Abstract
Streptococcus pneumoniae DnaJ is recognized as a virulence factor whose role in pneumococcal virulence remains unclear. Here, we attempted to reveal the contribution of DnaJ in pneumococcal virulence from the identification of its interacting proteins using co-immunoprecipitation method. dnaJ was cloned into plasmid pAE03 generating pAE03-dnaJ-gfp which was used to transform S. pneumoniae D39 strain. Then anti-GFP coated beads were used to capture GFP-coupled proteins from the bacterial lysate. The resulting protein mixtures were subjected to SDS-PAGE and those differential bands were determined by matrix-assisted laser desorption/ionization time of flight mass spectrometry. We finally obtained nine proteins such as DnaK, Gap, Eno, SpxB using this method. Furthermore, to confirm the interaction between DnaJ and these candidates, bacterial two-hybrid system was employed to reveal, for example, the interaction between DnaJ and DnaK, Eno, SpxB. Further protein expression experiments suggested that DnaJ prevented denaturation of Eno and SpxB at high temperature. These results help to understand the role of DnaJ in the pathogenesis of S. pneumoniae.
Introduction
The heat shock protein (HSP) genes are highly conserved in all eukaryotes and prokaryotes [9, 27]. Streptococcus pneumoniae DnaJ is a member of conserved HSP40 family, which has been reported to function in physiological and various stress processes. Recent studies supported the idea that DnaJ is a virulence factor. In Escherichia coli, dnaJ deficient mutants did not grow as well as the wild type at temperatures above 30 °C [33]; Takaya et al. [35] reported that Salmonella enterica serovar Typhimurium DnaJ was essential for invasion of epithelial cells and survival within macrophages. DjlA, a membrane-anchored DnaJ-like protein, functioned as chaperone was closely associated with pathogenesis in Legionella dumoffii [28]. djlA mutants displayed a reduced growth rate and showed a striking loss of cytotoxic activity against Ruditapes philippinarum hemocytes in vitro in Vibrio tapetis [21]. Besides, we noticed that D39ΔdnaJ mutant grew slowly in C+Y medium, and was defected in colonization of nasal and lung. Moreover, compared with mice infected with wild type D39, mice infected with D39ΔdnaJ survived much longer in murine models [6]. Nevertheless, it’s still unclear how DnaJ is implicated in the pneumococcal virulence.
In E. coli, DnaJ is a co-chaperone, together with DnaK and GrpE, composed of DnaK/DnaJ system, which functions in folding and assembly of newly synthesized proteins, aggregation prevention, dissolution and refolding of aggregated proteins, translocation of proteins across membranes, and protein degradation [23]. Generally, DnaJ recognizes and binds substrates first, then stimulates ATP hydrolysis by Hsp70/DnaK. Also, DnaJ can recognize and bind substrate in the way independent of DnaK, such as the TorI RDF in E. coli [5].
Although previous studies have reported the substrate proteins interacting with DnaJ in some bacteria [13, 18, 36], this kind of proteins have not been reported in S. pneumoniae. Therefore, it’s very important to find DnaJ interaction proteins in S. pneumoniae which may facilitate the understanding the underlying mechanism of DnaJ implicated in pneumococcal virulence.
Here, we constructed a recombinant plasmid pAE03-dnaJ-gfp, which was used to capture the interaction proteins in S. pneumoniae by the method of co-immunoprecipitation. Using this method, we finally obtained nine proteins that would possibly interact with DnaJ in S. pneumoniae strain D39 by MALDI-TOF MS. The interactions between DnaJ and DnaK, Eno, SpxB were confirmed with bacterial two-hybrid system. Further protein expression experiments showed that DnaJ prevented denaturation of Eno and SpxB at high temperature and might also facilitate the secretion to extracellular matrix of Eno. The results provide evidence for the understanding the role of DnaJ in the pathogenesis of S. pneumoniae.
Materials and Methods
Bacterial Strains and Growth Conditions
Streptococcus pneumoniae strain D39 (NCTC 7466, serotype 2) was obtained from the National Collection of Type Cultures (NCTC, London, UK). Pneumococci were routinely grown in C+Y medium or on blood agar (BA) plates under microaerophilic conditions at 37 °C. Erythromycin (0.25 μg/ml) and chloromycetin (0.25 μg/ml) were added to the culture medium as needed.
Construction of Recombinant Plasmids and Transformation into S. pneumoniae D39
Plasmid derivatives pAE03 were integrated to the S. pneumoniae chromosome at the native gene locus by single cross-over, which allowed the expression of C-terminal DnaJ-GFP+fusion protein.
The C-terminal part of target sequence was amplified by PCR from S. pneumoniae D39 with primers dnaJ 1 and dnaJ 2, which incorporated the flanking restriction enzyme sequences of Not I and Nhe I (TaKaRa, China) restriction sites (Table 1). After digestion with Not I and Nhe I, the resulting fragment was then cloned into plasmid pAE03and transformed into E. coli BL21 (TaKaRa, China). The recombinant plasmid was transformed into S. pneumoniae D39 with 100 ng/ml synthetic CSP-1 peptide [29]. Positive strains (D39-dnaJ-gfp) were selected on BA plates supplemented with 0.25 μg/ml erythromycin, and confirmed by PCR with primers gfp1 and gfp2. Primers gfp1 and gfp2 were designed from plasmid pAE03 itself. The full length of eno and spxB were amplified by PCR from S. pneumoniae D39 with primers eno 1, eno 2 and spxB 1, spxB 2 separately, which incorporated the flanking restriction enzyme sequences of Bgl II and Sma I (TaKaRa, China) restriction sites (Table 1). After digestion with Bgl II and Sma I, the resulting fragment was then cloned into plasmid pEVP3 and transformed into E. coli BL21 (TaKaRa, China). The recombinant plasmid was transformed into S. pneumoniae D39 and D39ΔdnaJ mutant with 100 ng/ml synthetic CSP-1 peptide [29]. Positive strains (D39-pEVP3-eno/spxB or D39ΔdnaJ-pEVP3-eno/spxB) were selected on BA plates supplemented with 0.25 μg/ml chloromycetin, and confirmed by PCR with primers chlo1 and chlo2. Primers chlo1 and chlo2 were designed from plasmid pEVP3 itself. All primers were listed in Table 1.
Table 1.
Bacterial strains, plasmids and primers used in this study
| Strains | Descriptions or sequences | Sources or references |
|---|---|---|
| S. pneumoniae strain D39 | NCTC 7466, serotype 2 | The National Collection of Type Cultures (NCTC, London, UK) |
| D39ΔdnaJ mutant | dnaJ deletion mutant of D39 | This study |
| D39-dnaJ-gfp | D39 containing pAE03-dnaJ-gfp, that is dnaJ:: gfp fusion | This study |
| D39-pEVP3-eno | D39 containing pEVP3-eno, that is pEVP3:: eno fusion | This study |
| D39-pEVP3-spxB | D39 containing pEVP3-spxB, that is pEVP3:: spxB fusion | This study |
| D39ΔdnaJ-pEVP3-eno | D39ΔdnaJ containing pEVP3-eno, that is pEVP3:: eno fusion | This study |
| D39ΔdnaJ-pEVP3-spxB | D39ΔdnaJ containing pEVP3-spxB, that is pEVP3:: spxB fusion | This study |
| E. coli BL21 | Carry recombinant plasmids | TaKaRa, China |
| Plasmids | ||
| Plasmid pAE03 | Ermr | Jan-Willem Veening |
| Plasmid pEVP3 | Camr | M. Donald |
| Primers(from 5′ to 3′) | ||
| Primer sets for co-IP | ||
| dnaJ 1 | ATAAGAATGCGGCCGCAGACAAGTTTGAACGTGAAGGAAC GA | This study |
| dnaJ 2 | CTAGCTAGCTTCTCCATCAAAGGCATCTT TAATA | This study |
| gfp1 | AAAGGAGAAGAACTTTTCACTGGAG | This study |
| gfp2 | AGTAGTGACAAGTGTTGGCCATGGA | This study |
| Primers for β-galactosidase reporter gene assay | ||
| eno 1 | GAAGATCTATGTCAATTATTACTGATGTTTACG | This study |
| eno 2 | TCCCCCGGGTTTTTTAAGGTTGTAGAATGATTTC | This study |
| spxB1 | GAAGATCTATGACTCAAGGGAAAATTACTGCAT | This study |
| spxB2 | TCCCCCGGGTTTAATTGCGCGTGATTGCAATCCT | This study |
| chlo1 | TTATAAAAGCCAGTCATTAGGCCTA | This study |
| chlo2 | ATGAACTTTAATAAAATTGATTTAG | This study |
| Primers for bacterial two-hybrid system | ||
| pBT-dnaJ 1 | ATAAGAATGCGGCCGCAATGAACAATACTGAATTT | This study |
| pBT-dnaJ 2 | CCGCTCGAGTTATTCTCCATCAAAGG | This study |
| pTRG-dnaK 1 | ATAAGAATGCGGCCGCAATGTCTAAAATTATCGGTATTGACT | This study |
| pTRG-dnaK 2 | CCGCTCGAGTTACTTTTCCGTAAACTCTCCGTCT | This study |
| pTRG-eno 1 | CGCGGATCCATGTCAATTATTACTGATGTTTACG | This study |
| pTRG-eno 2 | CCGCTCGAGTTATTTTTTAAGGTTGTAGAATGAT | This study |
| pTRG-spxB 1 | CCGGAATTCAGATGACTCAAGGGAAAATTACTGCAT | This study |
| pTRG-spxB 2 | CCGCTCGAGTTATTTAATTGCGCGTGATTGCAAT | This study |
Western Blot Analysis
Streptococcus pneumoniae strain D39 and D39-dnaJ-gfp were cultured in C+Y medium until an OD600 = 0.4–0.5. Bacteria were collected by centrifugation at 8,000 rpm for 10 min and washed twice with phosphate-buffered saline (PBS, pH 7.4). The pellet was then resuspended in 4 ml PBS, followed by adding appropriate protease inhibitor cocktail (BBI) to inhibit protein degradation. Then, the pellet was sonicated for 15–20 min and cell debris was removed by centrifugation at 12,000 rpm for 30 min. The supernatant was collected and used for further studies. S. pneumoniae strain D39 and D39ΔdnaJ were cultured in C+Y medium until an OD600 = 0.4–0.5. Erythromycin (0.25 μg/ml) was added to the latter culture medium. Bacteria were collected by centrifugation at 12, 000 rpm for 2 min and washed twice with PBS. The pellet was then resuspended in 200 μl 2× SDS loading buffer, followed by boiling 10 min and the supernatants were collected by centrifugation at 13,000 rpm for 2 min.
The proteins were electroblotted onto polyvinylidene difluoride (PVDF) membranes, blocked with 5 % skim milk (Sigma), and then probed with a 1:300–500 dilution of GFP antibody (Beyotime), 1:8,000 dilution of anti-DnaJ antiserum, 1:400 dilution of anti-CodY antiserum, 1:1,000 dilution of anti-Eno antiserum or 1:2,000 dilution of anti-SpxB antiserum. Polyclonal anti-DnaJ and anti-CodY/Eno/SpxB antiserum were raised in rabbit and mouse by routine immunogenic procedures separately [11, 38]. The secondary antibody was a 1:5,000 dilution of goat anti-rabbit or mouse immunoglobulin G conjugated to horseradish peroxidase (HRP, Promega). Chemiluminescence was used to detect HRP-conjugated secondary antibody used in western blots.
Co-Immunoprecipitation (co-IP)
The procedures were performed essentially described as the protocol for GFP antibody (A.v. Monoclonal Antibody, Clontech) with some modifications [4, 12, 32]. Briefly, 1 ml cleared lysate of D39 and D39-dnaJ-gfp were transferred to a 10-ml beaker separately. 500 μl protein G-agarose beads (GE) which were prewashed with PBS twice were added to the beaker and incubated together for 3 h at 4 °C on a rotating apparatus with magnetic stirrer to remove non-specifically bound proteins. The beads were spined down and the supernatants were transferred to another 10-ml beaker. 20–25 μg GFP antibody (Clontech) was added to the supernatants and incubated at 4 °C for 1 h beforehand. Then 800 μl protein G-agarose beads were added to the supernatants and incubated overnight at 4 °C on a rotating apparatus. The beads were spined down and the supernatants were removed. The beads were washed five times with 1 ml PBS for 2 min each wash. The supernatant was discarded from final wash and the pellet was resuspended in 50–80 μl 2× SDS sample buffer. The samples were boiled for 5 min and spined down. 10–15 μl of the supernatant was loaded on an SDS/polyacrylamide gel and continued with western blot by GFP antibody detection.
Matrix-Assisted Laser Desorption/Ionization Time of Flight Mass Spectrometry (MALDI-TOF MS) analysis
Samples from co-IP were analyzed by SDS-PAGE, and then stained with Coomassie brilliant blue (CBB G-250). Differential protein bands between D39 and D39-dnaJ-gfp were sent to BGI Tech Solutions Co., Ltd (Shenzhen, China) for MALDI-TOF MS analysis.
Bacterial Two-Hybrid System
A bacterial two-hybrid system utilizing bacteriophage λ repressor protein and RNA polymerase was used as previously described [7, 8, 15, 32]. The DnaJ was fused to the full-length bacteriophage λ repressor protein (λcI, 237 amino acids), containing the amino-terminal DNA-binding domain and the carboxylterminal dimerization domain, while DnaK or Eno, SpxB was fused to the N-terminal domain of the α-subunit of RNA polymerase (248 amino acids). pBT-dnaJ and pTRG-dnaK/spxB/eno were co-transformed into an XL1-Blue MRF’ Kan strain. Cells were plated on nonselective, selective, and dual selective screening plates all containing chloromycin (25 μg/ml) and tetracycline (30 μg/ml) and incubated at 37 °C. Activity of a reporter gene was monitored, as previously described [7, 8, 15, 32].
β-Galactosidase Reporter Gene Assay
Streptococcus pneumoniae strain D39 and D39-pEVP3-eno/spxB or D39 ΔdnaJ-pEVP3-eno/spxB were cultured in C+Y medium until an OD600 = 0.4–0.5. Then 107 CFU bacteria were collected by centrifugation at 12,000 rpm for 2 min, the supernatant was collected as needed, and washed twice with PBS. The pellet was then resuspended in 500 μl 0.1 % triton X-100 dissolved in PBS, followed by reacting 15 min at room temperature and 50 μl reaction product was taken for detection following the instructions of β-galactosidase reporter gene assay kit (Beyotime, China), finally the protein expressions were detected in Microplate reader at 450 nm.
Statistical Analysis
Statistical differences between groups were analyzed by either the Student’s t test or two-way ANOVA depending on the data. A probability level less than 0.05 was considered significant, P < 0.05 was indicated by *; P < 0.01 was indicated by **; P < 0.001 was indicated by ***.
Results
Recombinant Plasmid pAE03-dnaJ-gfp Is Successfully Constructed and Transformed into D39
PCR and sequencing were used to confirm the successful construction of pAE03-dnaJ-gfp (Fig. 1a). The resulting plasmid was transformed into pneumococcal D39 strain, GFP or DnaJ-GFP fusion protein was detected with western blot probed with a 1:300–500 dilution of GFP antibody (Fig. 1b) or a 1:8,000 dilution of anti-DnaJ antiserum (Fig. 1c). When detected with GFP antibody, there was one band with a molecular weight (Mw) ~67 kDa in S. pneumoniae strain D39-dnaJ-gfp; no band could be detected in WT D39 strain (Fig. 1b). The ~67 kDa protein is coincident with the Mw of DnaJ-GFP fusion protein, because DnaJ has the Mw of approximate 40 kDa and the Mw of GFP is approximate 27 kDa. We observed two bands in D39-dnaJ-gfp, but only one band in D39, no band in negative control pure protein DnaK (Fig. 1c) probed with anti-DnaJ antiserum. The lower band indicated DnaJ, and the higher represented fusion protein DnaJ-GFP. Together, these results showed that plasmid pAE03-dnaJ-gfp has been successfully constructed and transformed into D39 which can be used for further studies.
Fig. 1.
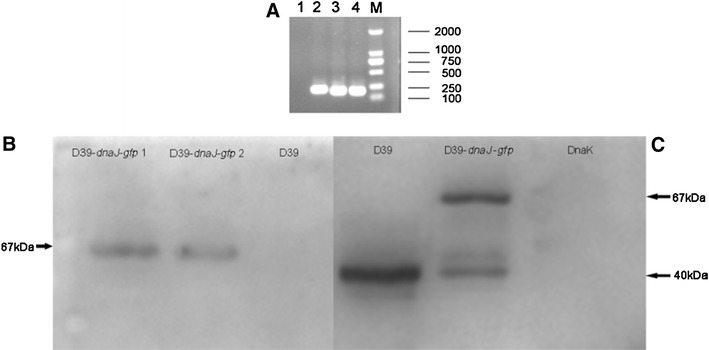
Identification of pAE03-dnaJ-gfp in S. pneumoniae strain D39-dnaJ-gfp. Two positive colonies (D39-dnaJ-gfp) were selected on BA plates supplemented with 0.25 μg/ml erythromycin, and confirmed by PCR with gfp primers (a). The samples were D39 (1), plasmid pAE03 (2), D39-dnaJ-gfp 2 (3), D39-dnaJ-gfp 1 (4) and marker (M).Then they were cultured in C+Y medium until OD600 0.4–0.5. Bacteria was collected by centrifugation at 12,000 rpm and washed twice with PBS. The pellet was then resuspended in 50–80 μl 2× SDS sample buffer. Samples were boiled for 30 min and spined down. 10–15 μl of supernatant was loaded on an SDS-PAGE gel and continued with western blot probed by GFP antibody (Beyotime) (b) and anti-DnaJ antiserum (c) detection. DnaK protein was purified from E. coli BL21 (TaKaRa, China)
Screening and Identification of DnaJ Interaction Proteins in S. pneumoniae D39 by co-IP and MALDI-TOF
The procedures of co-IP were mainly described as the protocol for GFP antibody with some modifications. As a result, we obtained 15 differential proteins attached on protein G-agarose beads between S. pneumoniae D39-dnaJ-gfp and WT D39 (Fig. 2).
Fig. 2.
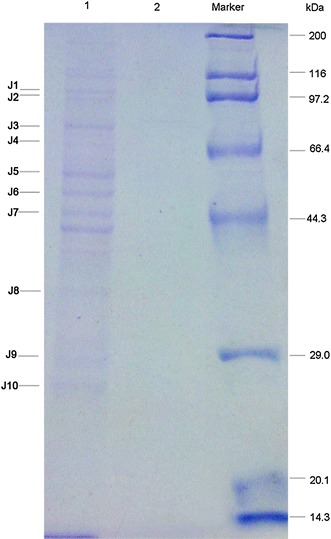
Differential DnaJ interaction proteins between S. pneumoniae strain D39 and D39-dnaJ-gfp by co-IP. S. pneumoniae strain D39 and D39-dnaJ-gfp were cultured in C+Y medium until OD600 0.4–0.5. Then the bacteria were collected and the pellet was sonicated. Cell debris was removed by centrifugation at 12,000 rpm for 30 min. The supernatant was collected and used for co-IP. Lanes 1 and 2 showed the proteins attached to protein G-agarose beads in S. pneumoniae D39-dnaJ-gfp or D39, respectively
To identify the differential DnaJ interaction proteins between D39 and D39-dnaJ-gfp and make sure the bands are reproductively distinguished from D39, we repeated co-IP experiment again and obtained 10 differential protein bands. The 10 bands were subjected to MALDI-TOF MS analysis. Those proteins included bi-functional acetaldehyde-CoA/alcohol dehydrogenase, fusA gene product/translation elongation factor G, chaperone protein DnaK, spxB gene product/pyruvate oxidase, tuf gene product/translation elongation factor Tu, eno gene product/phosphopyruvate hydratase, gap gene product/glyceraldehyde-3-phosphate dehydrogenase, type I, rplC gene product/ribosomal protein L3, and rplF gene product/ribosomal protein L6. Except for ribosomal proteins, the functions of seven proteins were basically defined, in particular the chaperone protein DnaK, which had been proved in many other bacteria as a DnaJ interaction partner [5, 21, 23, 28, 33, 35].
DnaJ Interacts with DnaK, Eno, and SpxB in XL1-Blue MRF’ Kan Strain
Previous studies demonstrated the interaction between DnaJ and DnaK, especially in E. coli [10]. Combined with the co-IP results, it’s reasonable that DnaJ may also interact with DnaK and Eno, SpxB in S. pneumoniae. To test this hypothesis, we checked the interaction between them in XL1-Blue MRF’ Kan strain using the bacterial two-hybrid system [7, 8, 15, 32]. DnaJ was fused to the full-length bacteriophage λ repressor protein, pBT; whereas, DnaK, Eno, and SpxB were fused to the N-terminal domain of the α-subunit of RNA polymerase, pTRG separately. DnaJ was tethered to the λ operator sequence upstream of the reporter promoter through the DNA-binding domain of λcI. If DnaJ interacts with the candidate protein, they recruit and stabilize the binding of RNA polymerase at the promoter and activate the transcription of the HIS3 reporter gene. aadA is a second reporter encoding a protein that conferred resistance against streptomycin to strengthen the evidence for the interaction between the bait and the target. pBT-dnaJ and pTRG-dnaK/eno/spxB were co-transformed into an XL1-Blue MRF’ Kan strain. Expression of fusion proteins was confirmed by western blot analysis. In control experiments, when the pBT and pTRG fragments alone were co-expressed, no colonies appeared on selective screening plates containing 5 mM 3-AT. When dnaJ and dnaK/eno/spxB were fused to both the pBT and pTRG fragments and co-expressed, colonies appeared on selective screening plates and dual selective screening plates containing 5 mM 3-AT and 12.5 μg/ml streptomycin.
Bacteria co-expressing DnaJ and DnaK/Eno/SpxB were viable on nonselective, selective, and dual selective screening plates as the positive control (Fig. 3a–i). In sharp contrast, there were no colonies on selective and dual selective screening plates when the pBT and pTRG-dnaK/eno/spxB, pBT-dnaJ and pTRG, or pBT and pTRG fragments were co-transformed respectively (Fig. 3b, c, e, f, h, i), although they could be seen on nonselective screening plates (Fig. 3a, d, g), indicating the fitness of the bacterial two-hybrid system. These results demonstrated the interaction between DnaJ and DnaK also works in S. pneumoniae and Eno/SpxB interacts with DnaJ in S. pneumoniae.
Fig. 3.
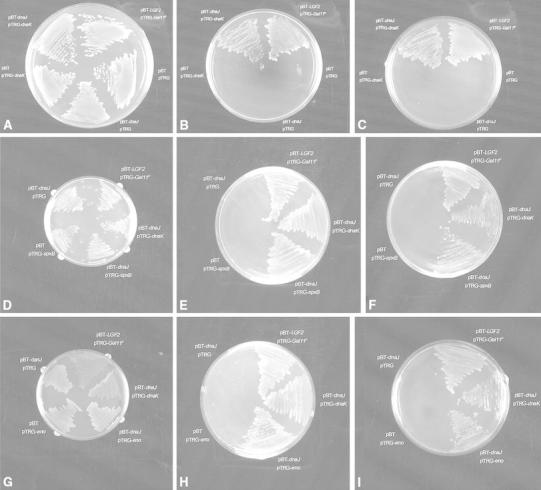
DnaJ interacts with DnaK, SpxB, and Eno in XL1-Blue MRF’ Kan strain in vivo. A bacterial two-hybrid system was used to confirm the interaction between DnaJ and DnaK/SpxB/Eno in vivo. Interaction between DnaJ and DnaK/SpxB/Eno was monitored by the expression of HIS3 and aadA reporter genes on nonselective (a, d, g), selective (b, e, h) and dual selective (c, f, i) screening plates
Prevention of Denaturation of SpxB and Eno by DnaJ at High Temperature
DnaJ is helpful to the correct functions of protein, we supposed whether the interacting partners were regulated by DnaJ in this way. Because the correct function of protein is necessary for the activity of β-galactosidase, β-galactosidase reporter assay was used to reflect the portion of functional proteins in the bacterial lysates and the supernatants, while western blot was employed to determine the total synthesized proteins in different bacterial lysates. We have known that DnaJ can facilitate some secretory proteins transport across the membrane, such as alkaline phosphatase (AP), ribose-binding protein (RBP), and β-lactamase (Bla) and metallo-β-lactamase (MβL) in E. coli [26, 36, 37]. And it has been reported that enolase can be secreted outside the bacteria [2], so we were also interested in the level of Eno in the supernatant. The amount of functional Eno was much more in wild type D39 strain than D39ΔdnaJ as revealed by β-galactosidase activity analysis (Fig. 4d), and the total synthesized Eno was also elevated with the increasing expression of DnaJ (Fig. 4a, b). Besides, we observed that the secretion of Eno was in parallel with the expression of DnaJ (Fig. 4b, d). It was suggested that besides its effect on the prevention of denaturation of Eno, DnaJ might also help to promote its outside secretion.
Fig. 4.
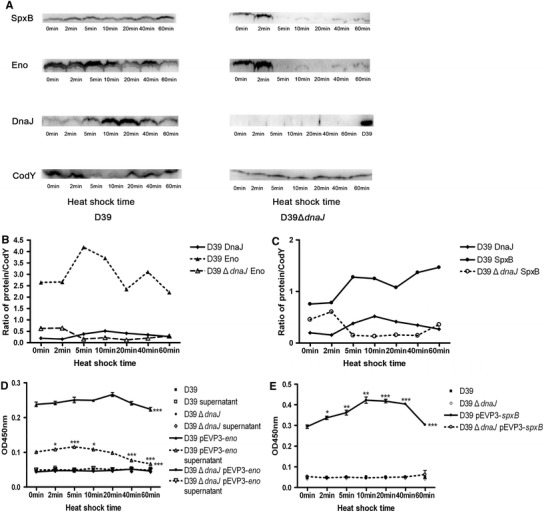
DnaJ prevents the denaturation of Eno and SpxB during heat shock. S. pneumoniae D39, D39ΔdnaJ, D39-pEVP3-eno/spxB, and D39ΔdnaJ-pEVP3-eno/spxB were cultured in C+Y medium and all the bacteria incubated in 42 °C water bath for 0, 2, 5, 10, 20, 40, and 60 min separately. DnaJ/SpxB/Eno expressions in the bacterial lysates of D39 and D39ΔdnaJ were analyzed by western blot separately. CodY served as the internal reference (a). Figure b and c were the ratios of DnaJ/Eno versus CodY and DnaJ/SpxB versus CodY. The results of β-galactosidase reporter gene assay were indicated by Fig. d, e, and the figures represented three independent experiments. Statistical differences were analyzed by two-way ANOVA between groups of D39-pEVP3-eno/spxB and D39, D39ΔdnaJ or D39ΔdnaJ-pEVP3-eno/spxB, respectively. Their P values were marked with *** located on the right of the spots. For D39-pEVP3-eno/spxB and D39-pEVP3-eno supernatant, statistical differences were compared by Student’s t test between samples at different time of heat shock and the initial time. Their P values were indicated above the spots. * indicates P < 0.05; ** indicates P < 0.01, and *** indicates P < 0.001
As to SpxB, the portion of functional SpxB was more in the amount in wild type D39 strain than D39ΔdnaJ (Fig. 4e), and decreased with the decline of DnaJ after heat shocked at 42 °C for 40 min (Fig. 4c, e), indicating that DnaJ was necessary to prevent the denaturation of SpxB despite the total synthesized SpxB was still increasing (Fig. 4c).
For D39ΔdnaJ, there were few SpxB and Eno expressions after heat shock for 5 min (Fig. 4a); while no functional proteins could be found through β-galactosidase reporter gene assays (Fig. 4d, e), indicating DnaJ might be important to protect SpxB and Eno from degradation.
Discussion
Streptococcus pneumoniae encounters heat stress upon penetration from the nasal mucosa into blood and/or meninges. Commonly, infection with S. pneumoniae leads to elevated temperature in host, which serves as a key trigger for the rapid, transient increase in the synthesis of HSPs instead [20]. Therefore, HSPs are essential for the pathogenesis of pneumococcal infections. As a HSP member, DnaJ is believed to be associated with this heat shock event and confers fitness for bacterial survival. Definitely, DnaJ is associated with bacterial virulence, including S. pneumoniae [6, 21, 28, 35]. To investigate the underlying mechanism, we attempted to screen DnaJ interaction proteins with the method of co-IP. Finally, we obtained nine proteins with MALDI-TOF MS analysis.
Using co-IP to screen the interactive proteins is reliable. In the present study, we noticed the interaction between DnaJ and DnaK/Eno/SpxB using co-IP, which were confirmed by using bacterial two-hybrid reporter system. This is the first time that we report the interaction between DnaK and DnaJ in S. pneumoniae, albeit their interaction in other bacteria.
Our results showed several ribosomal proteins could also interact with DnaJ, indicating the diverse roles for DnaJ in protein synthesis in pneumococci. Therefore, DnaJ may be an important virulence factor which confers fitness for pneumococci to adapt host stress by interacting with the ribosomal proteins. This result is supported by the observation on other bacteria, such as E. coli and Saccharomyces cerevisiae [19, 24, 39]. Thus, DnaJ-mediated host adaptation may be a common phenomenon among bacteria.
The elongation steps are the most highly conserved processes for living bacterial cells, which was learnt from the comparisons of the processes among the initiation, elongation, and termination stages of protein synthesis [14]. In bacteria, the elongation steps of protein synthesis require the sequential action of two different elongation factors (EF), EF-G and EF-Tu. EF-Tu is required for delivering the correct aminoacyl-tRNA to the A site on the ribosome and is, therefore, intimately involved in proofreading [31]. Translation elongation factor EF-G uses GTP to catalyze translocation of peptidyl-tRNA from the ribosomal A/P site to the P/P site [25, 30]. After GTP hydrolysis and translocation, EF-G·GDP leaves the ribosome and is regenerated by the spontaneous exchange of GDP for GTP off the ribosome [17, 30]. EF-G·GTP also plays a role with ribosome recycling factor in splitting the ribosome into its two subunits after translation termination [16]. And, EF-G in the virulence has also been reported in S. enterica serovar Typhimurium [22]. Together these evidences suggest the possible role of DnaJ in bacterial virulence, which may be partly attributed to its ability in interaction with EF-G and Tu.
In pneumococcus, enzyme pyruvate oxidase (SpxB) is responsible for the production of H2O2 under rich and aerobic conditions. S. pneumoniae SpxB was in relationship with the bacterial transformation. The contribution of SpxB in virulence has also been reported. Spellerberg B et al. [34] demonstrated that a spxB-deficient mutant exhibited reduced virulence for nasopharyngeal colonization, pneumonia and sepsis. D39ΔdnaJ mutant was defected in colonization of nasal and invasive infections [6].Our results suggested that DnaJ prevented the denaturation of SpxB during heat shock, so in D39ΔdnaJ mutant SpxB may cannot express correctly and contribute to the reduction of colonization.
eno is an essential gene in bacteria and enolase, eno designated, is the key enzyme in the glycolytic cycle [2]. Besides, enolase can be secreted outside the bacteria and bind to plasminogen and plasmin to activate fibrinolysis system facilitating pathogen invasion and dissemination in the infected host [2]. Moreover, it also binds to human complement inhibitor C4b-binding protein and contributes to complement evasion [1]. Pneumococcal enolase mutant also exhibits attenuation in a model of respiratory infection [2]. Besides, the mouse intranasal challenge studies indicate that defined amino acid substitutions of Eno affect the virulence of S. pneumoniae and contribute to the pathogenesis of diseases [3]. In our study, we found the secretion of Eno changed with DnaJ and the total expression of Eno increased during 42 °C heat shock. Therefore, in D39 DnaJ may help enolase translocate to the extracellular matrix leading to invasive infection. Together, the implications of DnaJ in virulence appear to correlate with its effects in controlling the correct expression or effective secretion of some known virulence factors.
In conclusion, we obtained several proteins that could interact with DnaJ to execute normal physiological or biological functions. Of them, SpxB, Eno, and some translation related factors could be used to interpret the contribution of DnaJ in pneumococcal virulence. Nevertheless, the precise mechanism is worth further investigation.
Acknowledgments
We thank Jan-Willem Veening professor of Newcastle University, Newcastle Upon Tyne, UK, for providing plasmid pAE03. We thank M. Donald for providing us plasmid pEVP3. This work was funded by a Grant from the excellent college and university talent foundation projects of Chongqing (the third group, No. 32, Yu Jiao Ren 2011 and No. 89, Chong Yi Da Ren 2011) and the Natural Science Foundation Project of CQ CSTC (No. 2012jjA10009).
References
- 1.Agarwal V, Hammerschmidt S, Malm S, Bergmann S, Riesbeck K, Blom AM. Enolase of Streptococcus pneumoniae binds human complement inhibitor C4b-binding protein and contributes to complement evasion. J Immunol. 2012;189:3575–3584. doi: 10.4049/jimmunol.1102934. [DOI] [PubMed] [Google Scholar]
- 2.Bergmann S, Rohde M, Chhatwal GS, Hammerschmidt S. α-Enolase of Streptococcus pneumoniae is a plasmin(ogen)-binding protein displayed on the bacterial cell surface. Mol Microbiol. 2001;40:1273–1287. doi: 10.1046/j.1365-2958.2001.02448.x. [DOI] [PubMed] [Google Scholar]
- 3.Bergmann S, Wild D, Diekmann O, Frank R, Bracht D, Chhatwal GS, Hammerschmidt S. Identification of a novel plasmin(ogen)-binding motif in surface displayed α-enolase of Streptococcus pneumoniae. Mol Microbiol. 2003;49:411–423. doi: 10.1046/j.1365-2958.2003.03557.x. [DOI] [PubMed] [Google Scholar]
- 4.Bjerrum OJ, Heegaard NHH. CRC handbook of immunoblotting of proteins. Florida: CRC Press; 1988. [Google Scholar]
- 5.Champ S, Puvirajesinghe TM, Perrody E, Menouni R, Genevaux P, Ansaldi M. Chaperone-assisted excisive recombination, a solitary role for DnaJ (Hsp40) chaperone in lysogeny escape. J Biol Chem. 2011;286:38876–38885. doi: 10.1074/jbc.M111.281865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cui J, Zhang Q, Dong J, Jiang H, Zhou AE, Dong SS, Zhang XM, Yin YB, Wang H. Construction of dnaJ-deficient mutant strain of Streptococcus pneumoniae and its preliminary study on virulence. J Third Mil Med Univ. 2011;33:2000–2003. [Google Scholar]
- 7.Dove SL, Hochschild A. Conversion of the omega subunit of Escherichia coli RNA polymerase into a transcriptional activator or an activation target. Genes Dev. 1998;12:745–754. doi: 10.1101/gad.12.5.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dove SL, Joung JK, Hochschild A. Activation of prokaryotic transcription through arbitrary protein–protein contacts. Nature. 1997;386:627–630. doi: 10.1038/386627a0. [DOI] [PubMed] [Google Scholar]
- 9.Feder ME, Hofmann GE. Heat-shock proteins, molecular chaperones, and the stress response: evolutionary and ecological physiology. Annu Rev Physiol. 1999;61:243–282. doi: 10.1146/annurev.physiol.61.1.243. [DOI] [PubMed] [Google Scholar]
- 10.Genevaux P, Georgopoulos C, Kelley WL. The Hsp70 chaperone machines of Escherichia coli: a paradigm for the repartition of chaperone functions. Mol Microbiol. 2007;66:840–857. doi: 10.1111/j.1365-2958.2007.05961.x. [DOI] [PubMed] [Google Scholar]
- 11.Gong Y, Cui YL, Niu SQ, Zhang XM, Xu WC, He YJ, Wang H. Expression and purification of putative protein SPD0414 from Streptococcus pneumoniae and analysis of its conservation. Zhong Hua Mian Yi Xue Za Zhi. 2010;26:824–827. [Google Scholar]
- 12.Harlow E, Lane E. Antibodies: a laboratory manual. New York: Cold Spring Harbor Laboratory; 1988. [Google Scholar]
- 13.Hendrick JP, Langer T, Davis TA, Hartl FU, Wiedmann M. Control of folding and membrane translocation by binding of the chaperone DnaJ to nascent polypeptides. Proc Natl Acad Sci USA. 1993;90:10216–10220. doi: 10.1073/pnas.90.21.10216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hou Y, Yaskowiak ES, March PE. Carboxyl-terminal amino acid residues in elongation factor G essential for ribosome association and translocation. J Bacteriol. 1994;176:7038–7044. doi: 10.1128/jb.176.22.7038-7044.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Joung JK, Ramm EI, Pabo CO. A bacterial two-hybrid selection system for studying protein-DNA and protein–protein interactions. Proc Natl Acad Sci USA. 2000;97:7382–7387. doi: 10.1073/pnas.110149297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karimi R, Pavlov MY, Buckingham RH, Ehrenberg M. Novel roles for classical factors at the interface between translation termination and initiation. Mol Cell. 1999;3:601–609. doi: 10.1016/S1097-2765(00)80353-6. [DOI] [PubMed] [Google Scholar]
- 17.Kaziro Y. The role of guanosine 50-triphosphate in polypeptide chain elongation. Biochim Biophys Acta. 1978;505:95–127. doi: 10.1016/0304-4173(78)90009-5. [DOI] [PubMed] [Google Scholar]
- 18.Kim SY, Sharma S, Hoskins JR, Wickner S. Interaction of the DnaK and DnaJ chaperone system with a native substrate, P1 RepA. J Biol Chem. 2002;277:44778–44783. doi: 10.1074/jbc.M206176200. [DOI] [PubMed] [Google Scholar]
- 19.Kudlicki W, Odom O, Kramer G, Hardesty B. Binding of an N-terminal rhodanase peptide to DnaJ and to ribosomes. J Biol Chem. 1996;271:31160–31165. doi: 10.1074/jbc.271.49.31160. [DOI] [PubMed] [Google Scholar]
- 20.Kwon HY, Kim SW, Choi MH, Ogunniyi AD, Paton JC, Park SH, Pyo SN, Rhee DK. Effect of heat shock and mutations in ClpL and ClpP on virulence gene expression in Streptococcus pneumoniae. Infect Immun. 2003;71:3757–3765. doi: 10.1128/IAI.71.7.3757-3765.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lakhal F, Bury-Mone’ S, Nomane Y, Le Goıc N, Paillard C, Jacq A. DjlA, a membrane-anchored DnaJ-like protein, is required for cytotoxicity of Clam pathogen Vibrio tapetis to hemocytes. Appl Environ Microbiol. 2008;74:5750–5758. doi: 10.1128/AEM.01043-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Macvanin M, Björkman J, Eriksson S, Rhen M, Andersson DI, Hughes D. Fusidic acid-resistant mutants of Salmonella enterica serovar Typhimurium with low fitness in vivo are defective in RpoS induction. Antimicrob Agents Chemother. 2003;47:3743–3749. doi: 10.1128/AAC.47.12.3743-3749.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mayer MP, Bukau B. Hsp70 chaperones: cellular functions and molecular mechanism. Cell Mol Life Sci. 2005;62:670–684. doi: 10.1007/s00018-004-4464-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meyer AE, Hung NJ, Yang P, Johnson AW, Craig EA. The specialized cytosolic J-protein, Jjj1, functions in 60S ribosomal subunit biogenesis. Proc Natl Acad Sci USA. 2007;104:1558–1563. doi: 10.1073/pnas.0610704104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moazed D, Noller HF. Intermediate states in the movement of transfer RNA in the ribosome. Nature. 1989;342:142–148. doi: 10.1038/342142a0. [DOI] [PubMed] [Google Scholar]
- 26.Morán-Barrio J, Limansky AS, Viale AM. Secretion of GOB metallo-beta-lactamase in Escherichia coli depends strictly on the cooperation between the cytoplasmic DnaK chaperone system and the Sec machinery: completion of folding and Zn(II) ion acquisition occur in the bacterial periplasm. Antimicrob Agents Chemother. 2009;53:2908–2917. doi: 10.1128/AAC.01637-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morimoto RI, Tissieres A, Georgopoulos C. Stress proteins in biology and medicine. Cold Spring Harbor: Cold Spring Harbor Press; 1990. [Google Scholar]
- 28.Ohnishi H, Mizunoe Y, Takade A, Tanaka Y, Miyamoto H, Harada M, Yoshida S. Legionella dumoffii DjlA, a member of the DnaJ family, is required for intracellular growth. Infect Immun. 2004;72:3592–3603. doi: 10.1128/IAI.72.6.3592-3603.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pozzi G, Masala L, Iannelli F, Manganelli R, Havarstein LS, Piccoli L, Simon D, Morrison DA. Competence for genetic transformation in encapsulated strains of Streptococcus pneumoniae: two allelic variants of the peptide pheromone. J Bacteriol. 1996;178:6087–6090. doi: 10.1128/jb.178.20.6087-6090.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rodnina MV, Savelsbergh A, Katunin VI, Wintermeyer W. Hydrolysis of GTP by elongation factor G drives tRNA movement on the ribosome. Nature. 1997;385:37–41. doi: 10.1038/385037a0. [DOI] [PubMed] [Google Scholar]
- 31.Rohrbach MS, Bodley JW. Selective chemical modification of Escherichia coli elongation factor G: butanedione modification of an arginine essential for nucleotide binding. J Biol Chem. 1977;251:930–933. [PubMed] [Google Scholar]
- 32.Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual. New York: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 33.Sell SM, Eisen C, Ang D, Zylicz M, Georgopoulos C. Isolation and characterization of dnaJ null mutants of Escherichia coli. J Bacteriol. 1990;172:4827–4835. doi: 10.1128/jb.172.9.4827-4835.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Spellerberg B, Cundell DR, Sandros J, Pearce BJ, Idanpaan-Heikkila I, Rosenow C, Masure HR. Pyruvate oxidase, as a determinant of virulence in Streptococcus pneumoniae. Mol Microbiol. 1996;19:803–813. doi: 10.1046/j.1365-2958.1996.425954.x. [DOI] [PubMed] [Google Scholar]
- 35.Takaya A, Tomoyasu T, Matsui H, Yamamoto T. The DnaK/DnaJ chaperone machinery of Salmonella enterica serovar Typhimurium is essential for invasion of epithelial cells and survival within macrophages, leading to systemic infection. Infect Immun. 2004;72:1364–1373. doi: 10.1128/IAI.72.3.1364-1373.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wild J, Altman E, Yura T, Gross CA. DnaK and DnaJ heat shock proteins participate in protein export in Escherichia coli. Genes Dev. 1992;6:1165–1172. doi: 10.1101/gad.6.7.1165. [DOI] [PubMed] [Google Scholar]
- 37.Wild J, Rossmeissl P, Walter WA, Gross CA. Involvement of the DnaK–DnaJ–GrpE chaperone team in protein secretion in Escherichia coli. J Bacteriol. 1996;178:3608–3613. doi: 10.1128/jb.178.12.3608-3613.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu KF, Zhang XM, Shi J, Li N, Li DR, Luo M, Cao J, Yin NL, Wang H, Xu WC, Yin YB. Immunization with a combination of three pneumococcal proteins confers additive and broad protection against Streptococcus pneumoniae infections in mice. Infect Immun. 2010;78:1276–1283. doi: 10.1128/IAI.00473-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yan W, Schilke B, Pfund C, Walter W, Kim S, Craig EA. Zuotin, a ribosome-associated DnaJ molecular chaperone. EMBO J. 1998;17:4809–4817. doi: 10.1093/emboj/17.16.4809. [DOI] [PMC free article] [PubMed] [Google Scholar]


