Abstract
In this study, a genetically diverse panel of 43 mouse strains was exposed to phosgene and genome-wide association mapping performed using a high-density single nucleotide polymorphism (SNP) assembly. Transcriptomic analysis was also used to improve the genetic resolution in the identification of genetic determinants of phosgene-induced acute lung injury (ALI). We prioritized the identified genes based on whether the encoded protein was previously associated with lung injury or contained a nonsynonymous SNP within a functional domain. Candidates were selected that contained a promoter SNP that could alter a putative transcription factor binding site and had variable expression by transcriptomic analyses. The latter two criteria also required that ≥10% of mice carried the minor allele and that this allele could account for ≥10% of the phenotypic difference noted between the strains at the phenotypic extremes. This integrative, functional approach revealed 14 candidate genes that included Atp1a1, Alox5, Galnt11, Hrh1, Mbd4, Phactr2, Plxnd1, Ptprt, Reln, and Zfand4, which had significant SNP associations, and Itga9, Man1a2, Mapk14, and Vwf, which had suggestive SNP associations. Of the genes with significant SNP associations, Atp1a1, Alox5, Plxnd1, Ptprt, and Zfand4 could be associated with ALI in several ways. Using a competitive electrophoretic mobility shift analysis, Atp1a1 promoter (rs215053185) oligonucleotide containing the minor G allele formed a major distinct faster-migrating complex. In addition, a gene with a suggestive SNP association, Itga9, is linked to transforming growth factor β1 signaling, which previously has been associated with the susceptibility to ALI in mice.
Keywords: ARDS, countermeasures, genetics, sodium absorption, lipoxygenase
Clinical Relevance
The capability to reliably predict survival in patients with acute lung injury (ALI) remains a major challenge to critical care. Previous genetics studies in humans have been limited. This study functionally assesses single-nucleotide polymorphism associations linked with survival during ALI in mice. Genetic associations were strengthened by an integrated assessment using transcriptomic profiling. The leading candidate genes associated with increased susceptibility to ALI in mice included Atp1a1, Alox5, Plxnd1, Ptprt, and Zfand4.
Acute lung injury (ALI) entails increased epithelial and endothelial permeability, disrupted surfactant associated protein function, and decreased clearance of edema fluid (1–3). It can result from direct (e.g., inhaled chemical) or indirect (e.g., sepsis) insults (4). The ability to reliably predict and enhance survival is a significant challenge to the treatment of ALI (5). Individual susceptibility varies greatly, with patients presenting with the same severity score having markedly different clinical outcomes (6). For this reason, studies have begun to investigate the role of genetics in determining survival during ALI (7–22).
Constant acquisition of information on the genetic variability among inbred mouse strains has made the mouse a powerful model to facilitate rapid evaluation of the genetic basis of human physiology and pathophysiology (23–31). Recently, next-generation whole genome sequencing information has been obtained on 17 mouse strains (29), which has strengthened the existing single nucleotide polymorphism (SNP) database for over 40 mouse strains (29–31). Using a genetically diverse panel of ≥40 inbred mouse strains, we previously conducted functional genomic analyses of acrolein-induced (32, 33) and chlorine-induced (34) ALI. In this study, we sought to compare these findings with an additional chemical, phosgene, also known to produce ALI after accidental or intentional terrorist exposures.
Phosgene (COCl2), a colorless gas with the odor of wet hay, was originally generated in 1812 by John Davy, who combined chlorine with carbon monoxide (35). Phosgene is a weak topical irritant to the eyes and upper respiratory tract, but, because phosgene readily penetrates into the alveolar region of the lung, it is a strong inducer of severe pulmonary edema, respiratory distress, hypoxia, and death due to respiratory failure (36, 37). The onset of pulmonary edema can be delayed and often follows a symptom-free period (37). Phosgene was used as a chemical warfare agent in World War I (38). Many countries, including the United States, have agreed to the Chemical Weapons Convention, which prohibits the deliberate release of this and other chemical compounds during warfare (39). However, Syria, Somalia, and the Democratic People’s Republic of Korea (North Korea), among others, have not signed this agreement (40). Phosgene (≥1 million tons per y produced worldwide) is used mainly in the manufacture of isocyanates, which are polymerized to produce polyurethane and polycarbonate resins (36, 41). Phosgene also can be generated accidentally by thermal degradation of several chemicals, including carbon tetrachloride, perchloroethylene (degreasing compound), and methylene chloride (paint remover) (41), and, because of its high toxic potency, phosgene is listed as a possible chemical terrorist agent (42). In this study, genome-wide association mapping using a high-density SNP assembly and transcriptomic analysis were combined to identify the genetic determinants of phosgene-induced ALI.
Materials and Methods
Experimental Design
This study was performed in accordance with the Institutional Animal Care and Use Committee of the University of Pittsburgh (Pittsburgh, PA). Forty-three inbred mouse strains (6- to 8-wk-old female mice; n = 10 mice per strain) (Jackson Laboratory, Bar Harbor, ME) were housed under specific pathogen–free conditions. Mice were exposed to filtered air (control) or phosgene (1.0 ppm for up to 24 h) (Matheson Tri-Gas, Montgomery, PA) in laminar-flow, dynamic 0.32-m3 stainless steel inhalation chambers. Phosgene concentrations were monitored (Chemguard Infrared Monitor; MSA Instruments Division, Pittsburgh, PA) during exposure. Survival time was recorded during exposure and after the mice had been returned to filtered room air. Genome-wide association analysis was performed using efficient mixed-models association corrected for confounding from population structure and genetic relatedness (43, 44). Previously, we determined that one statistical association by chance will occur in a genome-wide scan when the threshold is decreased to 1.6 × 10−5 or −log (P) = 4.8 (25, 32–34). Therefore, we used a significance threshold of –log (P) > 4.8 and a suggestive threshold of 4.8 ≥ –log (P) > 4.0. To examine phosgene-induced changes in lung transcripts (n = 8 mice per strain per group) or lung histology (n = 3 mice per strain per group), sensitive SM/J or resistant 129X1/SvJ mice were exposed to filtered air (0 h, control) or phosgene (1.0 ppm) for 6 or 12 hours. To examine neutrophils and protein in bronchoalveolar lavage fluid, additional groups (n = 5 mice per strain per group) of the sensitive SM/J and resistant 129X1/SvJ mice were exposed to filtered air (0 h, control) or phosgene (1.0 ppm) for 6 or 9 hours. In additional mice, lung transcript levels were measured by microarray analysis (n = 5 mice per strain per time), and real-time q-PCR (n = 8 mice per strain per time) was used to contrast transcript levels of identified candidate genes in the sensitive SM/J or resistant 129X1/SvJ mice. To determine consequences of rs215053185 variants in the Atp1a1 promoter on DNA-protein binding, an electrophoretic mobility shift assay was performed.
Selection of Candidate Genes
Because the next-generation genome-wide sequencing has been obtained directly (14 strains used in this study) or has been imputed (29 additional strains used in this study), all known SNPs in each of the identified candidate genes could be evaluated in our population for the functional consequences. This was done using a four-step process. The first two steps involved inclusion of genes previously associated with ALI and inclusion of genes that contained nonsynonymous SNP in a functional domain of the protein. In the second step, missense mutations were identified in the protein functional domain that could explain ≥ 10% of the phenotypic difference between the strains survival times and had a minor allelic frequency of ≥ 10%. The next two steps involved inclusion of genes that differed in baseline lung transcript levels in the SM/J compared with those of the 129X1/SvJ mouse and inclusion of genes that differed in lung transcript levels in the SM/J compared with those of the 129X1/SvJ mouse after phosgene exposure. These differences were evaluated by microarray and confirmed by real-time q-PCR. Once genes with differential expression were identified, then SNPs in 5′untranslated region (UTR) (promoter) that could alter putative transcription factor binding were evaluated using the same threshold criteria using ≥ 10% survival time and ≥ 10% allelic frequency of the 430 mice exposed. Additional details are provided in the online supplement.
Results
Assessment of Phosgene-Induced Lung Injury in Sensitive SM/J Mice and Resistant 129X1/SvJ Mice
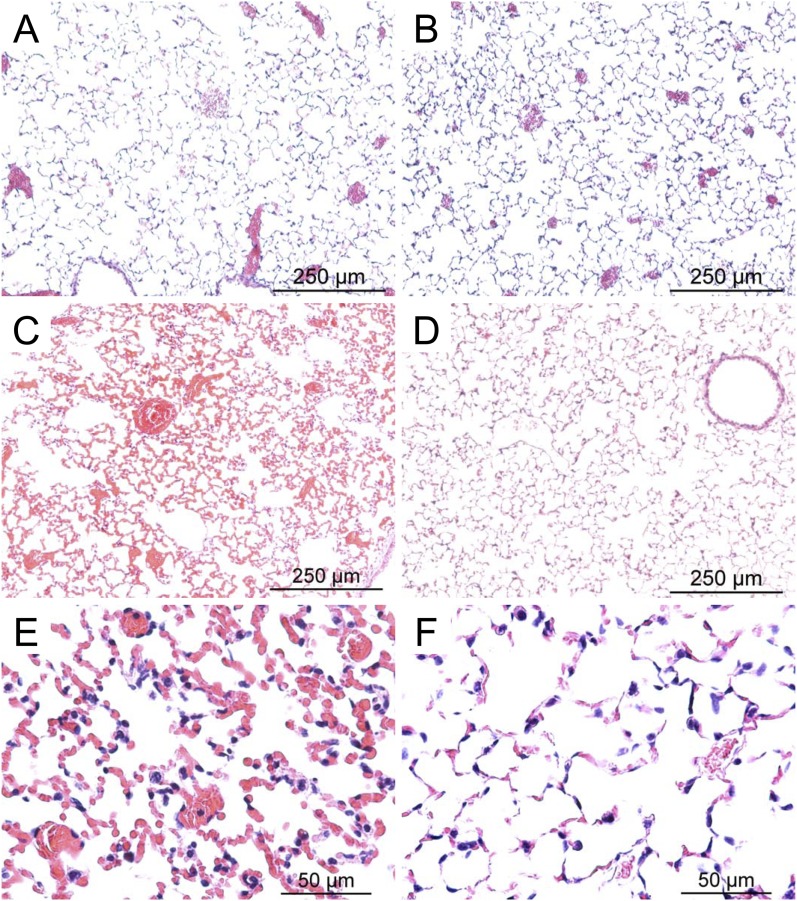
Survival time varied more than 4-fold among 43 mouse strains (Figure 1). To confirm that phosgene produced features consistent with ALI, sensitive SM/J and resistant 129X1/SvJ mice were exposed to filtered air (control) or to phosgene (1 ppm for 12 h) and anesthetized, and lung tissue was examined. Concordant with ALI, gross pathology indicated that hemorrhagic pulmonary edema was evident in the sensitive SM/J strain more than in the resistant 129X1/SvJ strain. Similarly, histological evidence of alveolar edema was more evident in the SM/J mouse at 12 hours (Figure 2). After 6 hours of exposure, bronchoalveolar lavage was performed, and the percentage of neutrophils increased in the sensitive SM/J strain and was significantly greater than the resistant 129X1/SvJ strain (Table 1). Protein in lavage increased at 6 hours in the SM/J and the 129X1/SvJ strains. Lavage was attempted at 12 hours; however, due to the extensive damage, recovery of lavage fluid was variable. Therefore, lavage was performed at 9 hours, and the percentage of neutrophils was found to be increased in both strains.
Figure 1.
Mouse strains vary in sensitivity to phosgene-induced acute lung injury. Survival time was determined for 43 mouse strains. Female mice were exposed to 1.0 ppm phosgene for up to 24 hours, and survival times were recorded hourly. Values are means ± SE (n = 10 mice per strain; 6–8 wk old).
Figure 2.
Histological assessment of lung tissue from control SM/J mice (A), control 129X1/SvJ mice (B), phosgene-exposed SM/J mice (C and E), or phosgene-exposed 129X1/SvJ mice (D and F). Consistent with phosgene-induced acute lung injury, hemorrhagic pulmonary edema was more evident in the sensitive SM/J strain than in the resistant 129X1/SvJ strain. Female mice were exposed to filtered air (control) or to phosgene (1 ppm for 12 h) and anesthetized, and lung tissue was obtained. Tissues were fixed in formaldehyde, and 5-μm sections were prepared with hematoxylin and eosin stain. Bars indicate magnification.
TABLE 1.
PHOSGENE INCREASED NEUTROPHILS AND PROTEIN IN BRONCHOALVEOLAR LAVAGE
| |
SM/J |
129X1/SvJ |
||||||
|---|---|---|---|---|---|---|---|---|
| Hours | Neutrophils | (%) | Protein | (μg/ml) | Neutrophils | (%) | Protein | (μg/ml) |
| |
Mean |
SE |
Mean |
SE |
Mean |
SE |
Mean |
SE |
| 0 |
0.15 |
0.11 |
274 |
44 |
0.13 |
0.15 |
204 |
47 |
| 6 |
3.68* |
1.12 |
1,838† |
634 |
1.67‡ |
0.54 |
1,300* |
183 |
| 9 | 2.58* | 0.41 | 1,871 | 1,114 | 3.48** | 0.80 | 1,065* | 251 |
Significantly different from strain-matched 0 hour control as determined by ANOVA followed by Holm-Sidak comparison test. P < 0.01.
Significantly different from strain-matched 0 hour control as determined by ANOVA followed by Holm-Sidak comparison test. P < 0.05.
Significantly different from SM/J at 6 hour as determined by ANOVA followed by Holm-Sidak comparison test. P < 0.05.
Genome-Wide Association Mapping of Mouse Strains Varies in Sensitivity to Phosgene-Induced ALI
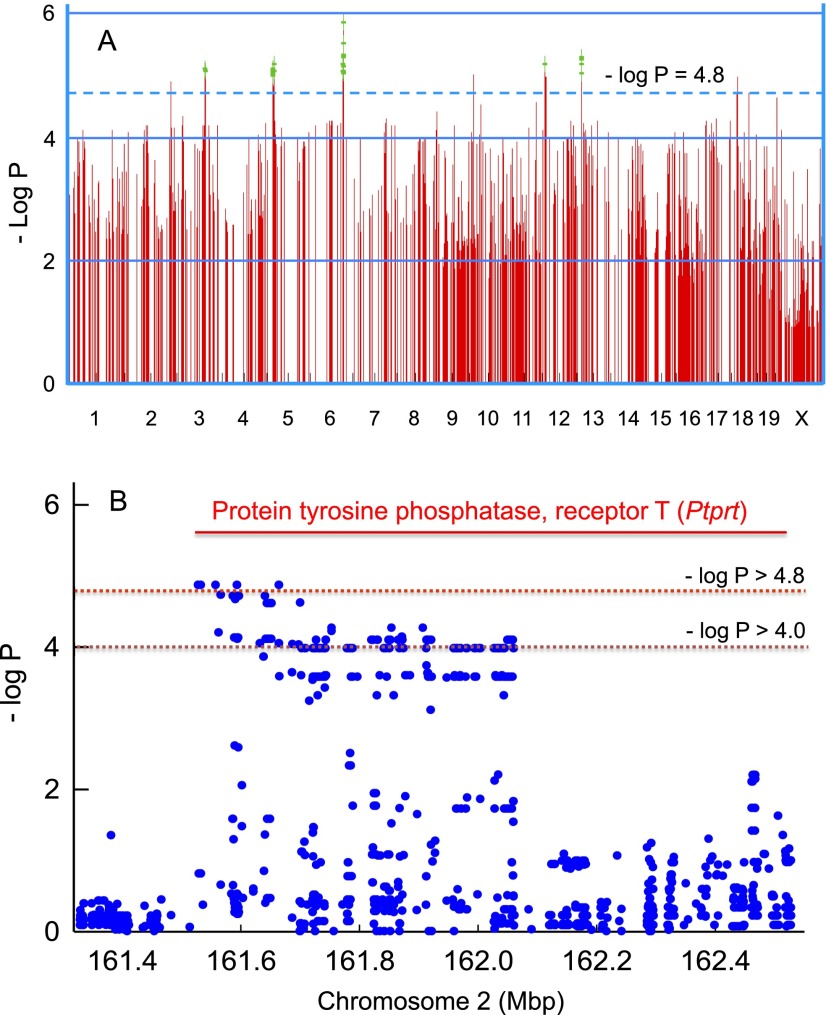
Survival time provided a quantitative trait for genome-wide association mapping for phosgene-induced ALI. A scatter (Manhattan) plot was generated by efficient mixed-models association corrected for confounding from population structure and genetic relatedness (Figure 3A). Figure 3A displays 1,000 SNPs with the highest –log P values of the > 4 million SNPs used in the analysis, and an exemplary genetic locus on chromosome 2 illustrates the overall SNP density (Figure 3B). Within the gene boundaries of protein tyrosine phosphatase, receptor T (Ptprt), four SNPs exceeded the significance threshold of −log P > 4.8, and 61 other SNPs exceeded the suggestive threshold of 4.8 > −log P > 4.0.
Figure 3.
Genome-wide association mapping of mouse strains that vary in sensitivity to phosgene-induced acute lung injury. (A) Genome-wide association map for phosgene-induced acute lung injury in mice. The scatter (Manhattan) plot was generated by efficient mixed-models association corrected for population structure and displays the corresponding –log (P) association probability for the top 1,000 region-wide single nucleotide polymorphisms (SNPs) at the indicated chromosomal locations. (B) Exemplary genetic locus on chromosome 2 illustrating SNP density. Within the gene boundaries of protein tyrosine phosphatase, receptor T, four SNPs exceeded the significance threshold of −log P > 4.8, and 61 other SNPs exceeded the suggestive linkage threshold of −log P > 4.0.
Thirty-one genes that contained significant (see Table E1 in the online supplement) and 70 genes that contained suggestive (Table E2) SNP associations were identified throughout the mouse genome. To prioritize the identified genes, we first determined whether a gene or the encoded protein previously had been associated with lung injury. Two genes with significant SNP associations (arachidonate 5-lipoxygenase [Alox5] [45] and ATPase, Na+/K+ transporting α1 polypeptide (Atp1a1) [3, 46]) and three genes with suggestive SNP associations (mannosidase, α, class 1A, member 2 [Man1a2] [47]; mitogen-activated protein kinase 14 [Mapk14, aka p38] [48, 49]; and von Willebrand factor [Vwf] [50, 51]) have been associated with lung injury.
Assessment of the Phenotypic Difference in Survival Time in Mice Carrying Nonsynonymous SNPs in Functional Domains of Candidate Genes and Comparison to Strains at the Phenotypic Extremes (Sensitive: PERA/EiJ and Resistant: NOD/ShiLtJ)
The genetic sequence for each of the 101 identified genes with significant (31 genes) or suggestive (70 genes) linkage was assessed for the presence of nonsynonymous SNPs in functional domains. The mean survival time was determined for mice carrying a nonsynonymous SNP predicted to alter amino acid sequence in a functional domain of the encoded protein. The difference between these groups was compared with the difference of the means of strains at the phenotypic extremes, PERA/EiJ (n = 10 mice) and resistant NOD/ShiLtJ (n = 10 mice) mouse strains exposed to 1 ppm phosgene. The resulting SNPs were evaluated as to whether each could explain ≥ 10% of the phenotypic difference between the survival time of strains at the phenotypic extremes and had a minor allelic frequency of ≥ 10% (Figure 4).
Figure 4.
Assessment of the phenotypic difference in survival time between the strains produced by nonsynonymous SNP associations in functional domains of candidate genes. For each SNP, the mean survival time was determined for mice in strains carrying one or the other of the two alleles (e.g., mean survival time of 320 mice with C for rs30121304 was compared with that for 110 mice with T for rs30121304). The difference between these allelic groups was then compared with the difference of the mean survival time of the mouse strains at the phenotypic extremes (i.e., sensitive PERA/EiJ [n = 10 mice] and resistant NOD/ShiLtJ [n = 10 mice] mouse strains exposed to 1 ppm phosgene; total = 410–430 female mice). The predicted amino acid substitutions in the corresponding protein are contained in the following domains: ALOX5 (V645I: valine 645 isoleucine): lipoxygenase domain; GALNT11 (S544L: serine 544 leucine): ricin-type β-trefoil lectin domain; ITGA9 (L353M leucine 353 methionine) VCBS (Vibrio, Colwellia, Bradyrhizobium, and Shewanella) putative adhesion domain; MBD4 (D128N: aspartic acid 128 asparagine): methyl CpG-binding domain; and MBD4 (A466T: alanine 466 threonine) endonuclease III domain, respectively. The predicted consequences of amino acid substitutions would alter side chain polarity (rs37913166, rs31503102), side chain charge (rs30840549), or hydropathy index (rs30121304, rs37913166, rs46653588). The SNP identification “rs” number is indicated on the abscissa with sample size within each bar. Values are means ± SE. P values indicate significance of the difference between the allele means as determined by ANOVA with an all-pairwise multiple comparison procedure (Holm-Sidak method). Alox5 = arachidonate 5-lipoxygenase; Galnt11 = UDP-N-acetyl-α-D-galactosamine: polypeptide N-acetylgalactosaminyltransferase 11; Itga9 = integrin α9; Mbd4 = methyl-CpG binding domain protein 4.
The predicted amino acid substitutions in the functional domains of the corresponding protein of four candidate genes included ALOX5: valine 645 isoleucine in a lipoxygenase domain; UDP-N-acetyl-α-D-galactosamine: polypeptide N-acetylgalactosaminyltransferase 11 (GALNT11): serine 544 leucine in a ricin-type β-trefoil lectin domain; integrin α9 (ITGA9): leucine 353 methionine in a VCBS (Vibrio, Colwellia, Bradyrhizobium, and Shewanella) putative adhesion domain; and methyl-CpG binding domain protein 4 (MBD4): aspartic acid 128 asparagine in a methyl CpG-binding domain and alanine 466 threonine in a endonuclease III domain (Table 2). The predicted consequences of amino acid substitutions could alter side chain polarity (Galnt11: rs37913166; Mbd4: rs31503102), side chain charge (Mbd4: rs30840549), or hydropathy index (Alox5: rs30121304; Galnt11: rs37913166; Itga9: rs46653588).
TABLE 2.
GENES WITH SINGLE NUCLEOTIDE POLYMORPHISM ASSOCIATIONS LINKED TO PHOSGEN ACUTE LUNG INJURY
| Chr | Position | − log (P) | SNPs | Symbol | ID | Description | Phenotype (%) | Allele freq. (%) | Consequence |
|---|---|---|---|---|---|---|---|---|---|
| Function has been associated with acute lung injury previously | |||||||||
| 6 |
116360826 |
5.33 |
3 |
Alox5 |
11689 |
Arachidonate 5-lipoxygenase |
27 |
12 |
V646I lipoxygenase |
| |
|
|
|
Alox5 |
11689 |
Arachidonate 5-lipoxygenase |
19 |
25 |
Loss CAC-binding gain Sp1 |
| |
|
|
|
Alox5 |
11689 |
Arachidonate 5-lipoxygenase |
21 |
27 |
Loss POU3F2 |
| 3 |
101398706 |
4.97 |
2 |
Atp1a1 |
11928 |
ATPase, Na+/K+ transporting, alpha 1 polypeptide |
13 |
17 |
Loss TCF/LEF1 site |
| 3 |
100453526 |
4.52 |
2 |
Man1a2* |
17156 |
Mannosidase, alpha, class 1A, member 2 |
— |
— |
ND |
| 17 |
28836181 |
4.18 |
5 |
Mapk14* |
26416 |
Mitogen-activated protein kinase 14 |
— |
— |
ND |
| 6 |
125601929 |
4.21 |
1 |
Vwf* |
22371 |
von Willebrand factor |
38 |
17 |
Regulation; splice site variant |
| Contains a nonsynonymous SNP in functional domain | |||||||||
| 5 |
24766046 |
4.83 |
1 |
Galnt11 |
231050 |
UDP-N-acetyl-alpha-D-galactosamine: polypep- tide N-acetylgalactosaminyltransferase 11 |
32 |
26 |
S544L Ricin-type beta-trefoil lectin |
| 9 |
118788857 |
4.16 |
1 |
Itga9* |
104099 |
Integrin alpha 9 |
14 |
18 |
L353M VCBS |
| 6 |
115790414 |
5.32 |
2 |
Mbd4 |
17193 |
Methyl-CpG binding domain protein 4 |
32 |
24 |
D128N Methyl-CpG binding |
| |
|
|
|
Mbd4 |
17193 |
Methyl-CpG binding domain protein 4 |
13 |
48 |
A446T Endonuclease III |
| Contains a promoter SNP that alters putative transcription factor binding site and transcript levels differed between strains | |||||||||
| 6 |
114358031 |
5.08 |
8 |
Hrh1 |
15465 |
Histamine receptor H1 |
37 |
33 |
Gain TCF/LEF1 site |
| 10 |
13010416 |
5.00 |
1 |
Phactr2 |
215789 |
Phosphatase and actin regulator 2 |
19 |
17 |
Loss AP-1 |
| 6 |
115911565 |
5.04 |
6 |
Plxnd1 |
67784 |
Plexin D1 |
14 |
31 |
Loss GCF gain TFCP2 |
| 2 |
161527626 |
4.87 |
65 |
Ptprt |
19624 |
Protein tyrosine phosphatase, receptor type, T |
14 |
31 |
Loss GCF gain TFCP2 |
| 5 |
21502773 |
4.83 |
54 |
Reln |
19699 |
Reelin |
45 |
17 |
Loss of CAC-binding, NF-E2 |
| 6 | 116267300 | 5.34 | 4 | Zfand4 | 67492 | Zinc finger, AN1-type domain 4 | 16 | 25 | Loss E2F+p107 |
Position is the base pair location of the single nucleotide polymorphism (SNP) in the gene. –log (P) is the highest negative log of probability for the SNP within the gene. SNPs is the total number of significant –log (P) > 4.8 and suggestive SNP 4.0 < –log (P) < 4.8 observed in the gene. Symbol is the official Entrez symbol. ID is the Entrez gene identification number. Description is the official full gene name provided to Entrez by Mouse Genome Informatics (MGI) Database. Phenotype (%) is the percent difference in mean survival time explained by the SNP. Allele (%) is the frequency of the minor allele in the population that has been genotyped for the functional SNP. Consequence is the amino acid substitution with the functional domain or the change produced in a putative DNA transcription factor binding site in the promoter.
Definition of Abbreviations: AP-1, activator protein 1; CAC-binding, CAC-binding protein transcription factor; Chr, chromosome; E2F+p107, E2F and p107 transcription factors; GCF, GC factor; ND, none detected; NF-E2, nuclear factor, erythroid derived 2; POU3F2, POU domain, class 3, transcription factor 2; Sp1: trans-acting transcription factor 1; TCF/LEF1, T cell-specific transcription factor/lymphoid enhancer-binding factor 1 transcription factor; TFCP2, transcription factor CP2; VCBS, Vibrio, Colwellia, Bradyrhizobium, and Shewanella protein domain.
Indicates genes with suggestive SNP association.
Transcriptomic Analysis
To assess the transcriptomic response during lung injury, microarray analysis was performed on SM/J and 129X1/SvJ mouse lung mRNA obtained at 0 (control), 6, and 12 hours during 1.0 ppm phosgene exposure. Transcripts that increased ≥ 2-fold or decreased ≤ 2-fold in SM/J mouse lung and not significantly different in 129X1/SvJ mouse lung as compared with SM/J control (0 h) transcripts were analyzed for enriched pathways using Database for Annotation, Visualization, and Integrated Discovery. The enriched terms in categories of Gene Ontogeny biological process, Gene Ontogeny molecular function, or Kyoto Encyclopedia of Genes and Genomes pathway were determined. Ten transcripts with the greatest difference between strains in each of these categories are displayed in Figure 5.
Figure 5.
Transcripts in enriched pathways that were altered in sensitive SM/J more than in resistant 129X1/J mice as determined by microarray analysis of lung mRNA after phosgene exposure. Microarray analysis was performed on mouse lung mRNA obtained at 0 (control), 6, and 12 hours during 1.0 ppm phosgene exposure (n = 5 arrays per strain per time), and significant differences were determined by Partek software (P < 0.05). Transcripts log 2 ≥ 1.0 (i.e., ≥ 2-fold) increased or log 2 ≤ −1.0 decreased in SM/J mouse lung but not significantly different in 129X1/SvJ mouse lung as compared with SM/J control (0 h) transcripts were analyzed using Database for Annotation, Visualization, and Integrated Discovery. The most enriched term in categories of Gene Ontogeny (GO) biological process, GO molecular function, or Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway were selected. Ten transcripts with the greatest difference between strains in these pathways are displayed. (A) At 6 hours, the enriched pathways with increased transcripts in SM/J mouse lung (n = 751) included apoptosis, unfolded protein response, and cytokine–cytokine receptor binding. The enriched pathways with decreased transcripts in SM/J mouse lung (n = 731) included positive regulation of angiogenesis, transmembrane receptor activity, and ribosome. (B) At 12 hours, the enriched pathways with increased transcripts in SM/J mouse lung (n = 898) included apoptosis, GTPase regulator activity, and mitogen activated protein kinase (MAPK) signaling. The enriched pathways with decreased transcripts in SM/J mouse lung (n = 989) included microtubule-based process, cytoskeletal protein binding, and axon guidance.
At 6 hours, the enriched pathways with increased transcripts in SM/J mouse lung (n = 751) but not in 129X1/SvJ mouse lung included apoptosis, unfolded protein response, and cytokine–cytokine receptor binding. Antioxidant response transcripts mediated by nuclear factor (erythroid derived 2)–like 2 (NFE2L2, aka Nrf2) were increased more in the SM/J than in the 129X1/SvJ mouse lung at 6 hours (Figure 6). At 12 hours, the enriched pathways with increased transcripts in SM/J mouse lung (n = 898) included apoptosis, GTPase regulator activity, and mitogen activated protein kinase signaling. The enriched pathways at 6 hours with decreased transcripts in SM/J mouse strain (n = 731) included positive regulation of angiogenesis, transmembrane receptor activity, and ribosome. The enriched pathways at 12 hours with decreased transcripts in SM/J mouse lung (n = 989) included microtubule-based process, cytoskeletal protein binding, and axon guidance.
Figure 6.
Transcripts in the enriched nuclear factor (erythroid-derived 2)–like 2 (NFE2L2, also known as Nrf2)–mediated oxidative stress response pathway in the lungs of sensitive SM/J and resistant 129X1/SvJ mice during phosgene-induced acute lung injury. Increased transcripts at 6 hours in SM/J but not in 129X1/SvJ mice were determined by microarray analysis. Transcripts in this pathway were similar between strains at 12 hours and included heme oxygenase (decycling) 1 (HMOX1); glutathione S-transferase α1 (Ya) (GSTA1); activating transcription factor 3 (ATF3); glutamate-cysteine ligase, catalytic subunit (GCLC); NAD(P)H dehydrogenase, quinone 1 (NQO1); glutathione reductase (GSR); v-maf musculoaponeurotic fibrosarcoma oncogene family, protein F (avian) (MAFF); thioredoxin reductase 1 (TXNRD1); activating transcription factor 4 (ATF4); and glutamate-cysteine ligase, modifier subunit (GCLM). Values represent means ± SE (n = 6–8 female mice per strain per time) normalized to SM/J control mice (0 h).
Transcripts ≥ 2-fold increased or ≤ 2-fold decreased in 129X1/SvJ mouse lung as compared with SM/J mouse lung control (0 h) but not significantly different in temporally matched SM/J as compared with the SM/J mouse lung control (0 h) are displayed in Figure 7. At 6 hours, the enriched pathways with increased transcripts in 129X1SvJ mouse lung (n = 1,016) included lymphocyte activation, calcium ion binding, and T-cell receptor signaling. The enriched pathways at 6 hours with decreased transcripts in 129X1/SvJ (n = 947) included positive regulation of cell death, peptidase inhibitor, and complement and coagulation. At 12 hours, the enriched pathways with increased transcripts in 129X1/SvJ mouse lung (n = 627) included defense mechanism, carbohydrate/pattern binding, and cell adhesion molecules. The enriched pathways at 12 hours with decreased transcripts in 129X1/SvJ (n = 954) included regulation of transcription, chemokine receptor, and DNA repair.
Figure 7.
Transcripts in enriched pathways that were altered in resistant 129X1/SvJ more than in sensitive SM/J mice as determined by microarray analysis of lung mRNA after phosgene exposure. Microarray analysis was performed on mouse lung mRNA obtained at 0 (control), 6, and 12 hours during 1.0 ppm phosgene exposure (n = 5 arrays per strain per time), and significance (P < 0.05) was determined by Partek software. Transcripts log 2 ≥ 1.0 (i.e., ≥2-fold) increased or log 2 ≤ −1.0 decreased in 129X1/SvJ mouse lung but not significantly different in SM/J mouse lung as compared with SM/J control (0 h) transcripts were analyzed using Database for Annotation, Visualization, and Integrated Discovery (DAVID). The most significant enriched term in the categories of Gene Ontogeny (GO) biological process, GO molecular function, and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway were selected. Ten transcripts with the greatest difference between strains in these pathways are displayed. (A) At 6 hours, the enriched pathways with increased transcripts in 129X1/SvJ mouse lung (n = 1,016) included lymphocyte activation, calcium ion binding, and T-cell receptor signaling. The enriched pathways with decreased transcripts in 129X1/SvJ mouse lung (n = 947) included positive regulation of cell death, peptidase inhibitor, and complement and coagulation. (B) At 12 hours, the enriched pathways with increased transcripts in 129X1/SvJ mouse lung (n = 627) included defense mechanism, carbohydrate/pattern binding, and cell adhesion molecules (CAMs). The enriched pathways with decreased transcripts in 129X1/SvJ mouse lung (n = 954) included regulation of transcription, chemokine receptor, and DNA repair.
At 6 hours, common enriched pathways with increased transcripts (n = 187) (Table E3) in SM/J and 129X1/SvJ mouse lung included the G-protein coupled receptor protein signaling pathway, cytokine activity, and the Jak-STAT signaling pathway. The enriched pathways at 6 hours with decreased transcripts in both strains (n = 584) included macromolecule catabolic process, endonuclease activity, and ribosome. At 12 hours, the enriched pathways with increased transcripts in both strains (n = 1,532) (Table E4) included inflammatory response, cytokine activity, and glutathione metabolism. The enriched pathways at 12 hours with decreased transcripts in both strains (n = 1,451) included cellular component morphogenesis, zinc ion binding, and focal adhesion.
Transcript Levels of a Candidate Gene that Differ between Strains at Baseline Control or that Change during Phosgene Exposure
To assess the identified candidate genes further, the microarray data were evaluated to determine whether lung transcripts differed between the sensitive SM/J and the resistant 129X1/SvJ mouse strain before or during phosgene exposure. Based on this initial analysis, transcript levels were determined by real-time q-PCR for 10 candidate genes with significant SNP associations and 10 candidate genes with suggestive SNP associations. Of these 20 genes, 8 of the 10 genes with significant SNP associations (Figure 8A) and 6 of the 10 genes with suggestive SNP associations (Table E5) had altered baseline transcripts in the 129X1/SvJ mouse lung as compared with the SM/J mouse lung. For example, the baseline lung levels of ATP1A1 transcript increased 1.8 ± 0.2-fold (log 2 = 0.85-fold) and ALOX5 transcript decreased 1.7 ± 0.1-fold (log 2 = −0.80-fold) in 129X1/SvJ as compared with the SM/J mice.
Figure 8.
Transcript levels of candidate genes in 129X1/SvJ as compared with SM/J mouse lung. (A) Baseline transcript levels for 10 candidate genes with significant SNP associations (−log P ≥ 4.8). Transcript levels were determined by real-time qPCR (n = 8 mice per strain). Values are means ± SE (n = 8 mice per strain) normalized to ribosomal protein L32 (RPL32). *Significant difference (P < 0.05) between the resistant 129X1/SvJ and sensitive SM/J mouse strains at baseline as determined by ANOVA with an all-pairwise multiple comparison procedure (Holm-Sidak method). (B) Transcript levels of a candidate gene, zinc finger, AN1-type domain 4 (Zfand4), that was changed during phosgene exposure. Transcript levels were determined by real-time qPCR (n = 8 mice per strain). Values are means ± SE (n = 8 mice/strain) normalized to ribosomal protein L32 (RPL32). *Significantly different (P < 0.05) from sensitive SM/J mouse strain at baseline (0 h control), as determined by ANOVA with an all pairwise multiple comparison procedure (Holm-Sidak method). †Significant difference (P < 0.05) between the resistant 129X1/SvJ and sensitive SM/J mouse strains at the same time as determined by ANOVA with an all-pairwise multiple comparison procedure (Holm-Sidak method). (C) Transcript levels of zinc finger, AN1-type domain proteins in the lung of sensitive SM/J or resistant 129X1/SvJ mice after phosgene exposure. Transcript levels were determined by microarray analysis. Values are means ± SE (n = 8 mice per strain). Three transcripts (ZFAND2A, ZFAND4, and ZFAND5) increased more in the lung of the sensitive SM/J as compared with the resistant 129X1/SvJ, whereas other ZFAND transcripts did not differ between strains (ZFAND1) or were unchanged (ZFAND2B, ZFAND3, and ZFAND6) in either strain after phosgene exposure. *Significantly different (P < 0.05) from sensitive SM/J mouse strain at baseline (0 h control) as determined by ANOVA with an all-pairwise multiple comparison procedure (Holm-Sidak method). †Significant difference (P < 0.05) between the resistant 129X1/SvJ and sensitive SM/J mouse strains at the same time as determined by ANOVA with an all-pairwise multiple comparison procedure (Holm-Sidak method). ALOX5 = arachidonate 5-lipoxygenase; ATP1A1 ATPase = Na+/K+ transporting α1 polypeptide; HRH1 = histamine receptor H1; MARCH8 = membrane-associated ring finger (C3HC4) 8; PHACTR2 = phosphatase and actin regulator 2; PTPRT = protein tyrosine phosphatase, receptor T; RELN = Reelin; 5033411D12Rik = RIKEN cDNA 5033411D12 gene (homolog to CaiB/baiF CoA-transferase protein family, C7ORF10); PLXND1: plexin D1; ZFAND1–6 = zinc finger, AN1-type domains 1 through 6.
Based on these strain differences in transcript levels, SNPs contained in the 5′UTR/promoter region of the 15 candidate genes were evaluated to determine whether the sequence variant would alter a putative transcription factor binding site. The resulting SNPs were evaluated to determine whether they could explain ≥10% of the phenotypic difference between the survival times of strains at the phenotypic extremes and had a minor allelic frequency of ≥10%; genes with such SNPs were included as candidate genes. Using these criteria, candidate genes remaining included histamine receptor H1 (Hrh1), phosphatase and actin regulator 2 (Phactr2), plexin D1 (Plxnd1), Ptprt, and reelin (Reln).
Transcripts for these candidate genes were measured by real-time q-PCR in these strains during phosgene exposure at 6 and 12 hours. Only one candidate gene, zinc finger, AN1-type domain 4 (Zfand4), had transcripts increase more in the sensitive SM/J mouse lung as compared with the 129X1/SvJ during phosgene exposure (Figure 8B). Because little is known about ZFAND4 function, changes in the transcripts for related ZFAND protein members in the microarray analysis were also compared between mouse strains after phosgene exposure (Figure 8C). Two related transcripts, ZFAND2A and ZFAND5, also increased more in the lung of the sensitive SM/J as compared with the resistant 129X1/SvJ, whereas other ZFAND transcripts did not differ between strains (ZFAND1) or were unchanged (ZFAND2B, ZFAND3, and ZFAND6) in either strain after phosgene exposure.
Protein Expression of Candidate Genes in the Lung
To assess whether the protein of the corresponding candidate gene can be detected in the lung, the Human Protein Atlas (52) was surveyed (Figure E1). Moderate to strong cytoplasmic or membranous immunostaining was present for HRH1, PHACTR2, PLXND1, PTPRT, and ZFAND4, and strong nuclear immunostaining was present for GALNT11 and MBD4 in the airway epithelium. In alveolar pneumocytes, moderate cytoplasmic or membranous immunostaining was present for ZFAND4, and strong nuclear immunostaining was present for GALNT11. In alveolar macrophages, moderate to strong cytoplasmic or membranous immunostaining was present for HRH1, ITGA9, PHACTR2, PLXND1, and RELN, and moderate nuclear immunostaining was present for GALNT11 and MBD4.
Atp1a1 Promoter Analysis
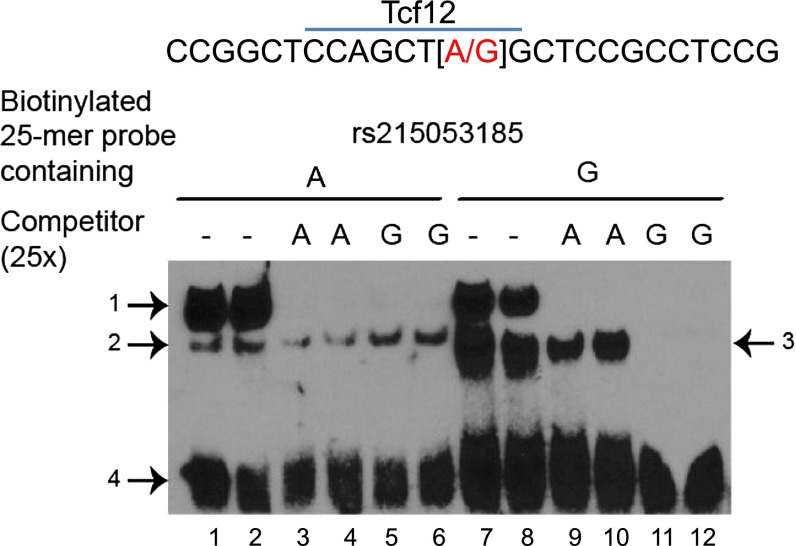
The mouse Atp1a1 gene proximal promoter region (−1,529 to +89 base pairs [bp] from the transcriptional start site) contains 35 SNPs and nine insertion/deletions. We evaluated these SNPs and insertion/deletions as to whether the change in DNA sequence was predicted to alter a putative transcription factor binding site and applied the phenotype and allele frequency criteria of ≥ 10%. One SNP, rs215053185 (−62 bp A/G), was identified that could explain 38% of the difference in survival time noted in the mouse strains at the phenotypic extremes (Table 2). Using the A or G allele oligonucleotide (“bait”) and nuclear protein extract from mouse MLE15 airway epithelial cells, a competitive electrophoretic mobility shift analysis was performed to assess the possible consequences of rs215053185. A slow migrating complex (Figure 9, arrow 1) could be competed with the unbiotinylated oligonucleotide A competitor or the G competitor. The biotinylated oligonucleotide containing the minor G allele (present in sensitive strains) also formed a major distinct faster-migrating complex, which is more effectively competed by unbiotinylated oligonucleotide containing the G variant (Figure 9, arrow 3).
Figure 9.
SNP (rs215053185) in the ATPase, Na+/K+ transporting α1 polypeptide gene promoter region influences nuclear protein binding capacity. Electrophoretic mobility shift assay of nuclear protein extract prepared from mouse lung epithelial cells (MLE-15) and 25-mers (−50 to −74 bp 5′ from the start site) containing the A or G allele in the middle of a biotinylated oligonucleotide. The biotinylated 25-mer oligonucleotide with the G allele (lanes 7–12), in contrast to the A allele (lanes 1–6), formed a major distinct faster-migrating complex (middle band arrow 3), which is more effectively competed by unbiotinylated oligonucleotide containing the G variant (lanes 11 and 12) compared with the unbiotinylated oligonucleotide containing the A variant (lanes 9 and 10). The G allele is the minor allele (present in sensitive strains) and could result in the gain of a putative DNA binding site for transcription factor 12 (Tcf12), which is a transcriptional repressor. Mice with the rs215053185 A allele were resistant (mean survival time, 24.8 ± 1.0 h), whereas mice with the G allele were sensitive (mean survival time, 11.3 ± 0.5 h; P < 0.001).
Discussion
In previous studies (53–62), increased bronchoalveolar lavage protein has been reported to be a sensitive endpoint in detecting early responses to phosgene. In this study, phosgene increased lavage protein at 6 hours from sensitive SM/J mice and resistant 129X1/SvJ mice (Table 1). Although temporal changes in lavage protein were similar between strains, neutrophil percent (Table 1) and lung histology (Figure 2) indicated that the SM/J mice developed injury sooner than the 129X1/SvJ mice. Previously, Sciuto and colleagues reported that glutathione S-transferase α1 (Ya) (GSTA1) and glutamate-cysteine ligase, catalytic subunit (GCLC) transcripts were among the most significantly changed in the microarray/real-time q-PCR analyses of mouse lung after phosgene exposure (63). Similarly, NFE2L2-mediated antioxidant response transcripts, including GSTA1 and GCLC, were increased in the SM/J more than in the 129X1/SvJ mouse lung at 6 hours, but these transcripts increased equally at 12 hours (Figure 6). The gross pathology revealed that lungs had marked focal hemorrhages at time of death in both strains. Thus, similarities in the pathology between strains suggest that the extent of injury was the same at time of death and that the phenotype being measured is extension of survival time.
Mean survival times varied by more than 4-fold among mouse strains, and genome-wide association analysis identified candidate genes with significant SNP associations on chromosomes 2, 3, 5, 6, and 10 and suggestive SNP associations on chromosomes 9 and 17. We prioritized the genes that had SNP associations within the gene borders or within 2,000 bp of the transcription start site by several criteria, including: 1) the encoded protein previously had been associated with adverse outcomes during lung injury, 2) the gene contained a nonsynonymous SNP in a functional domain of the encoded protein, or 3) the gene contained a 5′UTR (promoter) SNP that could alter a putative transcription factor binding site and was matched with variable expression by transcriptomic analyses. The latter two criteria also required that ≥ 10% of mice carried the minor allele and that this allele could explain ≥ 10% of the phenotypic difference noted between the sensitive PERA/EiJ and resistant NOD/ShiLtJ mouse strains. This integrative functional approach revealed 14 candidate genes that included 10 genes (Alox5, Atp1a1, Galnt11, Hrh1, Mbd4, Phactr2, Plxnd1, Ptprt, Reln, and Zfand4) that had significant SNP associations and four genes (Itga9, Man1a2, Mapk14, and Vwf) that had suggestive SNP associations (Table 2). Of the 10 genes with significant SNP associations, Atp1a1, Alox5, Plxnd1, Ptprt, and Zfand4 could be associated with ALI in several ways.
Although ALI is a complex trait governed by multiple genetic determinants, ATP1A1 is critical to the clearance of pulmonary edema fluid and thus to the outcome of patients with ALI (3). In this study, basal levels of lung ATP1A1 mRNA levels were increased in the resistant 129X1/SvJ as compared with the sensitive SM/J (Figure 8A), and we identified an Atp1a1 SNP rs215053185 that could alter the DNA-protein binding in a conserved promoter region (Table 2). Mice with the rs215053185 A allele were resistant (mean survival time, 24.8 ± 1.0 h), whereas mice with the G allele were sensitive (mean survival time, 11.3 ± 0.5 h; P < 0.001). Electrophoretic mobility shift assay of nuclear protein extract prepared from mouse lung epithelial cells (MLE-15) and biotinylated 25-mer oligonucleotide containing the A or G allele indicated that the G allele formed a major distinct faster-migrating complex, which was effectively competed by unbiotinylated oligonucleotide containing the G variant (Figure 9). The G allele is the minor allele (15.3% allelic frequency) and could result in the gain of a putative DNA binding site for transcription factor 12. Expressed in the lung (64), transcription factor 12 can repress E-cadherin (cadherin 1, type 1, E-cadherin [epithelial]) (65) and may modulate SMAD signaling (66), which is often active during ALI by transforming growth factor (TGF)β1 (32, 34).
Little is known about the functional significance of polymorphisms in human ATP1A1. The Dahl salt-sensitive (Dahl S) rat (67, 68) was thought to have a genetic variant in Atp1a1, and transgenic Dahl S rats bearing the Dahl salt-resistant Atp1a1 exhibited less salt-sensitive hypertension, less hypertensive renal disease, and longer life span when compared with nontransgenic Dahl S control rats (69–71). However, these findings are controversial (72, 73). In dairy cattle, an ATP1A1 polymorphism has been associated with heat tolerance and with respiratory rate (74).
A transmembrane protein, ATP1A1, is an α subunit of a heterodimeric Na,K-ATPase enzyme, which is expressed on the basolateral surface of alveolar type I and type II epithelial cells (3, 75, 76). In a process that consumes 40% of a cell’s energy, Na,K-ATPase maintains the sodium–potassium electrochemical gradient across the plasma membrane by exporting three Na+ and importing two K+ for each ATP hydrolyzed (77). In previous metabolomics analyses of acrolein- and chlorine-induced lung injury, we observed that the lung undergoes energetic stress and that resistant mice appear better able to use substrates for energy production (33, 34). The Na,K-ATPase–generated electrochemical gradient is essential for edema fluid absorption from the alveolus (1, 3). Na,K-ATPase also interacts with cytoskeletal and junction proteins to maintain epithelial integrity (78–80). Hypoxia- and mitochondrial-derived reactive oxygen species promote the protein kinase C-ζ–dependent phosphorylation of the Na,K-ATPase α1 subunit, triggering its endocytosis (81, 82). The phosphorylation occurs at a proximal serine in the N-terminus, and an induced mutation of this serine prevents endocytosis and a decrease in function (83, 84). Extended hypoxia causes the ubiquitination of four lysines on the N-terminus near the proximal serine and leads to degradation (84).
In examining the exiting sequence information for human ATP1A1, we identified a critical SNP (NCBI Reference Sequence: rs111860221) that results in a missense mutation that leads to a proline substitution for the proximal serine. This missense mutation (serine to proline conversion) is more frequent in persons of African ancestry and is predicted to produce an intolerant amino acid substitution by PolyPhen (85) or SIFT (86). Moreover, the resulting protein lacks the critical serine phosphorylation site and thereby would be resistant to protein kinase C-ζ phosphorylation mediated endocytosis ubiquitination (81–84). Thus, this mutation would be a gain of function preserving ATP1A1 during periods of hypoxia. In addition, ATP1A1 has two stop gained mutations (one somatic in cancer) and multiple human ATP1A1 protein isoforms exist (87, 88). One isoform, ATP1A1 isoform d (NCBI Reference Sequence: NP_001153706), encodes a 992–amino acid protein that has a truncated N-terminus compared with the full-length 1,024 amino acid isoform.
Another candidate gene with relevance to ALI is Alox5, which had significant SNP associations. The Alox5 encodes arachidonate 5-lipoxygenase, an enzyme that converts arachidonate (or eiosatetraenoic acid) to leukotriene A4, which can be converted to leukotrienes B4, C4, and D4. The resulting leukotrienes are elevated in ALI (45). In gene-targeted deficient mice or in animals treated with ALOX5 inhibitors, neutrophil infiltration and lavage protein are decreased after various forms of lung injury (89–94). Moreover, survival is increased after ventilator-induced lung injury (91). In this study we identified a SNP that was in the functional lipoxygenase domain and two SNPs in the 5′UTR region that were associated with the survival phenotype (Table 2). In humans, genetic variants in the ALOX5 promoter region are associated with diminished effectiveness of antileukotriene drugs used to treat asthma (95–98).
Plexin D1 (Plxnd1) also had significant SNPs associated with phosgene survival. Recently, we identified semaphorin 7a (Sema7a) as a candidate gene for susceptibility to chlorine-induced ALI in mice (34). Expressed in pulmonary vasculature, PLXND1 can function as a semaphorin receptor (99, 100), although its relationship to SEMA7A has not been evaluated. Plexin–semaphorin interactions are best known for their role in axon guidance (101, 102). However, these proteins also regulate diverse biological processes, including inhibition of lung branching morphogenesis (103, 104), pulmonary fibrosis (105), vascular development (100), and specific leukocyte functions (106, 107). In the microarray pathway analysis, angiogenesis and axon guidance were enriched in decreased transcripts in the sensitive SM/J mouse lung at 6 and 12 hours, respectively (Figure 4). In our analysis, we found a 5′UTR Plxnd1 polymorphism consistent with a loss of GC factor and a gain of a transcription factor CP2 putative binding site. Five 5′UTR, two frame-shift, and 230 missense variants have been identified in the human PLXND1 gene.
A member of the type IIB receptor protein tyrosine phosphatase family, PTPRT can dephosphorylate E-cadherin and paxillin, which stabilize junctional complexes and increase cell–cell adhesion (108). PTPRT can dephosphorylate and thereby inactivate signal transducer and activator of transcription 3 (acute-phase response factor) (109), which can reduce transcription of several target genes, including genes critical to surfactant phospholipid homeostasis (110) and cytokine activation (111) in ALI. In a genome-wide association study of subjects with asthma, a SNP in PTPRT (rs6016963) was among the top 50 significant SNPs associated with change in FEV1 in response to inhaled corticosteroids, but this SNP failed to reach significance in a replication population (112).
The transcript levels of one candidate gene, ZFAND4, increased more in the lung of the sensitive SM/J more than the resistant 129X1/SvJ mice during phosgene exposure (Figure 8B). ZFAND4 is a member of AN1-type zinc finger domain proteins, which are characterized by a metal (zinc)-bound α/β-fold domain that was first identified as a zinc finger at the C terminus of AN1, a ubiquitin-like protein in Xenopus laevis (113). To our knowledge, this is the first report in the literature on this member of the ZFAND protein family. Because very little is known about ZFAND4, we evaluated possible transcript changes of related AN1 domain–containing protein ZFAND members. In addition to ZFAND4, mouse lung ZFAND2A and ZFAND5 transcripts increased more in the SM/J than in the 129X1/SvJ (Figure 8C). Previous investigations have found that ZFAND2A expression is induced by arsenite (114, 115) and heat shock (116). Similarly in macrophages, ZFAND5 expression is induced by cell stress in response to TNF, IFNγ, LPS, and other toll-like receptor ligands (117–119). ZFAND5 also can compete with zinc finger protein 36 (aka tristetraprolin) and stabilizes class II adenylate/uridine-rich sequence element containing mRNA, a region common to inflammatory mediator transcripts including TNF mRNA. Thus, ZFAND5 is induced by and stabilizes TNF (119). However, the role of these proteins in ALI must be viewed with caution because structural similarity does not predict functional similarity.
Although phosgene-induced ALI in mice may have relevance to accidental or intentional human phosgene exposures, numerous other agents can produce ALI. The genetic and transcriptomic findings here may be limited to this compound and thus may not be relevant to other forms of ALI. We previously identified candidate genes associated with acrolein-induced (32, 33) and chlorine-induced ALI (34), and these genes differed from those identified with phosgene. Until several types of chemically induced ALI have been evaluated, generalizations to other forms may not be warranted. Nonetheless, a major candidate identified previously with acrolein was Activin A receptor type 1 (Acvr1), which mediates TGF-β1 signaling (32). TGF-β1 signaling was also implicated with chlorine-induced ALI by the identification of Sema7a (34). Integrin α9 (Itga9), a candidate gene with a suggestive SNP association in this study using phosgene, has been associated with TGF-β1 signaling in the lung (120). Partnering with integrin β1, integrin α9β1 can interact with vascular cell adhesion molecule 1 (121), vascular endothelial growth factors (122), and secreted phosphoprotein 1 (aka osteopontin) (123) and thus could have critical roles in ALI.
In summary, susceptibility to phosgene-induced ALI in mice was associated with 31 genes that contained significant SNP associations and many others with suggestive SNP associations. Candidate genes were selected based on previous relevance to ALI and whether genetic variants with putative functional consequences (amino acid substitution in a functional domain or sequence variants in promoters with transcript level changes) could explain a portion of the variation in survival time observed in 43 mouse strains. Of the genes with significant SNP associations, Atp1a1, Alox5, Plxnd1, Ptprt, and Zfand4 could be associated with ALI in several ways. In addition, one gene with a suggestive SNP association, Itga9, implicated TGF-β1 signaling with ALI, a relationship suggested by previous analyses.
Footnotes
This study was supported by the National Institutes of Health grants ES015675, HL077763, and HL085655 (G.D.L.); HL091938 (Y.P.P.D.); HL084932 and HL095397 (N.K.); HL080079 and DA024417 (E.E.); AT003203, AT005522, CA113793, and CA134433 (M.Y.); HL075562 (D.R.P.); and HL086981 (D.L.K.) and by NSF grants 0513612, 0731455, and 0729049 (E.E.).
This article has an online supplement, which is accessible from this issue's of contents at www.atsjournals.org
Originally Published in Press as DOI: 10.1165/rcmb.2012-0337OC on April 5, 2013
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Ware LB, Matthay MA. The acute respiratory distress syndrome. N Engl J Med. 2000;342:1334–1349. doi: 10.1056/NEJM200005043421806. [DOI] [PubMed] [Google Scholar]
- 2.Bein K, Wesselkamper SC, Liu X, Dietsch M, Majumder N, Concel VJ, Medvedovic M, Sartor MA, Henning LN, Venditto C, et al. Surfactant-associated protein B is critical to survival in nickel-induced injury in mice. Am J Respir Cell Mol Biol. 2009;41:226–236. doi: 10.1165/rcmb.2008-0317OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vadász I, Raviv S, Sznajder JI. Alveolar epithelium and Na,K-ATPase in acute lung injury. Intensive Care Med. 2007;33:1243–1251. doi: 10.1007/s00134-007-0661-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Matthay MA, Ware LB, Zimmerman GA. The acute respiratory distress syndrome. J Clin Invest. 2012;122:2731–2740. doi: 10.1172/JCI60331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ware LB, Koyama T, Billheimer DD, Wu W, Bernard GR, Thompson BT, Brower RG, Standiford TJ, Martin TR, Matthay MA NHLBI ARDS Clinical Trials Network. Prognostic and pathogenetic value of combining clinical and biochemical indices in patients with acute lung injury. Chest. 2010;137:288–296. doi: 10.1378/chest.09-1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Siempos II, Vardakas KZ, Kyriakopoulos CE, Ntaidou TK, Falagas ME. Predictors of mortality in adult patients with ventilator-associated pneumonia: a meta-analysis. Shock. 2010;33:590–601. doi: 10.1097/SHK.0b013e3181cc0418. [DOI] [PubMed] [Google Scholar]
- 7.Christie JD, Ma SF, Aplenc R, Li M, Lanken PN, Shah CV, Fuchs B, Albelda SM, Flores C, Garcia JG. Variation in the myosin light chain kinase gene is associated with development of acute lung injury after major trauma. Crit Care Med. 2008;36:2794–2800. doi: 10.1097/ccm.0b013e318186b843. [DOI] [PubMed] [Google Scholar]
- 8.Reddy AJ, Christie JD, Aplenc R, Fuchs B, Lanken PN, Kleeberger SR. Association of human NAD(P)H:quinone oxidoreductase 1 (NQO1) polymorphism with development of acute lung injury. J Cell Mol Med. 2009;13:1784–1791. doi: 10.1111/j.1582-4934.2008.00581.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reddy AJ, Kleeberger SR. Genetic polymorphisms associated with acute lung injury. Pharmacogenomics. 2009;10:1527–1539. doi: 10.2217/pgs.09.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gao L, Barnes KC. Recent advances in genetic predisposition to clinical acute lung injury. Am J Physiol Lung Cell Mol Physiol. 2009;296:L713–L725. doi: 10.1152/ajplung.90269.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sheu CC, Zhai R, Wang Z, Gong MN, Tejera P, Chen F, Su L, Thompson BT, Christiani DC. Heme oxygenase-1 microsatellite polymorphism and haplotypes are associated with the development of acute respiratory distress syndrome. Intensive Care Med. 2009;35:1343–1351. doi: 10.1007/s00134-009-1504-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fremerey C, Wiebe B, Feyen O, Lenski C, Pohlmann U, Ehlen M, Schofer O, Meindl A, Niehues T, Bartmann P. ARDS as presenting symptom in an infant with CD40L deficiency (Hyper-IgM syndrome Type 1) Klin Padiatr. 2009;221:302–304. doi: 10.1055/s-0029-1192041. [DOI] [PubMed] [Google Scholar]
- 13.Hu Z, Jin X, Kang Y, Liu C, Zhou Y, Wu X, Liu J, Zhong M, Luo C, Deng L, et al. Angiotensin-converting enzyme insertion/deletion polymorphism associated with acute respiratory distress syndrome among Caucasians. J Int Med Res. 2010;38:415–422. doi: 10.1177/147323001003800204. [DOI] [PubMed] [Google Scholar]
- 14.Flores C, Pino-Yanes MM, Casula M, Villar J. Genetics of acute lung injury: past, present and future. Minerva Anestesiol. 2010;76:860–864. [PubMed] [Google Scholar]
- 15.Jin X, Hu Z, Kang Y, Liu C, Zhou Y, Wu X, Liu J, Zhong M, Luo C, Deng L, et al. Association of interleukin-10–1082 G/G genotype with lower mortality of acute respiratory distress syndrome in a Chinese population. Genet Test Mol Biomarkers. 2011;15:203–206. doi: 10.1089/gtmb.2010.0180. [DOI] [PubMed] [Google Scholar]
- 16.Bajwa EK, Cremer PC, Gong MN, Zhai R, Su L, Thompson BT, Christiani DC. An NFKB1 promoter insertion/deletion polymorphism influences risk and outcome in acute respiratory distress syndrome among Caucasians. PLoS ONE. 2011;6:e19469. doi: 10.1371/journal.pone.0019469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rushefski M, Aplenc R, Meyer N, Li M, Feng R, Lanken PN, Gallop R, Bellamy S, Localio AR, Feinstein SI, et al. Novel variants in the PRDX6 gene and the risk of acute lung injury following major trauma. BMC Med Genet. 2011;12:77. doi: 10.1186/1471-2350-12-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang S, Cao S, Li J, Chang J. Association between vascular endothelial growth factor + 936 genotype and acute respiratory distress syndrome in a Chinese population. Genet Test Mol Biomarkers. 2011;15:737–740. doi: 10.1089/gtmb.2011.0054. [DOI] [PubMed] [Google Scholar]
- 19.Meyer NJ, Li M, Feng R, Bradfield J, Gallop R, Bellamy S, Fuchs BD, Lanken PN, Albelda SM, Rushefski M, et al. ANGPT2 genetic variant is associated with trauma-associated acute lung injury and altered plasma angiopoietin-2 isoform ratio. Am J Respir Crit Care Med. 2011;183:1344–1353. doi: 10.1164/rccm.201005-0701OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Christie JD, Wurfel MM, Feng R, O'Keefe GE, Bradfield J, Ware LB, Christiani DC, Calfee CS, Cohen MJ, Matthay M, et al. Trauma ALI SNP Consortium (TASC) investigators. Genome wide association identifies PPFIA1 as a candidate gene for acute lung injury risk following major trauma. PLoS ONE. 2012;7:e28268. doi: 10.1371/journal.pone.0028268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cruces P, Díaz F, Puga A, Erranz B, Donoso A, Carvajal C, Wilhelm J, Repetto GM. Angiotensin-converting enzyme insertion/deletion polymorphism is associated with severe hypoxemia in pediatric ARDS. Intensive Care Med. 2012;38:113–119. doi: 10.1007/s00134-011-2381-3. [DOI] [PubMed] [Google Scholar]
- 22.Kangelaris KN, Sapru A, Calfee CS, Liu KD, Pawlikowska L, Witte JS, Vittinghoff E, Zhuo H, Auerbach AD, Ziv E, et al. National Heart, Lung, and Blood Institute ARDS Network. The association between a DARC gene polymorphism and clinical outcomes in African American patients with acute lung injury. Chest. 2012;141:1160–1169. doi: 10.1378/chest.11-1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, Agarwal P, Agarwala R, Ainscough R, Alexandersson M, An P, et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 24.Pletcher MT, McClurg P, Batalov S, Su AI, Barnes SW, Lagler E, Korstanje R, Wang X, Nusskern D, Bogue MA, et al. Use of a dense single nucleotide polymorphism map for in silico mapping in the mouse. PLoS Biol. 2004;2:e393. doi: 10.1371/journal.pbio.0020393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu P, Wang Y, Vikis H, Maciag A, Wang D, Lu Y, Liu Y, You M. Candidate lung tumor susceptibility genes identified through whole-genome association analyses in inbred mice. Nat Genet. 2006;38:888–895. doi: 10.1038/ng1849. [DOI] [PubMed] [Google Scholar]
- 26.Kirby A, Kang HM, Wade CM, Cotsapas C, Kostem E, Han B, Furlotte N, Kang EY, Rivas M, Bogue MA, et al. Fine mapping in 94 inbred mouse strains using a high-density haplotype resource. Genetics. 2010;185:1081–1095. doi: 10.1534/genetics.110.115014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang H, Wang JR, Didion JP, Buus RJ, Bell TA, Welsh CE, Bonhomme F, Yu AH, Nachman MW, Pialek J, et al. Subspecific origin and haplotype diversity in the laboratory mouse. Nat Genet. 2011;43:648–655. doi: 10.1038/ng.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Keane TM, Goodstadt L, Danecek P, White MA, Wong K, Yalcin B, Heger A, Agam A, Slater G, Goodson M, et al. Mouse genomic variation and its effect on phenotypes and gene regulation. Nature. 2011;477:289–294. doi: 10.1038/nature10413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yalcin B, Wong K, Agam A, Goodson M, Keane TM, Gan X, Nellåker C, Goodstadt L, Nicod J, Bhomra A, et al. V Sequence-based characterization of structural variation in the mouse genome. Nature. 2011;477:326–329. doi: 10.1038/nature10432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang JR, de Villena FP, Lawson HA, Cheverud JM, Churchill GA, McMillan L. Imputation of single-nucleotide polymorphisms in inbred mice using local phylogeny. Genetics. 2012;190:449–458. doi: 10.1534/genetics.111.132381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leikauf GD, Concel VJ, Liu P, Bein K, Berndt A, Ganguly K, Jang AS, Brant KA, Dietsch M, Pope-Varsalona H, et al. Haplotype association mapping of acute lung injury in mice implicates activin a receptor, type 1. Am J Respir Crit Care Med. 2011;183:1499–1509. doi: 10.1164/rccm.201006-0912OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fabisiak JP, Medvedovic M, Alexander DC, McDunn JE, Concel VJ, Bein K, Jang AS, Berndt A, Vuga LJ, Brant KA, et al. Integrative metabolome and transcriptome profiling reveals discordant energetic stress between mouse strains with differential sensitivity to acrolein-induced acute lung injury. Mol Nutr Food Res. 2011;55:1423–1434. doi: 10.1002/mnfr.201100291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leikauf GD, Pope-Varsalona H, Concel VJ, Liu P, Bein K, Berndt A, Martin TM, Ganguly K, Jang AS, Brant KA, et al. Integrative assessment of chlorine-induced acute lung injury in mice. Am J Respir Cell Mol Biol. 2012;47:234–244. doi: 10.1165/rcmb.2012-0026OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Davy J. On a gaseous compound for carbonic oxide and chlorine. Philos Trans Royal Soc. 1812;102:144–151. [Google Scholar]
- 36.Pauluhn J, Carson A, Costa DL, Gordon T, Kodavanti U, Last JA, Matthay MA, Pinkerton KE, Sciuto AM. Workshop summary: phosgene-induced pulmonary toxicity revisited: appraisal of early and late markers of pulmonary injury from animal models with emphasis on human significance. Inhal Toxicol. 2007;19:789–810. doi: 10.1080/08958370701479133. [DOI] [PubMed] [Google Scholar]
- 37.Grainge C, Rice P. Management of phosgene-induced acute lung injury. Chem Toxicol (Phila) 2010;48:497–508. doi: 10.3109/15563650.2010.506877. [DOI] [PubMed] [Google Scholar]
- 38.Fitzgerald GJ. Chemical warfare and medical response during World War I. Am J Public Health. 2008;98:611–625. doi: 10.2105/AJPH.2007.11930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Organisation for the Prohibition of Chemical WeaponsBrief description of chemical weapons [accessed March 5, 2013]. Available from: http://www.opcw.org/about-chemical-weapons/what-is-a-chemical-weapon
- 40.Organisation for the Prohibition of Chemical WeaponsStatus of participation in the chemical weapons convention as AT 21 May 2009 [accessed March 5, 2013]. Available from: http://www.opcw.org/about-opcw/member-states/status-of-participation-in-the-cwc/
- 41.Ryan TA, Ryan C, Seddon EA, Seddon KR.ed). Topics in inorganic and organic chemistry. Vol. 24: phosgene and related carbonyl halides Amsterdam, The Netherlands: Elsevier; 1996 [Google Scholar]
- 42.Department of Homeland SecurityChemical attack warfare agents, industrial chemicals, and toxins: Table 1. Effects and treatment of some chemical weapons developed for military use [accessed August 15, 2012]. https://www.dhs.gov/files/publications/gc_1243884402361.shtm
- 43.Kang HM, Zaitlen NA, Wade CM, Kirby A, Heckerman D, Daly MJ, Eskin E. Efficient control of population structure in model organism association mapping. Genetics. 2008;178:1709–1723. doi: 10.1534/genetics.107.080101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bennett BJ, Farber CR, Orozco L, Kang HM, Ghazalpour A, Siemers N, Neubauer M, Neuhaus I, Yordanova R, Guan B, et al. A high-resolution association mapping panel for the dissection of complex traits in mice. Genome Res. 2010;20:281–290. doi: 10.1101/gr.099234.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moore EE, Organ CH., Jr. Memorial lecture: splanchnic hypoperfusion provokes acute lung injury via a 5-lipoxygenase-dependent mechanism. Am J Surg. 2010;200:681–689. doi: 10.1016/j.amjsurg.2010.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vadász I, Morty RE, Olschewski A, Königshoff M, Kohstall MG, Ghofrani HA, Grimminger F, Seeger W. Thrombin impairs alveolar fluid clearance by promoting endocytosis of Na+,K+-ATPase. Am J Respir Cell Mol Biol. 2005;33:343–354. doi: 10.1165/rcmb.2004-0407OC. [DOI] [PubMed] [Google Scholar]
- 47.Tremblay LO, Nagy Kovács E, Daniels E, Wong NK, Sutton-Smith M, Morris HR, Dell A, Marcinkiewicz E, Seidah NG, McKerlie C, et al. Respiratory distress and neonatal lethality in mice lacking Golgi alpha1,2-mannosidase IB involved in N-glycan maturation. J Biol Chem. 2007;282:2558–2566. doi: 10.1074/jbc.M608661200. [DOI] [PubMed] [Google Scholar]
- 48.Peng XQ, Damarla M, Skirball J, Nonas S, Wang XY, Han EJ, Hasan EJ, Cao X, Boueiz A, Damico R, et al. Protective role of PI3-kinase/Akt/eNOS signaling in mechanical stress through inhibition of p38 mitogen-activated protein kinase in mouse lung. Acta Pharmacol Sin. 2010;31:175–183. doi: 10.1038/aps.2009.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shao YR, Shen J, Yuan Z, He DK, Zhang L. (Expression and role of mitogen activated protein kinases signaling pathway in lung injury induced by phosgene) (in Chinese) Zhonghua Lao Dong Wei Sheng Zhi Ye Bing Za Zhi. 2012;30:278–283. [PubMed] [Google Scholar]
- 50.Ware LB, Eisner MD, Thompson BT, Parsons PE, Matthay MA. Significance of von Willebrand factor in septic and nonseptic patients with acute lung injury. Am J Respir Crit Care Med. 2004;170:766–772. doi: 10.1164/rccm.200310-1434OC. [DOI] [PubMed] [Google Scholar]
- 51.Meyer NJ, Christie JD. von Willebrand factor and angiopoietin-2: toward an acute lung injury endothelial endophenotype? Crit Care Med. 2012;40:1966–1967. doi: 10.1097/CCM.0b013e31824c8fad. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Uhlen M, Oksvold P, Fagerberg L, Lundberg E, Jonasson K, Forsberg M, Zwahlen M, Kampf C, Wester K, Hober S, et al. Towards a knowledge-based Human Protein Atlas. Nat Biotechnol. 2010;28:1248–1250. doi: 10.1038/nbt1210-1248. [DOI] [PubMed] [Google Scholar]
- 53.Hatch GE, Slade R, Stead AG, Graham JA. Species comparison of acute inhalation toxicity of ozone and phosgene. J Toxicol Environ Health. 1986;19:43–53. doi: 10.1080/15287398609530905. [DOI] [PubMed] [Google Scholar]
- 54.Sciuto AM. Assessment of early acute lung injury in rodents exposed to phosgene. Arch Toxicol. 1998;72:283–288. doi: 10.1007/s002040050503. [DOI] [PubMed] [Google Scholar]
- 55.Hatch GE, Kodavanti U, Crissman K, Slade R, Costa D. An “injury-time integral” model for extrapolating from acute to chronic effects of phosgene. Toxicol Ind Health. 2001;17:285–293. doi: 10.1191/0748233701th123oa. [DOI] [PubMed] [Google Scholar]
- 56.Duniho SM, Martin J, Forster JS, Cascio MB, Moran TS, Carpin LB, Sciuto AM. Acute changes in lung histopathology and bronchoalveolar lavage parameters in mice exposed to the choking agent gas phosgene. Toxicol Pathol. 2002;30:339–349. doi: 10.1080/01926230252929918. [DOI] [PubMed] [Google Scholar]
- 57.Sciuto AM, Carpin LB, Moran TS, Forster JS. Chronological changes in electrolyte levels in arterial blood and bronchoalveolar lavage fluid in mice after exposure to an edemagenic gas. Inhal Toxicol. 2003;15:663–674. doi: 10.1080/08958370390197308. [DOI] [PubMed] [Google Scholar]
- 58.Sciuto AM, Cascio MB, Moran TS, Forster JS. The fate of antioxidant enzymes in bronchoalveolar lavage fluid over 7 days in mice with acute lung injury. Inhal Toxicol. 2003;15:675–685. doi: 10.1080/08958370390197245. [DOI] [PubMed] [Google Scholar]
- 59.Sciuto AM, Clapp DL, Hess ZA, Moran TS. The temporal profile of cytokines in the bronchoalveolar lavage fluid in mice exposed to the industrial gas phosgene. Inhal Toxicol. 2003;15:687–700. doi: 10.1080/08958370390197254. [DOI] [PubMed] [Google Scholar]
- 60.Pauluhn J. Acute nose-only exposure of rats to phosgene. Part I: concentration x time dependence of LC50s, nonlethal-threshold concentrations, and analysis of breathing patterns. Inhal Toxicol. 2006;18:423–435. doi: 10.1080/08958370600563680. [DOI] [PubMed] [Google Scholar]
- 61.Pauluhn J. Acute nose-only exposure of rats to phosgene. Part II: concentration x time dependence of changes in bronchoalveolar lavage during a follow-up period of 3 months. Inhal Toxicol. 2006;18:595–607. doi: 10.1080/08958370600742771. [DOI] [PubMed] [Google Scholar]
- 62.Pauluhn J. Acute head-only exposure of dogs to phosgene. Part III: comparison of indicators of lung injury in dogs and rats. Inhal Toxicol. 2006;18:609–621. doi: 10.1080/08958370600742797. [DOI] [PubMed] [Google Scholar]
- 63.Sciuto AM, Phillips CS, Orzolek LD, Hege AI, Moran TS, Dillman JF., III. Genomic analysis of murine pulmonary tissue following carbonyl chloride inhalation. Chem Res Toxicol. 2005;18:1654–1660. doi: 10.1021/tx050126f. [DOI] [PubMed] [Google Scholar]
- 64.Hu JS, Olsen EN, Kingston RE. HEB, a helix-loop-helix protein related to E2A and ITF2 that can modulate the DNA-binding ability of myogenic regulatory factors. Mol Cell Biol. 1992;12:1031–1042. doi: 10.1128/mcb.12.3.1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lee CC, Chen WS, Chen CC, Chen LL, Lin YS, Fan CS, Huang TS. TCF12 protein functions as transcriptional repressor of E-cadherin, and its overexpression is correlated with metastasis of colorectal cancer. J Biol Chem. 2012;287:2798–2809. doi: 10.1074/jbc.M111.258947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yoon SJ, Wills AE, Chuong E, Gupta R, Baker JC. HEB and E2A function as SMAD/FOXH1 cofactors. Genes Dev. 2011;25:1654–1661. doi: 10.1101/gad.16800511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dahl LK, Heine M, Tassinari L. Role of genetic factors in susceptibility to experimental hypertension due to chronic excess salt ingestion. Nature. 1972;194:480–482. doi: 10.1038/194480b0. [DOI] [PubMed] [Google Scholar]
- 68.Dahl LK, Heine M, Thompson K. Genetic influence of the kidneys on blood pressure: evidence from chronic renal homografts in rats with opposite predispositions to hypertension. Circ Res. 1974;40:94–101. doi: 10.1161/01.res.40.4.94. [DOI] [PubMed] [Google Scholar]
- 69.Herrera VLM, Ruiz-Opazo N. Alteration of alpha-1 Na+,K(+)-ATPase 86R-beta(+) influx by a single amino acid substitution. Science. 1990;249:1023–1026. doi: 10.1126/science.1975705. [DOI] [PubMed] [Google Scholar]
- 70.Ruiz-Opazo N, Barany F, Hirayama K, Herrera VLM. Confirmation of mutant alpha-1 Na,K-ATPase gene and transcript in Dahl salt-sensitive/JR rats. Hypertension. 1994;24:260–270. doi: 10.1161/01.hyp.24.3.260. [DOI] [PubMed] [Google Scholar]
- 71.Herrera VLM, Xie HX, Lopez LV, Schork NJ, Ruiz-Opazo N. The alpha-1 Na,K-ATPase gene is a susceptibility hypertension gene in the Dahl salt-sensitive-HSD rat. J Clin Invest. 1998;102:1102–1111. doi: 10.1172/JCI3868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Barnard R, Kelly G, Manzetti SO, Harris EL. Neither the New Zealand genetically hypertensive strain nor Dahl salt-sensitive strain has an A1079T transversion in the alpha-1 isoform of the Na(+),K(+)-ATPase gene. Hypertension. 2001;38:786–792. doi: 10.1161/hy1001.091782. [DOI] [PubMed] [Google Scholar]
- 73.Mokry M, Cuppen E. The Atp1a1 gene from inbred Dahl salt sensitive rats does not contain the A1079T missense transversion. Hypertension. 2008;51:922–927. doi: 10.1161/HYPERTENSIONAHA.107.108415. [DOI] [PubMed] [Google Scholar]
- 74.Liu YX, Zhou X, Li DQ, Cui QW, Wang GL. Association of ATP1A1 gene polymorphism with heat tolerance traits in dairy cattle. Genet Mol Res. 2010;9:891–896. doi: 10.4238/vol9-2gmr769. [DOI] [PubMed] [Google Scholar]
- 75.Ridge KM, Rutschman DH, Factor P, Katz AI, Bertorello AM, Sznajder JL. Differential expression of Na-K-ATPase isoforms in rat alveolar epithelial cells. Am J Physiol Lung Cell Mol Physiol. 1997;273:L246–L255. doi: 10.1152/ajplung.1997.273.1.L246. [DOI] [PubMed] [Google Scholar]
- 76.Borok Z, Liebler JM, Lubman RL, Foster MJ, Zhou B, Li X, Zabski SM, Kim KJ, Crandall ED. Na transport proteins are expressed by rat alveolar epithelial type I cells. Am J Physiol Lung Cell Mol Physiol. 2002;282:L599–L608. doi: 10.1152/ajplung.00130.2000. [DOI] [PubMed] [Google Scholar]
- 77.Kaplan JH. Biochemistry of Na,K-ATPase. Annu Rev Biochem. 2002;71:511–535. doi: 10.1146/annurev.biochem.71.102201.141218. [DOI] [PubMed] [Google Scholar]
- 78.Lecuona E, Minin A, Trejo HE, Chen J, Comellas AP, Sun H, Grillo D, Nekrasova OE, Welch LC, Szleifer I, et al. Myosin-Va restrains the trafficking of Na+/K+-ATPase-containing vesicles in alveolar epithelial cells. J Cell Sci. 2009;122:3915–3922. doi: 10.1242/jcs.046953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Trejo HE, Lecuona E, Grillo D, Szleifer I, Nekrasova OE, Gelfand VI, Sznajder JI. Role of kinesin light chain-2 of kinesin-1 in the traffic of Na,K-ATPase-containing vesicles in alveolar epithelial cells. FASEB J. 2010;24:374–382. doi: 10.1096/fj.09-137802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Vagin O, Dada LA, Tokhtaeva E, Sachs G. The Na-K-ATPase α₁β₁ heterodimer as a cell adhesion molecule in epithelia. Am J Physiol Cell Physiol. 2012;302:C1271–C1281. doi: 10.1152/ajpcell.00456.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Dada LA, Chandel NS, Ridge KM, Pedemonte C, Bertorello AM, Sznajder JI. Hypoxia-induced endocytosis of Na,K-ATPase in alveolar epithelial cells is mediated by mitochondrial reactive oxygen species and PKC-zeta. J Clin Invest. 2003;111:1057–1064. doi: 10.1172/JCI16826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Comellas AP, Dada LA, Lecuona E, Pesce LM, Chandel NS, Quesada N, Budinger GR, Strous GJ, Ciechanover A, Sznajder JI. Hypoxia-mediated degradation of Na,K-ATPase via mitochondrial reactive oxygen species and the ubiquitin-conjugating system. Circ Res. 2006;98:1314–1322. doi: 10.1161/01.RES.0000222418.99976.1d. [DOI] [PubMed] [Google Scholar]
- 83.Dada LA, Welch LC, Zhou G, Ben-Saadon R, Ciechanover A, Sznajder JI. Phosphorylation and ubiquitination are necessary for Na,K-ATPase endocytosis during hypoxia. Cell Signal. 2007;19:1893–1898. doi: 10.1016/j.cellsig.2007.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lecuona E, Sun H, Vohwinkel C, Ciechanover A, Sznajder JI. Ubiquitination participates in the lysosomal degradation of Na,K-ATPase in steady-state conditions. Am J Respir Cell Mol Biol. 2009;41:671–679. doi: 10.1165/rcmb.2008-0365OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sim NL, Kumar P, Hu J, Henikoff S, Schneider G, Ng PC. SIFT web server: predicting effects of amino acid substitutions on proteins. Nucleic Acids Res. 2012;40:W452–W457. doi: 10.1093/nar/gks539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Glorioso N, Herrera VL, Bagamasbad P, Filigheddu F, Troffa C, Argiolas G, Bulla E, Decano JL, Ruiz-Opazo N. Association of ATP1A1 and dear single-nucleotide polymorphism haplotypes with essential hypertension: sex-specific and haplotype-specific effects. Circ Res. 2007;100:1522–1529. doi: 10.1161/01.RES.0000267716.96196.60. [DOI] [PubMed] [Google Scholar]
- 88.Sverdlov ED, Broude NE, Sverdlov VE, Monastyrskaya GS, Grishin AV, Petrukhin KE, Akopyanz NS, Modyanov NN, Ovchinnikov YA. Family of Na+,K+-ATPase genes: intra-individual tissue-specific restriction fragment length polymorphism. FEBS Lett. 1987;221:129–133. doi: 10.1016/0014-5793(87)80366-6. [DOI] [PubMed] [Google Scholar]
- 89.Guidot DM, Repine MJ, Westcott JY, Repine JE. Intrinsic 5-lipoxygenase activity is required for neutrophil responsivity. Proc Natl Acad Sci USA. 1994;91:8156–8159. doi: 10.1073/pnas.91.17.8156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Cuzzocrea S, Rossi A, Serraino I, Mazzon E, Di Paola R, Dugo L, Genovese T, Calabrò B, Caputi AP, Sautebin L. 5-Lipoxygenase knockout mice exhibit a resistance to pleurisy and lung injury caused by carrageenan. J Leukoc Biol. 2003;73:739–746. doi: 10.1189/jlb.1002477. [DOI] [PubMed] [Google Scholar]
- 91.Caironi P, Ichinose F, Liu R, Jones RC, Bloch KD, Zapol WM. 5-Lipoxygenase deficiency prevents respiratory failure during ventilator-induced lung injury. Am J Respir Crit Care Med. 2005;172:334–343. doi: 10.1164/rccm.200501-034OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ankermann T, Reisner A, Wiemann T, Koehler H, Krams M, Krause MF. Intrapulmonary application of a 5-lipoxygenase inhibitor using surfactant as a carrier reduces lung edema in a piglet model of airway lavage. Pediatr Pulmonol. 2006;41:452–462. doi: 10.1002/ppul.20392. [DOI] [PubMed] [Google Scholar]
- 93.Failla M, Genovese T, Mazzon E, Gili E, Muià C, Sortino M, Crimi N, Caputi AP, Cuzzocrea S, Vancheri C. Pharmacological inhibition of leukotrienes in an animal model of bleomycin-induced acute lung injury. Respir Res. 2006;7:137. doi: 10.1186/1465-9921-7-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Eun JC, Moore EE, Mauchley DC, Johnson CA, Meng X, Banerjee A, Wohlauer MV, Zarini S, Gijón MA, Murphy RC. The 5-lipoxygenase pathway is required for acute lung injury following hemorrhagic shock. Shock. 2012;37:599–604. doi: 10.1097/SHK.0b013e31824ee7bc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lima JJ, Zhang S, Grant A, Shao L, Tantisira KG, Allayee H, Wang J, Sylvester J, Holbrook J, Wise R, et al. Influence of leukotriene pathway polymorphisms on response to montelukast in asthma. Am J Respir Crit Care Med. 2006;173:379–385. doi: 10.1164/rccm.200509-1412OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Telleria JJ, Blanco-Quiros A, Varillas D, Armentia A, Fernandez-Carvajal I, Jesus Alonso M, Diez I. ALOX5 promoter genotype and response to montelukast in moderate persistent asthma. Respir Med. 2008;102:857–861. doi: 10.1016/j.rmed.2008.01.011. [DOI] [PubMed] [Google Scholar]
- 97.Tantisira KG, Lima J, Sylvia J, Klanderman B, Weiss ST. 5-lipoxygenase pharmacogenetics in asthma: overlap with Cys-leukotriene receptor antagonist loci. Pharmacogenet Genomics. 2009;19:244–247. doi: 10.1097/FPC.0b013e328326e0b1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Geiger EV, Doehring A, Kirchhof A, Lötsch J. Functional variants of the human 5-lipoxygenase gene and their genetic diagnosis. Prostaglandins Leukot Essent Fatty Acids. 2009;80:255–262. doi: 10.1016/j.plefa.2009.04.001. [DOI] [PubMed] [Google Scholar]
- 99.Gitler AD, Lu MM, Epstein JA. PlexinD1 and semaphorin signaling are required in endothelial cells for cardiovascular development. Dev Cell. 2004;7:107–116. doi: 10.1016/j.devcel.2004.06.002. [DOI] [PubMed] [Google Scholar]
- 100.Gu C, Yoshida Y, Livet J, Reimert DV, Mann F, Merte J, Henderson CE, Jessell TM, Kolodkin AL, Ginty DD. Semaphorin 3E and plexin-D1 control vascular pattern independently of neuropilins. Science. 2005;307:265–268. doi: 10.1126/science.1105416. [DOI] [PubMed] [Google Scholar]
- 101.Luo Y, Shepherd I, Li J, Renzi MJ, Chang S, Raper JA. A family of molecules related to collapsin in the embryonic chick nervous system. Neuron. 1995;14:1131–1140. doi: 10.1016/0896-6273(95)90261-9. [DOI] [PubMed] [Google Scholar]
- 102.Perälä N, Sariola H, Immonen T. More than nervous: the emerging roles of plexins. Differentiation. 2012;83:77–91. doi: 10.1016/j.diff.2011.08.001. [DOI] [PubMed] [Google Scholar]
- 103.Ito T, Kagoshima M, Sasaki Y, Li C, Udaka N, Kitsukawa T, Fujisawa H, Taniguchi M, Yagi T, Kitamura H, et al. Repulsive axon guidance molecule Sema3A inhibits branching morphogenesis of fetal mouse lung. Mech Dev. 2000;97:35–45. doi: 10.1016/s0925-4773(00)00401-9. [DOI] [PubMed] [Google Scholar]
- 104.Becker PM, Tran TS, Delannoy MJ, He C, Shannon JM, McGrath-Morrow S. Semaphorin 3A contributes to distal pulmonary epithelial cell differentiation and lung morphogenesis. PLoS ONE. 2011;6:e27449. doi: 10.1371/journal.pone.0027449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kang HR, Lee CG, Homer RJ, Elias JA. Semaphorin 7A plays a critical role in TGF-beta1-induced pulmonary fibrosis. J Exp Med. 2007;204:1083–1093. doi: 10.1084/jem.20061273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Hall KT, Boumsell L, Schultze JL, Boussiotis VA, Dorfman DM, Cardoso AA, Bensussan A, Nadler LM, Freeman GJ. Human CD100, a novel leukocyte semaphorin that promotes B-cell aggregation and differentiation. Proc Natl Acad Sci USA. 1996;93:11780–11785. doi: 10.1073/pnas.93.21.11780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Suzuki K, Kumanogoh A, Kikutani H. Semaphorins and their receptors in immune cell interactions. Nat Immunol. 2008;9:17–23. doi: 10.1038/ni1553. [DOI] [PubMed] [Google Scholar]
- 108.Scott A, Wang Z. Tumour suppressor function of protein tyrosine phosphatase receptor-T. Biosci Rep. 2011;31:303–307. doi: 10.1042/BSR20100134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zhang X, Guo A, Yu J, Possemato A, Chen Y, Zheng W, Polakiewicz RD, Kinzler KW, Vogelstein B, Velculescu VE, et al. Identification of STAT3 as a substrate of receptor protein tyrosine phosphatase T. Proc Natl Acad Sci USA. 2007;104:4060–4064. doi: 10.1073/pnas.0611665104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ikegami M, Falcone A, Whitsett JA. STAT-3 regulates surfactant phospholipid homeostasis in normal lung and during endotoxin-mediated lung injury. J Appl Physiol. 2008;104:1753–1760. doi: 10.1152/japplphysiol.00875.2007. [DOI] [PubMed] [Google Scholar]
- 111.Gao H, Guo RF, Speyer CL, Reuben J, Neff TA, Hoesel LM, Riedemann NC, McClintock SD, Sarma JV, Van Rooijen N, et al. Stat3 activation in acute lung injury. J Immunol. 2004;172:7703–7712. doi: 10.4049/jimmunol.172.12.7703. [DOI] [PubMed] [Google Scholar]
- 112.Tantisira KG, Damask A, Szefler SJ, Schuemann B, Markezich A, Su J, Klanderman B, Sylvia J, Wu R, Martinez F, et al. Genome-wide association identifies the T gene as a novel asthma pharmacogenetic locus. Am J Respir Crit Care Med. 2012;185:1286–1291. doi: 10.1164/rccm.201111-2061OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Linnen JM, Bailey CP, Weeks DL. Two related localized mRNAs from Xenopus laevis encode ubiquitin-like fusion proteins. Gene. 1993;128:181–188. doi: 10.1016/0378-1119(93)90561-g. [DOI] [PubMed] [Google Scholar]
- 114.Sok J, Calfon M, Lu J, Lichtlen P, Clark SG, Ron D. Arsenite-inducible RNA-associated protein (AIRAP) protects cells from arsenite toxicity. Cell Stress Chaperones. 2001;6:6–15. doi: 10.1379/1466-1268(2001)006<0006:airapa>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Stanhill A, Haynes CM, Zhang Y, Min G, Steele MC, Kalinina J, Martinez E, Pickart CM, Kong XP, Ron D. An arsenite-inducible 19S regulatory particle-associated protein adapts proteasomes to proteotoxicity. Mol Cell. 2006;23:875–885. doi: 10.1016/j.molcel.2006.07.023. [DOI] [PubMed] [Google Scholar]
- 116.Rossi A, Trotta E, Brandi R, Arisi I, Coccia M, Santoro MG. AIRAP, a new human heat shock gene regulated by heat shock factor 1. J Biol Chem. 2010;285:13607–13615. doi: 10.1074/jbc.M109.082693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Huang J, Teng L, Li L, Liu T, Li L, Chen D, Xu LG, Zhai Z, Shu HB. ZNF216 Is an A20-like and IkappaB kinase gamma-interacting inhibitor of NFkappaB activation. J Biol Chem. 2004;279:16847–16853. doi: 10.1074/jbc.M309491200. [DOI] [PubMed] [Google Scholar]
- 118.Hishiya A, Ikeda K, Watanabe K. A RANKL-inducible gene Znf216 in osteoclast differentiation. J Recept Signal Transduct Res. 2005;25:199–216. doi: 10.1080/10799890500240781. [DOI] [PubMed] [Google Scholar]
- 119.He G, Sun D, Ou Z, Ding A. The protein Zfand5 binds and stabilizes mRNAs with AU-rich elements in their 3′-untranslated regions. J Biol Chem. 2012;287:24967–24977. doi: 10.1074/jbc.M112.362020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Palmer EL, Rüegg C, Ferrando R, Pytela R, Sheppard D. Sequence and tissue distribution of the integrin alpha 9 subunit, a novel partner of beta 1 that is widely distributed in epithelia and muscle. J Cell Biol. 1993;123:1289–1297. doi: 10.1083/jcb.123.5.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Taooka Y, Chen J, Yednock T, Sheppard D. The integrin alpha9beta1 mediates adhesion to activated endothelial cells and transendothelial neutrophil migration through interaction with vascular cell adhesion molecule-1. J Cell Biol. 1999;145:413–420. doi: 10.1083/jcb.145.2.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Vlahakis NE, Young BA, Atakilit A, Hawkridge AE, Issaka RB, Boudreau N, Sheppard D. Integrin alpha9beta1 directly binds to vascular endothelial growth factor (VEGF)-A and contributes to VEGF-A-induced angiogenesis. J Biol Chem. 2007;282:15187–15196. doi: 10.1074/jbc.M609323200. [DOI] [PubMed] [Google Scholar]
- 123.Nishimichi N, Hayashita-Kinoh H, Chen C, Matsuda H, Sheppard D, Yokosaki Y. Osteopontin undergoes polymerization in vivo and gains chemotactic activity for neutrophils mediated by integrin alpha9beta1. J Biol Chem. 2011;286:11170–11178. doi: 10.1074/jbc.M110.189258. [DOI] [PMC free article] [PubMed] [Google Scholar]