Abstract
Fungi causes most plant disease. When fruits are stored at suboptimal conditions, fungi grows, and some produce mycotoxin which can be dangerous for human consumption. Studies have shown that the Penicillium and Monilinia species commonly cause spoilage of fruits, especially apples. Several other genera and species were reported to grow to spoil fruits. This study was conducted to isolate and identify fruit spoilage by fungi on apples collected in Riyadh, Saudi Arabia and conduct a molecular identification of the fungal isolates. Thus, we collected 30 samples of red delicious and Granny Smith apples with obvious spoilage from different supermarkets between February and March of 2012 in Riyadh, Saudi Arabia. Each apple was placed in a sterile plastic bag in room temperature (25–30 °C) for six days or until fungal growth was evident all over the sample. Growth of fungal colonies on PDA was counted and sent for molecular confirmation by PCR. Six fruit spoilage fungi were isolated, including Penicillium chrysogenum, Penicillium adametzii, Penicillium chrysogenum, Penicillium steckii, Penicillium chrysogenum, and Aspergillus oryzae. P. chrysogenum was the most frequent isolate which was seen in 14 of a total of 34 isolates (41.2%), followed by P. adametzii and A. oryzae with seven isolates each (20.6%) and the least was P. steckii with six isolates (17.6%). Penicillium species comprised 27 of the total 34 (79.4%) isolates. Sequence analysis of the ITS regions of the nuclear encoded rDNA showed significant alignments for P. chrysogenum, P. adametzii and A. oryzae. Most of these fungal isolates are useful and are rarely pathogenic; however they can still produce severe illness in immune-compromised individuals, and sometimes otherwise healthy people may also become infected. It is therefore necessary to evaluate the possible production of mycotoxins by these fungi to determine a potential danger and to establish its epidemiology in order to develop adequate methods of control.
Keywords: Apples, Saudi Arabia, Fungi, Penicillium, Aspergillus, Internal transcribed spacer, Genetic, Fruits
1. Introduction
Yeasts play a significant role in nutrition, medicine, and biocontrol of plant pathogens. Certain yeasts act as antagonists in postharvest infections on fruits such as apples and citrus (Blevea et al., 2006; Janisiewicz et al., 2001). However, fungi cause most plant disease, accounting for perhaps 70% of all major crop diseases (Doores and Splittstoesser, 1983; Janisiewicz et al., 2001). Apart from the effects of high temperature and relative humidity, fungi produce pectic enzymes that break down apple pectin to expose the nutrients of the cells to the fungi (Blevea et al., 2006; Garcia et al., 2011).
Fruits such as apples contain high levels of sugars and nutrients, making them desirable for fungal growth (Prasad, 2007). Fungi can take hold of apples, particularly through a puncture or other wound that breaks the skin of the apple. Toxigenic fungi have been isolated from spoiling fruits (Pose et al., 2004). When fruits are stored at suboptimal conditions, it promotes fungal growth and mycotoxin production (Tournas and Memon, 2009).
The most common causes of apple rot are from the fungi Penicillium expansum and Monilinia fructigena (Fiori et al., 2008; Holb and Scherm, 2008). Other fungal genera that were isolated from apples include Colletotrichum, Xylaria, Botryosphaeria (Camatti-Sartori et al., 2005) and Rhizopus oryzae (Kwon et al., 2011). Aspergillus spp. has also been isolated and known to cause infections or allergies (Mons, 2004). In some studies, Cladosporium spp. was found to be a frequent fungus found in stored apples, and also Penicillium, Acremonium, Aspergillus, Aureobasidium, Cryptococcus, Sporobolomyces and Alternaria spp. (Robiglio and Lopez, 1995; Watanabe, 2008).
Traditional practices for studies of fungi include conventional cultivation and microscopic identification (Advances in Food Mycology, 2005). Identification of the fungi species is based on mycelia (color, size and shape) and morphological characteristics (morphology, conidia size and morphology conidiophore) (Al-Hindi et al., 2011; Pitt and Hocking, 2009). These techniques require skilled taxonomists. Minor differences in medium composition can impair effective comparison of mycelia characters (Larone, 1995). Molecular techniques have been demonstrated as an effective and easy way to identify fungi. DNA-based assays are reliable to detect a variety of fungi. Various molecular approaches have been used for the detection of Aspergillus from environmental and clinical samples (Einsele et al., 1997; Leinberger et al., 2005; Liu, 2011). Targets for the genus level detection of Aspergillus have included the 18S rRNA gene, mitochondrial DNA, the intergenic spacer region and the internal transcribed spacer (ITS) regions The ITS regions are located between the 18S and 28S rRNA genes and offer distinct advantages over other molecular targets including sensitivity due to the existence of approximately 100 copies per genome. The sequency variation of the ITS regions has led to their use in phylogenetic studies of many different organisms (Anaissie et al., 2009). Other studies used ITS 1 and 2 nucleotide sequences of the clinically important Aspergillus species and determined whether sufficient variability existed for identification to species level (Henry et al., 2000; Hinrikson et al., 2005). We conducted this study to isolate and identify fruit spoilage by fungi on apples and conduct a molecular identification of the fungal isolates.
2. Materials and methods
2.1. Sample source
2.1.1. Red delicious apples
This apple variety has a deep red color and thick skin. The apple skin is bitter in taste and its edible portion has a crispy texture. Farmers prefer this popular cultivar over local varieties. Unlike the rounder varieties, the red delicious apples are thinner.
2.1.2. Granny Smith apples
The sweetness and tartness of Granny Smith apples are well-balanced. These green colored apples are durable and retain their freshness throughout the shipping journey. It is possible to store the Granny Smith apples for about 6 months in cold storage.
2.2. Sample collection
Thirty samples of apples were collected from different supermarkets between February and March of 2012 in Riyadh, Saudi Arabia. The samples were collected six at a time over five visits to various supermarkets. A total of fifteen apples of the red delicious variety and fifteen of the Granny Smith variety were collected. The collected apples had some obvious lesion or spoilage. Each apple was placed in a sterile plastic bag in room temperature (25–30 °C) for six days or until fungal growth was evident all over the sample (after approximately six days). Spoiled or diseased apples were identified by morphological examination using the method of Bukar et al. (2009).
2.3. Fungal isolation and purification
The apple fruits were divided into species. The samples, which were apparently diseased, were cut from the advancing edges of the lesion with a sterilized knife. The cut portion of the lesion was disinfected with ethanol of 85% concentration for 2 minutes. These were then rinsed in three different changes of distilled water. Each portion was then homogenized using a sterile glass rod and a test tube containing 10 ml of the homogenate (1 g + 9 ml) (101) was made and serially diluted down to 104. Plates of already prepared Potato Dextrose Agar (PDA) containing Chloramphenicol (30 mg/l) to prevent the growth of bacteria were inoculated with 0.1 ml aliquots of the serially diluted samples and incubated at ambient room temperature (25–30 °C) for seven days. After seven days, growth of fungal colonies on PDA was counted and recorded in a colony forming unit per milliliter (cfu/ml). Isolated species were sent for molecular confirmation.
2.4. DNA extraction and sequence analyses
DNA was extracted from isolates using the CTAB (N-cetyl-N,N,Ntrimethyl-ammonium bromide) method (Murray and Thompson, 1980). Small subunit ribosomal RNA (mtSSU rRNA) and β-tubulin were then amplified by PCR using primer pairs ITS1/ITS2 and the conditions described by O’Donnell et al. (1998), Borneman and Hartin (2000) and Glass and Donaldson (1995), respectively. PCR products were purified using the QIA quick PCR purification kit (QIAGEN, GmbH, Germany), and sequenced in both directions using the respective PCR primers. For this purpose, the Big Dye terminator sequencing kit (Version 3.1, Applied Biosystems) and an ABI PRISMTM 3100 DNA sequencer (Applied Biosystems) were used. All PCRs and sequencing reactions were performed on a GeneAmp PCR System 9700 (Applied Biosystems). Gene sequences were assembled using Sequence Navigator (Version 1.0.1, Applied Biosystems), and aligned using ClustalX; Version 1.8 (Thompson et al., 2002), after which the alignments were manually corrected where needed. The predicted sequences were then compared with the corresponding sequences in GenBank to determine the possible positions of introns. All characters were weighted equally and alignment gaps were treated as missing data. Bootstrap analysis was based on replications (Makarenkov et al., 2010).
3. Results
3.1. Fungal isolates
Six fruit spoilage fungi were isolated and identified as follows: P. chrysogenum, P. adametzii, P. chrysogenum, P. steckii, P. chrysogenum, and A. oryzae. Among red delicious apples, P. chrysogenum was isolated in six samples, P. adametzii and A. oryzae were isolated in four samples each and P. steckii was isolated in three samples. Among Granny Smith apples, P. chrysogenum was isolated in eight samples, whereas P. adametzii, A. oryzae and P. steckii were isolated in three samples each of Granny Smith apples (Table 1).
Table 1.
Frequency of occurrence of various fungal isolates and frequency percentage from 30 samples (total count: tfc/30 apples) on Potato Dextrose Agar (pda) containing Chloramphenicol (30 mg/l).
| Kinds of apples | TFC (cfu/g) | Fungi | TC |
|---|---|---|---|
| Red delicious apples | 4 × 10 | Penicillium steckii | 40 |
| 9 × 10 | Penicillium steckii | 90 | |
| 12 × 10 | Penicillium steckii | 120 | |
| 5 × 10 | Penicillium chrysogenum | 50 | |
| 64 × 102 | Penicillium chrysogenum | 6400 | |
| 12 × 10 | Penicillium chrysogenum | 120 | |
| 18 × 10 | Penicillium chrysogenum | 180 | |
| 25 × 10 | Penicillium chrysogenum | 250 | |
| 57 × 102 | Penicillium adametzii | 5700 | |
| 1 × 104 | Penicillium adametzii | 40 | |
| 4 × 10 | Penicillium adametzii | 40 | |
| 3 × 10 | Aspergillus oryzae | 30 | |
| 21 × 103 | Aspergillus oryzae | 21000 | |
| 50 × 10 | Aspergillus oryzaePenicillium adametzii | 500 | |
| 64 × 102 | Aspergillus oryzae/ Penicillium chrysogenum | 6400 | |
| Granny Smith apples | 20 × 102 | Penicillium steckii | 2000 |
| 23 × 103 | Penicillium steckii | 23000 | |
| 17 × 102 | Penicillium chrysogenum | 1700 | |
| 9 × 104 | Penicillium chrysogenum | 90000 | |
| 21 × 10 | Penicillium chrysogenum | 210 | |
| 23 × 10 | Penicillium chrysogenum | 230 | |
| 13 × 10 | Penicillium chrysogenum | 130 | |
| 15 × 10 | Penicillium chrysogenum | 150 | |
| 13 × 10 | Penicillium adametzii | 130 | |
| 23 × 10 | Penicillium adametzii | 230 | |
| 18 × 102 | Aspergillus oryzae | 1800 | |
| 24 × 103 | Aspergillus oryzae | 24000 | |
| 23 × 103 | Aspergillus oryzae/Penicillium chrysogenum | 2300 | |
| 34 × 103 | Penicillium chrysogenumPenicillium adametzii | 34000 | |
P. chrysogenum was the most frequent isolate, seen in 14 of a total of 34 isolates (41.2%), followed by P. adametzii and A. oryzae with seven isolates each (20.6%), and the least common was P. steckii with six isolates (17.6%). Penicillium species consisted of 27 of the total of 34 (79.4%) isolates. (Table 2)
Table 2.
Total count, frequency of occurrence of various fungal and frequency percentage from 30 samples on Potato Dextrose Agar (PDA) containing chloramphenicol (30 mg/l).
| Genera and species | Total count | Frequency | % |
|---|---|---|---|
| Penicillium chrysogenum | 4180 | 14 | 41.2 |
| Penicillium adametzii | 1195 | 7 | 20.6 |
| Penicillium steckii | 747 | 6 | 17.6 |
| Aspergillus oryzae | 1648 | 7 | 20.6 |
| Total count | 379 | 34 | 100 |
3.2. PCR identification
Sequence analysis of the ITS regions of the nuclear encoded rDNA showed significant alignments of 97–99% in sample 1 for P. chrysogenum with 550/557 (99%) identities, 96–100% alignment in sample 2 for P. adametzii with 710/715 (99%) identities, and 99% alignments in all 10 accessions for A. oryzae in sample 3 with 519/525 (99%) identities (Table 3). Similarities within each morphotype were relatively high, within the range of 96% to 100%. Inter-morphotype score was considerably less than this range (52–95). (Table 4) Fig. 1 shows the dendogram generated based on the similarity values of the three strains as well as reference strains obtained from GenBank. Isolates within each morphotype were clustered together and distinctly separated from each other. The generated phylogenetic tree (Fig. 1) shows that the studied isolates were clustered in three groups. The first group includes 3 isolates: numbers 1, 6 and 5. The second group includes the 2 isolates: numbers 2 and 3. The third group includes isolate number 4. Many authors have reported similar results. In this concern, Mansfield showed that ITS regions’ analyses detected a greater number of species then selective plating (Mansfield and Kuldau, 2007). Peterson also isolated 2 new Penicillin species from peanut field soils and used ITS to identify the novel species (Peterson and Horn, 2009).
Table 3.
Sequence and alignment of three isolates.
| Isolate no (MSA1) |
| 1 gaatccgagt gaggctctgg gtccacctcc cacccgtgtt tattttacct tgttgcttcg |
| 61 gcgggcccgc cttttctggc ccccgggggg cttacccccc cgggcccgcc ccccccgaag |
| 121 acaccctcga actctgtctg aagattgtag tctgagtgta aatataaatt atttaaaact |
| 181 ttcaacaacg gatctcttgg ttccggcatc gatgaagaac gcagcgaaat gcgatacgta |
| 241 atgtgaattg caaattcagt gaatcatcga gtctttgaac gcacattgcc ccccctggta |
| 301 ttccggccgg catgcctgtc cgagcgtcat ttctgccctc aagcacggct tgtgtgttgg |
| 361 gccccgtcct ccgatcccgg gggacgggcc cgaaaggcag cggcggcacc gcgtccggtc |
| 421 ctcgagcgta tggggctttg tcacccgctc tgtaggcccg gccggcgctt gccgatcaac |
| 481 ccaaattttt atccaggttg acctcggatc aggtaccgat acccgctgaa cttaagcata541 tcaataggac ggaggaag |
| Isolate no (MSA2) |
| 1 tcggccacgt cgattccggc aagtccacca ccaccggtaa gcttcaatcc ccccaacctt |
| 61 cgcatcccat cgaacaacag aactgattca tcaatttagg tcacttgatc tacaagtgtg |
| 121 gtggtatcga ctctcgtacc atcgagatgt tcgagaagga ggctgctgag ctcggtaagg |
| 181 gttccttcaa gtacgcttgg gttcttgaca agctcaaggc cgagcgtgag cgtggtatca |
| 241 ccatcgatat cgccctctgg aagttccaga cctccaagta tgaggtcacc gtcattggta |
| 301 agctttatcc ctgagccctc tttattctct ttcagccatt aaccctttta tttcagatgc |
| 361 ccccggtcac cgtgacttca tcaagaacat gatcactggt acctcccagg ctgactgcgc |
| 421 cattctcatc attgcctccg gtactggtga gttcgaggct ggtatctcca aggatggcct |
| 481 gacccgtgag cacgctctgc ttgctttcac ccttggtgtc cgccagctca tcgtcgccct |
| 541 gaacaagatg gatacctgca agtgggctga ggagcgttac atcgagattg tcaaggagac |
| 601 ctccaacttc atcaagaagg tcggctacaa ccccaagggt gtccccttcg tccccatctg |
| 661 cggtttcaac ggtgacaaca tgcttgagcc ctcccccaac tgcccctggt acaagc |
| Isolate no (MSA3) |
| 1 acggtgctgc tttctggtat gtctcaatgc cttcgagtta gtatgctttg gaccaaggaa |
| 61 ctcctcaaaa ccatgatctc ggatgtgtcc tgttatatct gccacatgtt cgctaacaac |
| 121 tttgcaggca aaccatctct ggcgagcacg gccttgacgg ctccggtgtg taagtacagc |
| 181 ctgtatacac ctcgaacgaa cgacgaccat atggcattag aagttggaat ggatctgacg |
| 241 gcaaggatag ttacaatggc tcctccgatc tccagctgga gcgtatgaac gtctacttca |
| 301 acgaggtgcg tacctcaaaa tttcagcatc tatgaaaacg ctttgcaact cgtgaccgct |
| 361 tctccaggcc agcggaaaca agtatgtccc tcgtgccgtc ctcgttgatc ttgagcctgg |
| 421 taccatggac gccgtccgtg gcggtccctt cggtcagctc ttccgtcccg acaacttcgt |
| 481 taacggccag tccggtgctg gtaacaactg ggccaagggt cactc |
| Isolate no (MSA4) |
| 1 cagcgctgcg ttgagaccaa ccgtgagatt tacttgaaca ttggtctgaa ggctgccact |
| 61 ttgactagtg gtcttaaata tgctcttgct acaggtaact ggggtgagca aaagaaggca |
| 121 gccagcgcca aggccggtgt ttctcaagtg ctgagtcgct atacatttgc ttcttcgtta |
| 181 tctcacttgc gtcggactaa tacgcccatt ggtcgtgatg gtaagattgc caaacctcgc |
| 241 cagctacata atacccactg gggtctggtt tgtccagccg aaacacctga aggacaggct |
| 301 tgtggtctcg tcaagaactt ggcactcatg tgctacatca ctgtcggtac cccaagcgag |
| 361 cctatcattg atttcatgat ccaaagaaac atggaagttc tggaagagtt cgagccccaa |
| 421 gtcacaccaa atgccacgaa ggtcttcgtc aatggtgtct gggttggtat ccaccgtgat |
| 481 ccatcgcatc tggtgaacac tatgcaatcg ctgcgtcgac gcaacatgat ttcccacgaa |
| 541 gtcagcttaa ttcgtgatat ccgtgagcga gagttcaaga tcttcaccga tactggacgt |
| 601 gtctgccgtc cattgttcgt cgttgacaat gatcctaaga gcgaaaatgc cggatctttg |
| 661 attctcaaca aggaacatat tcacaagctt gaacaagata aggacttgcc acttgatatg |
| 721 gatgtggaag agcgccggga gcgttacttc ggatgggatg gtcttgttcg atcaggtgcc |
| 781 gttgaatacg tcgatgctga agaagaagag acaattatga ttgtcatgac accagaagat |
| 841 cttgagatct ccaaacagct tcaggctggc tatgcactgc ccgaggagga gaccaatgat |
| 901 ccaaacaagc gagtc |
| Isolate no (MSA5) |
| 1 ttaccaagct tcgtagtgac tgcggaggac attaccgagt gagggccctc tgggtcctac |
| 61 ctcccacccg tgtttatttt accttgttgc ttcggcgggc ccgccttaac tggccgcggg |
| 121 ggggcttacg cccccgggcc cgcgcccgcc gaagacaccc tcgttctctg tctgaagatt |
| 181 ctagtctgag tgaaaatata aattatttaa aactttcaac aacgggtctc ttggttccgg |
| 241 gatcgatgaa gaacgcagcg aaatgcgata cgtaatgtga attgcaaatt cagtgaatca |
| 301 tcgagtcttt gaacgcacat tgcgccccct ggtattccgg ggggcatgcc tgtccgatcg |
| 361 tcatttctgc cctcaagcac ggcttgggtg tggggccccg ccctcggggg gggggggggg |
| 421 gggggcagaa cgccccgggg aaacatcggg gggccctccc aaagagtgat aattaaaaca |
| 481 aagggggggg gggggggggg ggccccccag ccccccccgc tttttttagt gcctcccgcc |
| 541 cggggggcca aaaggaaaaa cccttttttt tttttttttt tttgggaggg ggggggggag |
| 601 aaggg |
| Isolate no (MSA6) |
| 1 ggtttcgagt gagcccctct gggtccacct cccacccgtg tttattttac cttgttgctt |
| 61 cggcgggccc gccttaactg gccgccgggg ggcttacgcc cccgggcccg cgcccgccga |
| 121 agacaccctc gaactctgtc tgaagattgt agtctgagtg aaaatataaa ttatttaaaa |
| 181 ctttcaacaa cggatctctt ggttccggca tcgatgaaga acgcagcgaa atgcgatacg |
| 241 taatgtgaat tgcaaattca gtgaatcatc gagtctttga acgcacattg cgccccctgg |
| 301 tattccgggg ggcatgcctg tccgagcgtc atttctgccc tcatgcacgg cttgtgtgtt |
| 361 gggccccgtc ctccgatccc gggggacggg cccgaaaggc agcggcggca ccgcgtccgg |
| 421 tccacgagcg tatggggctt tgtcacccgc tctgtaggcc cggccggcgc ttgccgatca |
| 481 acccaaattt ttatccaggt tgtcctcgga tcaggtaggg atacccgctg aacttaagga |
| 541 tatcaaaggc cggggaggaa t |
Table 4.
The similarity between the sequences of isolates.
| SeqA | Name | Length | SeqB | Name | Length | Score |
|---|---|---|---|---|---|---|
| 1 | Isolate1 | 558 | 2 | Isolate2 | 716 | 54.0 |
| 1 | Isolate1 | 558 | 3 | Isolate3 | 525 | 52.0 |
| 1 | Isolate1 | 558 | 4 | Isolate4 | 915 | 60.0 |
| 1 | Isolate1 | 558 | 5 | Isolate5 | 605 | 78.0 |
| 1 | Isolate1 | 558 | 6 | Isolate6 | 561 | 95.0 |
| 2 | Isolate2 | 716 | 3 | Isolate3 | 525 | 58.0 |
| 2 | Isolate2 | 716 | 4 | Isolate4 | 915 | 54.0 |
| 2 | Isolate2 | 716 | 5 | Isolate5 | 605 | 46.0 |
| 2 | Isolate2 | 716 | 6 | Isolate6 | 561 | 53.0 |
| 3 | Isolate3 | 525 | 4 | Isolate4 | 915 | 66.0 |
| 3 | Isolate3 | 525 | 5 | Isolate5 | 605 | 52.0 |
| 3 | Isolate3 | 525 | 6 | Isolate6 | 561 | 53.0 |
| 4 | Isolate4 | 915 | 5 | Isolate5 | 605 | 55.0 |
| 4 | Isolate4 | 915 | 6 | Isolate6 | 561 | 60.0 |
| 5 | Isolate5 | 605 | 6 | Isolate6 | 561 | 80.0 |
Figure 1.
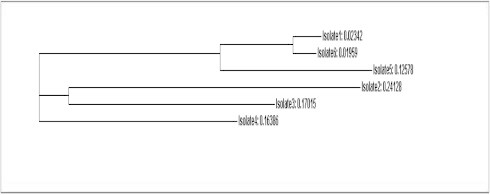
The cluster analysis and distance of tested isolates.
4. Discussion
Penicillin and Aspergillus species have been reported before as pathogens of fruit spoilage (Fiori et al., 2008; Holb and Scherm, 2008; Mons, 2004; Robiglio and Lopez, 1995; Watanabe, 2008). However, in this study, we found a different species specifically P. chrysogenum, P. adametzii and A. oryzae. P. chrysogenum, previously known as P. notatum can be found on salted food products and in damp buildings (Andersen et al., 2011; Houbraken et al., 2012). It rarely causes human disease; in fact it is the source of β-lactam antibiotics, most significantly penicillin (Anaissie et al., 2009). Transcription of genes involved in the biosynthesis of valine, cysteine and α-aminoadipic acid—precursors for penicillin biosynthesis—as well as of genes encoding microbody proteins, was increased in the high-producing strain of P. chrysogenum (Berg et al., 2008). However, despite its highly useful traits, P. chrysogenum has been implicated in several diseases. There have been reports of Intestinal invasion and disseminated disease associated with P. chrysogenum identified in immunosuppressed patients, either due to human immunodeficiency virus or from immunosuppressant medications post-transplantation (Barcus et al., 2005). In other reports, P. chrysogenum was identified to cause invasive pulmonary mycosis in transplant patients (Geltner et al., 2013).
P. adametzii on the other hand, is a glucose oxidase producing fungi (Mikhailova et al., 2007). Glucose oxidase per se, acts a natural preservative acting as bactericide in many cells. It has become commercially important in the last few years, gaining a multitude of different uses in the chemical, pharmaceutical, food, beverage, and other industries (Guimarães et al., 2006).
The genus Aspergillus, particularly A. oryzae, is a filamentous fungus used in Chinese and Japanese cuisine to ferment soybeans, saccharify rice and potatoes. Its most popular use is in making the alcoholic Japanese beverage sake and the production of rice vinegars (Rokas, 2008). Pathogenecity for A. oryzae is quite rare, although there are reported cases of fungal peritonitis caused by A. oryzae reported in the literature (Schwetz et al., 2007). Other reports showed cultures of A. oryzae in nasal mucosa and sinuses of patients undergoing chemotherapy (Pagella et al., 2007).
Although most of these fungal isolates are useful, whether in the production of penicillin or in the food industry, they can still produce severe illness in immune-compromised individuals, and sometimes otherwise healthy people may also become infected. They can be an occupational hazard, especially among people who work on farms. It is therefore necessary to evaluate the possible production of mycotoxins by these funguses to determine a potential danger and to establish its epidemiology in order to develop adequate methods of control.
Footnotes
Peer review under responsibility of King Saud University.
References
- Advances in Food Mycology. (2005). (vol. 571): Springer.
- Al-Hindi R.R., Al-Najada A.R., Mohamed S.A. Isolation and identification of some fruit spoilage fungi: screening of plant cell wall degrading enzymes. African Journal of Microbiology. 2011;5(4):443–448. [Google Scholar]
- Anaissie E.J., McGinnis M.R., Pfaller M.A. second ed. Churchill Livingstone; 2009. Clinical Mycology. [Google Scholar]
- Andersen B., Frisvad J.C., Søndergaard I., Rasmussen S., Larsen L.S. Associations between fungal species and water-damaged building materials. Applied and Environmental Microbiology. 2011;77(12):4180–4188. doi: 10.1128/AEM.02513-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barcus A.L., Burdette S.D., Herchline T.E. Intestinal invasion and disseminated disease associated with Penicillium chrysogenum. Annals of Clinical Microbiology and Antimicrobials. 2005;4(21) doi: 10.1186/1476-0711-4-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg M.A.v.d., Albang R., Albermann K., Badger J.H., Daran e.M., Driessen A.J.M. Genome sequencing and analysis of the filamentous fungus Penicillium chrysogenum. Nature Biotechnology. 2008;26(10):1161–1168. doi: 10.1038/nbt.1498. [DOI] [PubMed] [Google Scholar]
- Blevea G., Griecoa F., Cozzib G., Logrieco A., Viscontib A. Isolation of epiphytic yeasts with potential for biocontrol of Aspergillus carbonarius and A. niger on grape. International Journal of Food Microbiology. 2006;108(2):204–209. doi: 10.1016/j.ijfoodmicro.2005.12.004. [DOI] [PubMed] [Google Scholar]
- Borneman J., Hartin R.J. PCR primers that amplify fungal rRNA genes from environmental samples. Applied and Environmental Microbiology. 2000;66(10):4356. doi: 10.1128/aem.66.10.4356-4360.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukar A., Mukhtar M., Adamu S. Isolation and identification of postharvest spoilage fungi associated with sweet oranges (Citrus sinensis) traded in Kano metropolis. Bayero Journal of Pure and Applied Sciences. 2009;2(1):122–124. [Google Scholar]
- Camatti-Sartori V., Silva-Ribeiro R.T.d., Valdebenito-Sanhueza R.M., Pagnocca F.C., Echeverrigaray S., Azevedo J.L. Endophytic yeasts and filamentous fungi associated with southern Brazilian apple (Malus domestica) orchards subjected to conventional, integrated or organic cultivation. Journal of Basic Microbiology. 2005;45(5):397–402. doi: 10.1002/jobm.200410547. [DOI] [PubMed] [Google Scholar]
- Doores S., Splittstoesser D.F. The microbiology of apples and apple products. Critical Reviews in Food Science and Nutrition. 1983;19(2):133–149. doi: 10.1080/10408398309527372. [DOI] [PubMed] [Google Scholar]
- Einsele H., Hebart H., Roller G., Löffler J., Rothenhofer I., Müller C.A. Detection and identification of fungal pathogens in blood by using molecular probes. Journal of Clinical Microbiology. 1997;35(6):1353–1360. doi: 10.1128/jcm.35.6.1353-1360.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiori S., Fadda A., Giobbe S., Berardi E., Migheli Q. Pichia angusta is an effective biocontrol yeast against postharvest decay of apple fruit caused by Botrytis cinerea and Monilia fructicola. FEMS Yeast Research. 2008;8(6):961–963. doi: 10.1111/j.1567-1364.2008.00424.x. [DOI] [PubMed] [Google Scholar]
- Garcia D., Ramos A.J., Sanchis V., Marín S. Intraspecific variability of growth and patulin production of 79 Penicillium expansum isolates at two temperatures. International Journal of Food Microbiology. 2011;151(2):195–200. doi: 10.1016/j.ijfoodmicro.2011.08.021. [DOI] [PubMed] [Google Scholar]
- Geltner C., Lass-Flörl C., Hugo Bonatti, Müller L., Stelzmüller I.( Invasive pulmonary mycosis due to Penicillium chrysogenum: a new invasive pathogen. Transplantation. 2013;95(4) doi: 10.1097/TP.0b013e31827ff214. e21-e23. [DOI] [PubMed] [Google Scholar]
- Glass N.L., Donaldson G.C. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Applied and Environmental Microbiology. 1995;61(4):1323–1330. doi: 10.1128/aem.61.4.1323-1330.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guimarães L.H.S., Peixoto-Nogueira S.C., Michelin M., Rizzatti A.C.S., Sandrim V.r.C., Zanoelo F.F. Screening of filamentous fungi for production of enzymes of biotechnological interest. Brazilian Journal of Microbiology. 2006;37(4):474–480. [Google Scholar]
- Henry T., Iwen P.C., Hinrichs S.H. Identification of Aspergillus species using internal transcribed spacer regions 1 and 2. Journal of Clinical Microbiology. 2000;38(4):1510–1515. doi: 10.1128/jcm.38.4.1510-1515.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinrikson H.P., Hurst S.F., Lott T.J., Warnock D.W., Morrison C.J. Assessment of ribosomal large-subunit D1–D2, internal transcribed spacer 1, and internal transcribed spacer 2 regions as targets for molecular identification of medically important Aspergillus Species. Journal of Clinical Microbiology. 2005;43(5):2092–2103. doi: 10.1128/JCM.43.5.2092-2103.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holb I.J., Scherm H. Quantitative relationships between different injury factors and development of brown rot caused by Monilinia fructigena in integrated and organic apple orchards. Phytopathology. 2008;98(1):79–86. doi: 10.1094/PHYTO-98-1-0079. [DOI] [PubMed] [Google Scholar]
- Houbraken J., Frisvad J.C., Seifert K.A., Overy D.P., Tuthill D.M., Valdez J.G. New penicillin-producing Penicillium species and an overview of section Chrysogena. Persoonia – molecular phylogeny and evolution of fungi. 2012;29:78–100. doi: 10.3767/003158512X660571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janisiewicz W.J., Tworkoski T.J., Kurtzman C.P. Biocontrol potential of Metchnikowia pulcherrima strains against blue mold of apple. Phytopathology. 2001;91(11):1098–1108. doi: 10.1094/PHYTO.2001.91.11.1098. [DOI] [PubMed] [Google Scholar]
- Kwon J.-H., Kim J., Kim W.-I. First report of Rhizopus oryzae as a postharvest pathogen of apple in Korea. Mycobiology. 2011;39(2):140–142. doi: 10.4489/MYCO.2011.39.2.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larone D.H. fourth ed. ASM Press; 1995. Medically Important Fungi: A Guide to Identification. [Google Scholar]
- Leinberger D.M., Schumacher U., Autenrieth I.B., Bachmann T.T. Development of a DNA microarray for detection and identification of fungal pathogens involved in invasive mycoses. Journal of Clinical Microbiology. 2005;43(10):4943–4953. doi: 10.1128/JCM.43.10.4943-4953.2005. 4943-3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D. CRC Press; 2011. Molecular Detection of Human Fungal Pathogens. [Google Scholar]
- Makarenkov V., Boc A., Xie J., Peres-Neto P., Lapointe F.-J., Legendre P. Weighted bootstrapping: a correction method for assessing the robustness of phylogenetic trees. BMC Evolutionary Biology. 2010;10(250) doi: 10.1186/1471-2148-10-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansfield M.A., Kuldau G.A. Microbiological and molecular determination of mycobiota in fresh and ensiled maize silage. Mycologia. 2007;99(2):269–278. doi: 10.3852/mycologia.99.2.269. [DOI] [PubMed] [Google Scholar]
- Mikhailova R.V., Zhukovskaya L.A., Lobanok A.G. Spontaneous variability of glucose oxidase-producing fungus Penicillium adametzii LF F-2044. Applied Biochemistry and Microbiology. 2007;43(2):207–210. [PubMed] [Google Scholar]
- Mons E. Occupational asthma in greenhouse workers. Current Opinion in Pulmonary Medicine. 2004;10(2):147–150. doi: 10.1097/00063198-200403000-00010. [DOI] [PubMed] [Google Scholar]
- Murray M., Thompson W. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Research. 1980;8(19):4321–4326. doi: 10.1093/nar/8.19.4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donnell K., Kistler C., Cigelnik E., Ploetz R.C. Multiple evolutionary origins of the fungus causing Panama disease of banana: concordant evidence from nuclear and mitochondrial gene genealogies. Proceedings of the National Academy of Sciences of the United States of America. 1998;95(5):2044–2049. doi: 10.1073/pnas.95.5.2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagella F., Matti E., Bernardi F.D., Semino L., Cavanna C., Marone P. Paranasal sinus fungus ball: diagnosis and management. Mycosis. 2007;50(6):451–456. doi: 10.1111/j.1439-0507.2007.01416.x. [DOI] [PubMed] [Google Scholar]
- Peterson S.W., Horn B.W. Penicillium parvulum and Penicillium georgiense, sp. nov., isol‘ated from the conidial heads of Aspergillus species. Mycologia. 2009;101(1):71–83. doi: 10.3852/08-036. [DOI] [PubMed] [Google Scholar]
- Pitt J.I., Hocking A.D. third ed. SpringerLink: Springer e-Books; 2009. Fungi and Food Spoilage. [Google Scholar]
- Pose G., Ludemann V., Segura J., Pinto V.F. Mycotoxin production by Alternaria strains isolated from tomatoes affected by Blackmold in Argentina. Mycotoxin Research. 2004;20(2):80–86. doi: 10.1007/BF02946738. [DOI] [PubMed] [Google Scholar]
- Prasad D. Daya Publishing House; New Delhi: 2007. Sustainable Pests Management. [Google Scholar]
- Robiglio A.L., Lopez S.E. Mycotoxin production by Alternaria alternata strains isolated from red delicious apples in Argentina. International Journal of Food Microbiology. 1995;24(3):413–417. doi: 10.1016/0168-1605(94)00035-5. [DOI] [PubMed] [Google Scholar]
- Rokas A. The effect of domestication on the fungal proteome. Trends in Genetics. 2008;25(2):60–63. doi: 10.1016/j.tig.2008.11.003. [DOI] [PubMed] [Google Scholar]
- Schwetz I., Horina J., Buzina W., Roob J., Olschewski H., Krause R. Aspergillus oryzae peritonitis in CAPD: case report and review of the literature. American Journal of Kidney Diseases. 2007;49(5):701–704. doi: 10.1053/j.ajkd.2007.02.260. [DOI] [PubMed] [Google Scholar]
- Thompson J.D., Gibson T.J., Higgins D.G. UNIT 2.3 multiple sequence alignment using ClustalW and ClustalX. Current Protocols in Bioinformatics. 2002;5(1):6. doi: 10.1002/0471250953.bi0203s00. [DOI] [PubMed] [Google Scholar]
- Tournas V.H., Memon S.U. Internal contamination and spoilage of harvested apples by patulin-producing and other toxigenic fungi. International Journal of Food Microbiology. 2009;133(1–2):206–209. doi: 10.1016/j.ijfoodmicro.2009.05.025. [DOI] [PubMed] [Google Scholar]
- Watanabe M. Production of mycotoxins by Penicillium expansum inoculated into apples. Journal of Food Protection. 2008;71(8):1714–1719. doi: 10.4315/0362-028x-71.8.1714. [DOI] [PubMed] [Google Scholar]



