Abstract
Planarian adult stem cells (pASCs) or neoblasts represent an ideal system to study the evolution of stem cells and pluripotency as they underpin an unrivaled capacity for regeneration. We wish to understand the control of differentiation and pluripotency in pASCs and to understand how conserved, convergent or divergent these mechanisms are across the Bilateria. Here we show the planarian methyl-CpG Binding Domain 2/3 (mbd2/3) gene is required for pASC differentiation during regeneration and tissue homeostasis. The genome does not have detectable levels of 5-methylcytosine (5mC) and we find no role for a potential DNA methylase. We conclude that MBD proteins may have had an ancient role in broadly controlling animal stem cell pluripotency, but that DNA methylation is not involved in planarian stem cell differentiation.
Keywords: Methyl binding domain, Pluripotency, Differentiation, Regeneration
Highlights
-
•
A single ancestral MBD2/3 protein is present in the planarian Schmidtea mediterranea.
-
•
The genome of S. mediterranea does not have pervasive cytosine methylation.
-
•
MBD2/3 is required for pluripotent stem cell differentiation down multiple but not all cell lineages.
-
•
MBD2/3 may have had an ancestral role in regulating stem cell pluripotency.
Introduction
The precise features that stem cells of different animals have in common are unknown. Of particular interest is whether or not the mechanisms that control pluripotency are deeply conserved across animal stem cells. The life history of planarian flatworms provides a model system within which to assess the extent to which conserved genetic networks might control pluripotency across the Bilateria. These animals contain large numbers of adult stem cells that account for their ability for profound regenerative capacity. Recent genome wide expression studies have suggested that the mechanisms controlling pluripotency in planarian adult stem cells (pASC) may have much in common with both mammalian embryonic stem cells (ESCs) and germ stem cells (Aboobaker, 2011; Aboobaker and Kao, 2012; Labbe et al., 2012; Önal et al., 2012; Solana et al., 2012; Wagner et al., 2012; Zeng et al., 2013). This suggests that the intriguing possibility of both deep conservation of molecular mechanisms and a blurring of the barrier between germ line and somatic stem cells (Aboobaker and Kao, 2012; Solana, 2013).
While pASCs are known to be pluripotent (Wagner et al., 2011), there are currently few detailed studies that demonstrate potential roles for conserved genes in pluripotency and differentiation (Bonuccelli et al., 2010; Hubert et al., 2013; Önal et al., 2012; Scimone et al., 2010; Zeng et al., 2013). The existence of markers of many differentiated cell types and the elucidation of at least some stem cell progeny markers provides the tools necessary to look for genes that control the epigenetic and transcriptional mechanisms involved in pASC differentiation (Eisenhoffer et al., 2008)
Two recent studies reported roles for conserved epigenetic regulatory proteins in pASC biology, the Schmidtea mediterranea Mi2/CHD4 ortholog Smed-CHD4 and the Dugesia japonica RbAp48 ortholog (Bonuccelli et al., 2010; Scimone et al., 2010). While both of these proteins are present in more than one chromatin related complex, both are components of the widely conserved nucleosome remodeling and histone deacetylase (NuRD) complex which is required for correct epigenetic control of cell fates in multiple tissues in many organisms. This complex is formed by at least 7 proteins in mammals: HDAC1 and 2, RbAp46 and 48, MTA1/2, Mi-2 and MBD3/MBD2 (Denslow and Wade, 2007; Xue et al., 1998; Zhang et al., 1999). By altering chromatin structure, the NuRD complex contributes to the capacity of embryonic stem cells to preserve their undifferentiated proliferative state while maintaining capacity to differentiate (Denslow and Wade, 2007; McDonel et al., 2009; Xue et al., 1998; Zhang et al., 1999). MBD3 is one member of the NuRD complex that has not yet been described as being active in other conserved chromatin associated complexes and thus can be studied to investigate specific effects of NuRD. MBD3 belongs to a family of proteins that contain a conserved methyl-CpG binding domain (MBD) and include MeCP2, MBD1, MBD2, MBD3 and MBD4 in vertebrates, with the first four being thought to play a role in transcriptional repression (Hendrich and Bird, 1998; Jaenisch and Bird, 2003). Although a member of this family MBD3 does not itself bind methylated DNA (Hendrich and Bird, 1998; Ohki et al., 2001), which is a key genome wide component of in the control of both pluripotency and differentiation programs in mammals (Boyer et al., 2005; Feldman et al., 2006; Meissner et al., 2008; Mohn et al., 2008). To date there is no direct data to address how or whether either MBD proteins or DNA methylation play a role in pASCs or planarian regeneration.
Here we investigated whether methylation or genes associated with the methylation machinery have a role in planarians. At least a single ancestral MBD encoding gene MBD2/3, orthologous to MBD2 and MBD3, is thought to have been present in the ancestral bilaterian (Albalat, 2008; Gutierrez and Sommer, 2004, 2007; Marhold et al., 2004a, 2004b; Matsumoto and Toraya, 2008; Tweedie et al., 1999) and we have identified one of these in S. mediterranea. Our results demonstrate that the single Smed-mbd2/3 present in planarians is required to allow pASCs to differentiate correctly. Without Smed-mbd2/3 pASCs are able to proliferate despite the loss of differentiated tissues. We observe that early post-mitotic stem cell progeny are formed but they do not differentiate further. We also show that Smed-mbd2/3 identified in planarians is unlikely to function by binding methylated DNA as it does not have the conserved residues for binding methylated DNA (Ohki et al., 2001), we do not detect methylation in the genome and we find no function for the single potential DNA methylase found in the genome. Together our findings uncover a potentially ancient role for MBD2/3 proteins in sustaining the pluripotency of animal stem cells independently of DNA methylation.
Materials and methods
Planarian culture
S. mediterranea of both the asexual and sexual strains originally provided by Professor Emili Salo from the University of Barcelona were maintained at 20 °C in tap water filtered through activated charcoal and buffered with 0.5 ml/L 1 M NaHCO3. Planarians were fed veal liver and starved for at least one week prior to experiments or amputation as previously described (Gonzalez-Estevez et al., 2009).
Irradiation to remove proliferative cells
Whole worms were starved for 1 week prior to gamma irradiation as previously described (Gonzalez-Estevez et al., 2009).
Cloning and identification of Smed-mbd2/3 and Smed-dnmt2
To identify planarian homologues of MBD and DNA methyl-transferase proteins we searched a local database of Version 3.1 of the S. mediterranea Genome Project (http://genome.wustl.edu/genomes) (Robb et al., 2008) as well as local transcriptome data (Blythe et al., 2010). Sequence data was supplemented by using RACE (Ambion RLM Race Kit) to generate full-length cDNAs. S. mediterranea genes were aligned with MBD (see Supplementary Table 1) and DNMT proteins from other species and phylogenetic reconstruction was conducted using maximum likelihood. The Smed-mbd2/3 and Smed-dnmt-2 sequences have been submitted to GenBank with accession numbers KF664582 and KF664583.
RNAi Experiments and phenotypic scoring
Primers used to generate fragments used for dsRNA production were Smed-mbd2/3F and Smed-mbd2/3R for mbd2/3(RNAi); Smed-dnmt2F and Smed-dnmt2R for Smed-dnmt2(RNAi); DsRed F and DsRed R for Control(RNAi). For mbd2/3(RNAi) specificity experiment, F1mbd-Spec and R1mbd-Spec were used for the first fragment (mbdF1R1(RNAi)); F2mbd-Spec and Smed-mbd2/3R2 were used for the second fragment (mbdF2Rex(RNAi)). All primer sequences are in Supplementary Table 2.
dsRNAs were produced and used as previously described (Felix and Aboobaker, 2010). DsRNA from the DsRed gene at matched concentration was used as a control. Animals were either observed for tissue homeostasis or amputated to observe regeneration. One round of injection resulted in weaker penetrance and was not pursued.
In situ hybridizations and cell counting
Primers used to generate fragments used for probe production are Smed-mbd2/3F and Smed-mbd2/3Rex for mbd2/3 probe; Smed-dnmt2F and Smed-dnmt2R2 for Smed-dnmt2 probe; nb2111ePWR F and nb2111ePWR R for Smed-nb.21.11e probe; F-nb.32.1g-fj5′ and R-nb.32.1g-fj3′ for Smed-nb.32.1g probe; F-cytP450-fj5′ and R-cytP450-fj3′ for Smed-CYP1A1-1 probe. Sequences of these primers are listed in Supplementary Table 1. Primers used to make other in situ hybridisation probes were taken from their corresponding references.
Whole mount in situ hybridization was carried out on intact non-irradiated, irradiated worms and regenerating pieces which were fixed and stained using methods previously described (Gonzalez-Estevez et al., 2009). The following probes were used: Smedwi-2, Smed-HistoneH2B, Smed-GluR, Smed-cintillo, Smed-eye53, Smed-TCEN49, Smed-laminin, Smed-porcn-1, Smed-CAVII-1, Smed-NB.21.11e, Smed-AGAT-1, Smed-NB.32.1g and Smed-CYP1A1-1 (Cebria and Newmark, 2005, 2007; Eisenhoffer et al., 2008; Gurley et al., 2008; Inoue et al., 2004a; Rink et al., 2011). Bright field images were taken on a Zeiss Discovery V8 from CarlZeiss using Axio Cam MRC from Carl Zeiss. In situ hybridisation on paraffin sections was performed as previously described (Cardona et al., 2005). Fluorescence in situ hybridization was performed as described elsewhere (Gonzalez-Estevez et al., 2009). For double fluorescence in situ hybridisation worms were incubated with DIG-labeled Smed-AGAT-1 probe and fluorescein-labeled Smed-NB.21.11e probes. After developing the first probe, peroxidase activity was quenched with 1% H2O2 in PBS-0.1% Triton-100 (TPBS) for 1.5 h. The number of cells expressing either Smed-NB.21.11e or Smed-AGAT-1 was counted from the region anterior to the pharynx on the dorsal side and normalized to the area of that region in pixels using imaging with a Leica SP2 confocal microscope (CLSM, Leica Lasertechnik, Heidelberg).
Quantiative real time PCR (qRT-PCR) analysis of gene expression levels
cDNA was synthesized from RNA extracts by using SuperScriptIII RT enzyme (Invitrogen). A total of 18 ng of cDNA per data point was used to perform Real-time RT-PCR using the Absolute QPCR SYBR Green master mix (Thermo Scientific) and the gene Smed-cystatin was used for normalization. All experiments were performed in triplicate. Primers used are listed in Supplementary Table 3. A Student's t-test was performed to test for significance at specific time points.
Immunohistochemistry
For immunostaining animals were killed, fixed and processed as previously described (Cebria and Newmark, 2007). The worms were stained with: anti-SYNORF1 3C11 (Developmental Studies Hybridoma Bank, dilution 1:100), anti-acetylated tubulin (Developmental Studies Hybridoma Bank, dilution 1:100), anti-arrestin VC-1 (kindly provided by Hidefumi Orii, dilution 1:15,000), anti-phosphotyrosine (Cell Signaling), and anti-phospho-Serine10 Histone H3 (H3P) (Upstate, dilution 1:1000). Secondary antibodies used were: goat anti-mouse antibody conjugated to Alexa 488 or Alexa 568 (Molecular Probes, used at 1:400 dilution) and goat anti-rabbit antibody conjugated to Alexafluor 568 (Molecular Probes, 1:1000). Fluorescent images were taken on a Leica MZ16F fluorescence stereomicroscope using Leica DFC 300Fx camera (Leica Lasertechnik, Heidelberg). Confocal laser scanning microscopy was performed with a Leica SP2 confocal microscope (CLSM, Leica Lasertechnik, Heidelberg).
Analysis of proliferation
To assess proliferation, worms were stained with anti-H3P. For each regenerating piece or homeostatic worm the number of mitotic cells and the area in mm2 was determined using Adobe Photoshop CS4, and used to calculate the individual mitotic index. The standard error was calculated for the pooled regeneration and homeostasis time points and a Student's t-test was performed to test for significance at specific time points.
Immunohistochemistry for 5mC
Paraffin-embedded formaldehyde-fixed sections of adult wild type Drosophila and S. mediterranea were used for immunohistochemistry (Almeida et al., 2012). Tissue sections were de-waxed according to standard procedures. Cells and tissue sections were permeabilised for 15 min with PBS containing 0.5% Triton X-100, incubated in 4 N HCl for 1 h at 37 °C and then neutralised in 100 mM Tris–HCl (pH 8.5) for 10 min, followed by a standard immunostaining protocol. Anti-5-mC (Eurogentec) antibody was used at 1:200 dilution followed by anti-mouse HRP (Roche, 1:300) and reacted with FITC tyramide (Perkin Elmer). DNA was visualized using propidium iodide (PI) at 1 µg/ml.
Methylation-dependent restriction digestion
Digestions were performed using the manufacturer's guidelines for McrBC (NEB) using 1 µg of gDNA. Human gDNA from the AF11 lymphoblastoid cell line was used as a positive control.
Methylation-sensitive and insensitive restriction digests
A panel of methylation sensitive and methylation insensitive isoshizomers (NEB), were use to digest between 2 and 2.5 μg of S. medierranea gDNA (see Supplementary Table 4 for details).
Enzymatic digestion of Schmidtea mediterranea gDNA
Genomic DNA was prepared from S. mediterranea, which had been starved for at least 2 weeks weeks to minimize any potential dietary source of methylated DNA. Worms were disrupted by homogenisation and DNA purified by two rounds of phenol-chloroform extraction and ethanol precipitation. An RNase treatment was included between the two extractions. Up to 5 mg of purified DNA was digested to nucleosides for subsequent LC–MS analysis. Each 20 ml reaction contained S. mediterranea DNA, 5 units of Antarctic Phosphatase (NEB) and 2 mU Snake venom Phosphodiesterase (Phosphodiesterase I from Crotalus adamentus venom; Sigma) in a buffer that was 0.5×Micrococcal Nuclease Buffer (NEB) and 0.5×Antarctic Phosphatase Buffer (NEB). The digestions were performed at 37 °C for 16 h then stored at −20 °C. Control digests were performed on methylated DNA samples (human Jurkat cell line DNA, lambda DNA and pBR322; all from NEB), as well as an un-methylated DNA (pUC19 DNA prepared after propagation in dcm-, dam- Escherichia coli; a gift from A. Formenkov). Mock digests lacking any DNA substrate were also performed.
Nucleoside analysis
Samples of nucleosides prepared as described above were analyzed by reverse phase liquid chromatography (LC) and electrospray ionization time-of-flight mass spectrometry (ESI-TOF MS). A reverse phase column, 1×150 mm, Develosil RP-Aqueous C30, 3 mm particles, 140 A pore size (Phenomenex), was developed at a flow rate of 20 ml/min at 30 °C using an Agilent 1100 capillary LC connected directly to an Agilent 6210 series ESI-TOF MS. The column was equilibrated with 50 mM ammonium acetate, pH 6 in water. Up to eight ml of nucleoside sample was injected onto the column that was developed after 2 min at initial conditions with a 15-min linear gradient from 0% to 22.5% acetonitrile and then held at 22.5% acetonitrile for an additional 5 min. Nucleosides were found to elute at approximately 16 to 23 min after injection. The mass spectra were acquired from 100 to 400 m/z, one cycle/s and 10,000 transients per scan at 4000 V, 300 °C and drying gas 7.0 l/min and nebulizer 15 psi and a fragmentor voltage of 215. The acquired spectra were extracted with Agilent MassHunter Qualitative Analysis Software (with Bioconfirm) B 2.0.2 software using mass ranges of 136.0–136.1, 112.0–112.1, 152.0–152.1, 127.0–127.1, 126.0–126.2, 142.0–142.2, 305.1–305.2 for the liberated base fragments of dA, dC, dG, dT, d(5m)C, d(5hm)C and d(5Glucose,hm)C, respectively.
Results
A single methyl binding domain protein expressed in planarian stem cells
In S. mediterranea, we identified a single MBD containing gene most closely related to the MBD2/3 family of proteins in available genomic and transcriptomic data {Blythe, 2010 #52}{Robb, 2008 #56}. Supported by phylogenetic analysis (Supplementary Fig. 1A) we have called this gene Smed-mbd2/3 (in short mbd2/3). The sequenced genomes of pre-bilaterian lineages represented by Trichoplax adherens, Amphimedon queenslandica and Nematostella vectensis all had a single methyl binding domain protein in the MBD2/3 family, showing that at least single gene was present in the metazoan ancestor (Supplementary Table 1, Supplementary Fig. 1A). We found no members of other methyl binding domain protein families in these genomes. Smed-MBD2/3 had sequence similarity with the vertebrate MBD2 and MBD3 proteins at the C-terminal outside the MBD domain, as expected (Supplementary Fig. 1B). We found that Smed-MBD2/3 is unlikely to bind 5mC due to the presence of a lysine residue instead of an arginine residue at position 17 in its MBD (Ohki et al., 2001) (Supplementary Fig. 1C). Significantly this is a different residue substitution to the one thought to result in the inability of mouse MBD3 to bind methylated DNA.
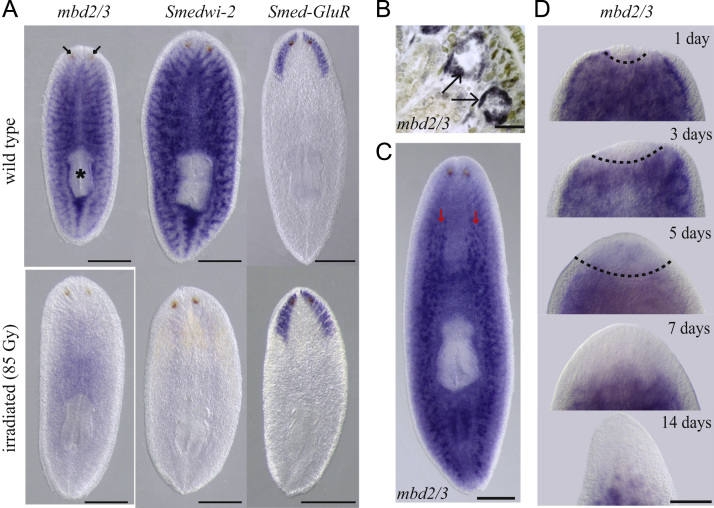
We observed irradiation sensitive parenchymal mbd2/3 expression in intact animals, absent from regions in front of the eyes and from the pharyngeal region, as described for genes expressed in pASCs such as Smedwi-2 (Reddien et al., 2005) (Fig. 1A). The expression of Smed-GluR expressed in the differentiated brain cells was unaffected by irradiation (Fig. 1A). Additionally we were also able to detect mbd2/3 expression in the germline, a feature shared with many genes expressed in pASCs (Fig. 1B and C). During regeneration we noted a lack of transcript expression in the early blastema suggesting that mbd2/3 is not expressed in stem cell progeny (Fig. 1D). Blastemas remain clear of mbd2/3 expression through to 5 days when faint expression is detectable as pASCs repopulate the differentiated anterior region of the regenerating animal. At seven days of regeneration and later the pattern observed in intact planarians is restored (Fig. 1D). Overall these data suggest that mbd2/3 transcript expression is confined to proliferating pASCs and germ line cells.
Fig. 1.
mbd2/3 Is expressed in stem cells and the germline. (A) mbd2/3 is expressed cells throughout the parenchyma but absent from the pharynx (black asterisk) and anterior to the eyes (black arrows) and is lost by irradiation similar to the pASC marker gene Smedwi-2. Smed-GluR (glutamate receptor) expression is unaffected by irradiation. Smed-mbd2/3 is expressed in the germline, shown is expression in testes primordia of juvenile animals in paraffin sections (B) and in the testes whole mount in mature sexual animals (red arrows). Scale bars, 50 µm in (B) 500 µm. in (C). (D) Expression of mbd2/3 is absent from the regeneration blastema through 3 days of regeneration, but faint expression can be detected in 5 day old blastema and later the worms restore normal mbd2/3 distribution. Dashed lines show the boundary between old and new tissue. Scale bar, 500 µm.
mbd2/3 Is required for the correct differentiation during regeneration
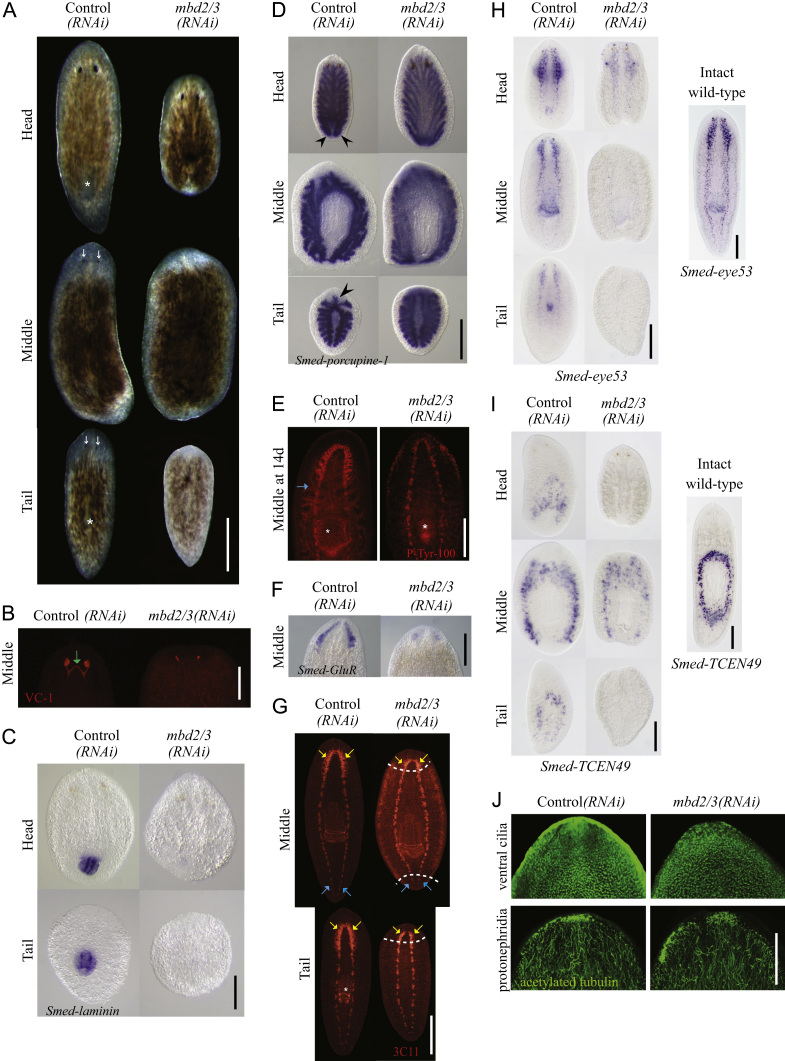
We performed mbd2/3(RNAi) and checked the efficacy with qRT-PCR (Supplementary Fig. 2A–C). We observed mbd2/3 transcripts in mbd2/3(RNAi) animals levels were reduced to between 2 and 9% of control(RNAi) levels. A summary of phenotypic progression during both regeneration and homeostasis is presented in Supplementary Tables 5 and 6. We found that animals were able to form both anterior and posterior blastemas (Fig. 2A, Supplementary Fig. 2D). However, both trunks (77%) and tail fragments (91%) failed to make any visible eyes after 7 days of regeneration (Fig. 2A, Supplementary Fig. 2D). After 14 days of regeneration some eye pigment cells were observed in 60% of middle pieces and 38% of tail pieces in the still un-pigmented head region, but in all cases these were greatly reduced (when present at all) compared to conrol(RNAi) controls. To confirm the specificity of this phenotype we also performed RNAi with a second region of the gene and observed the same phenotype (Supplementary Fig. 2E). We confirmed our observation of only partial eye regeneration by staining with the VC-1 monoclonal antibody, which labeled just small set of eye specific cells (7/15 regenerating middle pieces, no staining in remainder) in mbd2/3(RNAi) animals but labeled completely regenerated visual neurons in control(RNAi) animals (Fig. 2B).
Fig. 2.
mbd2/3(RNAi) Leads to the disruption of various differentiated tissues during regeneration. (A) mbd2/3(RNAi) effect at 7 days of regeneration showing defects in pharynx (white asterisk) and eye (white arrows) formation. (B)–(F) mbd2/3(RNAi) affects the formation of the eyes (B), pharynx (C), anterior and posterior gut branches (black arrow heads) (D) and (E), brain ganglia (F) and (G); yellow arrows in (G), cells in and around the CNS (H) and secretory gland cells around the pharynx (I) during regeneration. However, the VNCs (G), blue arrows), cilia and secretory cell types are formed during regeneration. White dashed lines represent the boundary between the old and new tissue. Immunostaining was performed in (B), (E), (G) and (J), in situ hybridisation in (C), (D), (F), (H) and (I). 3C11: anti-SYNORF-1.
Head and tail fragments showed pharynx regeneration defects (Fig. 2A, Supplementary Fig. 2D). To confirm these defects we used the pharyngeal marker Smed-laminin. We were able to detect clear expression in control(RNAi) animals (10/10) but no expression in mbd2/2(RNAi) animals (10/10) (Fig. 2C). We also observed that mbd2/3(RNAi) animals failed to correctly remodel and regenerate the gut (Fig. 2D and E). Whereas control(RNAi) animals correctly regenerated gut branches labeled with Smed-porcupine-1 (6/6) and diverticulae labeled with an anti-phosphotyrosine antibody (5/5), mbd2/3(RNAi) animals were unable to do either (5/5 and 5/5 animals, respectively, Fig. 2D and E).
We found that in both regenerating trunks and tails, mbd2/3(RNAi) led to greatly reduced or entirely absent brain ganglia (Fig. 2A and E–G, 20/20 regenerating middles, 20/20 regenerating tails, anti-SYNORF1 staining, 20/20 animals both middles and tails, anti-phosphotyrosine staining 5/5 middles, Smed-GluR 10/10 middles). Significantly, we were able to detect regeneration of the ventral nerve cords using both anti-phosphotyrosine staining (Fig. 2E 5/5 animals) and anti-SYNORF 3C11 (Fig. 2G, 20/20 animals middles and tails).
With Smed-eye53, which labels a subset of cells in and around the planarian CNS (Inoue et al., 2004b) and Smed-TCEN49, which labels a set of secretory gland cells positioned around the pharynx (Iglesias et al., 2008), and we observed that mbd2/3(RNAi) animals failed to reconstitute these cells during regeneration (Fig. 2H and I, 20/20 trunks and tails for Smed-TCEN-49, 20/20 heads and tails for Smed-eye53).
Taken together, our data suggest that that mbd2/3 is required to allow pASCs to produce many different differentiated cell types. We observe that mbd2/3(RNAi) animals can form blastemas, albeit smaller than controls. This predicts that stem cell progeny are produced and are able to migrate to the wound site, but that this population fails to correctly differentiate. Since we observed that animals are able to make VNCs we wished to know if other cell types and tissues are still produced by mbd2/3(RNAi) animals. To do this we used anti acetylated tubulin to look at both ciliated cells and protonephridia (excretory organs). We found that both these cell types are regenerated in mbd2/3(RNAi) animals (Fig. 2J). As anti-acetylated tubulin labels many types of secretory cells (which do regenerate in mbd2/3(RNAi) animals) we also used the protonephridia specific marker Smed-CAVII-1. This marker also supported that protonephridia can be regenerated (Supplementary Fig. 2F, 7/7 animals). Thus although mbd2/3(RNAi) has broad affects on regeneration, it appears some cell types do differentiate correctly.
mbd2/3 Is required for the maintenance of differentiated tissues during tissue homeostasis
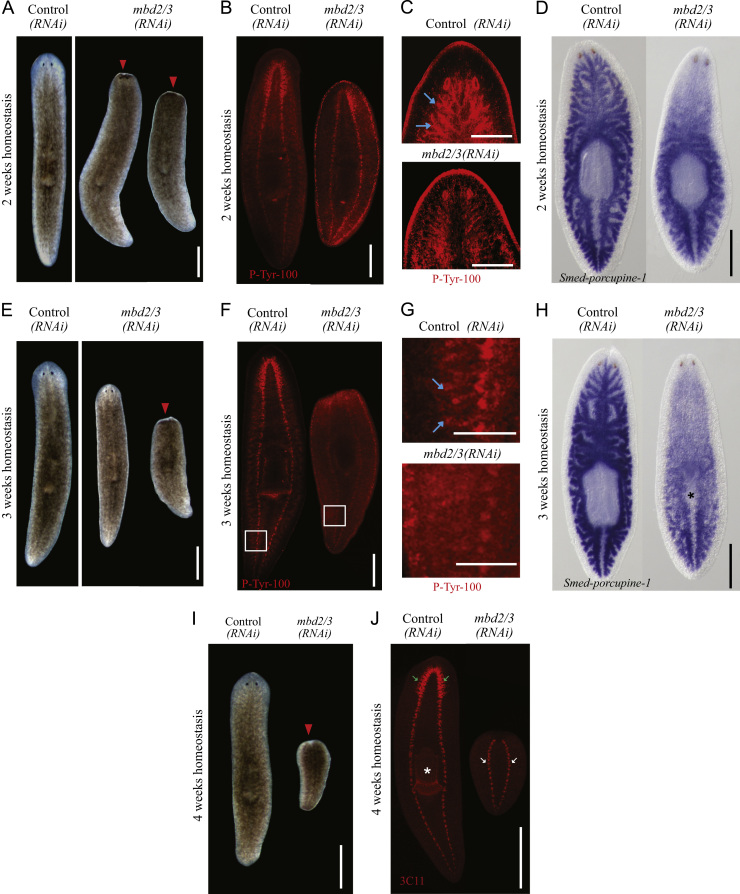
The widespread differentiation defects observed in mbd2/3(RNAi) worms during regeneration were also apparent in worms that were left to undergo normal tissue homeostasis. After 2 weeks of homeostasis (equivalent to 14 days of regeneration) most animals started to show some degree of anterior regression, starting with the anterior tip and anterior lateral margins (Fig. 3A). We found that head regression correlated with a loss of brain ganglia cells (Fig. 3B). At 2 weeks of homeostasis we also observed loss of anterior gut branches, which correlated with the regression of medial tissue between the brain ganglia (Fig. 3C, 9/10 animals and Fig. 3D, 5/5 animals).
Fig. 3.
mbd2/3(RNAi) Leads to disruption of tissue homeostasis. Most mbd2/3(RNAi) animals start to regress anterior regions by 2 weeks varying from mild to more extreme phenotypes (A). Head regression correlates with loss of cephalic ganglia (B) and the anterior gut branches (C) and (D). At 3 weeks almost all animals have completely lost anterior regions (red arrow-head), although a few display only mild anterior regression (E). By three weeks posterior gut branches have also started to be lost (F)–(H). While gut branches (blue arrows in (G)) and the pharynx (stars in (H) and (J)) are lost, the VNCs (white arrows in (J)) are maintained (F)–(H) and (J). Animals at 4 weeks have regressed further, but are mostly still alive (I). Scale bars 500 µm, except in C and G 200 µm. Panels in G represent regions in white boxes depicted in panel F.
We observed a complete loss of the head and eyes in 37% of worms by 3 weeks, at which time small pieces resembling amputated tails were observed (Fig. 3E). Some animals displayed only very slight signs of head regression even by 3 weeks of homeostasis (Fig. 3E). mbd2/3(RNAi) animals continued to decrease in size (and eventually started to die at 4 weeks of homeostasis, 40 days after the first RNAi injections.
We also observed the gradual loss of the major posterior and peripheral gut branches, with all animals losing gut branches by 3 weeks of homeostasis (Fig. 3F–H 25/25 animals). The strength of the homeostatic phenotypes observed increased with time and we observed loss of the pharynx as animals regressed to tail stumps by 4 weeks (Fig. 3I and J). In animals that had regressed to small tail stumps we were still able to see clear labeling of VNCs, suggesting that these cells are maintained or can still be produced by stem cells. From these experiments we conclude that mbd2/3 is required for both regeneration and maintenance of numerous differentiated cell types and tissues.
mbd2/3 Is not required for ongoing stem cell proliferation or maintenance
We wished to explain the underlying cause of regenerative and homeostatic defects in mbd2/3(RNAi) worms. One possibility would be the pASC cell population is either lost or unable to proliferate correctly.
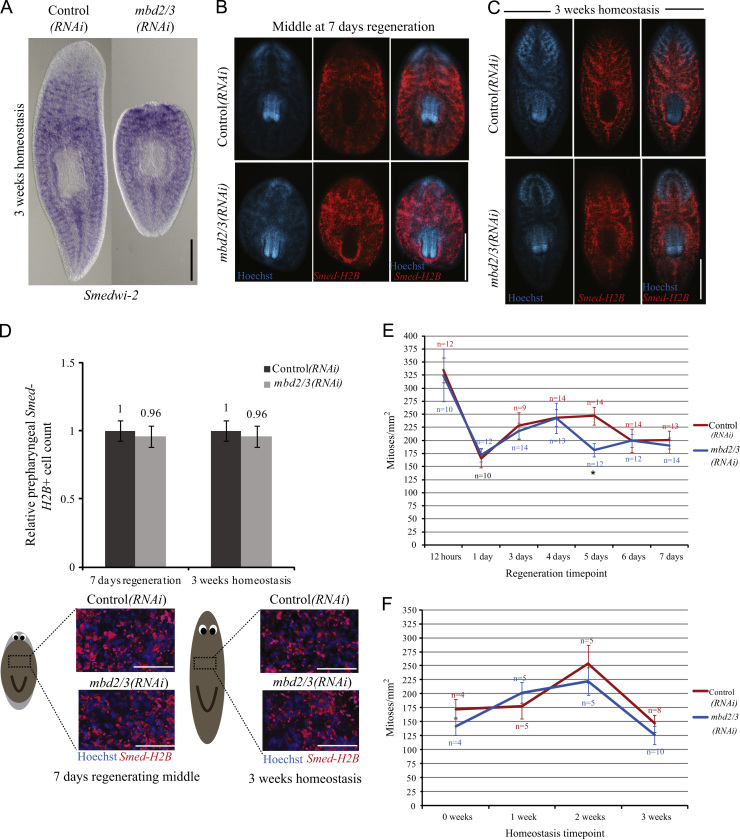
Previous studies in planarians have described genes that are required for stem cell differentiation and in each case stem cell maintenance and proliferation are also affected (Bonuccelli et al., 2010; Reddien et al., 2005; Scimone et al., 2010). We assessed pASC maintenance and proliferation using markers of pASCs Smedwi-2 (Reddien et al., 2005) and Smed-H2B (Guo et al., 2006; Solana et al., 2012) both expressed in proliferating stem cells and by counting mitotic cells.
We observed no effect on the distribution, proliferation or relative numbers of pASCs during tissue homeostasis or regeneration in mbd2/3(RNAi) animals relative to their controls (Fig. 4A–F; Supplementary Fig. 3A–C). Even animals that had regressed to tail stumps after 3 weeks of homeostasis had normal Smedwi-2 expression in pASCs and normal levels of mitosis (Fig. 4A and Supplementary Fig. 3A). We subjected mbd2/3(RNAi) animals to a further round of standard RNAi injections and regeneration and also observed no defect in proliferation (Supplementary Fig. 3C).
Fig. 4.
mbd2/3(RNAi) Does not affect pASC maintenance or proliferation. (A) The distribution of pASCs assessed by Smedwi-2 expression found to be consistent between mbd2/3(RNAi) animals and their controls during homeostasis even in animals with regressed anterior regions. Smed-H2B expressing cells are also maintained in regenerating and animals undergoing normal homeostasis (B) and (C). Numbers of Smed-H2B expressing cells are not significantly different from controls in these animals (D). Numbers of mitotic cells are not significantly different from controls during regeneration or homeostasis, except at 5 days of regeneration (E) and (F). Scale bars 500 µm, accept in (C) and (D) 100 µm. ⁎ Indicates p<0.05, determined by Student's t-test. Standard error bars are shown.
We conclude that pASC proliferation and maintenance do not require mbd2/3. Taken together our data suggest that the mbd2/3(RNAi) phenotype uncouples the two processes of pASC maintenance and differentiation.
The expression profiles of stem cell progeny markers in mbd2/3(RNAi) animals highlight a failure to progress through the differentiation process
The observation that pASCs are maintained but fail to produce and replace differentiated cells suggests defects in stem cell differentiation. We therefore used the previously described early progeny marker Smed-NB.21.11.e and the later progeny marker Smed-AGAT-1 to investigate the dynamics of progeny (Eisenhoffer et al., 2008).
We observed that Smed-NB.21.11e+ cells were produced during regeneration in mbd2/3(RNAi) animals, and accumulated in regeneration blastemas when compared to control animals (Fig. 5A–C). However, the later Smed-AGAT-1+ stem cell progeny were depleted in mbd2/3(RNAi) animals during regeneration (Fig. 5A, B and D. These data correlated with increased transcript levels for Smed-NB.21.11e and decreased transcript levels of Smed-AGAT-1 (Fig. 5E). Also during regeneration we observed a reduction in the expression of Smed-CYP1A1-1, another marker of late pASC progeny (Eisenhoffer et al., 2008) (Fig. 5F).
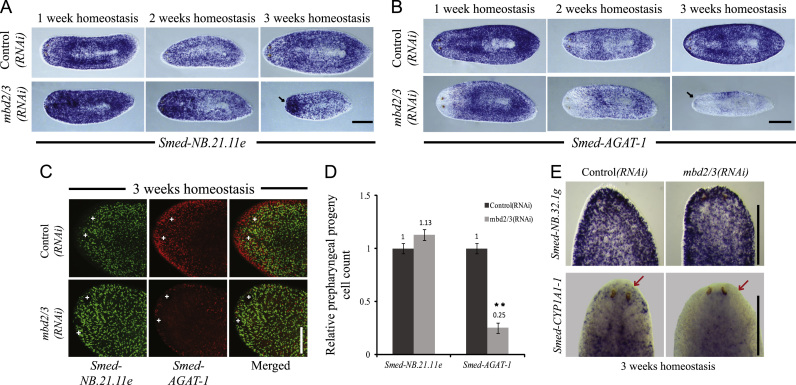
Fig. 5.
mbd2/3(RNAi) Leads to an accumulation of NB.21.11e+ cells and a decline Smed-AGAT-1+ during regeneration. Double FISH on middle pieces shows maintenance of Smed-NB.21.11e+ cells and loss of Smed-AGAT-1+ cells during regeneration in mbd2/3(RNAi) animals (A). Anterior to the left. Scale bar, 200 µm. mbd2/3(RNAi) leads a significant increase in NB.21.11e+ cells and a significant decrease in Smed-AGAT-1+ cells (B), which is quantification of (A); n=3. Standard error bars are shown. ⁎ Indicates p<0.05. Accumulation of NB.21.11e+ cells and a decline in AGAT-1+ cells is apparent throughout regenerating head, middle and tail pieces (C) and (D) Scale bar 500 µm. RT-qPCR analyses of both progeny marker transcripts correlates with the accumulation and loss of early and late stem cell progeny (E). The alternate progeny marker Smed-CYP1A1-1 is also depleted during regeneration by mbd2/3(RNAi) during regeneration (F). Scale bar, 200 µm.
During tissue homeostasis in intact animals Smed-NB.21.11e+ cells were maintained in mbd2/3(RNAi) animals and accumulated at the sites of anterior regression (Fig. 6A, C and D). On the other hand, later Smed-AGAT-1+ stem cell progeny were depleted in mbd2/3(RNAi) animals (Fig. 6B–D). This resulted in animals with accumulated Smed-NB.21.11e+ cells and depleted Smed-AGAT-1+ cells by 3 weeks of homeostasis (Fig. 6C). The data observed for Smed-NB.21.11e+ and Smed-AGAT-1+ cells was supported by an increase in Smed-NB.21.11e transcript levels and a decrease in Smed-AGAT-1 transcript levels (Supplementary Fig. 4A and B). An alternate early progeny marker Smed-NB.32.1g (Eisenhoffer et al., 2008) was also maintained at the third week of homeostasis following mbd2/3(RNAi) (Fig. 6E). The loss of late neoblast progeny in mbd2/3(RNAi) animals during homeostasis was confirmed using the Smed-CYP1A1 marker gene (Fig. 6F). These data suggest that pASCs lacking mbd2/3 are able to give rise to early stem cell progeny, but that these progeny are unable to differentiate correctly, with later stem cell progeny failing to form.
Fig. 6.
mbd2/3(RNAi) Leads to an accumulation of NB.21.11e+ cells and a decline Smed-AGAT-1+ during homeostasis. Smed-NB.21.11e expressing early progeny cells during homeostasis are maintained and they accumulate at sites of anterior regression (black arrow) in mbd2/3(RNAi) animals (A). Reduction in the number of Smed-AGAT-1 expression late pASC progeny cells during tissue homeostasis in mbd2/3(RNAi) animals, with loss in an anterior to posterior direction (B). Scale bars, 500 µm. (C) Double FISH on intact animals during homeostasis to visualize neoblast progeny cell formation. +: eyes. Scale bar, 200 µm. Quantification of (C) with n=3. ⁎⁎ Indicates p<0.01 as determined by Student's t-test. (D) The alternate early progeny marker Smed-NB.32.1g is also maintained while the alternate late progeny marker Smed-CYP1A1-1 is depleted (E). Scale bars, 500 µm.
The genome of Schmidtea mediterranea is not broadly methylated and Smed-dnmt2 is not required for regeneration or homeostasis
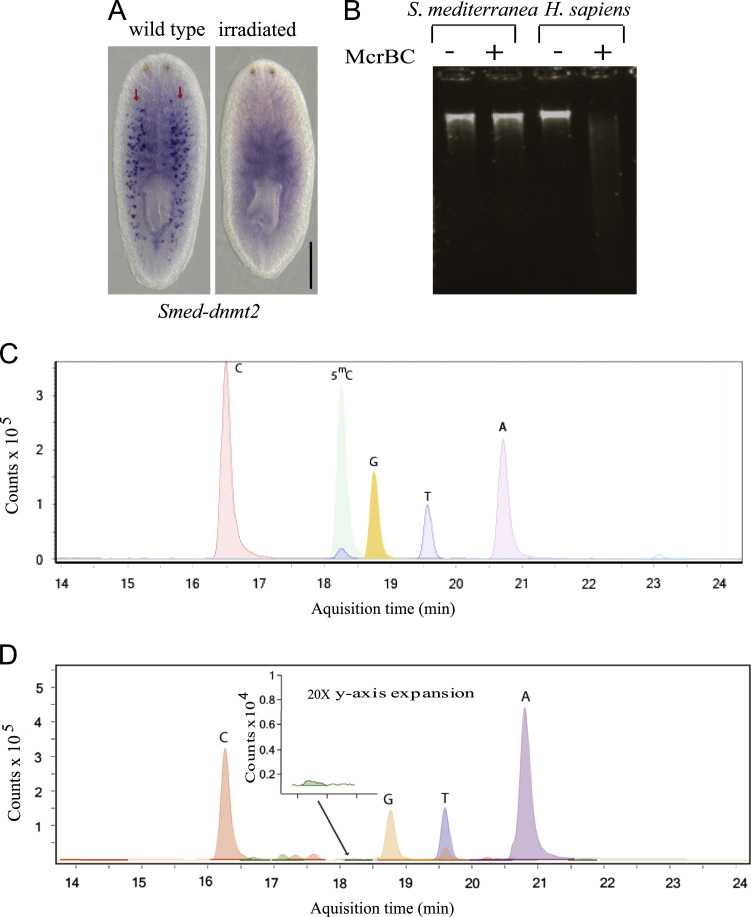
Some MBD proteins are thought to act through binding 5mC and affect gene expression (Hendrich and Bird, 1998; Jaenisch and Bird, 2003). We looked for evidence for the presence 5mC in genomic DNA (gDNA) and for genes encoding potential DNA methyltransferases. We identified a single gene with identity to known DNA methyltransferases, Smed-dnmt2 (Supplementary Fig. 5A and B). Neither the observed expression, which was confined to the germline (Wang et al., 2007) (Fig. 7A), nor RNAi experiments indicated any role in regeneration (Supplementary Fig. 5C and D). Another recent study reported a DMNT1 gene in S. mediterranea (Zeng et al., 2013), but we have found only a DMNT2 ortholog in line with a recent study that only found DMNT2 encoding genes across the platyhelminthes (Geyer et al., 2013).
Fig. 7.
Lack of evidence for a role for DNA methylation in pASC biology Smed-dnmt-2 is expressed in the germ line and germ line stem cells (A) Scale bars, 500 µm. gDNA digestion with the methylation dependent restriction enzyme McrBC does not indicate any DNA cytosine methylation in the Schmidtea mediterranea genome as compared with that of humans. −: no enzyme; +: with enzyme (B). Control chromatogram after HPLC-MS displaying the sensitivity of nucleosides detection, in particular for 5mC. (C) A representative chromatogram of S. mediterranea nucleoside analysis indicating the lack of 5mC (D).
Within the phylum platyhelminthes Schistosoma mansoni was the first member to be described as methylated (Geyer et al., 2011), and more recently another study detected methylation throughout the phylum (Geyer et al., 2013). However, a recent study based on bisulfite sequencing found no evidence for methylation in S. mansoni (Raddatz et al., 2013). We took several approaches to look for 5mC in the asexual and sexual S. mediterranea genome. gDNA digestion with the methylation sensitive and insensitive isoschizomers PleI/MlyI, HpaII/MspI, AvaI/BsoBI and Acc65I/KpnI (Lindsay and Bird, 1987; McClelland et al., 1994) showed similar patterns indicating the lack of abundant cytosine methylation (Supplementary Table 4; Supplementary Fig. 6A–E). The lack of abundant 5mC was also confirmed using the methylation dependent restriction enzyme McrBC (Panne et al., 1999) (Fig. 7B). In addition, a monoclonal antibody to 5mC (Ruzov et al., 2011) could detect putative low levels of methylation in nerve and muscle cells of Drosophila melanogaster but not in asexual S. mediterranea (Supplementary Fig. 6F, sexual animals were not assayed by this approach). Finally, 5mC was undetectable using High Performance Liquid Chromatography coupled Mass Spectrometry (HPLC-MS) (Fig. 7C and D). Together our results provide strong evidence for the absence of abundant 5mC from S. mediterranea genome, at levels below our limit of HPLC-MS detection of 0.1%. We conclude that this aspect of epigenetic regulation is not conserved between pASCs and mammalian stem cells. These data indicates that mbd2/3, in agreement with not having the correct amino acid sequence to bind methylated DNA (Supplementary Fig. 1C) (Ohki et al., 2001), is unlikely to be binding 5mC in vivo.
Discussion
In this study we have shown that the single MBD containing gene in planarians is required for the correct differentiation of pASCs. Despite pASCs being able to proliferate correctly and produce a regeneration blastema consisting of stem cell progeny, these cells fail to terminally differentiate and some early progeny cells accumulate. Significantly, the loss of mbd2/3 does not affect pASC proliferation and pASCs are maintained. Nematode MBD2/3 proteins lack the methyl-binding domain and functional studies have described relatively minor and variable phenotypes (Gutierrez and Sommer, 2007). MBD2/3 mutants in Drosophila have described problems in chromosome segregation in early embryogenesis in a fraction of embryos but with no effect on viability or fertility (Marhold et al., 2004b). A requirement of planarian mbd2/3 for stem cell differentiation reveals a significant role for this gene in invertebrates and it may be that the MBD2/3 family has an ancestral role in stem cell differentiation in metazoans.
We observe the expression of mbd2/3 is limited to radiation sensitive cells and the germline, suggesting that it is expressed pASCs and germ line stem cells. Absence of expression from blastemas suggests that mbd2/3 is not expressed in stem cell progeny although this is the cellular compartment primarily affected by mbd2/3(RNAi). Two alternate possibilities may explain this. First, the MBD2/3 protein may perdure in progeny while transcripts are only present in pASCs. Alternatively MBD2/3 may have a role in controlling heritable epigenetic regulatory changes that have subsequent effects in allowing correct differentiation programs to unfold. Future detailed work on epigenetic mechanisms and modifications will allow these possibilities to be investigated.
The finding that mbd2/3 does not have the full complement of conserved residues known to contact methylated DNA (Ohki et al., 2001) suggested that a potential regulation of planarian pASC differentiation through the 5mC epigenetic modification would be unlikely (Supplementary Fig. 1). We find no evidence of DNA methylation in S. mediterranea from multiple analyses (Supplementary Figs. 6 and 7) and observe no role for Smed-dnmt2 in regeneration (Supplementary Figs. 5 and 7). Overall our data supports the recent finding from whole genome bisulfite sequencing of animals with only a single DNA methylase protein in the DNMT-2 family, that these genomes do not contain DNA methylation (Raddatz et al., 2013). Future work will have to resolve these findings with the reports of DNA methylation detection in S. mansoni and other Platyhelminthes (Geyer et al., 2011, 2013).
In vertebrates Xenopus laevis MBD3 shows significant sequence homology to mbd2/3 and mouse MBD2 (Suplementary Fig. 1A and B), and like mouse MBD2 is able to bind methylated DNA in vitro (Iwano et al., 2004). In contrast mouse MBD3 does not bind specifically to methylated DNA in vitro (Hendrich and Bird, 1998) but instead mediates the function of the NuRD complex (Kaji et al., 2006). It therefore seems likely that mbd2/3 regulates pASC differentiation through interaction with a planarian version of the conserved NuRD complex, analogous to the role of mouse MBD3 in stable formation of the Mi-2/NuRD complex in pre-implantation embryos (Kaji et al., 2006).
The function of MBD2/3 has always been associated with the NuRD complex (Kaji et al., 2007) Binding of the MBD3/NuRD complex at target genes in ESCs can lead to further histone modifications and therefore transcriptional repression. Polycomb-repressive complex 2 (PRC2) is a transcriptional repressor that leads to di- and tri-methylation of H3K27, and it gets recruited by the MBD3/NuRD complex to target promoters in ESCs to maintain them in a methylated, deacetylated state (Reynolds et al., 2012). MBD3/NuRD repressor also regulates JNK signaling, a pathway known to be important in tissue morphogenesis, cell polarity, stem cell regeneration and apoptosis (Aguilera et al., 2011). While phosphorylation of c-Jun leads to the activation of target gene expression, MBD3 specifically interacts with the unphosphorylated form of c-Jun. This leads to repression of target gene expression by reducing histone acetylation at promoter regions at target genes, such as the intestinal stem cell marker lgr5, affecting intestinal epithelial homeostasis. Further study of epigenetic modifications of histones in planarians and transcriptional changes resulting from loss of mbd2/3 will shed further light on how pASC differentiation and pluripotency is controlled. Experiments defining the transcriptional differences caused by mbd2/3 loss may also help to broadly identify differentiation factors and assess the level of underlying conservation across animals.
The functions of two other planarian NuRD components, Smed-CHD4/Mi2 and an ortholog of RbAp48 in the planarian D. japonica, suggested that NuRD may also be required for stem cell maintenance as well as differentiation (Bonuccelli et al., 2010; Scimone et al., 2010). Abrogation of these genes led to both defects in stem cell differentiation and depletion of pASCs. Comparing the initial loss of Smed-AGAT-1 cells and then the loss of Smed-NB.21.11e by 18–20 days after initial RNAi in Smed-CHD4(RNAi) animals (equivalent of 1 week homeostasis in our experiments) with the maintenance of Smed-NB.21.11e positive cells and loss of Smed-AGAT-1 in mbd2/3 animals highlights a significant difference between the functions of mbd2/3 and Smed-CHD4/Mi2. If early and late progeny are related by lineage this would suggest that loss of mbd2/3 causes a defect at a distinct and later point in the differentiation process. Also mbd2/3 is not required for neoblast maintenance to the same extent as Smed-CHD4(RNAi) and DjRbAp48(RNAi) animals, in which neoblasts and proliferation are ultimately depleted whereas mbd2/3(RNAi) animals are able to form a blastema and initiate an appropriate proliferative response to amputation. Significantly, both RbAp48 and CHD4/Mi2 are known to have broad molecular roles as active components of other complexes such as the Sin3, CAF-1 and dMec complexes (Ahringer, 2000; Kunert et al., 2009; Verreault et al., 1996). It is possible that the observed effects of Smed-CHD4 and DjRbAp48 loss on proliferation, blastema formation and pASC maintenance are the result of the loss of their activities in the context of these alternate complexes. Alternatively the planarian NuRD complex may have forms that include and exclude mbd2/3, with functions independent of the MBD2/3 component required for pASC maintenance. Further work will shed light on which of these scenarios is true.
In summary our data supports a model where the planarian NuRD complex functions to allow pASCs to differentiate correctly. In MBD3-null ES cells it appears that the inability to silence pluripotency genes results in the inability to properly differentiate, indirectly implicating the rest of the NuRD complex in this process (Kaji et al., 2006). While the role of the conserved NuRD complex component CHD4/Mi2 and family members in differentiation in defining cell fates in Caenorhabditis elegans (von Zelewsky et al., 2000), D. melanogaster (Kehle et al., 1998) and even Arabidopsis (Ogas et al., 1999) has been previously described, a central role for a conserved MBD containing protein in these processes outside of the vertebrates has not been previously described. Like other invertebrates it appears that planarians have lost components of a more complex DNA methylation system, including DNA methylases and other MBD containing proteins (Albalat, 2008). However, the findings that an ortholog of mbd2/3 in a simple bilaterian is required for correct stem cell differentiation, and acts independently of DNA methylation, suggest that the mode of action observed for MBD3 in mammals may in fact be ancient.
Acknowledgements
This work funded by grants from the MRC (G0601133) and BBSRC (BB/K007564/1) awarded to AAA. We are grateful to Mr. Jamie Jowett, Ms. Sabrina Lam and Ms. Virginie Lemay for technical support and Dr. Karen Smeets for qPCR guidance.
Footnotes
This is an open-access article distributed under the terms of the Creative Commons Attribution-NonCommercial-No Derivative Works License, which permits non-commercial use, distribution, and reproduction in any medium, provided the original author and source are credited.
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.ydbio.2013.09.020.
Appendix A. Supplementary materials
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
References
- Aboobaker A.A. Planarian stem cells: a simple paradigm for regeneration. Trends in Cell Biology. 2011;21:304–311. doi: 10.1016/j.tcb.2011.01.005. [DOI] [PubMed] [Google Scholar]
- Aboobaker A.A., Kao D. A lack of commitment for over 500 million years: conserved animal stem cell pluripotency. EMBO Journal. 2012;31:2747–2749. doi: 10.1038/emboj.2012.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilera C., Nakagawa K., Sancho R., Chakraborty A., Hendrich B., Behrens A. c-Jun N-terminal phosphorylation antagonises recruitment of the Mbd3/NuRD repressor complex. Nature. 2011;469:231–235. doi: 10.1038/nature09607. [DOI] [PubMed] [Google Scholar]
- Ahringer J. NuRD and SIN3 histone deacetylase complexes in development. Trends in Genetics. 2000;16:351–356. doi: 10.1016/s0168-9525(00)02066-7. [DOI] [PubMed] [Google Scholar]
- Albalat R. Evolution of DNA-methylation machinery: DNA methyltransferases and methyl-DNA binding proteins in the amphioxus Branchiostoma floridae. Development Genes and Evolution. 2008;218:691–701. doi: 10.1007/s00427-008-0247-7. [DOI] [PubMed] [Google Scholar]
- Almeida R.D., Sottile V., Loose M., De Sousa P.A., Johnson A.D., Ruzov A. Semi-quantitative immunohistochemical detection of 5-hydroxymethyl-cytosine reveals conservation of its tissue distribution between amphibians and mammals. Epigenetics. 2012;7:137–140. doi: 10.4161/epi.7.2.18949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blythe M.J., Kao D., Malla S., Rowsell J., Wilson R., Evans D., Jowett J., Hall A., Lemay V., Lam S., Aboobaker A.A. A dual platform approach to transcript discovery for the planarian Schmidtea mediterranea to establish RNAseq for stem cell and regeneration biology. PLoS One. 2010;5:e15617. doi: 10.1371/journal.pone.0015617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonuccelli L., Rossi L., Lena A., Scarcelli V., Rainaldi G., Evangelista M., Iacopetti P., Gremigni V., Salvetti A. An RbAp48-like gene regulates adult stem cells in planarians. Journal of Cell Science. 2010;123:690–698. doi: 10.1242/jcs.053900. [DOI] [PubMed] [Google Scholar]
- Boyer L.A., Lee T.I., Cole M.F., Johnstone S.E., Levine S.S., Zucker J.P., Guenther M.G., Kumar R.M., Murray H.L., Jenner R.G., Gifford D.K., Melton D.A., Jaenisch R., Young R.A. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardona A., Fernandez J., Solana J., Romero R. An in situ hybridization protocol for planarian embryos: monitoring myosin heavy chain gene expression. Development Genes and Evolution. 2005;215:482–488. doi: 10.1007/s00427-005-0003-1. [DOI] [PubMed] [Google Scholar]
- Cebria F., Newmark P.A. Planarian homologs of netrin and netrin receptor are required for proper regeneration of the central nervous system and the maintenance of nervous system architecture. Development. 2005;132:3691–3703. doi: 10.1242/dev.01941. [DOI] [PubMed] [Google Scholar]
- Cebria F., Newmark P.A. Morphogenesis defects are associated with abnormal nervous system regeneration following roboA RNAi in planarians. Development. 2007;134:833–837. doi: 10.1242/dev.02794. [DOI] [PubMed] [Google Scholar]
- Denslow S.A., Wade P.A. The human Mi-2/NuRD complex and gene regulation. Oncogene. 2007;26:5433–5438. doi: 10.1038/sj.onc.1210611. [DOI] [PubMed] [Google Scholar]
- Eisenhoffer G.T., Kang H., Sanchez Alvarado A. Molecular analysis of stem cells and their descendants during cell turnover and regeneration in the planarian Schmidtea mediterranea. Cell Stem Cell. 2008;3:327–339. doi: 10.1016/j.stem.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman N., Gerson A., Fang J., Li E., Zhang Y., Shinkai Y., Cedar H., Bergman Y. G9a-mediated irreversible epigenetic inactivation of Oct-3/4 during early embryogenesis. Nature Cell Biology. 2006;8:188–194. doi: 10.1038/ncb1353. [DOI] [PubMed] [Google Scholar]
- Felix D.A., Aboobaker A.A. The TALE class homeobox gene Smed-prep defines the anterior compartment for head regeneration. PLoS Genetics. 2010;6:e1000915. doi: 10.1371/journal.pgen.1000915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geyer K.K., Rodriguez Lopez C.M., Chalmers I.W., Munshi S.E., Truscott M., Heald J., Wilkinson M.J., Hoffmann K.F. Cytosine methylation regulates oviposition in the pathogenic blood fluke Schistosoma mansoni. Nature Communications. 2011;2:424. doi: 10.1038/ncomms1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geyer K.K., Chalmers I.W., Mackintosh N., Hirst J.E., Geoghegan R., Badets M., Brophy P.M., Brehm K., Hoffmann K.F. Cytosine methylation is a conserved epigenetic feature found throughout the phylum Platyhelminthes. BMC Genomics. 2013;14:462. doi: 10.1186/1471-2164-14-462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Estevez C., Arseni V., Thambyrajah R.S., Felix D.A., Aboobaker A.A. Diverse miRNA spatial expression patterns suggest important roles in homeostasis and regeneration in planarians. International Journal of Developmental Biology. 2009;53:493–505. doi: 10.1387/ijdb.082825cg. [DOI] [PubMed] [Google Scholar]
- Guo T., Peters A.H., Newmark P.A. A Bruno-like gene is required for stem cell maintenance in planarians. Developmental Cell. 2006;11:159–169. doi: 10.1016/j.devcel.2006.06.004. [DOI] [PubMed] [Google Scholar]
- Gurley K.A., Rink J.C., Sanchez Alvarado A. Beta-catenin defines head versus tail identity during planarian regeneration and homeostasis. Science. 2008;319:323–327. doi: 10.1126/science.1150029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez A., Sommer R.J. Evolution of dnmt-2 and mbd-2-like genes in the free-living nematodes Pristionchus pacificus, Caenorhabditis elegans and Caenorhabditis briggsae. Nucleic Acids Research. 2004;32:6388–6396. doi: 10.1093/nar/gkh982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez A., Sommer R.J. Functional diversification of the nematode mbd2/3 gene between Pristionchus pacificus and Caenorhabditis elegans. BMC Genetics. 2007;8:57. doi: 10.1186/1471-2156-8-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrich B., Bird A. Identification and characterization of a family of mammalian methyl-CpG binding proteins. Molecular and Cellular Biology. 1998;18:6538–6547. doi: 10.1128/mcb.18.11.6538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubert A., Henderson J.M., Ross K.G., Cowles M.W., Torres J., Zayas R.M. Epigenetic regulation of planarian stem cells by the SET1/MLL family of histone methyltransferases. Epigenetics. 2013;8:79–91. doi: 10.4161/epi.23211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iglesias M., Gomez-Skarmeta J.L., Salo E., Adell T. Silencing of Smed-betacatenin1 generates radial-like hypercephalized planarians. Development. 2008;135:1215–1221. doi: 10.1242/dev.020289. [DOI] [PubMed] [Google Scholar]
- Inoue T., Kumamoto H., Okamoto K., Umesono Y., Sakai M., Sanchez Alvarado A., Agata K. Morphological and functional recovery of the planarian photosensing system during head regeneration. Zoological Science. 2004;21:275–283. doi: 10.2108/zsj.21.275. [DOI] [PubMed] [Google Scholar]
- Inoue T., Kumamoto H., Okamoto K., Umesono Y., Sakai M., Sanchez Alvarado A., Agata K. Morphological and functional recovery of the planarian photosensing system during head regeneration. Zoological Science. 2004;21:275–283. doi: 10.2108/zsj.21.275. [DOI] [PubMed] [Google Scholar]
- Iwano H., Nakamura M., Tajima S. Xenopus MBD3 plays a crucial role in an early stage of development. Developmental Biology. 2004;268:416–428. doi: 10.1016/j.ydbio.2003.12.032. [DOI] [PubMed] [Google Scholar]
- Jaenisch R., Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nature Genetics. 2003;33(Suppl):245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- Kaji K., Caballero I.M., MacLeod R., Nichols J., Wilson V.A., Hendrich B. The NuRD component Mbd3 is required for pluripotency of embryonic stem cells. Nature Cell Biology. 2006;8:285–292. doi: 10.1038/ncb1372. [DOI] [PubMed] [Google Scholar]
- Kaji K., Nichols J., Hendrich B. Mbd3, a component of the NuRD co-repressor complex, is required for development of pluripotent cells. Development. 2007;134:1123–1132. doi: 10.1242/dev.02802. [DOI] [PubMed] [Google Scholar]
- Kehle J., Beuchle D., Treuheit S., Christen B., Kennison J.A., Bienz M., Muller J. dMi-2, a hunchback-interacting protein that functions in polycomb repression. Science. 1998;282:1897–1900. doi: 10.1126/science.282.5395.1897. [DOI] [PubMed] [Google Scholar]
- Kunert N., Wagner E., Murawska M., Klinker H., Kremmer E., Brehm A. dMec: a novel Mi-2 chromatin remodelling complex involved in transcriptional repression. EMBO Journal. 2009;28:533–544. doi: 10.1038/emboj.2009.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labbe R.M., Irimia M., Currie K.W., Lin A., Zhu S.J., Brown D.D., Ross E.J., Voisin V., Bader G.D., Blencowe B.J., Pearson B.J. A comparative transcriptomic analysis reveals conserved features of stem cell pluripotency in planarians and mammals. Stem Cells. 2012;30:1734–1745. doi: 10.1002/stem.1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsay S., Bird A.P. Use of restriction enzymes to detect potential gene sequences in mammalian DNA. Nature. 1987;327:336–338. doi: 10.1038/327336a0. [DOI] [PubMed] [Google Scholar]
- Marhold J., Brehm A., Kramer K. The Drosophila methyl-DNA binding protein MBD2/3 interacts with the NuRD complex via p55 and MI-2. BMC Molecular Biology. 2004;5:20. doi: 10.1186/1471-2199-5-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marhold J., Kramer K., Kremmer E., Lyko F. The Drosophila MBD2/3 protein mediates interactions between the MI-2 chromatin complex and CpT/A-methylated DNA. Development. 2004;131:6033–6039. doi: 10.1242/dev.01531. [DOI] [PubMed] [Google Scholar]
- Matsumoto M., Toraya T. cDNA cloning, expression, and characterization of methyl-CpG-binding domain type 2/3 proteins from starfish and sea urchin. Gene. 2008;420:125–134. doi: 10.1016/j.gene.2008.05.007. [DOI] [PubMed] [Google Scholar]
- McClelland M., Nelson M., Raschke E. Effect of site-specific modification on restriction endonucleases and DNA modification methyltransferases. Nucleic Acids Research. 1994;22:3640–3659. doi: 10.1093/nar/22.17.3640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonel P., Costello I., Hendrich B. Keeping things quiet: roles of NuRD and Sin3 co-repressor complexes during mammalian development. International Journal of Biochemistry and Cell Biology. 2009;41:108–116. doi: 10.1016/j.biocel.2008.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner A., Mikkelsen T.S., Gu H., Wernig M., Hanna J., Sivachenko A., Zhang X., Bernstein B.E., Nusbaum C., Jaffe D.B., Gnirke A., Jaenisch R., Lander E.S. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–770. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohn F., Weber M., Rebhan M., Roloff T.C., Richter J., Stadler M.B., Bibel M., Schubeler D. Lineage-specific polycomb targets and de novo DNA methylation define restriction and potential of neuronal progenitors. Molecular Cells. 2008;30:755–766. doi: 10.1016/j.molcel.2008.05.007. [DOI] [PubMed] [Google Scholar]
- Ogas J., Kaufmann S., Henderson J., Somerville C. PICKLE is a CHD3 chromatin-remodeling factor that regulates the transition from embryonic to vegetative development in Arabidopsis. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:13839–13844. doi: 10.1073/pnas.96.24.13839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohki I., Shimotake N., Fujita N., Jee J., Ikegami T., Nakao M., Shirakawa M. Solution structure of the methyl-CpG binding domain of human MBD1 in complex with methylated DNA. Cell. 2001;105:487–497. doi: 10.1016/s0092-8674(01)00324-5. [DOI] [PubMed] [Google Scholar]
- Önal P., Grun D., Adamidi C., Rybak A., Solana J., Mastrobuoni G., Wang Y., Rahn H.P., Chen W., Kempa S., Ziebold U., Rajewsky N. Gene expression of pluripotency determinants is conserved between mammalian and planarian stem cells. EMBO Journal. 2012;31:2755–2769. doi: 10.1038/emboj.2012.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panne D., Raleigh E.A., Bickle T.A. The McrBC endonuclease translocates DNA in a reaction dependent on GTP hydrolysis. Journal of Molecular Biology. 1999;290:49–60. doi: 10.1006/jmbi.1999.2894. [DOI] [PubMed] [Google Scholar]
- Raddatz G., Guzzardo P.M., Olova N., Fantappie M.R., Rampp M., Schaefer M., Reik W., Hannon G.J., Lyko F. Dnmt2-dependent methylomes lack defined DNA methylation patterns. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:8627–8631. doi: 10.1073/pnas.1306723110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddien P.W., Oviedo N.J., Jennings J.R., Jenkin J.C., Sanchez Alvarado A. SMEDWI-2 is a PIWI-like protein that regulates planarian stem cells. Science. 2005;310:1327–1330. doi: 10.1126/science.1116110. [DOI] [PubMed] [Google Scholar]
- Reynolds N., Salmon-Divon M., Dvinge H., Hynes-Allen A., Balasooriya G., Leaford D., Behrens A., Bertone P., Hendrich B. NuRD-mediated deacetylation of H3K27 facilitates recruitment of polycomb repressive complex 2 to direct gene repression. EMBO Journal. 2012;31:593–605. doi: 10.1038/emboj.2011.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rink J.C., Vu H.T., Sanchez Alvarado A. The maintenance and regeneration of the planarian excretory system are regulated by EGFR signaling. Development. 2011;138:3769–3780. doi: 10.1242/dev.066852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robb S.M., Ross E., Sanchez Alvarado A. SmedGD: the Schmidtea mediterranea genome database. Nucleic Acids Research. 2008;36:D599–606. doi: 10.1093/nar/gkm684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruzov A., Tsenkina Y., Serio A., Dudnakova T., Fletcher J., Bai Y., Chebotareva T., Pells S., Hannoun Z., Sullivan G., Chandran S., Hay D.C., Bradley M., Wilmut I., De Sousa P. Lineage-specific distribution of high levels of genomic 5-hydroxymethylcytosine in mammalian development. Cell Research. 2011;21:1332–1342. doi: 10.1038/cr.2011.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scimone M.L., Meisel J., Reddien P.W. The Mi-2-like Smed-CHD4 gene is required for stem cell differentiation in the planarian Schmidtea mediterranea. Development. 2010;137:1231–1241. doi: 10.1242/dev.042051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solana J. Closing the circle of germline and stem cells: the primordial stem cell hypothesis. EvoDevo. 2013;4:2. doi: 10.1186/2041-9139-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solana J., Kao D., Mihaylova Y., Jaber-Hijazi F., Malla S., Wilson R., Aboobaker A. Defining the molecular profile of planarian pluripotent stem cells using a combinatorial RNAseq, RNA interference and irradiation approach. Genome Biology. 2012;13:R19. doi: 10.1186/gb-2012-13-3-r19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tweedie S., Ng H.H., Barlow A.L., Turner B.M., Hendrich B., Bird A. Vestiges of a DNA methylation system in Drosophila melanogaster? Nature Genetics. 1999;23:389–390. doi: 10.1038/70490. [DOI] [PubMed] [Google Scholar]
- Verreault A., Kaufman P.D., Kobayashi R., Stillman B. Nucleosome assembly by a complex of CAF-1 and acetylated histones H3/H4. Cell. 1996;87:95–104. doi: 10.1016/s0092-8674(00)81326-4. [DOI] [PubMed] [Google Scholar]
- von Zelewsky T., Palladino F., Brunschwig K., Tobler H., Hajnal A., Muller F. The C. elegans Mi-2 chromatin-remodelling proteins function in vulval cell fate determination. Development. 2000;127:5277–5284. doi: 10.1242/dev.127.24.5277. [DOI] [PubMed] [Google Scholar]
- Wagner D.E., Ho J.J., Reddien P.W. Genetic regulators of a pluripotent adult stem cell system in planarians identified by RNAi and clonal analysis. Cell Stem Cell. 2012;10:299–311. doi: 10.1016/j.stem.2012.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner D.E., Wang I.E., Reddien P.W. Clonogenic neoblasts are pluripotent adult stem cells that underlie planarian regeneration. Science. 2011;332:811–816. doi: 10.1126/science.1203983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Zayas R.M., Guo T., Newmark P.A. nanos function is essential for development and regeneration of planarian germ cells. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:5901–5906. doi: 10.1073/pnas.0609708104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y., Wong J., Moreno G.T., Young M.K., Cote J., Wang W. NURD, a novel complex with both ATP-dependent chromatin-remodeling and histone deacetylase activities. Molecular Cells. 1998;2:851–861. doi: 10.1016/s1097-2765(00)80299-3. [DOI] [PubMed] [Google Scholar]
- Zeng A., Li Y.Q., Wang C., Han X.S., Li G., Wang J.Y., Li D.S., Qin Y.W., Shi Y., Brewer G., Jing Q. Heterochromatin protein 1 promotes self-renewal and triggers regenerative proliferation in adult stem cells. Journal of Cell Biology. 2013;201:409–425. doi: 10.1083/jcb.201207172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Ng H.H., Erdjument-Bromage H., Tempst P., Bird A., Reinberg D. Analysis of the NuRD subunits reveals a histone deacetylase core complex and a connection with DNA methylation. Genes Development. 1999;13:1924–1935. doi: 10.1101/gad.13.15.1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data