Abstract
iNKT cells can produce high levels of IL-4 early during infection. However, indirect evidence suggests they may produce this immunomodulatory cytokine in the steady state. Through intracellular staining for transcription factors, we define 3 subsets of iNKT cells that produce distinct cytokines (NKT1, NKT2, and NKT17), which represent diverse lineages and not developmental stages as previously thought. These subsets exhibit substantial inter-strain variation in numbers. In several strains, including BALB/c, NKT2 cells are abundant and stimulated by self-ligands to produce IL-4. In these strains, steady state IL-4 conditions CD8 T cells to become “memory-like”, increases serum IgE levels, and causes dendritic cells to produce chemokines. Thus iNKT cell derived IL-4 alters immune properties under normal steady state conditions.
Introduction
It has become increasingly apparent that many gene deficient and transgenic mice display a unique population of memory-like CD8 T cells in the thymus1. These cells have also been referred to as “innate CD8 T cells” because they behave like memory T cells and rapidly produce high levels of IFN-γ, yet antigen recognition was not required for their differentiation2. Studies on the differentiation mechanism revealed a common pathway, whereby various genetic alterations all led to the increased development of promyelocytic leukemia zinc finger (PLZF) expressing αβ or γδ NKT cells3–6. In all of these models, IL-4, presumably produced by iNKT cells in the steady state, was required for CD8 T cells to express Eomesodermin (Eomes) and adopt the characteristic features of memory CD8 T cells. Notably, not only these gene deficient mice, but also wild type BALB/c mice have a distinct population of memory-like CD8 T cells, which were shown to be dependent on iNKT cells and IL-43, 7. Even C57BL/6 mice have subsequently been shown to have a much smaller, but defined, population of memory-like CD8 T cells8. These data all imply that iNKT cells must have been activated to produce IL-4 continuously, or at some point in their development.
In this study, we sought to identify the iNKT cells that produce IL-4 and give rise to memory-like CD8 T cells. Our data strongly support a newly proposed classification of iNKT cells into NKT1, NKT2 and NKT17 lineages, according to their function and transcription factor expression9, analogous to CD4 T helper cells. In several strains of mice, iNKT precursors efficiently differentiate into NKT2 cells, and secret abundant IL-4 in the steady state. This IL-4 not only conditioned bystander CD8 T cells to become memory-like, but also stimulated thymic dendritic cells (DCs) to secrete CCL17 and CCL22, and peripheral B cells to produce immunoglobulin E (IgE).
Results
iNKT cells produce IL-4 in the steady state in BALB/c mice
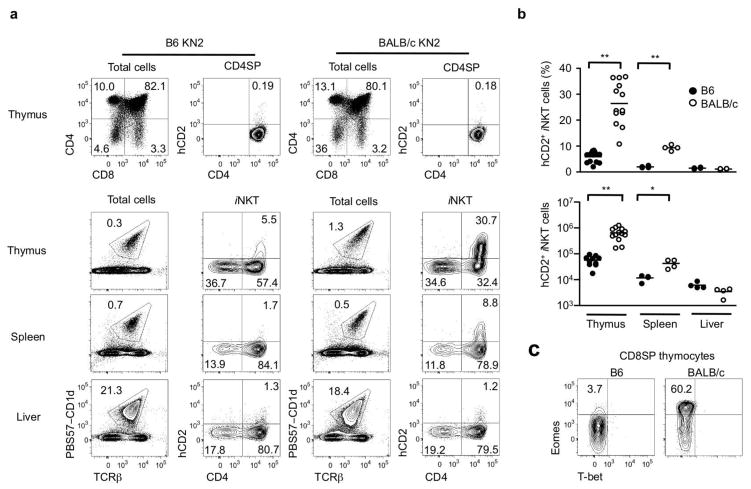
The thymus of young BALB/c mice contains 3 to 5 times more iNKT cells than B6 mice and these cells were hypothesized to secrete IL-4, which conditions bystander CD8 single positive (SP) thymocytes to become memory-like CD8 T cells3. Based on this finding, we sought to further investigate the subset of iNKT cells secreting IL-4 in the steady state. For this purpose, we used KN2 mice, where the first two exons of the endogenous IL-4 gene were replaced by human CD2 (hCD2) and its expression on the cell surface reports IL-4 secretion in vivo10. We found that up to 40% of thymic iNKT cells in BALB/c-KN2 mice expressed hCD2 on the cell surface, whereas B6 iNKT cells from B6-KN2 mice were largely hCD2 negative (Fig. 1a), consistent with a recent report11. In young BALB/c-KN2 mice, there was a >10 fold increase in the number of hCD2 positive cells in the thymus compared to B6-KN2 mice (Fig. 1b). Peripheral iNKT cells also expressed hCD2 in BALB/c-KN2 mice, albeit not as striking as in the thymus (Fig. 1a). In contrast, hardly any conventional CD4 SP cells were positive for hCD2, either in the thymus (Fig. 1a) or periphery (data not shown). These results indicate that iNKT cells are a major source of IL-4 in vivo in the steady state. As shown previously3, this increased IL-4 correlated with an increase in the percentage and number of Eomes positive “memory-like CD8 T cells” in BALB/c mice (Fig. 1c).
Figure 1. BALB/c iNKT cells produce IL-4 in the steady state.
(a) Flow cytometric analysis shows hCD2 expression in conventional CD4 SP thymocytes (top row) and CD1d tetramer binding iNKT cells from thymus, spleen and liver (bottom three rows) of 7 week-old B6 and BALB/c KN2+/− mice. (b) Percentages and numbers of hCD2+ iNKT cells in thymus, spleen and liver of 7–8 week old B6-KN2 (N=4~10) and BALB/c-KN2 (N=4~13) mice. Horizontal bars indicate mean values. Unpaired two tailed t-tests were used to compare B6 and BALB/c mice. ***p<0.0001, **p=0.0095 (c) Representative FACS data of T-bet and Eomes expression in CD8 SP thymocytes of indicated mouse strains.
PLZF, ROR-γt, and T-bet differentiate NKT1, NKT2 and NKT17 cells
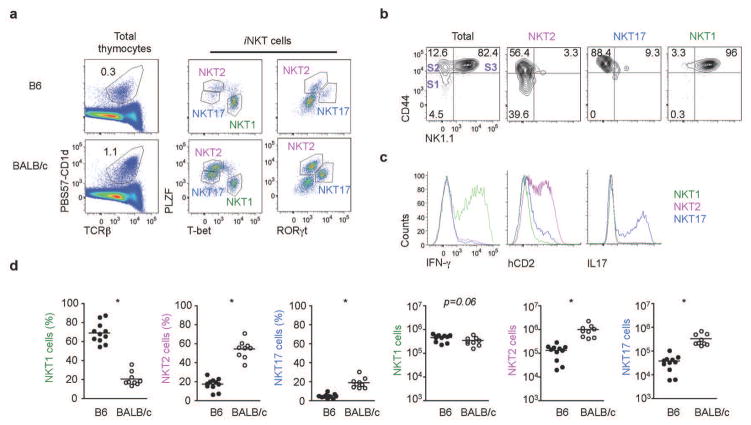
To further characterize the IL-4 producing iNKT cells in BALB/c mice, we compared the developmental profile of thymic iNKT cells in B6 and BALB/c mice. In the standard iNKT cell classification, a combination of CD24 (HSA), CD44 and NK1.1 are used to discriminate iNKT cells as stage 0, 1, 2 and 312. However, NK1.1, which has been considered to be a marker of terminal maturation of iNKT cells, is neither expressed in BALB/c mice nor correlated with functional capacity13. Therefore, instead of surface markers, we performed intracellular staining for transcription factors, which are recognized equivalently in different mouse strains, and more closely linked to function. PLZF is an essential factor for the development and innate function of iNKT cells14, 15, and T-bet, GATA-3 and ROR-γt are transcription factors regulating Th1, Th2 and Th17 lineages in conventional CD4 T cells respectively16. As shown in Fig. 2a, the combination of PLZF, T-bet and ROR-γt separated iNKT cells into three distinctive subsets and, analogous to T helper lineage nomenclature, we designated these cells as NKT1, NKT2 and NKT17 cells. Th2 specific transcription factors, including GATA-3 and IRF-4, were highly expressed in both NKT2 and NKT17 cells (Supplementary Fig. 1a). NKT1 cells, expressing a high level of T-bet, were low for GATA-3 expression, consistent with a previous report that showed all T cells including Th1 and iNKT cells express variably low levels of GATA-317. This classification roughly correlates with the conventional staging system in B6 mice as NKT1 cells are predominantly stage 3 and NKT2 cells are stage 1 and 2 (Fig. 2b) although NKT17 cells can not be distinguished from NKT2 with the conventional classification.
Figure 2. PLZF, ROR-γt and T-bet differentiate NKT1, NKT2 and NKT17 cells.
(a) Thymic iNKT cells from 7 week-old B6 and BALB/c mice were stained for intracellular PLZF, T-bet and ROR-γt. We designated 3 distinct populations as NKT1, NKT2 and NKT17 cells. (b) NK1.1 and CD44 expression on each iNKT subset is shown. Historical stages are indicated by S1, S2, and S3. (c) Thymocytes of BALB/c KN2+/− mice were depleted of CD8 and CD24 positive cells by MACS, stimulated with PMA and ionomycin for 4 hours and stained for intracellular cytokines and hCD2. (d) Frequencies and numbers of each iNKT subset in thymi of 7–8 week-old B6 (N=11) and BALB/c (N=9) mice were compared. Horizontal bars indicate mean values. Unpaired two tailed t-tests were used to compare B6 and BALB/c mice. ***p<0.001.
Importantly, this method of defining iNKT cell subsets correlated precisely with cytokine production: thymic NKT1, NKT2 and NKT17 cells produced IFN-γ, IL-4, and IL-17 respectively (Fig. 2c), when stimulated with PMA and ionomycin. NKT2 cells were abundant in BALB/c mice, whereas NKT1 cells were predominant in B6 mice (Fig. 2d). Numerically, 7–8 week old BALB/c mice produced 7.5 times more NKT2 cells, and 9 times more NKT17 compared to B6 mice. However, the absolute number of NKT1 cells was not significantly different between these two strains (Fig. 2d). Peripheral NKT cells were similarly identified by this transcription factor staining profile (Supplementary Fig. 2a). As in the thymus, NKT2 cells were predominant in the spleen of BALB/c mice, whereas NKT1 cells were predominant in B6 mice (Supplementary Fig. 2a and 2b). Liver iNKT cells were mostly NKT1 in both strains. NKT2 cells produced IL-4, but not IFN-γ, after strong agonistic stimulation in vivo with the synthetic lipid α-galactosylceramide (αGalCer); while peripheral NKT1 cells secreted IFN-γ and also IL-4 (albeit to lower levels) (Supplementary Fig. 2c).
We also investigated other surface markers that can be used to discriminate these subsets. Supplementary Fig. 1b shows that CD122 (and NK1.1 in B6 mice) are reasonable markers of NKT1 cells in the thymus and, among the CD122/NK1.1 negative population, CD4 and CD27 could differentiate NKT2 and NKT17 cells. As a result, a combination of CD122 (or NK1.1 in B6 mice) together with CD4 or CD27 can differentiate thymic iNKT cells consistent with their transcription factor profile. IL17RB was described as a surface marker capable of distinguishing NKT cell subsets18. NKT2 cells are IL17RB+CD4+ and NKT1 cells are IL17RB− CD4+/− (Supplementary Fig. 1c and 1d). However, NKT17 cells are IL17RB+CD4− in B6 mice, but IL17RB− CD4− in BALB/c mice. The phenotypic and functional properties of iNKT subsets are summarized in Table 1.
Table 1.
Transcription factors differentiate iNKT cells into NKT1, NKT2 and NKT17 lineages
| PLZF | T-bet | ROR-γt | GATA-3 | IRF-4 | major cytokine | surface markers | conventional classification | |
|---|---|---|---|---|---|---|---|---|
| NKT2 | high | − | − | + | + | IL-4 | CD4+CD27+CD122− | stage 1 and 2 |
| NKT17 | intermediate | − | + | + | + | IL-17 | CD4−CD27−CD122− | stage 2 |
| NKT1 | low | + | − | +/− | − | IFN-γ | CD4+/−CD27+CD122+ | stage 3 |
IL4 producing NKT2 cells do not give rise to NKT1 cells
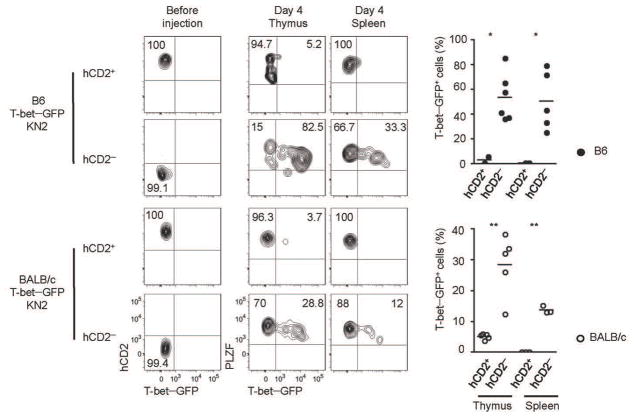
The conventional classification of iNKT cells defined in B6 mice proposed a linear differentiation model, where CD44loNK1.1− (stage 1) cells upregulate CD44 (stage 2) before finally expressing NK1.1 (stage 3), and that stage 1 cells primarily produce IL-4, while stage 3 cells produce both IFN-γ and IL-412. However, we show here that iNKT cells with these markers are heterogenous and express distinct transcription factors, which is more suggestive of a model where multiple lineages differentiate from a common progenitor9. Thus, we wished to address whether NKT2 cells, which fall within the CD44hiNK1.1− (stage 2) gate, give rise to NK1.1+ NKT1 cells (stage 3) or not. It has been firmly established that NK1.1− CD44hi cells can give rise to NK1.1+ cells19, 20. However, a recent report showed that the fraction of NK1.1− cells that express IL17RB do not give rise to NK1.1+ cells18. We investigated expression of IL-17RB in BALB/c and B6 KN2 mice and found that hCD2 expression was confined to an IL17RB+ population (Supplementary Fig. 1c) but not vice versa (Supplementary Fig. 1d), suggesting that the NKT2 cells secreting IL-4 in the steady state are not developmental intermediates that would give rise to NKT1 cells. To more rigorously test the precursor – progenitor relationship, we sorted hCD2+ and hCD2− NKT2 from B6 and BALB/c T-betGFPKN2+/− mice, labeled them violet cell tracer (VCT) and intrathymically injected them into congenic hosts (Fig. 3). Four days later, a significant fraction of hCD2− cells had converted into NKT1 cells expressing T-betGFP in thymus and spleen, whereas hCD2+ NKT2 cells did not. This result indicates that NKT2 cells are not precursors of NKT1 cells. Also these data show that NK1.1− iNKT cells are heterogeneous, containing terminally differentiated IL-4 producers (hCD2+ NKT2 cells), NKT17 cells, as well as the progenitors for NKT1 cells.
Figure 3. IL4 producing NKT2 cells do not give rise to NKT1 cells.
Thymocytes from B6 and BALB/c T-betGFP KN2+/− mice were enriched iNKT cells by depleting CD8 and CD24 positive cells and sorted hCD2+ or hCD2− NKT2 cells. Cells were labeled with VCT, injected into congenic host mice and analyzed four days later for T-betGFP expression from donor cells. Pooled data of 3 independent experiments are shown.
NKT2 cells produce IL-4 in the steady state
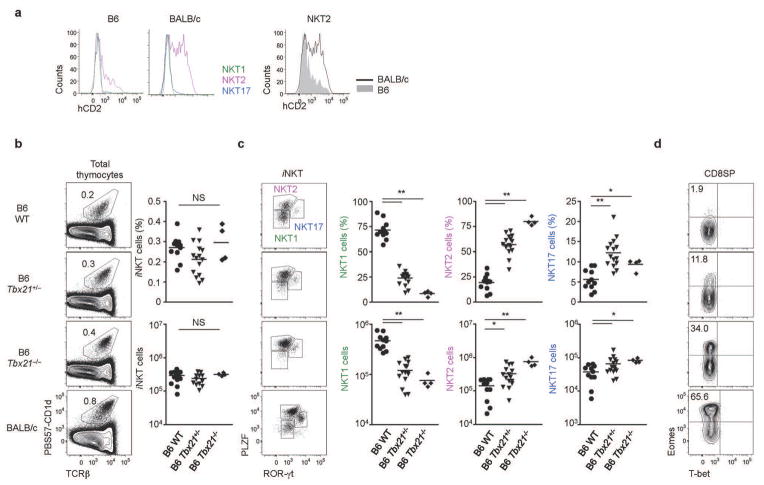
To determine which NKT subset produces IL-4 in the steady state, we examined hCD2 expression in KN2 mice directly ex vivo. Ex vivo hCD2 expression was observed exclusively in NKT2 cells (Fig. 4a). In addition to the greater abundance of NKT2 cells in BALB/c mice (Fig. 2d), the mean fluorescence intensity (MFI) of hCD2 was higher in BALB/c iNKT cells (Fig. 4a), suggesting that not only do more cells with the potential to produce IL-4 exist in the steady state in BALB/c mice, but more of them are actively producing IL-4 in vivo.
Figure 4. Steady state IL-4 is produced by NKT2 cells, and is enhanced in the absence of T-bet.
(a) hCD2 staining histograms of each iNKT cell subset in B6 and BALB/c mice in the steady state are shown. In the panels on the right, hCD2 expresion on NKT2 cells of B6 and BALB/c mice are overlaid. Data are representative of at least three independent experiments. (b and c) Representative profiles of iNKT cell transcription factor expression are shown (left) with statistical analysis (right) for six week old mice. One-way ANOVA was used to compare B6 WT (N=10), B6 T-bet+/− (N=15), and B6 T-bet−/ − (N=4). ***p<0.001, *p<0.05. Horizontal bars indicate mean values. (d) Representative FACS profiles of T-bet and Eomes staining in CD8 SP thymocytes are shown.
Since both NK1.1− NKT2 and NK1.1+ NKT1 have the potential to produce IL-420 (Supplementary Fig. 2c), we wished to definitively test if steady state IL-4 was produced by NKT2 or NKT1 cells. Thus, we analyzed T-bet KO mice, as these mice were previously shown to have a drastic loss of NK1.1+ iNKT cells21. Although the previous report suggested that T-bet deficiency reduced total iNKT cells number, we did not observe a loss of total iNKT cells in young (6 week old) T-bet deficient mice (Fig. 4b). Rather, while NKT1 cells were drastically reduced, higher percentages and numbers of NKT2 and NKT17 cells were observed in compensation (Fig. 4c). The difference between our results and the previous report may have to do with age, since in older mice (10–14 weeks), the overall frequency and total number of iNKT cells was decreased in T-bet KO mice, due to the major reduction of NKT1 cells, but not NKT2 or NKT17 cells (Supplementary Fig. 3a and 3b). Furthermore, this was a cell intrinsic effect, since in mixed BM chimeras, T-bet deficient BM cells generated more NKT2 and NKT17 and fewer NKT1 than WT BM cells irrespective of donor cell frequencies (Supplementary Fig. 3c and 3d). Collectively, these findings suggest that T-bet directs the dominance of NKT1 in B6 mice and that T-bet deficiency redirects cellular fates to facilitate the development of NKT2 and NKT17 cells. To test for evidence of developmental exposure to IL-4, we examined CD8 SP thymocytes. Strikingly, up to 30% of CD8 SP thymocytes in T-bet deficient B6 mice expressed high levels of Eomes, the hallmark of IL-4 dependent memory-like CD8 T cell development (Fig. 4d and Supplementary Fig. 3e). This level approaches that seen in BALB/c mice. We also showed the source of IL-4 in T-bet deficient mice is iNKT cells as T-bet and CD1d double deficient mice failed to generate any Eomes+ CD8 SP thymocytes (Supplementary Fig. 3f and 3g). This analysis of T-bet deficient mice demonstrates that steady state IL-4 does not require NK1.1+ NKT1 cells, and indeed is enhanced by a compensating increase in NKT2 cell numbers in such mice.
Likewise, we also found that KLF2 deficiency facilitated a 10-fold increase in NKT2 cells in B6 mice, but did not increase the number of NKT1 or NKT17 cells (Supplementary Fig. 4). Memory-like CD8 T cells were dramatically increased in KLF2 deficient mice (Supplementary Fig. 4) as previously reported22, providing further evidence that NKT2 cells are the source of IL-4 produced in the steady state.
This genetic evidence together with the intrathymic injection data strongly support a lineage diversification model of iNKT cells, where a common progenitor gives rise to NKT1, NKT2 and NKT17 lineages instead of linear conversion of IL-4 producing iNKT cells into IFN-γ secreting ones (Supplementary Fig. 5).
iNKT cell lineage diversification in different inbred strains
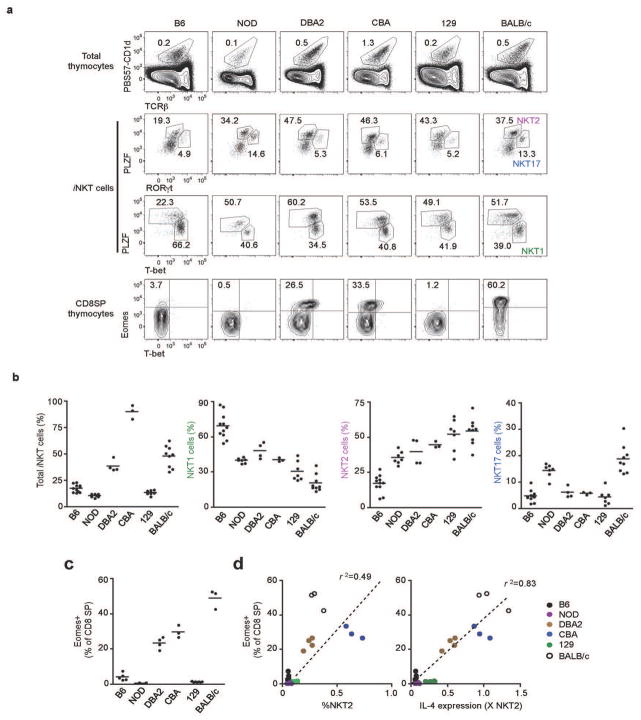
Having confirmed that iNKT lineage diversification happens differently in B6 and BALB/c mice (Fig. 2d) we wished to determine which pattern is representative amongst inbred mouse strains. We compared six different commonly used inbred strains of mice (Fig. 5). Mice of a similar age showed variability in the percentage (Fig. 5a and 5b) and number (data not shown) of total thymic iNKT cells. Using intracellular straining for transcription factors, we determined the relative proportion of NKT1, NKT2, and NKT17 in each. B6 mice were at one end of the spectrum, with high NKT1 and low NKT2, whereas BALB/c were at the other. The six strains showed a general inverse relationship between the proportion of NKT1 and NKT2 (Fig. 5b). Interestingly, three strains showed a marked population of Eomes expressing memory-like CD8 thymocytes: CBA, DBA/2, and BALB/c (Fig. 5a and 5c). Indeed, there was a positive correlation between the strains that have high percentages of NKT2 cells, and those that have Eomes expressing memory-like CD8 thymocytes (Fig. 5d, left panel, R2=0.49). Because IL-4 production by NKT2 cells was shown to be important for the development of memory-like CD8 thymocytes in BALB/c mice, we sorted NKT2 cells from all strains and measured gene expression of IL-4 by RT-PCR. The level of IL-4 produced by NKT2 cells correlated even more very tightly with the abundance of memory-like CD8 thymocytes in different inbred strains of mice (Fig. 5d, right panel, R2=0.83) suggesting that a universal mechanism of IL-4 dependent induction of memory-like CD8 thymocytes applies regardless of genetic background.
Figure 5. Interstrain comparison of iNKT subsets: NKT2 cell production of IL-4 is associated with memory-like CD8 T cells.
(a) Representative FACS profiles of thymic iNKT cells and CD8 SP thymocytes in 7–8 week-old indicated strains. (b) Percentage of iNKT cells among total thymocytes (left panel) and percentage of each subset (NKT1, NKT2 and NKT17) among total thymic iNKT cells (right panels), N=3~9. (c) Percentages of thymic CD8 SP thymocytes that express Eomes as detected by intracellular staining. Horizontal bars indicate mean values. (d) Correlation of Eomes expression in CD8 SP thymocytes with the frequency of NKT2 cells among total thymocytes (left) or IL-4 mRNA in NKT2 cells (right). Arbituary units represent the IL-4 message level in sorted NKT2 cells, normalized to GAPDH, and multiplied by the frequency of NKT2 cells among total thymocytes. Linear regression was used to calculate goodness of fit (R2).
IL-4 producing NKT2 cells are stimulated by self-ligands
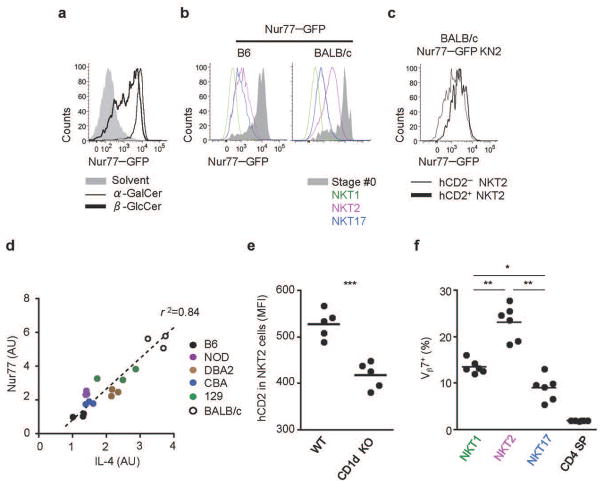
In T helper cells, IL-4 production is stimulated through the antigen receptor16. However, it was recently shown in various “innate lymphoid cells” that IL-4 can be stimulated by cytokines such as IL-25 and IL-3323. To understand what type of signals are stimulating IL-4 in NKT2 cells, we employed Nur77GFP mice, in which GFP expression is driven by antigen receptor signaling, and is independent of cytokine or inflammatory signals24. In these mice, GFP is rapidly upregulated in peripheral iNKT cells upon injection with the αGalCer or with the putative self-lipids β-glucosylceramide β GlcCer) (Fig. 6a) or isoglobotrihexosylceramide (iGb3) (data not shown). With this mouse, we sought to compare TCR signal perception of NKT2 cells to that of other subsets, hypothesizing that a difference in self-lipid antigen presentation might regulate IL-4 secretion in the steady state. We fully backcrossed Nur77GFP mice onto the BALB/c background and compared GFP expression of iNKT cells in B6 and BALB/c mice. Interestingly, NKT2 cells expressed a higher level of GFP than NKT17 and NKT1 cells in both strains (Fig. 6b) and NKT2 cells from BALB/c mice expressed a higher level of GFP than B6 (Fig. 6b), suggesting that such cells are being stimulated through the antigen receptor in vivo. We also generated BALB/c Nur77GFPKN2+/− mice, and found that hCD2+ NKT2 cells expressed a higher level of GFP than hCD2 negative NKT2 cells (Fig. 6c). Furthermore, endogenous Nur77 expression correlated well with IL-4 expression in sorted NKT2 cells amongst different inbred strains (Fig. 6d), consistent with the idea that iNKT cell stimulation through the TCR drives IL-4 production.
Figure 6. IL-4 producing iNKT cells are stimulated by self ligands.
(a) BM derived DCs pulsed with solvent (gray), βGlcCer (thick) or αGalCer (thin) were injected into B6 Nur77GFP mice and splenic iNKT cells were analyzed 16 hours later. Data are representative of two independent analyses with a total of 4 mice per group. (b) Thymocytes from B6 and BALB/c Nur77GFP mice were analyzed for expression of GFP in each iNKT subset. Data are representative of 3 independent experiments. (c) GFP expression in BALB/c Nur77GFP KN2+/− mice shows IL-4 producing NKT2 cells (hCD2+) express a higher level of GFP than hCD2 negative NKT2 cells. Representative data from 3 independent experiments is shown. (d) Gene expression of Nur77 was compared to IL-4 in sorted NKT2 cells (mRNA arbitrary units normalized to Gapdh). Linear regression was used to calculate goodness of fit(R2). (e) hCD2 expression is dependent on sustained TCR signaling. Thymic iNKT cells of BALB/c KN2+/− mice were MACS enriched by depleting CD8+ and CD24+ cells, labeled with VCT and injected into WT BALB/c (N=5) or BALB/c CD1d KO (N=5) mice. Six days later, hCD2 expression in NKT2 cells was analyzed. MFI, mean fluorescence intensity. An unpaired two tailed t-test was used to compare hCD2 MFI in WT or CD1d KO hosts. ***p=0.0004. (f) iNKT subsets have distinct TCR repertoires. TCR Vβ7+ usage is shown among NKT1, NKT2 and NKT17 cells in B6 mice (N=6). One-way ANOVA was used to compare the frequency of NKT1, NKT2 and NKT17 cells. ***p<0.001, *p<0.05.
To further test if antigen receptor signaling in the steady state is important for IL-4 expression by NKT2 cells, we intrathymically transferred thymic KN2+/− iNKT cells into WT or CD1d KO hosts (Supplementary Fig. 6a). Consistent with data from Nur77GFP mice, we observed a decreased mean fluorescence intensity of hCD2 on NKT2 cells (Fig. 6e) as well as a reduced frequency of hCD2+ NKT2 cells in CD1d deficient hosts after 6 days of transfer (Supplementary Fig. 6a). We also found that the NKT2 population has a significantly altered frequency of TCR Vβ2 and Vβ7+ cells in both B6 and BALB/c mice (Fig. 6f and Supplementary Fig. 6b) indicating that NKT2 cells have a distinct TCR repertoire. Collectively, these results indicate that IL-4 production is associated with TCR stimulation.
If self- or commensal lipid ligands were driving NKT2 cell activation and/or differentiation in the steady state, it is possible that this might vary with age. Thus we examined iNKT cell subsets and IL-4 production in B6 and BALB/c mice of different ages. At all ages, BALB/c mice produced more intrathymic iNKT than B6 mice (Supplementary Fig. 7a). Consistent with previous reports20, B6 mice were heavily skewed toward NKT1 at all ages, with the exception of very young (3 week old) mice, which had a modest proportion (30–35%) of NKT2 cells (Supplementary Fig. 7b). BALB/c mice, in contrast, were heavily skewed toward NKT2 up to 8 to 10 weeks of age; after that NKT1 and NKT17 cells predominated in older BALB/c mice. Interestingly, NKT2 percentages in BALB/c mice of different ages closely paralleled the percent producing IL-4 (hCD2+) and the development of memory-like CD8 T cells (Supplementary Fig. 7c). Although we do not understand what molecular factors drive these age dependent differences, it does suggest the potential for environmental influences on iNKT cell differentiation.
Steady state IL-4 influences CD8 T cells, B cells, and thymic dendritic cells
We speculated that if iNKT cells provide sufficient IL-4 for generation of memory-like CD8 T cells in some strains of mice, it might have other systemic effects. IL-4 stimulates B cells to secrete IgE25 and BALB/c mice have higher serum IgE levels than B6 mice26. As shown in Fig. 7a, we confirmed that BALB/c mice had more than 10 times higher serum IgE concentrations compared to age-matched B6 mice. This difference was attenuated in BALB/c CD1d KO mice, suggesting that at least some of the IL-4 that stimulates B cells to secrete IgE are generated by iNKT cells. T-bet heterozygous null B6 mice showed significantly increased serum IgE concentrations compared to WT B6 mice, which is further compatible with increased NKT2 cell IL-4 production (Fig. 7a).
Figure 7. Steady state IL-4 produced by iNKT cells influences B cells and thymic DCs.
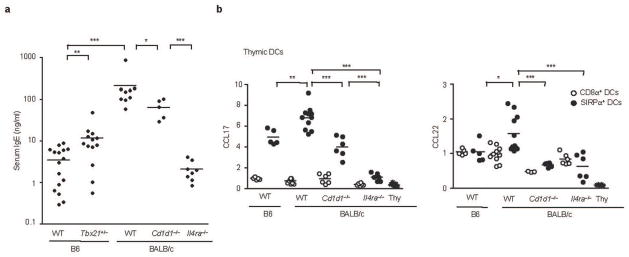
(a) Serum IgE concentrations measured by ELISA in indicated mice are shown. Unpaired two tailed t-tests were used to compare indicated two groups. N=5~16, *** p<0.001, **p<0.01, *p<0.05 (b) Thymic CD8α+ or SIRPα+− DCs were sorted as described in experimental procedures and analyzed for the expression of CCL17 and CCL22 by real time RT-PCR. One-way ANOVA was used to compare CCL17 and CCL22 levels in SIRPα+ DCs. Pooled data from four independent experiments using 6 to 12 mice total in each group. *** p<0.001, ** p<0.01, *p<0.05. Horizontal bars indicate mean values. Thy, thymocytes.
It has also been shown that IL-4 can cause DCs to secrete the “Th2 chemokines” CCL17 and CCL2227. Based on this finding, we investigated CCL17 and CCL22 expression in thymic DCs (Fig. 7b). We purified the two major thymic DC subtypes: CD8α+ and SIRPα+, and performed RT-PCR for CCL17 and CCL22. These chemokines, produced primarily from the SIRPα+ DCs as previously reported28, were expressed higher in BALB/c mice. Interestingly, DCs from CD1d deficient BALB/c mice showed reduced levels of CCL17 and CCL22, similar to that of their B6 counterpart, and IL-4 deficiency abrogated their expression entirely. Collectively, the above results show that IL-4 produced by iNKT cells not only influences the generation of memory CD8 T cells but also influences B cells and thymic DCs, possibly contributing to Th2 dominance in BALB/c mice.
Discussion
In this report, we defined subsets of iNKT cells according to their transcription factor expression and cytokine production, and showed that NKT2 cells are a source of steady state IL-4 production. This IL-4 influences the size of the memory CD8 T cell pool, stimulates thymic DC to secrete CCL17/CCL22, and increases serum IgE levels.
Until recently, the NKT field has embraced a sequential lineage developmental model20, in which “developmental intermediates” produce Th2 type cytokines and “mature” NK1.1+ iNKT cells produce Th1 cytokines. Dickgreber and colleagues suggested that “innate IL-4” is secreted from immature iNKT cells11. However, based on our findings of distinct transcription factors expressed in iNKT subsets, we considered an alternative “lineage diversification” model for iNKT cells9, analogous to effector T helper cell16 and innate lymphoid cell29 differentiation (Supplementary Fig. 5). In support of such a model, we observed that iNKT subsets exist in different ratios in different inbred strains of mice, that NKT2 and NKT1 lack a precursor-progeny relationship, and that T-bet deficiency blocks development of NKT1 while increasing NKT17 and NKT2. In a complementary fashion to T-bet, GATA-3 deficiency was reported to block the differentiation of NK1.1− CD4+ (NKT2) or NK1.1− CD4− (NKT17) lineages, while allowing NK1.1+ cells (NKT1) to mature30. Although the phenotype of iNKT cells from ROR-γt KO mice is not available as its deficiency abrogates iNKT cell development at the DP stage31, data from T-bet and GATA-3 deficient mice strongly support a model where development of NKT1 and NKT2 cells are controlled by lineage diversification rather than a linear maturation process. The complex interplay between T-bet, GATA-3 and ROR-γt described in conventional CD4 T cells lineages16 likely regulates the differentiation of NKT1, NKT2 and NKT17 cells from a PLZFhi common progenitor. Considering the analogous T helper cell differentiation, we also looked for a Bcl-6 expressing iNKT subset, which together with CXCR5 and PD-1 are hallmarks of follicular helper T cells. Cells with this phenotype have been described amongst peripheral iNKT cells of immunized mice32. However, we did not find significant numbers of such cells in the thymus of young adult B6 or BALB/c mice in the steady state (data not shown).
In several strains of mice, a prominent population of NKT2 cells produces an IL-4 rich environment and results in the development of large numbers of innate CD8 T cells among other IL-4 mediated effects described here. Our data suggest that NKT2 cells continually perceive stimulation through their TCR. Multiple self-lipids have been proposed as endogenous ligands for iNKT cells33–35 and it is possible that distinct lipids could be presented by distinct thymic APCs. The recent finding that mice with an inability to produce ether lipids have a preferential defect in NK1.1+ iNKT (NKT1) cells34 supports the idea that distinct endogenous lipids influence iNKT cell positive selection and differentiation. Interestingly, we observed differences in the Vβ repertoire in NKT2 compared to NKT1 or NKT17 cells, further supporting the idea that distinct iNKT subsets may be stimulated by different TCR ligands.
Nonetheless, certain cytokines can also cause cells to produce IL-4. A previous report showed that CD4+ NK1.1− iNKT cells (similar to our NKT2) express IL-17RB, a receptor for IL-2536. IL-25 is a cytokine produced by stromal cells that can promote the Th2 immune response23. Indeed, hCD2 positive (IL-4 producing) cells were found within the IL17RB positive population both in B6 and BALB/c mice. Watarai and colleagues showed that IL-17RB+ iNKT cells could produce IL-4 when exposed to exogenous IL-25 and that IL-25 receptor deficiency partially prevented the development of NK1.1 negative iNKT cells18. However, B6 NKT2 cells produce less IL-4 than BALB/c despite comparable level of IL17RB expression, and B6 NKT17 cells also express IL17RB indicating that IL17RB is not sufficient to drive IL-4 secretion (Supplementary Fig. 1c). It remains to be determined if a combination of TCR stimulation provided by self ligands and stromal cells secreting IL-25 might determine either the frequency of NKT2 cells or their ability to secrete IL-4 in BALB/c mice.
The relative abundance of NKT2 and NKT17 cells in BALB/c mice raises questions as to whether NKT subset skewing is due to a defective generation of NKT1 cells or an enhanced generation of NKT2 and NKT17 cells. The absolute number of NKT1 cells in the thymus was not different between B6 and BALB/c mice, whereas the number of NKT2 and NKT17 cells was 3 to 20 fold higher in BALB/c mice depending on the age of the mice (data not shown), suggesting it may be NKT2 and NKT17 lineages that are differently regulated between BALB/c and B6 mice. BALB/c mice lacking the transcription factor KLF13 do not have memory-like CD8 T cells7. This suggests that KLF13 may be important for the differentiation of NKT2 cells. Although they did not examine the specific transcription factors we used here, they showed IL-4 producing PLZFhi iNKT cells decreased in the thymus, which is consistent with this interpretation. Nonetheless, the level of KLF13 itself does not differ between B6 and BALB/c mice7, so the contributing genetic differences between these two strains remain undefined. Amongst six different inbred strains, a reciprocal relationship was generally observed between NKT1 and NKT2, but not necessarily NKT17. Notably, B6 mice were much more highly “NKT1 skewed” than other strains. Since NK1.1 is expressed only on NKT1 cells, NK1.1 expression identifies a substantial fraction of iNKT cells in B6 mice, but this is not the case in other inbred strains.
Our studies also uncovered a substantial age dependence of iNKT subset skewing, with B6 mice having 35% NKT2 cells at 3 weeks of age, but dropping to less than 5% by adulthood, while BALB/c mice showed up to 50% NKT2, which also dropped to less than 5%, but not until 18 weeks of age. We do not understand the factors that cause this. One possibility, suggested by a recent study, relates to Lin28b, a protein that regulates the Let7 family of microRNAs37. Lin28b was hypothesized to direct fetal like lymphopoiesis based in part on the observation that retroviral expression of Lin28b in adult hematopoietic progenitors led to an increase in NK1.1 negative iNKT and γδ T cells, and a dramatic increase in memory-like CD8 T cells37. Indeed, we found a modestly prolonged expression of Lin28b in thymi of BALB/c mice during the neonatal period compared to B6 mice (data not shown). However, BALB/c chimeras made using fetal hematopoietic cells that expressed a high level of Lin28b generated approximately the same NKT1, NKT2 and NKT17 lineages as chimeras made with adult BALB/c marrow cells that expressed a low level of Lin28b (data not shown), ruling out that NKT subsets are skewed differently by intrinsic differences in fetal and adult progenitors. Another possibility is that the thymic microenvironment of BALB/c mice supports iNKT precursors to differentiate into NKT2 and NKT17 lineages, possibly owing to differences in lipid antigen, SLAM expression on APC, or production of inductive cytokines, and some of these factors may differ with age. Finally, distinct microbiota were shown to influence the frequency and TCR Vβ usage of iNKT cells38. However, T-bet KO, heterozygous, and WT mice from the same cages still showed differences in iNKT phenotype, and (B6XBALB/c) F1 mice showed the same iNKT phenotype whether they were of BALB/c or B6 maternal origin (data not shown). Therefore, it is unlikely that our results came from a biased source of microbiota in different strains.
B6 and BALB/c mice are the prototypes of strains dominating Th1 and Th2 responses respectively. In a murine model of human asthma, ovalbumin specific Th2 responses induce airway hyper-responsiveness (AHR) and lung inflammation in BALB/c mice39. As T-bet deficient B6 mice became susceptible40 and BALB/c mice with a dominant negative form of GATA-3 were resistant to AHR induction41, it was suggested that Th2 skewing of conventional CD4 T cells was responsible for the strain specificity of disease induction. However CD1d KO BALB/c mice are resistant to asthma induction42 and T-bet or GATA-3 deficiency was shown to facilitate the development of NKT2 or NKT1 cells respectively (this study and ref30). Moreover, T-bet deficient BALB/c mice still required iNKT cells for AHR induction43. Therefore, NKT2 cells may play a role in strain differences of murine models of asthma.
iNKT cells influence the outcome of pathogenic infection in various ways. They can both directly and indirectly recognize pathogens, and the cytokines they produce can subsequently skew adaptive immune responses44. Some evidence suggests that iNKT cells can secrete IL-4 to enhance CD8 T cell responses. In an RSV infection model, iNKT cells activated virus specific CD8 T cells and CD1d KO BALB/c mice showed less severe response at the expense of delayed viral clearance45. In a malaria infection model, BALB/c mice were more resistant than B6 mice and the protective response was dependent on IL-4 receptors on CD8 T cells46. Furthermore, IL-4 or CD1d KO BALB/c mice were more susceptible to parasite infection and exhibited severe pathology47. These findings suggest that iNKT cells provide IL-4 required for proper CD8 T cell activation upon infection with malarial parasites. To this point, however, it is not clear whether IL-4 and iNKT cells are acting during the infection or developmentally (i.e. by inducing Eomes+ memory-like CD8 T cells). More sophisticated experimental systems will be required to properly address these issues in the future.
In this report, we provide evidence for a new transcription factor based classification system of iNKT cells that encompasses different strains of mice and correlates more directly with function. Surprisingly, B6 mice were one extreme of iNKT cell development, with NKT1 cells dominating. NKT2 cells were found to dominate in many other inbred strains of mice, and the consequence of this was higher basal IL-4 production with broad effects of immune response potential.
Online methods
Mice
B6 (C57BL/6NCr) and B6.CD45.1 (CD45.1 congenic B6) (B6-LY5.2/Cr) mice were purchased from the National Cancer Institute. Other mouse strains were purchased from JAX: T-bet deficient B6 (B6.129S6-Tbx21tm1Glm/J), B6 Cd1d−/− (B6.129S6-Cd1d1/Cd1d2tm1Spb/J), 129 (129X1/SvJ), DBA2 (DBA/2J), CBA (CBA/J), NOD (NOD/ShiLtJ), BALB/c (BALB/cByJ or BALB/cJ), and BALB/cJ Cd1d−/− (C.129S2-Cd1tm1Gru/J). CB6 T-bet+/− Cd1d−/− mice were generated by crossing B6 T-bet+/− Cd1d+/− and BALB/cJ Cd1d−/− mice. KN2 mice in B6 and BALB/c background were obtained from Markus Mohrs (Trudeau Institute). C57BL/6 Nur77GFP mice were generated previously 24 and backcrossed onto BALB/cByJ. T-bet reporter with ZsGreen (T-betGFP) mice were described previously48 and backcrossed onto a BALB/cByJ background. All the mice experiments were performed under protocols approved by the Institutional Animal Care and Use Committee of the University of Minnesota and the NIAID Animal Care and Use Committee.
Flow cytometry and antibodies
Single-cell suspensions were prepared from thymi and spleens and hepatic mononuclear cells were separated by percoll gradient. Biotinylated PBS57 loaded or unloaded CD1d monomers were obtained from the tetramer facility of the US National Institutes of Health. For hCD2 and PLZF staining, cells were surface stained with hCD2 (S5.2, mIgG2a; BD Bioscience), fixed with Foxp3 staining buffer set (eBioscience) and were incubated with antibody to PLZF (D-9, mIgG1; Santa Cruz) followed by staining with anti mouse immunoglobulin G1 (A85-1 or X-56; BD Biosciences). T-bet (4B10, mIgG1; eBioscience) was stained after blocking of free arm of anti-mouse IgG1 with mouse sera or PLZF antibody. In some staining, AF488 labeled (kind gift from Derek B. Saint’Angelo) or PE conjugated PLZF antibody (ebioscience) were used. Anti mouse IL17RB antibody was obtained from R&D Systems (752101, rat IgG1) or MBL (B5F6, rat IgG2a). All other antibodies were from eBioscience, BD or Biolegend. Cells were analyzed on an LSR II (Becton Dickinson) and data were processed with FlowJo software (TreeStar).
In vitro and in vivo cytokine production
For in vitro stimulation with PMA and ionomycin, total thymocytes or thymocytes depleted of CD8 and CD24 positive cells using MACS, were plated at a density of 1 × 106 cells per ml in RPMI medium plus 10% (vol/vol) FCS. Cells were incubated for 4 hours with PMA (50 ng/ml) and ionomycin (1.5 μM) with Monensin (eBioscience) for last 2 hours and analyzed for intracellular cytokines by FACS. For in vivo stimulation of iNKT cells, mice were intravenously injected 5 ug of α-galactosylceramide (KRN7000, Avanti Polar Lipids) and spleen and liver analyzed 3 hours later.
Mixed bone marrow chimera
Total BM cells were prepared from the femurs and tibias of B6 WT (CD45.1/1) or T-bet−/− (CD45.1/2) donor mice, and depleted of mature T cells with anti-Thy1.2 antibody and complement. Recipient mice (CD45.2/2) were lethally irradiated (1,000 rad) and received 107 T cell–depleted adult BM cells. Chimeras were analyzed at 6 weeks after transplant.
Intrathymic injection
Thymocytes from B6 or BALB/c T-betGFPKN2+/− mice were MACS depleted for CD8+ and CD24+ cells to enrich thymic iNKT cells. hCD2+CD4+T-betGFP NKT2 and hCD2− CD4+T-betGFP NKT2 cells were sorted using FACS Aria. Host (B6 or BALB/c) mice were anesthetized with an intraperitoneal injection of ketamine (90mg/kg) and xylazine (9mg/kg). Sorted cells were mixed with splenic CD8 T cells from B6 or BLAB/c CD1d KO mice, labeled with violet cell tracer (VCT) (Invitrogen) and injected into thymi of B6 or BALB/c recipients. In some experiments, enriched iNKT cells were labeled with VCT and injected into WT or CD1d KO BALB/c mice together with CFSE (Sigma) labeled splenic CD8 T cells.
Cell sorting and real-time RT-PCR
CD11c+ cells were enriched from thymocytes using MACS and CD8α+ or SIRPα+ DCs were purified by FACSAria (Becton Dickinson) sorter after gating CD11c+MHCII+Thy1.2− CD3− B220− cells. Cells with more than 95% purity of the target population were used for experiments. An RNeasy mini or micro kit (Qiagen) and a SuperScript III first Strand Synthesis SuperMix for qRT-PCR (Invitrogen) were used for the isolation of RNA and production of cDNA. FastStart Universal SYBR Green Master (Roche) and an ABI PRISM 7000 sequence detection system (Applied Bioscience) were used for amplification and detection. Primers were as follows: GAPDH, for 5′-TGGCCTACATGGCCTCCA -3′ and rev 5′-TCCCTAGGCCCCTCCTGTTAT -3′; IL-4, for 5′-GAGACTCTTTCGGGCTTTTC -3′ and rev 5′-TGATGCTCTTTAGGCTTTCCA -3′; Nur77, for 5′-ATGCTTCGTGTCAGCACTAT -3′ and rev 5′-GTACTTGGCGCTTTTCTGTA -3′. CCL17 and CCL22 primers were described previously27.
Lipid Pulsed BMDCs
Bone marrow cells from femurs of mice were cultured for 7 days (5×106 cells/well) in 6-well cell-culture dishes with complete RPMI medium in the presence of recombinant murine GM-CSF (50 ng/ml, PeproTech) and IL-4 (10 ng/ml, PeproTech). On day six, cells were pulsed with either 100 ng/ml of α-galactosylceramide (KRN7000, Avanti Polar Lipids), 1 ug/ml of β-glucosylceramide (C24:1 Glucosyl(β) Ceramide (d18:1/24:1(15Z)), Avanti Polar Lipids), or 1 ul/ml solvent (2:1 methanol:chloroform) for 12–15 hours. BMDCs were intravenously injected into Nur77GFP mice (0.5–1.0×106cells/mouse) and endogenous splenic iNKT cells were analyzed 16 hours later.
Statistical analysis
Prism software (Graphpad) was used for statistical analysis. Unpaired or paired two-tailed t-tests, linear regression, and ANOVA were used for data analysis and the generation of P or R2 values.
ELISA
A mouse IgE ELISA MAX kit (Biolegend) was used for quantifying serum IgE. All samples were serially diluted three times to get optimal range of dilution.
Supplementary Material
Acknowledgments
The authors wish to thank Jane Ding and Stephen Perry for excellent technical support. Markus Mohrs (Trudeau Institute) provided KN2 mice on a B6 and BALB/c background and Derek B. Sant’Angelo (MSKCC) provided AF488 conjugated anti-PLZF antibody. We thank Markus Mohrs and Mitch Kronenberg for helpful discussions, and Sara Hamilton for critical review of the manuscript. This research was supported by NIH grants R37-AI39560 (to K.A.H.) and RO1-AI075168 (to S.C.J.), an Irvington Institute Fellowship from the Cancer Research Institute (to Y.J.L), and T32 HD060536 training grant (to K.L.H). Jinfang Zhu is supported by the Division of Intramural Research, National Institute of Allergy and Infectious Diseases, National Institutes of Health, USA.
Footnotes
Author contribution
Y.J.L designed and performed experiments, analyzed data and wrote the manuscript; K.L.H. performed experiments and provided input on interpretation; J.Z. provided reagents and research interpretation; S.C.J. provided input to the research design and interpretation; and K.A.H. conceptualized the research, directed the study, analyzed data, and edited the manuscript.
Competing financial interests
The authors declare no competing financial interests.
References (up to 50)
- 1.Lee YJ, Jameson SC, Hogquist KA. Alternative memory in the CD8 T cell lineage. Trends Immunol. 2011;32:50–56. doi: 10.1016/j.it.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berg LJ. Signalling through TEC kinases regulates conventional versus innate CD8(+) T-cell development. Nat Rev Immunol. 2007;7:479–485. doi: 10.1038/nri2091. [DOI] [PubMed] [Google Scholar]
- 3.Weinreich MA, Odumade OA, Jameson SC, Hogquist KA. T cells expressing the transcription factor PLZF regulate the development of memory-like CD8+ T cells. Nat Immunol. 2010;11:709–716. doi: 10.1038/ni.1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alonzo ES, et al. Development of promyelocytic zinc finger and ThPOK-expressing innate gamma delta T cells is controlled by strength of TCR signaling and Id3. J Immunol. 2010;184:1268–1279. doi: 10.4049/jimmunol.0903218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Verykokakis M, Boos MD, Bendelac A, Kee BL. SAP protein-dependent natural killer T-like cells regulate the development of CD8(+) T cells with innate lymphocyte characteristics. Immunity. 2010;33:203–215. doi: 10.1016/j.immuni.2010.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Min HS, et al. MHC class II-restricted interaction between thymocytes plays an essential role in the production of innate CD8+ T cells. J Immunol. 2011;186:5749–5757. doi: 10.4049/jimmunol.1002825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lai D, et al. KLF13 sustains thymic memory-like CD8(+) T cells in BALB/c mice by regulating IL-4-generating invariant natural killer T cells. J Exp Med. 2011;208:1093–1103. doi: 10.1084/jem.20101527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rafei M, et al. Development and function of innate polyclonal TCRalphabeta+ CD8+ thymocytes. J Immunol. 2011;187:3133–3144. doi: 10.4049/jimmunol.1101097. [DOI] [PubMed] [Google Scholar]
- 9.Constantinides MG, Bendelac A. Transcriptional regulation of the NKT cell lineage. Curr Opin Immunol. 2013 doi: 10.1016/j.coi.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mohrs K, Wakil AE, Killeen N, Locksley RM, Mohrs M. A two-step process for cytokine production revealed by IL-4 dual-reporter mice. Immunity. 2005;23:419–429. doi: 10.1016/j.immuni.2005.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dickgreber N, et al. Immature murine NKT cells pass through a stage of developmentally programmed innate IL-4 secretion. J Leukoc Biol. 2012;92:999–1009. doi: 10.1189/jlb.0512242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bendelac A, Savage PB, Teyton L. The biology of NKT cells. Annu Rev Immunol. 2007;25:297–336. doi: 10.1146/annurev.immunol.25.022106.141711. [DOI] [PubMed] [Google Scholar]
- 13.McNab FW, et al. Peripheral NK1.1 NKT cells are mature and functionally distinct from their thymic counterparts. J Immunol. 2007;179:6630–6637. doi: 10.4049/jimmunol.179.10.6630. [DOI] [PubMed] [Google Scholar]
- 14.Savage AK, et al. The transcription factor PLZF directs the effector program of the NKT cell lineage. Immunity. 2008;29:391–403. doi: 10.1016/j.immuni.2008.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kovalovsky D, et al. The BTB-zinc finger transcriptional regulator PLZF controls the development of invariant natural killer T cell effector functions. Nat Immunol. 2008;9:1055–1064. doi: 10.1038/ni.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhu J, Yamane H, Paul WE. Differentiation of effector CD4 T cell populations (*) Annu Rev Immunol. 2010;28:445–489. doi: 10.1146/annurev-immunol-030409-101212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wei G, et al. Genome-wide analyses of transcription factor GATA3-mediated gene regulation in distinct T cell types. Immunity. 2011;35:299–311. doi: 10.1016/j.immuni.2011.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Watarai H, et al. Development and function of invariant natural killer T cells producing T(h)2- and T(h)17-cytokines. PLoS Biol. 2012;10:e1001255. doi: 10.1371/journal.pbio.1001255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Berzins SP, McNab FW, Jones CM, Smyth MJ, Godfrey DI. Long-term retention of mature NK1.1+ NKT cells in the thymus. J Immunol. 2006;176:4059–4065. doi: 10.4049/jimmunol.176.7.4059. [DOI] [PubMed] [Google Scholar]
- 20.Benlagha K, Kyin T, Beavis A, Teyton L, Bendelac A. A thymic precursor to the NK T cell lineage. Science. 2002;296:553–555. doi: 10.1126/science.1069017. [DOI] [PubMed] [Google Scholar]
- 21.Townsend MJ, et al. T-bet regulates the terminal maturation and homeostasis of NK and Valpha14i NKT cells. Immunity. 2004;20:477–494. doi: 10.1016/s1074-7613(04)00076-7. [DOI] [PubMed] [Google Scholar]
- 22.Weinreich MA, et al. KLF2 transcription-factor deficiency in T cells results in unrestrained cytokine production and upregulation of bystander chemokine receptors. Immunity. 2009;31:122–130. doi: 10.1016/j.immuni.2009.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saenz SA, Noti M, Artis D. Innate immune cell populations function as initiators and effectors in Th2 cytokine responses. Trends Immunol. 2010;31:407–413. doi: 10.1016/j.it.2010.09.001. [DOI] [PubMed] [Google Scholar]
- 24.Moran AE, et al. T cell receptor signal strength in Treg and iNKT cell development demonstrated by a novel fluorescent reporter mouse. J Exp Med. 2011;208:1279–1289. doi: 10.1084/jem.20110308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Coffman RL, et al. B cell stimulatory factor-1 enhances the IgE response of lipopolysaccharide-activated B cells. J Immunol. 1986;136:4538–4541. [PubMed] [Google Scholar]
- 26.Pochanke V, Hatak S, Hengartner H, Zinkernagel RM, McCoy KD. Induction of IgE and allergic-type responses in fur mite-infested mice. Eur J Immunol. 2006;36:2434–2445. doi: 10.1002/eji.200635949. [DOI] [PubMed] [Google Scholar]
- 27.Semmling V, et al. Alternative cross-priming through CCL17-CCR4-mediated attraction of CTLs toward NKT cell-licensed DCs. Nat Immunol. 2010;11:313–320. doi: 10.1038/ni.1848. [DOI] [PubMed] [Google Scholar]
- 28.Proietto AI, et al. Dendritic cells in the thymus contribute to T-regulatory cell induction. Proc Natl Acad Sci U S A. 2008;105:19869–19874. doi: 10.1073/pnas.0810268105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Spits H, et al. Innate lymphoid cells--a proposal for uniform nomenclature. Nat Rev Immunol. 2013;13:145–149. doi: 10.1038/nri3365. [DOI] [PubMed] [Google Scholar]
- 30.Kim PJ, et al. GATA-3 regulates the development and function of invariant NKT cells. J Immunol. 2006;177:6650–6659. doi: 10.4049/jimmunol.177.10.6650. [DOI] [PubMed] [Google Scholar]
- 31.Egawa T, et al. Genetic evidence supporting selection of the Valpha14i NKT cell lineage from double-positive thymocyte precursors. Immunity. 2005;22:705–716. doi: 10.1016/j.immuni.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 32.Chang PP, et al. Identification of Bcl-6-dependent follicular helper NKT cells that provide cognate help for B cell responses. Nat Immunol. 2012;13:35–43. doi: 10.1038/ni.2166. [DOI] [PubMed] [Google Scholar]
- 33.Zhou D, et al. Lysosomal glycosphingolipid recognition by NKT cells. Science. 2004;306:1786–1789. doi: 10.1126/science.1103440. [DOI] [PubMed] [Google Scholar]
- 34.Facciotti F, et al. Peroxisome-derived lipids are self antigens that stimulate invariant natural killer T cells in the thymus. Nat Immunol. 2012;13:474–480. doi: 10.1038/ni.2245. [DOI] [PubMed] [Google Scholar]
- 35.Brennan PJ, et al. Invariant natural killer T cells recognize lipid self antigen induced by microbial danger signals. Nat Immunol. 2011;12:1202–1211. doi: 10.1038/ni.2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Terashima A, et al. A novel subset of mouse NKT cells bearing the IL-17 receptor B responds to IL-25 and contributes to airway hyperreactivity. J Exp Med. 2008;205:2727–2733. doi: 10.1084/jem.20080698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yuan J, Nguyen CK, Liu X, Kanellopoulou C, Muljo SA. Lin28b reprograms adult bone marrow hematopoietic progenitors to mediate fetal-like lymphopoiesis. Science. 2012;335:1195–1200. doi: 10.1126/science.1216557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wingender G, et al. Intestinal microbes affect phenotypes and functions of invariant natural killer T cells in mice. Gastroenterology. 2012;143:418–428. doi: 10.1053/j.gastro.2012.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim HY, DeKruyff RH, Umetsu DT. The many paths to asthma: phenotype shaped by innate and adaptive immunity. Nat Immunol. 2010;11:577–584. doi: 10.1038/ni.1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Finotto S, et al. Development of spontaneous airway changes consistent with human asthma in mice lacking T-bet. Science. 2002;295:336–338. doi: 10.1126/science.1065544. [DOI] [PubMed] [Google Scholar]
- 41.Zhang DH, et al. Inhibition of allergic inflammation in a murine model of asthma by expression of a dominant-negative mutant of GATA-3. Immunity. 1999;11:473–482. doi: 10.1016/s1074-7613(00)80122-3. [DOI] [PubMed] [Google Scholar]
- 42.Akbari O, et al. Essential role of NKT cells producing IL-4 and IL-13 in the development of allergen-induced airway hyperreactivity. Nat Med. 2003;9:582–588. doi: 10.1038/nm851. [DOI] [PubMed] [Google Scholar]
- 43.Kim HY, et al. The development of airway hyperreactivity in T-bet-deficient mice requires CD1d-restricted NKT cells. J Immunol. 2009;182:3252–3261. doi: 10.4049/jimmunol.0803339. [DOI] [PubMed] [Google Scholar]
- 44.Tupin E, Kinjo Y, Kronenberg M. The unique role of natural killer T cells in the response to microorganisms. Nat Rev Microbiol. 2007;5:405–417. doi: 10.1038/nrmicro1657. [DOI] [PubMed] [Google Scholar]
- 45.Johnson TR, Hong S, Van Kaer L, Koezuka Y, Graham BS. NK T cells contribute to expansion of CD8(+) T cells and amplification of antiviral immune responses to respiratory syncytial virus. J Virol. 2002;76:4294–4303. doi: 10.1128/JVI.76.9.4294-4303.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Morrot A, Hafalla JC, Cockburn IA, Carvalho LH, Zavala F. IL-4 receptor expression on CD8+ T cells is required for the development of protective memory responses against liver stages of malaria parasites. J Exp Med. 2005;202:551–560. doi: 10.1084/jem.20042463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hansen DS, Siomos MA, Buckingham L, Scalzo AA, Schofield L. Regulation of murine cerebral malaria pathogenesis by CD1d-restricted NKT cells and the natural killer complex. Immunity. 2003;18:391–402. doi: 10.1016/s1074-7613(03)00052-9. [DOI] [PubMed] [Google Scholar]
- 48.Zhu J, et al. The transcription factor T-bet is induced by multiple pathways and prevents an endogenous Th2 cell program during Th1 cell responses. Immunity. 2012;37:660–673. doi: 10.1016/j.immuni.2012.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.