Highlights
-
•
The cytoplasmic dynein complex is the main retrograde motor in all eukaryotic cells.
-
•
This complex is built around a dimer of cytoplasmic dynein heavy chains (DYNC1H1).
-
•
Mouse DYNC1H1 mutants have sensory defects, but motor defects have been controversial.
-
•
Now human DYNC1H1 mutations with sensory, motor, and cognitive deficits are being found.
-
•
The study of these mutations will give us new insight into DYNC1H1 function in the nervous system.
Keywords: amyotrophic lateral sclerosis, axonal transport, Cramping 1, Legs at odd angles, motor neurons, neurodegeneration, Sprawling
Abstract
Cytoplasmic dynein is the main retrograde motor in all eukaryotic cells. This complex comprises different subunits assembled on a cytoplasmic dynein heavy chain 1 (DYNC1H1) dimer. Cytoplasmic dynein is particularly important for neurons because it carries essential signals and organelles from distal sites to the cell body. In the past decade, several mouse models have helped to dissect the numerous functions of DYNC1H1. Additionally, several DYNC1H1 mutations have recently been found in human patients that give rise to a broad spectrum of developmental and midlife-onset disorders. Here, we discuss the effects of mutations of mouse and human DYNC1H1 and how these studies are giving us new insight into the many critical roles DYNC1H1 plays in the nervous system.
Introduction
Cytoplasmic dynein 1 is a large (∼1.5 MDa), multisubunit motor complex (Figure 1A) that moves towards the minus end of microtubules in eukaryotic cells [1]. It belongs together with the axonemal dyneins and cytoplasmic dynein 2 to the dynein superfamily. Axonemal dyneins are responsible for the movement of cilia and flagella, whereas cytoplasmic dynein 2 has a role in intraflagellar transport and is required for cilia and flagella assembly [1].
Figure 1.
The cytoplasmic dynein complex. (A) Diagram of the cytoplasmic dynein motor complex including the heavy chain (HC) dimer and its associated subunits. A model of the motor domain [5] built from yeast cytoplasmic dynein (PDB ID 4AKG) and the mouse microtubule-binding domain (MTBD) (PDB ID 3ERR) assembled by Dr A.P. Carter has been overlapped with the schematic of the dynein HC in its apo or post-power stroke form [5,93,94]. Adapted, with permission, from The Company of Biologists (J. Cell Sci. 126, 705–713; [4]). The electron micrograph of an isolated molecule of monomeric dynein from Chlamydomonas reinhardtii flagella in its pre-power stroke form is shown for comparison on the right. Adapted with permission from Macmillan Publishers (Nature 421, 715–718; [93]). Conformational changes driven by ATP hydrolysis in the motor domain, which alter the relative position of the stem and the tail/linker, are hypothesised to lead to the power stroke and progression on microtubules [5,94]. The HCs (in dark violet) contain the six AAA ATPase domains (in red), the stalk region, which includes the MTBD (in dark yellow and yellow, respectively), the buttress (in orange), and the linker region. HCs are associated with light intermediate chains (LICs) (in green), intermediate chains (ICs) (in cyan), and light chains (LCs) (in light yellow). (B) Domain composition of the cytoplasmic dynein HC. In addition to the functional domains shown in (A), this scheme displays the homodimerisation region and linker (in white). The positions on the dynein HC of the three mouse mutations (Loa, Legs at odd angles; Cra, Cramping 1; Swl, Sprawling; bottom) and the human mutations discussed in this review (top) are indicated.
The core of the cytoplasmic dynein 1 complex is the heavy chain (DYNC1H1) dimer (Figure 1A). Each heavy chain is enormous – a half-megadalton protein – and, perhaps unsurprisingly, serves multiple purposes. Towards the N terminal it has a long tail domain with binding sites for other structural and regulatory components of the dynein complex and docking sites for cargoes including adaptor proteins (Figure 1B and Table 1); at the C terminal, DYNC1H1 folds into a daisy-like structure comprising six ATPase domains associated with diverse cellular activities (AAA+) and a microtubule-interacting stalk region [2,3] (Figure 1A,B). This motor domain drives the entire complex and its cargoes along microtubules [2,4], although it is not completely understood how ATP hydrolysis is coupled to force generation or even the total number of ATP molecules bound to DYNC1H1 at any given time [4–6].
Table 1.
Cytoplasmic dynein-interacting proteinsa
| Protein | Type of binding | Site/subunit | Refs |
|---|---|---|---|
| LIS1 | Direct | DYNC1H1 (AAA3/AAA4 junction) DYNC1H1 (AAA4 arginine finger) |
[83] |
| NudE | Indirect | Intermediate chain Light chain – LC8 |
[84] |
| Dynactin | Indirect | Intermediate chain | [85] |
| Snapin | Indirect | Intermediate chain | [86,87] |
| Htt | Indirect | Intermediate chain | [88] |
| HAP1 | Indirect | Dynactin | [88] |
| BICD1 BICD2 |
Indirect Indirect |
Dynactin Intermediate chain |
[89,90] [91] |
The table shows some of the interactors with the cytoplasmic dynein motor complex involved in CNS development and homeostasis.
DYNC1H1 is highly conserved and is an essential protein in higher eukaryotes because the dynein complex has housekeeping functions in all cells, including orientation of the mitotic spindle, nuclear positioning, Golgi maintenance, and endosomal dynamics [7–9]. In the nervous system, the complex takes on additional roles specific to neurons; driven by the heavy-chain motor, it transports cargoes within dendrites and is the sole motor supporting retrograde transport in axons – carrying signalling complexes that affect gene expression, development, and regeneration, misfolded proteins, and organelles from the synapse back to the cell body. Interestingly, the analysis of motor function in axons revealed that motor proteins, including cytoplasmic dynein, strictly rely for axonal transport on the local synthesis of ATP by glycolytic enzymes, which are bound to transported organelles [10,11], and not on mitochondrial ATP.
In mammals, new insight into DYNC1H1 function came from characterising a set of dynein heavy chain mouse mutations. Now mutations in humans are also being found. Both mouse and human data show that the dynein heavy chain is essential for normal function of the nervous system and even single conservative amino acid substitutions in this > 500-kDa protein can result in neurological abnormalities.
An allelic series of DYNC1H1 mutants for understanding the role of cytoplasmic dynein in the nervous system
Mouse Dync1h1 mutant strains
A phenotype arising from a single point mutation gives a snapshot of protein function. By working with multiple mutations of one protein, an ‘allelic series’, we gain a much richer picture – particularly helpful when trying to dissect the function of a large protein like the cytoplasmic dynein heavy chain.
Although fly and worm laboratories have worked at the molecular level with multiple mutant alleles of the dynein heavy chain for many years, the first mammalian DYNC1H1 mutation came in 1998 with a knockout mouse (Table 2) [12]. In mouse and human, DYNC1H1 appears to be a single isoform encoded by 78 exons [13]. Heterozygous knockout mice had no reported phenotype, but homozygous nulls died early in gestation (<8.5 days) with Golgi apparatus, endosome, and lysosome abnormalities, underlining that DYNC1H1 is an essential protein with housekeeping roles.
Table 2.
Mouse mutations in the cytoplasmic dynein heavy chain gene
| Approved allele name | Allele | Phenotype of heterozygotes | Refs |
|---|---|---|---|
| Mouse Dync1h1tm1Noh | Knockout, created by gene targeting of the first exon. | Heterozygotes reported normal. Nulls die by embryonic day (E)8.5 with Golgi and other abnormalities. | [12] |
| Mouse Dync1h1Loa | Point mutation resulting in missense change F580Y in homodimerisation and dynein intermediate chain-binding site; created by chemical mutagenesis. | Motor, sensory, and other abnormalities. Loss of 50% motor neurons in E18.5 embryos. By 13 weeks, muscle spindles are reduced by 86% in hind limbs. Homozygotes dead by 1 day after birth. | [14,15] |
| Mouse Dync1h1Swl | 9-bp deletion resulting in loss of three amino acids from 1040–1043 (GIVT to A); created by radiation mutagenesis. | Sensory early-onset neuropathy, with reduction of 88% of muscle spindles in hind-limb muscles compared with wild type. Homozygotes die in utero before E8.5. | [14,19] |
| Mouse Dync1h1Cra1 | Point mutation resulting in missense change Y1055C in homodimerisation domain, created by chemical mutagenesis. | Motor, sensory, and other abnormalities. Loss of 20% of motor neurons in E18.5 embryos. Heterozygotes display progressive mitochondrial dysfunction in muscle, hyperinsulinemia, and hyperglycaemia, progressing to glucose intolerance with age. Homozygotes dead by 1 day after birth. |
[15,58] |
Insight into the nervous system function of Dync1h1 came with three further mutants: Legs at odd angles (Dync1h1Loa), Cramping 1 (Dync1h1Cra1), and Sprawling (Dync1h1Swl) mice [14,15] (Table 2 and Figure 1B). Dync1h1Loa mice present a single point mutation from a phenylalanine to a tyrosine (F580Y) in the dynein tail domain. Surprisingly, the addition of a single hydroxyl group onto the 532-kDa protein is enough to give rise to a dominant phenotype [15]. This residue lies within the binding site for dynein intermediate chains (ICs) [16] (Figure 1B) and the homodimerisation region [16,17]. Dync1h1Cra1 mice have instead a single point mutation at the opposite end of the homodimerisation domain (Y1055C) (Figure 1B) that causes a neurological phenotype overlapping that of Dync1h1Loa [15–17] (Table 2). Dync1h1Loa and Dync1h1Cra1 were generated by independent chemical mutagenesis experiments (Box 1); a radiation-induced mutant, Dync1h1Swl, has a small deletion (G1040–T1043delinsA) in the proximity of the Dync1h1Cra1 mutation (Table 2 and Figure 1B) and also produces a dominant phenotype that primarily affects the nervous system [14,18,19].
Box 1. Dync1h1Loa, Dync1h1Cra1, and Dync1h1Swl mutations and the Harwell connection.
In 1994, one of the authors was talking to a member of Professor J. Peters’ Laboratory of the MRC Mammalian Genetics Unit in Harwell, UK, a world centre for mouse genetics since 1946. The Peters laboratory produced novel mouse mutants with the chemical mutagen N-ethyl-N-nitrosourea (ENU), which typically causes point mutations, particularly A-to-T changes [95]. A discussion arose about a new mutation that had disrupted locomotion. The first mouse described was an obligate heterozygote female born in 1984 from a C3H/HeH male previously treated with ENU and crossed to a PTP-strain female. This ‘Legs at odd angles’ (Loa) mouse was noticed because of her unusual locomotion and named because she held her legs at a strange angle (limb grasping, a generic sign of neuromuscular/motor system deficits) when rapidly suspended by the tail. This mouse and her progeny were crossed to several inbred strains, but mainly the 101 strain. During the following behavioural characterisation [96], it was noticed that heterozygous animals, which otherwise lived to a normal age compared with wild type littermates, developed a slowly progressive locomotor deficit that manifested as a low-based ‘reptilian’ gait and Rotarod test and grip-strength defects.
In that pre-genome project era, positional cloning of an unknown mutant gene was a gruelling undertaking of several years. Having found linkage of the Loa trait to mouse chromosome 12, the physical maps spanning the critical region were published [97]. In 2001, the critical region was displayed at the 15th International Mouse Genome Conference, Edinburgh, UK. Dr Andreas Russ, then working for the German Biotech company Ingenium saw the map and realised that the description of the mutant and the position of the unknown mutation were almost identical to that of a mouse mutation, Cramping 1, created by the German ENU project [98]. The groups in the UK and Germany cooperated and in 2003 found that the Loa and Cra1 strains have point mutations in Dync1h1. The Loa mutation is a transversion resulting in a change from phenylalanine to tyrosine at position 580 (F580Y), whereas the Cra1 mutation changes tyrosine to cysteine at position 1055 (Y1055C) [15]. The non-complementation found in the compound heterozygotes (Dync1h1Loa/Cra) added considerable weight of evidence that the correct mutations had been identified.
The first Sprawling mouse, described in 1967, had been produced from an irradiation experiment [18,19]. However, the causative mutation was published only in 2007 by Popko and colleagues, who determined that the phenotype arose from a 9-bp deletion in exon 12 of Dync1h1 that changed four amino acids (GIVT) to a single alanine (A) residue in a cargo-binding domain [14]. The Sprawling mouse strain originated at MRC Harwell.
Homozygous Dync1h1Loa and Dync1h1Cra1 mice die by 1 day after birth. This could potentially be due to a large reduction in spinal cord motor neurons, limiting their ability to feed [15]. By contrast, Dync1h1Swl homozygotes die at the late implantation–early gastrulation stage [14,18,19].
Human DYNC1H1 mutations
In 2010, the first human DYNC1H1 mutation (DYNC1H1H3822P; Figure 1B and Table 3) was found in an individual with developmental delay, hypotonia, and brain malformations [20]. These phenotypic manifestations are of great interest because cytoplasmic dynein is essential for neuronal migration. A second mutation (DYNC1H1E1518K) has also been found to cause severe intellectual disability and other features [20,21]. Recently, Poirer and colleagues reported seven novel de novo mutations (two of them overlapping) and a small deletion, in addition to a familial mutation, in individuals with malformations of cortical development (MCD)22. MCD is characterised by alterations of the cerebral cortex and is often associated with severe intellectual disability. The mutations causing MCD are located in different domains of DYNC1H1, but several of them (DYNC1H1K3241T, DYNC1H1K3336N, DYNC1H1R3344Q, DYNC1H1R3384Q) cluster in the stalk region and on the surface of its microtubule-binding domain (MTBD) (Figure 1B) and have been shown to weaken binding of the dynein complex to microtubules [22]. The remaining mutations lie in the first AAA ATPase domain (AAA1) (DYNC1H1R1962C), in an area proximal to the linker region (DYNC1H1R1567Q) and at the N terminus (DYNC1H1K129I) of the tail domain, whereas the deletion (DYNC1H1Δ659-662) was found within the binding sites for ICs and Light Intermediate Chains (LICs) [16] (Figure 1B and Table 3). Although the N terminus of the tail domain is not involved in in vitro motility or in the binding of structural and regulatory components, the dramatic effect of the DYNC1H1K129I mutation (Table 3), together with mutagenesis analyses in Aspergillus [23], suggest that this region of DYNC1H1 is critical for cytoplasmic dynein functions in vivo, possibly for the coordination of force production and movement under heavy load [23].
Table 3.
Human mutations in the cytoplasmic dynein heavy chain gene
| Approved allele name | Allele | Phenotype of heterozygotes | Refs |
|---|---|---|---|
| Human DYNC1H1K129I | De novo point mutation resulting in missense change K129I at the N terminus of the tail domain. | Normocephaly. Pathological thick convolutions of the posterior cerebral cortex (posterior pachygyria) and severe intellectual disability. Late-onset epilepsy. | [22] |
| Human DYNC1H1H306R | Point mutation resulting in missense change H306R in homodimerisation domain. | Early-onset, slowly progressive distal lower-limb weakness and wasting (similar to CMT2 neuropathy) or SMA with lower-extremity predominance. Learning difficulties in some individuals. | [24,25] |
| Human DYNC1H1I584L | Point mutation resulting in missense change I584L in homodimerisation and dynein intermediate chain-binding site. | SMA with lower-extremity predominance. Normal upper-extremity strength. | [26,33] |
| Human DYNC1H1Δ659-662 | De novo 12-bp deletion resulting in loss of four amino acids from 659–662 in homodimerisation and dynein intermediate chain-binding site. | Microcephaly associated with posterior pachygyria. Early-onset epilepsy and spastic tetraplegia. | [22] |
| Human DYNC1H1K671E | Point mutation resulting in missense change K671E in homodimerisation and dynein intermediate chain-binding site. | Early-onset, slowly progressive distal lower-limb weakness and wasting. May have a ‘waddling’ gait. No known sensory involvement. | [26] |
| Human DYNC1H1Y970C | Point mutation resulting in missense change Y970C in homodimerisation domain. | Significant motor delay, no known sensory involvement, mild cognitive impairment. | [26] |
| Human DYNC1H1E1518K | De novo point mutation resulting in missense change E1518K. | Severe mental retardation, unable to walk or talk, hypertonia and club feet; untested reflexes. Epilepsy. Cortical malformation. Lack of sensory data. | [20,21] |
| Human DYNC1H1R1567Q | De novo point mutation resulting in missense change R1567Q. | Normocephaly. Excessive number of small convolutions of the frontal cortex (frontal polymicrogyria) and severe intellectual disability. Foot deformities. | [22] |
| Human DYNC1H1R1962C | De novo point mutation resulting in missense change R1962C in AAA1. | Normocephaly. Posterior pachygyria with severe intellectual disability and awkwardness. Transient focal epilepsy at early age. | [22] |
| Human DYNC1H1K3241T | Familial point mutation resulting in missense change K3241T in the stalk domain. | Normocephaly. Posterior pachygyria with mild or absent intellectual disability and variable awkwardness. Focal epilepsy. | [22] |
| Human DYNC1H1K3336N | De novo point mutation resulting in missense change K3336N in the stalk domain (microtubule-binding domain [MTBD]). | Microcephaly associated with posterior pachygyria, frontal polymicrogyria, and other CNS malformations. Early-onset epilepsy. Spastic tetraplegia with foot deformities. | [22] |
| Human DYNC1H1R3344Q | Point mutation resulting in missense change R3344Q in the stalk domain (MTBD). This de novo mutation was reported in two unrelated individuals. | Mild microcephaly associated with posterior pachygyria with moderate intellectual disability and awkwardness. Focal epilepsy. | [22] |
| Human DYNC1H1R3384Q | De novo point mutation resulting in missense change R3384Q in the stalk domain (MTBD). | Microcephaly associated with posterior pachygyria, frontal polymicrogyria and other CNS malformations. Early-onset epilepsy. Spastic tetraplegia with foot deformities. | [22] |
| Human DYNC1H1H3822P | De novo point mutation resulting in missense change H3822P in a linker region between AAA5 and AAA6 within the motor domain. | Hypotonia, moderately severe mental retardation, broad-based, waddling gait, reduced reflexes. Bilateral deficient gyration of the frontal lobes. Lack of sensory data. | [20,21] |
DYNC1H1 mutations have also now been reported in motor neuropathies, which may be accompanied by cognitive impairment. The first such family presented with the dominant hereditary distal motor neuropathy Charcot–Marie–Tooth (CMT) type 2, with delayed motor milestones and/or abnormal gait. Affected individuals have early-onset lower-limb weakness and wasting caused by a mutation, DYNC1H1H306R, within the homodimerisation domain of the dynein heavy chain [24] (Figure 1B and Table 3). However, an independent family with an identical mutation manifests symptoms of spinal muscular atrophy (SMA) [25]. Other DYNC1H1 mutations have been found by exome sequencing in families with a similar SMA presentation [26]. The key clinical features in individuals with three different mutations in the tail domain (DYNC1H1I584L, DYNC1H1K671E, DYNC1H1Y970C) were congenital or very early-onset weakness in the proximal legs and other areas and a static or mildly progressive disease course. Clinical findings support a diagnosis of motor neuron disease without sensory involvement [26,27]. Interestingly, DYNC1H1K671E and the DYNC1H1Δ659-662 deletion are only 10 residues apart, yet their main clinical outcome, MCD associated with spastic tetraplegia, and motor neuron disease, respectively, are dramatically different.
The human and mouse mutations described so far are all autosomal-dominant disorders with high penetrance; it is unknown whether the phenotypes arise from gain of function or hemizygous loss of function or, perhaps more likely in such a large protein, a combination of both. There is, however, evidence that the Dync1h1Loa mutation results in reduced affinity between cytoplasmic dynein and the p150 subunit of dynactin (DCTN1), a binding partner of dynein (Table 1) [28] and in impaired motor processivity [28,29], although it is unclear whether this is the sole cause of the phenotype.
Consequences of DYNC1H1 mutations in the nervous system
Locomotor and motor system deficits
Dync1h1Loa, Cra1, Swl heterozygous mouse mutants have gait deficits that manifest by approximately 1 month of age (Table 2). They limb-clench when held by the tail and have grip-strength deficits, which occur as early as 1 week after birth in Dync1h1+/Swl mice [14,15]. Dync1h1+/Cra1 mice have early-onset reduction in grip strength, which does not progress [30] or mildly [31] progresses with age. Forelimb strength and movement are preserved in Dync1h1+/Loa mice [32].
Interestingly, all of the human DYNC1H1 mutation cases have gait abnormalities ranging from spastic tetraplegia, inability to walk at all, and ‘waddling gait’ to mild distal limb weakness; pes cavus or other foot deformities are often a feature, indicating an underlying neuromuscular disorder [20,21,24–27,33] (Table 3). The striking similarity between the locomotor phenotypes observed in humans and mice suggest that DYNC1H1 plays crucial roles in the physiological control of the motor system in both species.
The first description of heterozygous Dync1h1+/Loa and Dync1h1+/Cra1 mice reported a progressive loss of motor neurons with a predominance of large type I fibres in specific muscles [15]. However, later reports found no loss of alpha motor neurons in Dync1h1+/Loa [14,34] or Dync1h1+/Cra1 [30,35] mice. Although these contradictory results are complicated by possible methodological differences (e.g., how different laboratories count large alpha motor neurons), they may also give insights for unravelling the effect of genetic factors in dynein function, because one possible source of variation is the genetic background of the mice examined in different papers. Our original report [15] described motor neuron loss in Dync1h1+/Loa mice with a mixed genetic background that included at least C3H, PTP, 101, and C57BL/6, whereas subsequent studies were all on congenic C57BL/6 [14,34]. It is therefore possible that genetic background plays a modifying role on this phenotype.
This would not be the first case in which genetic background affects phenotype severity. In an established mouse model of amyotrophic lateral sclerosis (ALS) (SOD1G93A), the C57BL/6 background extends life compared with a C3H background [36]. Extrapolating to the Dync1h1 mutants, it is possible that alleles in the mixed genetic background used in the original study enhanced motor neuron loss whereas the C57BL/6 background was protective.
However, genetic effects are not an explanation of the discrepancies found in Dync1h1+/Cra1 and Dync1h1+/Swl mice [14,15,35], because these studies were performed on mice of the same (C3H) or very similar (C3H/101) background. Curiously, neuromuscular junctions (NMJs) have been found to be normal in Dync1h1+/Cra1 mice on a congenic C3H background [35], whereas on a mixed genetic background that includes C3H, NMJs were reported as abnormal [30].
As expected, embryos bearing homozygous mutations in Dync1h1 (e.g., Dync1h1Loa/Loa and Dync1h1Cra1/Cra1) display augmented phenotypes with severe motor neuron loss, although at present it is unknown whether these are developmental or neurodegenerative defects.
How do these results help in deciphering the clinical manifestations of human patients bearing mutations in DYNC1H1? Motor neuropathy in human patients displays different degrees of severity and includes distal wasting and weakness with reduced or absent reflexes and chronic distal denervation [27]. Distal motor neuron loss has been suggested in DYNC1H1H306R, DYNC1H1I584L, DYNC1H1K671E, and DYNC1H1Y970C patients [24–26,33], whereas the DYNC1H1E1518K individual has hypertonia and did not learn to walk [20,21]. Two interesting observations arise from the human data that strengthen the correspondence with the results obtained in the mouse studies and may relate to the modifier effects of genetic background found with the mouse mutations. First, the human DYNC1H1I584L mutation lies almost on top of the Dync1h1Loa mutation and is also very conservative (Ile to Leu). The functional characterisation of this human mutation indicates loss or dysfunction of motor neurons, akin to the original observation in the Dync1h1+/Loa mice [15]. Henceforth, the Dync1h1+/Loa mouse may represent an ideal system to model the pathophysiological effects of the DYNC1H1I584L human mutation. Second, DYNC1H1H306R manifests with distinct symptoms in different human pedigrees. In one (UK pedigree), patients display a predominant axonal CMT2B disease [24], whereas in the other (Japanese pedigree) they present with SMA with lower extremity predominance [25], potentially demonstrating the strong effects of genetic background also in the human cases.
Sensory system defects
In addition to motor system deficits, mutations in the dynein heavy chain also cause impairments in other areas of the nervous systems, including sensory neurons. An overt sensory phenotype is found in Dync1h1+/Swl mice, which have early-onset sensory neuropathy probably due to degeneration during late embryonic development [14,18] (Table 2). These mice have proprioceptive and nociceptive sensory neuron loss compared with wild types [14]. Sensory nerve conduction velocities are reduced and the mice have moderate sensory neuropathy arising from the proprioceptive defect [14]. Similarly, Dync1h1+/Loa mice have progressive loss of sensory axons from at least 1.5 months after birth, mostly from proprioceptive nerves [34]. Branching and elongation of primarily sensory limb nerves is impaired in E13.5 and E14.5 Dync1h1Loa homozygotes and heterozygotes [15]. Dync1h1+/Cra1 mice have milder loss of sensory axons than other Dync1h1 mutants and, perhaps surprisingly, no difference in proprioceptive neuron number or morphology compared with wild types at 6 months of age [30].
All three mutant strains of mice were identified because of hind-limb clenching and low-based gait (Table 2). In Dync1h1+/Loa and Dync1h1+/Swl, these features may be due to proprioceptive defects. However, in Dync1h1+/Cra1 they may arise from dysfunction of the NMJ rather than motor or sensory neurodegeneration, which fits with findings from Drosophila and other model organisms in which dynein disruption results in NMJ abnormalities [30].
This sensory phenotype is also found in human patients with DYNC1H1 mutations, but with important differences that may be explained at least in part by modifier genes associated with a different genetic background. Individuals bearing the DYNC1H1H306R mutation range from having mild reduction of proprioception to deficits in all sensory modalities in the UK pedigree [24], but normal sensory nerve conduction and no obvious sensory abnormalities in the Japanese pedigree [25]. Surprisingly, several human patients with DYNC1H1 mutations have no obvious sensory loss [22,26,33] (Table 3), raising the possibility that this is due to a permissive genetic background in these patients. An alternative explanation is that the human sensory system is more robust than in rodents and is protected from the deleterious effects of DYNC1H1 mutation by compensatory mechanisms. In this regard, the resulting sensory phenotype may be too subtle to be clinically relevant and detected by current protocols.
Abnormalities in brain morphology and function
Dync1h1 mutations affect brain function at multiple levels. Brain imaging showed striatal atrophy and lateral ventricle enlargement in Dync1h1+/Cra1 mice, which also had altered dopamine signalling. However, neuronal loss was found only in the substantia nigra [31]. In humans, Perry syndrome, an atypical Parkinson-like disease, can be caused by point mutations in the p150 subunit of dynactin [37] (Table 1). Furthermore, cytoplasmic dynein directly binds huntingtin (Htt) and huntingtin-associated protein 1 (HAP1), two proteins involved in the physiology of the dopaminergic system, via dynactin [38] (Table 1).
Correct cortical development also requires cytoplasmic dynein. MCD has been detected in several cases with DYNC1H1 mutations [21,22] and is often associated with microcephaly, pachygyria (presence of pathological thick convolutions of the cerebral cortex), polymicrogyria (excessive number of small convolutions of the cortex), and other central nervous system (CNS) malformations. Patients often display early-onset or focal epilepsy and mild-to-severe intellectual disability (Table 3). Interestingly, two cases (DYNC1H1K3336N and DYNC1H1R3384Q) display both pachygyria and polymicrogyria patterns, which are normally regarded as distinct clinical entities and have been found associated only in patients with WDR62-related brain abnormalities [39].
These malformations probably arise because the cytoplasmic dynein complex is important for neuronal migration [40]. This may also explain the abnormal migration of facial motor neuron cell bodies to the hindbrain found in E10.5 Dync1h1Loa/Loa embryos [15]. This striking phenotype was not observed for cranial and spinal nerves in the neural tube, suggesting that the susceptibility of distinct neuronal subtypes to the same Dync1h1 mutations varies within the same genetic background. This possibility is strengthened by the finding that alpha rather than gamma trigeminal motor neurons or proprioceptive neurons are affected in Dync1h1+/Loa mice, showing abnormal dendrites, aberrant mitochondria, and other defects consistent with altered trafficking of organelles [41]. Uncovering the mechanistic basis of this differential sensitivity will greatly further our understanding of the factors determining type-specific neuronal cell death as a response to generic insults or mutations of ubiquitously expressed proteins, a major unsolved problem in basic and clinical neuroscience.
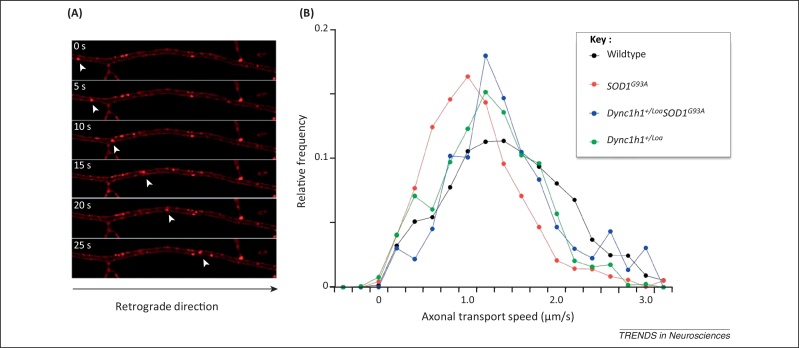
Based on the importance of cytoplasmic dynein in microtubule-dependent movement [42], the effects of Dync1h1 mutations on axonal transport were tested. Motor neuron cultures from Dync1h1Loa/Loa but not Dync1h1+/Loa embryos show a highly significant reduction in axonal retrograde transport [15,43]. The lack of an overt phenotype in heterozygous neurons is unexpected because mutant DYNC1H1 acts as a dimer [15,28]. The DYNC1H1Loa protein displays impaired run-lengths on microtubules [29] and the human DYNC1H1I584L protein has decreased binding to microtubules in the presence of ATP [26]. Both mutations have a similar biochemical profile, which is interesting given their proximity and highly conserved nature. The lack of an axonal transport phenotype in cultured Dync1h1+/Loa embryonic motor neurons was confirmed by assessing the retrograde transport of signalling endosomes in the sciatic nerve of adult Dync1h1+/Loa mice by intravital microscopy [44]. This technique allows the quantitative real-time assessment of the retrograde movement of transport organelles in the intact sciatic nerve of living mice and revealed only a minor drop in high transport speed in Dync1h1+/Loa mice (Figure 2), suggesting that only a subset of the specific functions of dynein is affected in these mice, causing a mild phenotype compatible with life.
Figure 2.
Quantitative analysis of axonal retrograde transport by intravital microscopy. (A) Axonal retrograde transport of signalling endosomes containing a fluorescently labelled atoxic fragment of tetanus neurotoxin was monitored in single axons in the intact sciatic nerve by time-lapse confocal microscopy and shown as a time series [44]. (B) The deficit in axonal retrograde transport observed in early symptomatic SOD1G93A transgenic mice (74 ± 1.7 days; in red) is almost completely rescued by the Dync1h1Loa allele in Dync1h1+/LoaSOD1G93A double-mutant mice (in blue). The speed distribution profile displayed by Dync1h1+/LoaSOD1G93A mice overlaps with that observed in Dync1h1+/Loa animals (in green) and is slightly shifted towards lower speed values compared with wild type mice (in black) of similar age.
Mutations in dynein heavy chain and neurodegenerative diseases
Although human DYNC1H1 mutations have been described causing CMT2 neuropathy (DYNC1H1H306R) [24], SMA (DYNC1H1H306R) [25], (DYNC1H1I584L, K671E, Y970C) [26], and severe cognitive/neuronal migration deficits (DYNC1H1E1518K, H3822P) [20,21], to date some of the most interesting results for dissecting disease mechanisms come from the mouse mutants. In this section, we focus on a subset of breeding experiments testing the genetic interactions of Dync1h1 mutations with known genes important for neurodegenerative diseases.
ALS
ALS is a neurodegenerative disease that usually manifests in midlife and results in progressive loss of upper and lower motor neurons, leading to paralysis and death within 3–5 years of diagnosis. Around 10–20% of ALS is an inherited autosomal-dominant disease and of these familial cases, about 10% are due to mutations in the superoxide dismutase 1 (SOD1) gene. Mice carrying human mutant SOD1 transgenes model ALS [36,45] and develop deficits including defective axonal retrograde transport (for example, see [43,44,46]).
SOD1G93A transgenic ALS mice were crossed to Dync1h1+/Loa mice and double-mutant progeny (i.e., SOD1G93ADync1h1Loa) surprisingly lived 28% longer than their SOD1G93A transgenic littermates, with a significant increase in survival of spinal cord motor neurons at a 120-day late-disease time point compared with SOD1G93A littermates [43] (Table 4). Curiously, double mutants had an increased rate of axonal retrograde transport in embryonic motor neuron cultures compared with their single-mutant and wild type littermates [43]. Interestingly, the Dync1h1Loa allele is able to rescue almost completely the axonal transport deficits observed in early symptomatic SOD1G93A mice, as shown by intravital microscopy (Figure 2). This restoration of axonal retrograde transport may determine the amelioration of the ALS disease phenotype observed in Dync1h1LoaSOD1G93A double mutants, including significant life extension (see below).
Table 4.
Genetic interactions between SOD1 and the cytoplasmic dynein complexa
| Dync1h1 mutant mouse parent | SOD1-ALS mutant mouse parent | Effect on lifespan of double mutants compared with SOD1-ALS littermates (time to humane end point shown for SOD1-ALS double mutants compared with their SOD1-ALS littermates) | Refs |
|---|---|---|---|
| Dync1h1+/Loa on C57BL/6 congenic background | SOD1G93A transgenic on hybrid SJL, C57BL/6 background | 28% significant increase in lifespan (160 days, n = 18, compared with 125 days, n = 20) | [43] |
| Dync1h1+/Loa on C57BL/6 congenic background | SOD1G93A transgenic on hybrid SJL, C57BL/6 background | 21% significant increase in lifespan (156 days, n = 12, compared with 129 days, n = 16) | [14] |
| Dync1h1+/Loa on C57BL/6 congenic background | SOD1G93A transgenic on C57BL/6 congenic background | 9% significant increase in lifespan (165 days, n = 9, compared with 152 days, n = 9) | [34] |
| Dync1h1+/Cra1 on C3H background | SOD1G93A transgenic on congenic C57BL/6 background | 14% significant increase in lifespan (167 days, compared with 147 days, n = 14/15) | [47] |
| Dync1h1+/Swl on C3H/101 background | SOD1G93A transgenic on hybrid SJL, C57BL/6 background | No significant differences in lifespan (124 days double mutants, n = 15, compared with 122 days single mutants, n = 15) | [14] |
| BICD2GFP-N on FVB background | SOD1G93A low-copy transgenic on FVB/N background | 14% increase in lifespan (271 days, n = 12, versus 237 days, n = 12 | [48,92] |
| Dync1h1+/Loa on C57BL/6 congenic background | SOD1G37R transgenic on C57BL/6 congenic background | No significant differences in lifespan (192 days, n = 10, compared with 189 days, n = 28) | [34] |
| Dync1h1+/Loa on C57BL/6 congenic background | SOD1G85R transgenic on C57BL/6 congenic background | 6% increase in lifespan (386 days, n = 14, compared with 364 days, n = 43) | [34] |
Effects on lifespan (to humane end point) of double-mutant progeny from crossing SOD1-ALS mutant mice to animals with mutations in Dync1h1 or other strains discussed in this review.
This phenomenon was unexpected and therefore similar experiments were performed by different laboratories by crossing Dync1h1+/Loa mice to various mutant SOD1 strains. Although the increase in lifespan is reproducible (21% life extension was found in an identical cross [14]), changing the genetic background of SOD1G93A mice to C57BL/6 reduced this increase to only 9% [34] (Table 4). This effect was confirmed using a different Dync1h1 mutation (Dync1h1+/Cra1). Indeed,,a 14% increase in time to humane end point was found in Dync1h1+/Cra1SOD1G93A double mutants compared with SOD1G93A littermates [47]. Further support for a functional interaction between SOD1 and the dynein motor is provided by work using a transgenic mouse with chronically impaired cytoplasmic dynein and dynactin function because of neuronal expression of Bicaudal D2 N terminus (BICD2GFP-N) [48]. BICD2 is a conserved motor-adaptor protein that is involved in dynein-mediated transport by linking the dynein motor complex to various cargoes (Table 1). It plays a major role in organelle trafficking in various cells, including neurons, and its mutation causes autosomal-dominant SMA and hereditary spastic paraplegia (HSP) [49–51]. Double mutants of BICD2GFP-N with ‘low-copy’ SOD1G93A mice also show an increased lifespan (+14%), further supporting a genetic interaction between SOD1 and cytoplasmic dynein and suggesting that the expression level of the SOD1G93A transgene is not likely to play a role in this phenomenon.
What is the molecular mechanism at the basis of the amelioration of the ALS phenotype by Dync1h1 mutations? Expression of SOD1G93A in primary motor neurons alters the cellular localisation of cytoplasmic dynein [52]. As a consequence, SOD1G93A mice have axonal transport defects and a change of cargoes in the axon from survival signals to stress/death signals [53]. Thus, inhibiting a specific subset of retrograde organelles may help delay the activation of cell stress pathways. This could help motor neurons to survive SOD1G93A toxicity [53] and may determine the rescue of axonal retrograde transport deficits observed in Dync1h1LoaSOD1G93A double mutants by intravital microscopy (Figure 2). Additionally, differences in the expression of the main anterograde motor kinesin 1 have also been noted in Dync1h1+/Cra1SOD1G93A mice [54].
Although the presence of a direct interaction between DYNC1H1 and SOD1 remains controversial [55,56], Dync1h1LoaSOD1G93A mice have significant reductions in mutant SOD1 protein in the mitochondrial matrix [57] that determine amelioration of mitochondrial respiration and restoration of membrane potential in embryonic motor neurons. Therefore, these neurons are more resistant to the toxic effects of mutant SOD1 on mitochondria [57]. This phenotype is surprising given that mouse fibroblasts derived from Dync1h1+/Cra1 and Dync1h1Cra/Cra1 embryos display profound alterations of mitochondrial morphology and that Dync1h1+/Cra1 mice develop, hyperinsulinemia, hyperglycaemia, and progressive mitochondrial dysfunction [58]. Similar mitochondrial abnormalities have also been reported in fibroblasts isolated from SMA patients carrying the DYNC1H1I584L and DYNC1H1K671E mutations [58].
Tau redistribution has also been called on to explain the increased lifespan found in double-mutant animals. Dync1h1+/Cra1SOD1G93A mice display some restoration of tau isoform ratios compared with single-mutant animals, although the main effects were in the cortex and cerebellum rather than the spinal cord [59]. Dync1h1Cra1SOD1G93A mice also show increased systemic expression of insulin-like growth factor 1 (IGF-1), which may contribute to the amelioration of the SOD1G93A disease phenotype [60]. Indeed, viral delivery of IGF-1 to the CNS has been shown to delay motor neuron death in SOD1G93A mice [61], although the efficacy of IGF-1-based therapy in ALS patients remains controversial [62]. An improved disease outcome may also result from reduced excitotoxicity caused by loss of glutamatergic proprioceptive sensory neurons [34]. However, this seems unlikely because the Dync1h1Swl mutation, which causes a greater loss of such neurons, does not rescue the mutant SOD1G93A phenotype [14] (see below).
Despite these intriguing results, no significant increase in lifespan was observed when the Dync1h1+/Loa mouse was crossed to two other SOD1-ALS models: dismutase-active SOD1G37R and dismutase-inactive SOD1G85R transgenic mice [34]. Similarly, a comparable Dync1h1+/Swl cross with SOD1G93A mice showed no effect of this dynein mutation on lifespan [14] (Table 4). Despite active research in this area, the molecular basis for these discrepancies is presently unknown, although the slower progression, lower penetrance, and severity of the SOD1G37R and SOD1G85R mutant phenotypes may explain some of the experimental differences found using these strains.
Huntington's disease (HT) and other neurological diseases
Functional interactions with other genes implicated in human neurodegenerative diseases, in addition to ALS, have also been inferred. Treatment of cells expressing mutant Htt or alpha-synuclein, which are mutated in HD and Parkinson's disease (PD), respectively, with a cytoplasmic dynein inhibitor caused inhibition of the clearance of both aggregate-prone proteins by autophagy [63]. When Dync1h1+/Loa mice were crossed to an HD model, tremor onset and severity were greatly enhanced by the dynein mutation in the double-mutant progeny, which had significantly shorter lifespans [63]. This was thought to arise from the deleterious effects of the Dync1h1Loa mutation on autophagy, which relies on cytoplasmic dynein for the intracellular transport and fusion of autophagosomes with lysosomes [63]. Interestingly, Htt and HAP1 help to control vesicle transport by interacting with the dynein–dynactin complex [38] and mutant Htt adversely affects dynein transport of brain-derived neurotrophic factor (BDNF) [64,65]. Crucially, Htt protein levels are decreased in dynein-mutant mice [64]. Interestingly, defects in brown and white adipose tissues were found in Dync1h1+/Loa and Dync1h1+/Cra1 mice, reminiscent of those seen in human HD patients, potentially because of the functional role of dynein in lipid droplet trafficking [66].
Enhancement of the severity of the disease phenotype was also observed in other animal models of neurodegeneration crossed with Dync1h1+/Loa mice. Mutations in the enzyme glycyl-tRNA synthetase (GARS) cause motor and sensory axon loss in humans due to dose-dependent gain of function and give rise to clinical phenotypes that range from forms of CMT neuropathy to severe infantile SMA. A mouse with a missense mutation in the Gars gene that has locomotor and sensory deficits was crossed to Dync1h1+/Loa mice and, as expected, the double mutants were more severely affected than either parent [67]. However, it is presently unclear whether this increase in severity is linked to a defect in the known role of GARS in translation, amino acid mischarging, or a still unknown mechanism [68].
Several pathogens and virulence factors exploit cytoplasmic dynein and other molecular motors to reach their site of action or replication, harness the biosynthetic machinery towards their replication compartment, or leave their host and spread [69] to other cells or tissues. In agreement with this view, vesicles containing mammalian prion protein (PrPC) have been shown to engage both cytoplasmic dynein and kinesin 1 for their intracellular movement [70] and clinical prion disease correlates with impaired axonal transport in motor neurons [71]. However, no differences in prion disease incubation times were found in Dync1h1+/Loa and wild type littermates inoculated intraperitoneally and intracerebrally with mouse-adapted scrapie protein [72]. This result suggests that, although PrPC undergoes dynein-mediated intracellular trafficking, PrPSc toxicity relies on alternative transport mechanisms independent of this molecular motor. This conclusion is in agreement with recent evidence showing that the plasma membrane is a primary site for prion conversion [73,74] and that the transfer of PrPSc among adjacent cells could occur via tunnelling nanotubes [75].
Concluding remarks and future perspectives
The cytoplasmic dynein heavy chain is involved in housekeeping and neuron-specific cellular processes. The first allelic series of Dync1h1 mutations in mice provided important insight into the function of this motor complex, but at the same time highlighted some phenotyping differences between investigating laboratories that remain incompletely understood. Crucially, several mutations now found in human DYNC1H1 are enriching the picture and clearly show pivotal roles for this motor complex in nervous system function. Disruption of DYNC1H1 results in developmental and degenerative defects, which may be modulated by genetic background. It is highly likely that more human and mouse mutations will be found, which will provide us with a better understanding of the roles played by this very large protein. Many intriguing questions remain (Box 2), such as why the known mutations have overlapping phenotypes but remain distinct from each other. This could be partly explained by potential disruption of binding by different cargoes and adaptors [42], for example, and we note the strong correlation in phenotype between the mouse Dync1h1Loa and the human DYNC1H1I584L mutations. Newly identified variants in regions of Dync1h1 that are still poorly understood, such as the buttress, the linker region, and the C-terminal end of the molecule (Figure 1B), will provide us with novel information on how these domains contribute to the force-generating process and determine the step size of this molecular motor. Similarly, we have only a partial understanding of the coordination of the six AAA ATPase subunits. Novel mutations in this region of DYNC1H1, such as the recently discovered DYNC1H1R1962C in AAA1, will greatly help us to dissect the precise mechanism of ATP hydrolysis and how chemical energy is translated into mechanical force.
Box 2. Outstanding questions.
-
•
How do mutations in Dync1h1 and DYNC1H1 generate such a broad spectrum of phenotypes ranging from motor neuropathies to malformations of the cerebral cortex?
-
•
Why do these mutations have cell specificity?
-
•
How many different types of cytoplasmic dynein complex exist in a eukaryotic cell?
-
•
How is the assembly of cytoplasmic dynein complexes of specific composition regulated in different cell types?
-
•
What is the basis of the functional interaction between SOD1 and Dync1h1 in mice and humans?
-
•
Why do the known Dync1h1 and DYNC1H1 mutations have overlapping phenotypes but remain distinct from each other?
-
•
Why do nearly overlapping mutations in Dync1h1 and DYNC1H1 give rise to dramatically different phenotypes and clinical outcomes?
-
•
What is the precise role of genetic background in the clinical phenotypes of Dync1h1 and DYNC1H1 mutations?
-
•
What are the roles of the novel domains identified by the mutations recently found in DYNC1H1 in dynein function and regulation?
With respect to neurodegeneration, defects in axonal retrograde transport, which is mainly driven by cytoplasmic dynein, have been reported in several neurodegenerative diseases [42,46,76] and cytoplasmic dynein trafficking is involved in processes that are often aberrant, including stress granule formation and trophic factor signalling. The intriguing finding of extended lifespan in Dync1h1+/LoaSOD1G93A double-mutant mice may inspire new avenues of research into novel potential drugs modulating axonal retrograde transport rate. Sadly, even a 10% increase in lifespan in humans with ALS would currently be a triumph. Thus, the interplay of mutant SOD1 and DYNC1H1 deserves further investigation as well as the reason why the lifespan-enhancing effect of Dync1h1+/Loa has been found, to date, only with a specific human SOD1 mutation.
With respect to development, a recent study has shown that DYNC1H1 is of fundamental importance for cell-size sensing, especially in neurons, and indeed there are defects in neuronal length in Dync1h1+/Loa mice [77]. The nature of the signals transported by cytoplasmic dynein regulating cell size and the pathways responsible for this process are presently unknown. However, their discovery holds promises for the exploitation of this machinery to sustain axonal regeneration and homeostasis in pathologies in which axonal integrity is compromised, such as CMT2 and HSP [78].
We have confined this review to data obtained from studies in mice and humans, but several results from other model organisms also support the key role of cytoplasmic dynein in neuronal physiology. These include studies in Drosophila, where the creation of an allelic series of cytoplasmic dynein heavy-chain (cDhc64C) mutants provided an early demonstration that disruption of cDhc64C (and the dynactin orthologue p150Glued) causes bidirectional disruption of axonal transport [79], thus suggesting that retrograde and anterograde transport are tightly coordinated. More recently, the zebrafish dync1h1 cannonball mutant has disrupted photoreceptor morphogenesis, revealing multiple roles for the protein in retinal development [80]. Importantly, the general principles learned by studying cytoplasmic dynein in metazoans can be transferred to lower organisms. The Dync1h1Loa mutation has been recreated in Neurospora, allowing further dissection of the molecular effects including alterations in dynein localisation and impaired speed of vesicle transport [81]. Future studies using protocols allowing the reconstitution of human cytoplasmic dynein from recombinant subunits [82] will allow the in vitro assembly of cytoplasmic dynein complexes containing specific heavy-chain mutants, such as those described in mouse and human animal models. These mutant complexes may then be tested in in vitro motility assays, allowing us to uncover the secrets of this fascinating molecular motor and its complex network of interacting proteins and regulators. Integration of in vitro studies addressing how cytoplasmic dynein works at the molecular level with future genetic investigations in mice and humans will be key to understanding how cytoplasmic dynein juggles such a range of functions and its so many physiological masters.
Acknowledgements
The authors thank Professor Josephine Peters and Dr Simon Ball for the original Dync1h1Loa mouse, Dr Lynsey Bilsland for the intravital axonal retrograde transport data, Dr S Burgess for the electron micrograph of dynein from Chlamydomonas reinhardtii flagella, and Dr Andrew P. Carter for the structural model of the cytoplasmic dynein motor domain. G.S. is funded by Cancer Research UK, L.G. is the Graham Watts Senior Research Fellow, funded by The Brain Research Trust, E.M.C.F. is funded by the UK Medical Research Council (MRC), G.S., L.G., M.H., and E.M.C.F are funded by the UK Motor Neuron Disease Association, and L.G. and E.M.C.F. are supported by the European Community Seventh Framework Programme (FP7/2007-2013).
References
- 1.Pfister K.K. Genetic analysis of the cytoplasmic dynein subunit families. PLoS Genet. 2006;2:e1. doi: 10.1371/journal.pgen.0020001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kikkawa M. Big steps toward understanding dynein. J. Cell Biol. 2013;202:15–23. doi: 10.1083/jcb.201304099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vallee R.B. Multiple modes of cytoplasmic dynein regulation. Nat. Cell Biol. 2012;14:224–230. doi: 10.1038/ncb2420. [DOI] [PubMed] [Google Scholar]
- 4.Kon T. The 2.8 angstrom crystal structure of the dynein motor domain. Nature. 2012;484:345–351. doi: 10.1038/nature10955. [DOI] [PubMed] [Google Scholar]
- 5.Carter A.P. Crystal clear insights into how the dynein motor moves. J. Cell Sci. 2013;126:705–713. doi: 10.1242/jcs.120725. [DOI] [PubMed] [Google Scholar]
- 6.Schmidt H. Insights into dynein motor domain function from a 3.3-angstrom crystal structure. Nat. Struct. Mol. Biol. 2012;19:492–497. doi: 10.1038/nsmb.2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bader J.R., Vaughan K.T. Dynein at the kinetochore: timing, interactions and functions. Semin. Cell Dev. Biol. 2010;21:269–275. doi: 10.1016/j.semcdb.2009.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Riera J., Lazo P.S. The mammalian NudC-like genes: a family with functions other than regulating nuclear distribution. Cell. Mol. Life Sci. 2009;66:2383–2390. doi: 10.1007/s00018-009-0025-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vallee R.B. Emerging roles for myosin II and cytoplasmic dynein in migrating neurons and growth cones. Trends Cell Biol. 2009;19:347–355. doi: 10.1016/j.tcb.2009.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schiavo G., Fainzilber M. Alternative energy for neuronal motors. Nature. 2013;495:178–180. doi: 10.1038/495178a. [DOI] [PubMed] [Google Scholar]
- 11.Zala D. Vesicular glycolysis provides on-board energy for fast axonal transport. Cell. 2013;152:479–491. doi: 10.1016/j.cell.2012.12.029. [DOI] [PubMed] [Google Scholar]
- 12.Harada A. Golgi vesiculation and lysosome dispersion in cells lacking cytoplasmic dynein. J. Cell Biol. 1998;141:51–59. doi: 10.1083/jcb.141.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ahmad-Annuar A. No association with common Caucasian genotypes in exons 8, 13 and 14 of the human cytoplasmic dynein heavy chain gene (DNCHC1) and familial motor neuron disorders. Amyotroph. Lateral Scler. 2003;4:150–157. doi: 10.1080/14660820310011737. [DOI] [PubMed] [Google Scholar]
- 14.Chen X.J. Proprioceptive sensory neuropathy in mice with a mutation in the cytoplasmic dynein heavy chain 1 gene. J. Neurosci. 2007;27:14515–14524. doi: 10.1523/JNEUROSCI.4338-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hafezparast M. Mutations in dynein link motor neuron degeneration to defects in retrograde transport. Science. 2003;300:808–812. doi: 10.1126/science.1083129. [DOI] [PubMed] [Google Scholar]
- 16.Tynan S.H. Distinct but overlapping sites within the cytoplasmic dynein heavy chain for dimerization and for intermediate chain and light intermediate chain binding. J. Biol. Chem. 2000;275:32769–32774. doi: 10.1074/jbc.M001537200. [DOI] [PubMed] [Google Scholar]
- 17.Habura A. Interaction mapping of a dynein heavy chain - identification of dimerization and intermediate-chain binding domains. J. Biol. Chem. 1999;274:15447–15453. doi: 10.1074/jbc.274.22.15447. [DOI] [PubMed] [Google Scholar]
- 18.Duchen L.W. A dominant hereditary sensory disorder in the mouse with deficiency of muscle spindles: the mutant Sprawling. J. Physiol. (Lond.) 1974;237:10P–11P. [PubMed] [Google Scholar]
- 19.Morris T. Sprawling (Swl) Mouse News Lett. 1967;37:29. [Google Scholar]
- 20.Vissers L.E.L.M. A de novo paradigm for mental retardation. Nat. Genet. 2010;42:1109–1112. doi: 10.1038/ng.712. [DOI] [PubMed] [Google Scholar]
- 21.Willemsen M.H. Mutations in DYNC1H1 cause severe intellectual disability with neuronal migration defects. J. Med. Genet. 2012;49:179–183. doi: 10.1136/jmedgenet-2011-100542. [DOI] [PubMed] [Google Scholar]
- 22.Poirier K. Mutations in TUBG1, DYNC1H1, KIF5C and KIF2A cause malformations of cortical development and microcephaly. Nat. Genet. 2013;45:639–647. doi: 10.1038/ng.2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qiu R. Identification of a novel site in the tail of dynein heavy chain important for dynein function in vivo. J. Biol. Chem. 2013;288:2271–2280. doi: 10.1074/jbc.M112.412403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weedon M.N. Exome sequencing identifies a DYNC1H1 mutation in a large pedigree with dominant axonal Charcot-Marie-Tooth disease. Am. J. Hum. Genet. 2011;89:308–312. doi: 10.1016/j.ajhg.2011.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tsurusaki Y. A DYNC1H1 mutation causes a dominant spinal muscular atrophy with lower extremity predominance. Neurogenetics. 2012;13:327–332. doi: 10.1007/s10048-012-0337-6. [DOI] [PubMed] [Google Scholar]
- 26.Harms M.B. Mutations in the tail domain of DYNC1H1 cause dominant spinal muscular atrophy. Neurology. 2012;78:1714–1720. doi: 10.1212/WNL.0b013e3182556c05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rossor A.M. The distal hereditary motor neuropathies. J. Neurol. Neurosurg. Psychiatry. 2012;83:6–14. doi: 10.1136/jnnp-2011-300952. [DOI] [PubMed] [Google Scholar]
- 28.Deng W.H. Neurodegenerative mutation in cytoplasmic dynein alters its organization and dynein-dynactin and dynein-kinesin interactions. J. Biol. Chem. 2010;285:39922–39934. doi: 10.1074/jbc.M110.178087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ori-McKenney K.M. A cytoplasmic dynein tail mutation impairs motor processivity. Nat. Cell Biol. 2010;12:1228–1234. doi: 10.1038/ncb2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Courchesne S.L. Neuromuscular junction defects in mice with mutation of dynein heavy chain 1. PLoS ONE. 2011;6:e16753. doi: 10.1371/journal.pone.0016753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Braunstein K.E. A point mutation in the dynein heavy chain gene leads to striatal atrophy and compromises neurite outgrowth of striatal neurons. Hum. Mol. Genet. 2010;19:4385–4398. doi: 10.1093/hmg/ddq361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tucci V. Reaching and grasping phenotypes in the mouse (Mus musculus): a characterization of inbred strains and mutant lines. Neuroscience. 2007;147:573–582. doi: 10.1016/j.neuroscience.2007.04.034. [DOI] [PubMed] [Google Scholar]
- 33.Harms M.B. Dominant spinal muscular atrophy with lower extremity predominance linkage to 14q32. Neurology. 2010;75:539–546. doi: 10.1212/WNL.0b013e3181ec800c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ilieva H.S. Mutant dynein (Loa) triggers proprioceptive axon loss that extends survival only in the SOD1 ALS model with highest motor neuron death. Proc. Natl. Acad. Sci. U.S.A. 2008;105:12599–12604. doi: 10.1073/pnas.0805422105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dupuis L. Mice with a mutation in the dynein heavy chain 1 gene display sensory neuropathy but lack motor neuron disease. Exp. Neurol. 2009;215:146–152. doi: 10.1016/j.expneurol.2008.09.019. [DOI] [PubMed] [Google Scholar]
- 36.Acevedo-Arozena A. A comprehensive assessment of the SOD1(G93A) low-copy transgenic mouse, which models human amyotrophic lateral sclerosis. Dis. Model. Mech. 2011;4:686–700. doi: 10.1242/dmm.007237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Farrer M.J. DCTN1 mutations in Perry syndrome. Nat. Genet. 2009;41:163–165. doi: 10.1038/ng.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Caviston J.P. Huntingtin facilitates dynein/dynactin-mediated vesicle transport. Proc. Natl. Acad. Sci. U.S.A. 2007;104:10045–10050. doi: 10.1073/pnas.0610628104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bilguvar K. Whole-exome sequencing identifies recessive WDR62 mutations in severe brain malformations. Nature. 2010;467:207–210. doi: 10.1038/nature09327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tsai J.W. Dual subcellular roles for LIS1 and dynein in radial neuronal migration in live brain tissue. Nat. Neurosci. 2007;10:970–979. doi: 10.1038/nn1934. [DOI] [PubMed] [Google Scholar]
- 41.Wiggins L.M. A novel phenotype for the dynein heavy chain mutation Loa: altered dendritic morphology, organelle density, and reduced numbers of trigeminal motoneurons. J. Comp. Neurol. 2012;520:2757–2773. doi: 10.1002/cne.23085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Franker M.A.M., Hoogenraad C.C. Microtubule-based transport - basic mechanisms, traffic rules and role in neurological pathogenesis. J. Cell Sci. 2013;126:2319–2329. doi: 10.1242/jcs.115030. [DOI] [PubMed] [Google Scholar]
- 43.Kieran D. A mutation in dynein rescues axonal transport defects and extends the lifespan of ALS mice. J. Cell Biol. 2005;169:561–567. doi: 10.1083/jcb.200501085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bilsland L.G. Deficits in axonal transport precede ALS symptoms in vivo. Proc. Natl. Acad. Sci. U.S.A. 2010;107:20523–20528. doi: 10.1073/pnas.1006869107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Joyce P.I. SOD1 and TDP-43 animal models of amyotrophic lateral sclerosis: recent advances in understanding disease toward the development of clinical treatments. Mamm. Genome. 2011;22:420–448. doi: 10.1007/s00335-011-9339-1. [DOI] [PubMed] [Google Scholar]
- 46.Perlson E. Retrograde axonal transport: pathways to cell death? Trends Neurosci. 2010;33:335–344. doi: 10.1016/j.tins.2010.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Teuchert M. A dynein mutation attenuates motor neuron degeneration in SOD1(G93A) mice. Exp. Neurol. 2006;198:271–274. doi: 10.1016/j.expneurol.2005.12.005. [DOI] [PubMed] [Google Scholar]
- 48.Teuling E. A novel mouse model with impaired dynein/dynactin function develops amyotrophic lateral sclerosis (ALS)-like features in motor neurons and improves lifespan in SOD1-ALS mice. Hum. Mol. Genet. 2008;17:2849–2862. doi: 10.1093/hmg/ddn182. [DOI] [PubMed] [Google Scholar]
- 49.Neveling K. Mutations in BICD2, which encodes a Golgin and important motor adaptor, cause congenital autosomal-dominant spinal muscular atrophy. Am. J. Hum. Genet. 2013;92:946–954. doi: 10.1016/j.ajhg.2013.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Oates E.C. Mutations in BICD2 cause dominant congenital spinal muscular atrophy and hereditary spastic paraplegia. Am. J. Hum. Genet. 2013;92:965–973. doi: 10.1016/j.ajhg.2013.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Peeters K. Molecular defects in the motor adaptor BICD2 cause proximal spinal muscular atrophy with autosomal-dominant inheritance. Am. J. Hum. Genet. 2013;92:955–964. doi: 10.1016/j.ajhg.2013.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ligon L.A. Mutant superoxide dismutase disrupts cytoplasmic dynein in motor neurons. Neuroreport. 2005;16:533–536. doi: 10.1097/00001756-200504250-00002. [DOI] [PubMed] [Google Scholar]
- 53.Perlson E. A switch in retrograde signaling from survival to stress in rapid-onset neurodegeneration. J. Neurosci. 2009;29:9903–9917. doi: 10.1523/JNEUROSCI.0813-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kuzma-Kozakiewicz M. Changes in kinesin expression in the CNS of mice with dynein heavy chain 1 mutation. Acta Biochim. Pol. 2013;60:51–55. [PubMed] [Google Scholar]
- 55.Strom A.L. Interaction of amyotrophic lateral sclerosis (ALS)-related mutant copper-zinc superoxide dismutase with the dynein-dynactin complex contributes to inclusion formation. J. Biol. Chem. 2008;283:22795–22805. doi: 10.1074/jbc.M800276200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang F.J. Interaction between familial amyotrophic lateral sclerosis (ALS)-linked SOD1 mutants and the dynein complex. J. Biol. Chem. 2007;282:16691–16699. doi: 10.1074/jbc.M609743200. [DOI] [PubMed] [Google Scholar]
- 57.El Kadi A.M. The Legs at odd angles (Loa) mutation in cytoplasmic dynein ameliorates mitochondrial function in SOD1(G93A) mouse model for motor neuron disease. J. Biol. Chem. 2010;285:18627–18639. doi: 10.1074/jbc.M110.129320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Eschbach J. Dynein mutations associated with hereditary motor neuropathies impair mitochondrial morphology and function with age. Neurobiol. Dis. 2013;58:220–230. doi: 10.1016/j.nbd.2013.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kuzma-Kozakiewicz M. Mice with mutation in dynein heavy chain 1 do not share the same Tau expression pattern with mice with SOD1-related motor neuron disease. Neurochem. Res. 2011;36:978–985. doi: 10.1007/s11064-011-0436-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fergani A. A mutation in the dynein heavy chain gene compensates for energy deficit of mutant SOD1 mice and increases potentially neuroprotective IGF-1. Mol. Neurodegener. 2011;6:26. doi: 10.1186/1750-1326-6-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dodge J.C. Delivery of AAV-IGF-1 to the CNS extends survival in ALS mice through modification of aberrant glial cell activity. Mol. Ther. 2008;16:1056–1064. doi: 10.1038/mt.2008.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Beauverd M. Recombinant human insulin-like growth factor I (rhIGF-I) for the treatment of amyotrophic lateral sclerosis/motor neuron disease. Cochrane Database Syst. Rev. 2012;11:CD002064. doi: 10.1002/14651858.CD002064.pub3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ravikumar B. Dynein mutations impair autophagic clearance of aggregate-prone proteins. Nat. Genet. 2005;37:771–776. doi: 10.1038/ng1591. [DOI] [PubMed] [Google Scholar]
- 64.Gauthier L.R. Huntingtin controls neurotrophic support and survival of neurons by enhancing BDNF vesicular transport along microtubules. Cell. 2004;118:127–138. doi: 10.1016/j.cell.2004.06.018. [DOI] [PubMed] [Google Scholar]
- 65.Liot G. Mutant huntingtin alters retrograde transport of TrkB receptors in striatal dendrites. J. Neurosci. 2013;33:6298–6309. doi: 10.1523/JNEUROSCI.2033-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bostrom P. SNARE proteins mediate fusion between cytosolic lipid droplets and are implicated in insulin sensitivity. Nat. Cell Biol. 2007;9:1286–1293. doi: 10.1038/ncb1648. [DOI] [PubMed] [Google Scholar]
- 67.Banks G.T. Mutant glycyl-tRNA synthetase (Gars) ameliorates SOD1(G93A) motor neuron degeneration phenotype but has little affect on Loa dynein heavy chain mutant mice. PLoS ONE. 2009;4:e6218. doi: 10.1371/journal.pone.0006218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Motley W.W. Charcot-Marie-Tooth-linked mutant GARS is toxic to peripheral neurons independent of wild-type GARS levels. PLoS Genet. 2011;7:e1002399. doi: 10.1371/journal.pgen.1002399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Salinas S. A hitchhiker's guide to the nervous system: the complex journey of viruses and toxins. Nat. Rev. Microbiol. 2010;8:645–655. doi: 10.1038/nrmicro2395. [DOI] [PubMed] [Google Scholar]
- 70.Encalada S.E. Stable kinesin and dynein assemblies drive the axonal transport of mammalian prion protein vesicles. Cell. 2011;144:551–565. doi: 10.1016/j.cell.2011.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ermolayev V. Impaired axonal transport in motor neurons correlates with clinical prion disease. PLoS Pathog. 2009;5:e1000558. doi: 10.1371/journal.ppat.1000558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hafezparast M. Prion disease incubation time is not affected in mice heterozygous for a dynein mutation. Biochem. Biophys. Res. Commun. 2005;326:18–22. doi: 10.1016/j.bbrc.2004.10.206. [DOI] [PubMed] [Google Scholar]
- 73.Goold R. Rapid cell-surface prion protein conversion revealed using a novel cell system. Nat. Commun. 2011;2:281. doi: 10.1038/ncomms1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Goold R. Alternative fates of newly formed PrPSc upon prion conversion on the plasma membrane. J. Cell Sci. 2013;126:3552–3562. doi: 10.1242/jcs.120477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Costanzo M. Transfer of polyglutamine aggregates in neuronal cells occurs in tunneling nanotubes. J. Cell Sci. 2013;126:3678–3685. doi: 10.1242/jcs.126086. [DOI] [PubMed] [Google Scholar]
- 76.Liu X.A. Pathologies of axonal transport in neurodegenerative diseases. Transl. Neurosci. 2012;3:355–372. doi: 10.2478/s13380-012-0044-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rishal I. A motor-driven mechanism for cell-length sensing. Cell Rep. 2012;1:608–616. doi: 10.1016/j.celrep.2012.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Holzbaur E.L.F. Motor neurons rely on motor proteins. Trends Cell Biol. 2004;14:233–240. doi: 10.1016/j.tcb.2004.03.009. [DOI] [PubMed] [Google Scholar]
- 79.Martin M. Cytoplasmic dynein, the dynactin complex, and kinesin are interdependent and essential for fast axonal transport. Mol. Biol. Cell. 1999;10:3717–3728. doi: 10.1091/mbc.10.11.3717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Insinna C. Analysis of a zebrafish dync1h1 mutant reveals multiple functions for cytoplasmic dynein 1 during retinal photoreceptor development. Neural Dev. 2010;5:12. doi: 10.1186/1749-8104-5-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sivagurunathan S. A mouse neurodegenerative dynein heavy chain mutation alters dynein motility and localisation in Neurospora crassa. Cytoskeleton. 2012;69:613–624. doi: 10.1002/cm.21049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Trokter M. Reconstitution of the human cytoplasmic dynein complex. Proc. Natl. Acad. Sci. U.S.A. 2012;109:20895–20900. doi: 10.1073/pnas.1210573110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Huang J. Lis1 acts as a “Clutch” between the ATPase and microtubule-binding domains of the dynein motor. Cell. 2012;150:975–986. doi: 10.1016/j.cell.2012.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.McKenney R.J. Mutually exclusive cytoplasmic dynein regulation by NudE-Lis1 and dynactin. J. Biol. Chem. 2011;286:39615–39622. doi: 10.1074/jbc.M111.289017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Schroer T.A. Dynactin. Annu. Rev. Cell Dev. Biol. 2004;20:759–779. doi: 10.1146/annurev.cellbio.20.012103.094623. [DOI] [PubMed] [Google Scholar]
- 86.Cai Q.A. Snapin-regulated late endosomal transport is critical for efficient autophagy-lysosomal function in neurons. Neuron. 2010;68:73–86. doi: 10.1016/j.neuron.2010.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhou B. Snapin recruits dynein to BDNF-TrkB signaling endosomes for retrograde axonal transport and is essential for dendrite growth of cortical neurons. Cell Rep. 2012;2:42–51. doi: 10.1016/j.celrep.2012.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Caviston J.P., Holzbaur E.L.F. Huntingtin as an essential integrator of intracellular vesicular trafficking. Trends Cell Biol. 2009;19:147–155. doi: 10.1016/j.tcb.2009.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Liu Y. Bicaudal-D uses a parallel, homodimeric coiled coil with heterotypic registry to coordinate recruitment of cargos to dynein. Genes Dev. 2013;27:1233–1246. doi: 10.1101/gad.212381.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Matanis T. Bicaudal-D regulates COPI-independent Golgi-ER transport by recruiting the dynein-dynactin motor complex. Nat. Cell Biol. 2002;4:986–992. doi: 10.1038/ncb891. [DOI] [PubMed] [Google Scholar]
- 91.Hoogenraad C.C. Mammalian Golgi-associated Bicaudal-D2 functions in the dynein-dynactin pathway by interacting with these complexes. EMBO J. 2001;20:4041–4054. doi: 10.1093/emboj/20.15.4041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Jaarsma D. CuZn superoxide dismutase (SOD1) accumulates in vacuolated mitochondria in transgenic mice expressing amyotrophic lateral sclerosis-linked SOD1 mutations. Acta Neuropathol. 2001;102:293–305. doi: 10.1007/s004010100399. [DOI] [PubMed] [Google Scholar]
- 93.Burgess S.A. Dynein structure and power stroke. Nature. 2003;421:715–718. doi: 10.1038/nature01377. [DOI] [PubMed] [Google Scholar]
- 94.Roberts A.J. AAA plus ring and linker swing mechanism in the dynein motor. Cell. 2009;136:485–495. doi: 10.1016/j.cell.2008.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Nolan P.M. A systematic, genome-wide, phenotype-driven mutagenesis programme for gene function studies in the mouse. Nat. Genet. 2000;25:440–443. doi: 10.1038/78140. [DOI] [PubMed] [Google Scholar]
- 96.Rogers D.C. SHIRPA, a protocol for behavioral assessment: validation for longitudinal study of neurological dysfunction in mice. Neurosci. Lett. 2001;306:89–92. doi: 10.1016/s0304-3940(01)01885-7. [DOI] [PubMed] [Google Scholar]
- 97.Witherden A.S. An integrated genetic, radiation hybrid, physical and transcription map of a region of distal mouse chromosome 12, including an imprinted locus and the ‘Legs at odd angles’ (Loa) mutation. Gene. 2002;283:71–82. doi: 10.1016/s0378-1119(01)00853-8. [DOI] [PubMed] [Google Scholar]
- 98.de Angelis M.H. Genome-wide, large-scale production of mutant mice by ENU mutagenesis. Nat. Genet. 2000;25:444–447. doi: 10.1038/78146. [DOI] [PubMed] [Google Scholar]