Abstract
Aims/hypothesis
In humans, disruption of the gene BSCL2, encoding the protein seipin, causes congenital generalised lipodystrophy (CGL) with severe insulin resistance and dyslipidaemia. While the causative gene has been known for over a decade, the molecular functions of seipin are only now being uncovered. Most pathogenic mutations in BSCL2 represent substantial disruptions including significant deletions and frameshifts. However, several more subtle mutations have been reported that cause premature stop codons or single amino acid substitutions. Here we have examined these mutant forms of seipin to gain insight into how they may cause CGL.
Methods
We generated constructs expressing mutant seipin proteins and determined their expression and localisation. We also assessed their capacity to recruit the key adipogenic phosphatidic acid phosphatase lipin 1, a recently identified molecular role of seipin in developing adipocytes. Finally, we used atomic force microscopy to define the oligomeric structure of seipin and to determine whether this is affected by the mutations.
Results
We show that the R275X mutant of seipin is not expressed in pre-adipocytes. While the other premature stop mutant forms fail to bind lipin 1 appropriately, the point mutants T78A, L91P and A212P all retain this capacity. We demonstrate that wild-type human seipin forms oligomers of 12 subunits in a circular configuration but that the L91P and A212P mutants of seipin do not.
Conclusions/interpretation
Our study represents the most comprehensive analysis so far of mutants of seipin causing lipodystrophy and reveals several different molecular mechanisms by which these mutations may cause disease.
Electronic supplementary material
The online version of this article (doi:10.1007/s00125-013-3029-3) contains peer-reviewed but unedited supplementary material, which is available to authorised users.
Keywords: Adipogenesis, Adipose tissue, BSCL2, Lipin, Lipodystrophy, Seipin
Introduction
Mutations in the gene BSCL2 cause Berardinelli–Seip congenital lipodystrophy type 2 (BSCL2), the most severe form of lipodystrophy. Affected individuals almost completely lack adipose tissue [1, 2] and typically suffer from severe insulin resistance, dyslipidaemia, hypertriglyceridaemia and muscular hypertrophy [2–4]. BSCL2 encodes the protein seipin, which can be either 398 or 462 amino acids long as a result of an alternative start codon that leads to the addition of a 64 amino acid extension to the N-terminus, thereby generating the longer form. The seipin protein has two transmembrane domains and resides in the endoplasmic reticulum (ER) membrane, with both termini in the cytoplasm [5, 6].
Studies in yeast and non-adipose cells have suggested that seipin may affect lipid droplet (LD) size, morphology and inheritance, although the molecular mechanism underlying these phenomena remains unclear. Yeast cells lacking fld1p (fldΔ1), the orthologue of seipin, form irregular LDs with proliferation of the ER and impaired lipolysis [7–11]. The fldΔ1 cells also show alterations in phospholipid composition and in levels of phosphatidic acid (PA) [8, 9]. Failure to identify any seipin-interacting proteins in yeast has led to speculation about a structural role for seipin in the budding of LDs from the ER membrane [7].
Seipin has been shown to play an essential cell-autonomous role in adipogenesis [12, 13]. A lack of seipin causes failure to sustain the expression of key adipogenic factors, including peroxisome proliferator-activated receptor γ (PPARγ), which may explain the significantly reduced expression of adipogenic genes [12, 13]. Mouse embryonic fibroblasts lacking seipin undergo unrestrained lipolysis at a relatively early stage of adipogenesis, causing the loss of lipid initially stored in these cells and loss of adipogenic gene expression [14]. This observation provided the first indications as to how the lack of seipin may contribute to failure of adipogenesis; however, the molecular details of this phenomenon remain uncertain. Recently, we demonstrated that seipin can bind the PA phosphatase lipin 1, which itself is required for adipogenesis and adipose tissue development in mice [15–18]. Seipin knockdown increases levels of the lipin 1 substrate PA and decreases the association of lipin 1 with the ER membrane in differentiating adipocytes [17]. Lipin 1 binding requires the C-terminal cytoplasmic domain of seipin [17], which Yang and colleagues have shown is critical for the adipogenic function of the protein [19]. In contrast, the LD modifying function of seipin appears to be mediated by a conserved core seipin sequence present in the yeast orthologue, which lacks this cytoplasmic domain [8, 10, 19].
Previously published studies regarding mutations in seipin that cause congenital generalised lipodystrophy (CGL) have focused mainly on the A212P mutant form of seipin [13, 20]. Here, we perform a more comprehensive analysis of the effects of several mutations in seipin that cause lipodystrophy. We have examined four point mutations that result in premature truncation of the seipin protein when translated (E113X, R138X, R275X, Q391X) and three point mutations that cause single amino acid substitutions (T78A, L91P, A212P) [2, 3, 21]. Our data indicate that several pathogenic mechanisms may underlie the failure of adipocyte development in BSCL2 patients carrying these mutations.
Methods
Constructs
Constructs to express haemagglutinin (HA)-tagged lipin 1β, Myc- and FLAG-tagged long forms of seipin were as previously described [5, 13, 17, 22]. All Myc- and FLAG-tagged seipin mutants were generated from wild-type constructs using the QuikChange II Site-Directed Mutagenesis kit (Agilent Technologies, Wokingham, UK) and appropriate changes were confirmed by sequencing.
Cell culture
Human HEK293 and murine C3H10T1/2 cells were routinely cultured, and C3H10T1/2 cells were differentiated, as previously described [13, 17]. Confluent C3H10T1/2 cells were transiently transfected using Lipofectamine LTX (Invitrogen, Paisley, UK) and immediately induced to differentiate. Cells were lysed 48 h later. HEK293 cells were cultured in DMEM (Sigma, Dorset, UK) containing 10% (vol./vol.) FBS and transiently transfected using Fugene6 Transfection Reagent (Promega, Southampton, UK). HEK293 cells were harvested 48 h after transfection. For atomic force microscopy (AFM) experiments tsA 201 cells, a sub-clone of HEK293 cells stably expressing the SV40 large T antigen, were grown in DMEM supplemented with 10% (vol./vol.) FBS, 100 U/ml penicillin and 100 μg/ml streptomycin in an atmosphere of 5% CO2/air. Transfection was carried out using calcium phosphate precipitation. A total of 250 μg of DNA was used to transfect cells in 5 × 162 cm2 culture flasks and cells were harvested 48 h later.
Immunoprecipitations and immunoblotting
Cells were lysed in 50 mmol/l Tris, pH 7.4, 150 mmol/l NaCl, 1 mmol/l EDTA containing 50 mmol/l n-octyl-β-d-glucopyranoside and protease inhibitors (Complete EDTA free; Roche, Burgess Hill, UK). Samples were sonicated, and centrifuged at 16,000 g for 10 min at 4°C. Supernatant fractions containing 1 mg of protein were incubated with anti-FLAG antibody conjugated to agarose beads (Santa Cruz, Dallas, TX, USA) for 2 h at 4°C. Immunoprecipitated samples were eluted using triple-FLAG peptide (Sigma). Samples were separated by SDS-PAGE, and immunoblotted using antibodies to Myc (clone 4A6; Millipore, Watford, UK), FLAG (Sigma), HA (Y-11, Santa Cruz), lipin 1 (generously provided by Symeon Siniossoglou, CIMR, Cambridge, UK) or calnexin (Abcam, Cambridge, UK). Following incubation with horseradish peroxidase-linked secondary antibodies, bands were visualised by enhanced chemi-luminescence (ECL). All immunoblots are representative of at least three independent experiments. Blots were quantified using ImageJ 1.45 s (http://imagej.nih.gov/ij).
Immunofluorescence
Cells were grown and transfected on glass coverslips and then fixed, permeabilised, blocked and probed with antibodies as previously described [17, 23]. Antibodies used were as described for immunoblotting. Highly cross-adsorbed Alexa Fluor anti-mouse 488 or anti-rabbit 594 secondary antibodies were used for detection (Invitrogen). Coverslips were mounted with Prolong Gold antifade reagent with DAPI (Invitrogen) and analysed on a Carl Zeiss LSM510 META Confocal microscope (Carl Zeiss, Cambridge, UK). Fluorescence from 405 nm excitation of DAPI was collected through a 420–480 nm bandpass filter; 488 nm excitation emission was collected through a 505–575 nm bandpass filter, and emission from 594 nm through a long-pass 615 nm filter. Images were recorded and analysed using Zeiss ZEN software with a 63x objective.
RNA extraction and real-time quantitative PCR
Total RNA was extracted using RNeasy kits (Qiagen, Hilden, Germany), following the manufacturer’s instructions. cDNA was synthesised, and real-time PCR was performed with Taqman or Sybr Green reagents (ABI, Paisley, UK) as previously described [23].
AFM imaging
Wild-type and mutant seipin constructs were expressed in tsA 201 cells. Transfected cells were solubilised in 1% Triton X-100 for 1 h, centrifuged at 61,740 g to remove insoluble material and then incubated with anti-FLAG-agarose beads for 3 h. Proteins were eluted with triple-FLAG peptide (0.15 mg/ml). All purification steps were carried out at 4°C.
Isolated proteins were imaged either alone or following overnight incubation with anti-Myc, anti-HA or anti-V5 antibodies. Various dilutions of each protein sample were made, and 45 μl of each sample was allowed to adsorb onto freshly cleaved mica disks. After a 5 min incubation, the sample was washed with Biotechnology Performance Certified-grade water (Sigma) and dried in a stream of nitrogen gas. Imaging was performed with a Veeco Digital Instruments Multimode atomic force microscope controlled by a Nanoscope IIIa controller (Veeco, Cambridge, UK). Samples were imaged in air, using tapping mode. The silicon cantilevers used had a drive frequency of ∼300 kHz and a specified spring constant of 40 N/m (Olympus, Southend, UK). The applied imaging force was kept as low as possible (AS/A0 ∼0.85). Disks with approximately equal protein densities for the various constructs were chosen for subsequent analysis.
The molecular volumes of protein particles were determined from particle dimensions based on AFM images. After adsorption of the proteins onto the mica support, the particles adopted the shape of a spherical cap. Molecular volume was calculated using the equation
| 1 |
where h is the particle height and r is the radius. Molecular volume based on molecular mass was calculated using the equation
| 2 |
where M 0 is the molecular mass, N 0 is Avogadro’s number, V 1 and V 2 are the partial specific volumes of particle (0.74 cm3/g) and water (1 cm3/g), respectively, and d is the extent of protein hydration (taken as 0.4 g water/g protein). It has been shown previously [24] that the molecular volumes of proteins measured by imaging in air are similar to the values obtained by imaging under fluid; hence, the process of drying does not significantly affect the measured molecular volume. It has also been shown by us [25] and by others [24] that there is a close correspondence between the measured and predicted molecular volumes for various proteins over a wide range of molecular masses; hence, molecular volume is measured reasonably accurately by AFM imaging.
Data were separated into appropriate bin widths and histograms plotted. Gaussian curves were fitted and the mean value of the peaks calculated.
Results
The R275X mutation in seipin, but not other premature stop mutations, leads to a lack of protein expression in pre-adipocytes
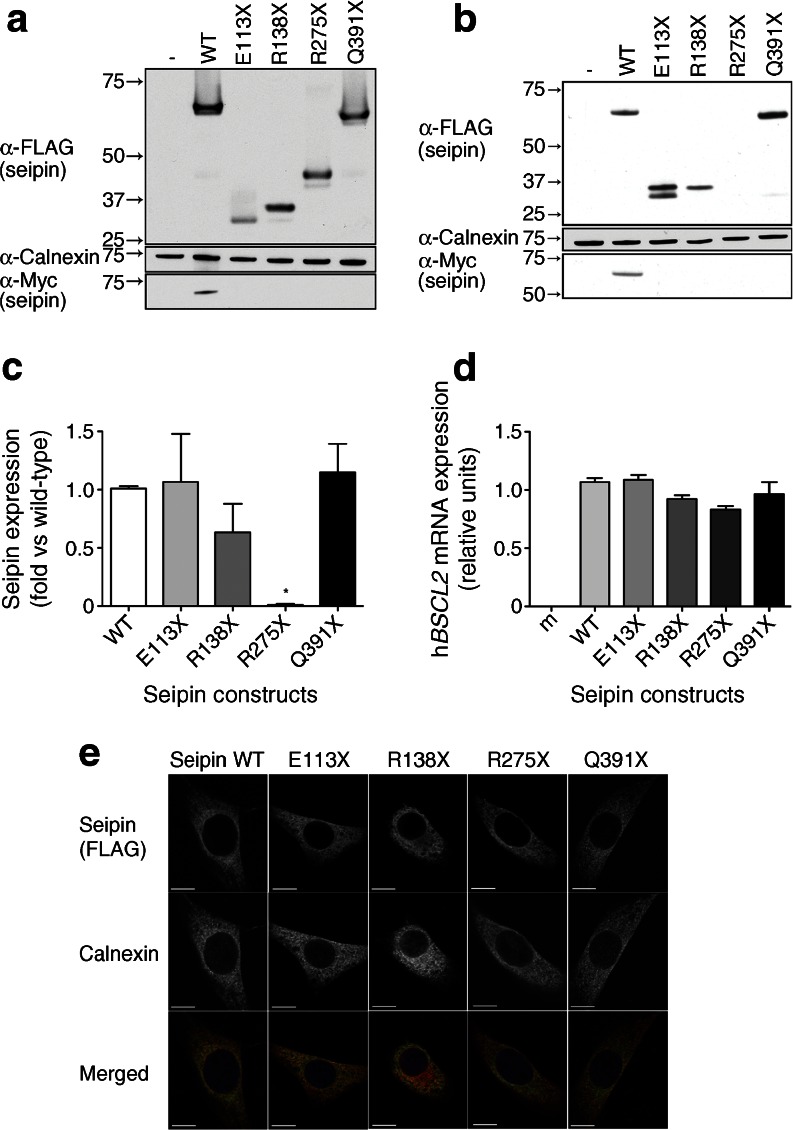
To examine how premature stop mutations may affect the function of seipin, we generated constructs in which the E113X, R138X, R275X and Q391X pathogenic mutations [2, 3, 21] were introduced by site-directed mutagenesis into the full-length cDNA encoding the long form of seipin bearing an N-terminal triple-FLAG tag and a C-terminal Myc tag. These constructs were transfected into HEK293 cells to ensure maximal transfection efficiency and protein abundance and each resulted in the expression of proteins of the expected sizes when probed with antibodies to detect the triple-FLAG tag at their N-termini (Fig. 1a). As predicted, only wild-type seipin resulted in the expression of a protein that could be detected via the C-terminal Myc tag, confirming that each pathogenic mutation caused the expected premature stop in translation.
Fig. 1.
The R275X mutation in seipin leads to loss of protein expression in differentiating adipocyte precursors. (a) HEK293 cells were transfected with empty vector (−), the long form of wild-type seipin bearing N-terminal triple-FLAG and C-terminal Myc tags (WT) or identically tagged forms with the point mutations E113X, R138X, R275X or Q391X. Lysates were separated by SDS-PAGE and immunoblotted for FLAG (α-FLAG), Myc (α-Myc) and calnexin (α-Calnexin). (b) Confluent C3H10T1/2 cells were transfected with empty vector (−) or with epitope-tagged WT or mutant forms of seipin as in (a). Cells were induced to differentiate for 2 days; lysates were separated by SDS-PAGE and immunoblotted for FLAG, Myc and calnexin. In (a) and (b) blots are representative of at least three independent experiments. (c) Quantitative analysis of WT, E113X, R138X, R275X or Q391X forms of seipin protein in C3H10T1/2 cells. FLAG–seipin bands from replicated blots as shown in (b) were normalised to calnexin expression in the same samples and expressed as means ± SEM, n = 3. *p < 0.05 vs WT. (d) C3H10T1/2 cells were transfected with empty vector (m) or with epitope-tagged WT or mutant forms of seipin as in (a). Cells were induced to differentiate for 2 days, and expression of mRNA encoding transfected human seipin (hBSCL2 mRNA) was determined by real-time PCR. Data are normalised to cyclophilin A and expressed as means ± SEM, n = 4. (e) Subconfluent C3H10T1/2 pre-adipocytes were transfected with epitope-tagged WT or mutant forms of seipin, fixed and immunostained for seipin using anti-FLAG antibody or with anti-calnexin antibody to reveal the ER. Individual images are shown in greyscale, and merged images show overlay of FLAG–seipin (green) and calnexin (red). Scale bars, 10 μm
To examine expression in a more relevant pre-adipocyte cell line the above constructs were expressed in C3H10T1/2 cells. Consistent with our previous studies using an artificially truncated version of the short form of human seipin [13], we were unable to detect the R275X mutant seipin protein (Fig. 1b). However, the other three premature stop seipin mutants tested were robustly expressed. Quantified data are shown in Fig. 1c. We observed a doublet for E113X when expressed in C3H10T1/2 cells and suspect that the lower band represents the monomer and the upper a partially denatured form of the protein as the samples were not heated before separation on SDS-PAGE. Real-time PCR analysis revealed equivalent expression of mRNA encoding wild-type or mutant forms of seipin, confirming that the inability to detect the R275X seipin did not result from altered levels of mRNA expression (Fig. 1d). Despite the high sequence similarity between murine and human seipin we observed no signal in mock-transfected cells (m, Fig. 1d), demonstrating that this assay did not detect endogenous murine Bscl2 mRNA.
When analysed by confocal microscopy, wild-type seipin showed the typical reticular distribution and co-localisation with calnexin, consistent with the previously reported ER localisation (Fig. 1e) [5, 13, 20]. Although very few cells transfected with the R275X mutant seipin showed any staining for the protein, in occasional cells showing weak expression the R275X seipin co-localised with the ER marker calnexin, as did the other premature stop mutants examined.
The T78A, L91P and A212P mutations do not dramatically affect seipin protein expression, but L91P and A212P seipin show altered localisation
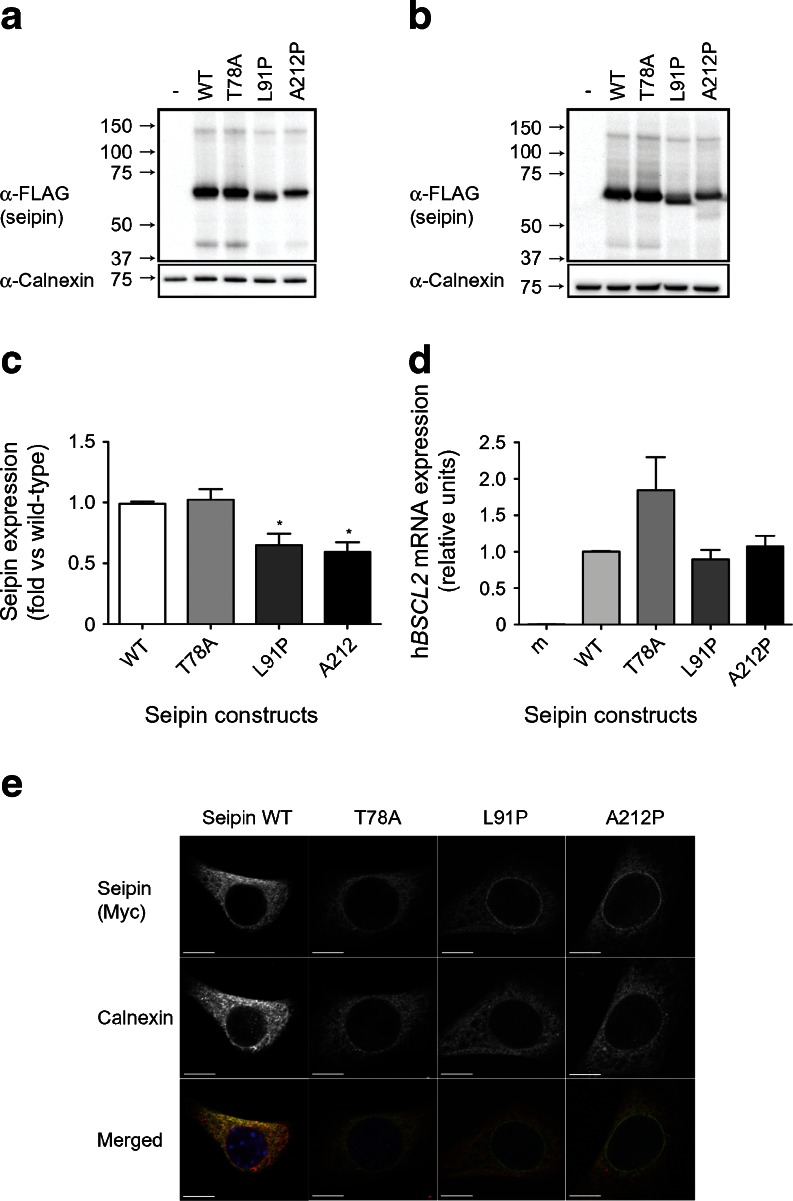
We next examined the effect of the T78A, L91P and A212P mutations on seipin expression. When each of these constructs was transfected into either HEK293 cells (Fig. 2a) or C3H10T1/2 cells (Fig. 2b), seipin was robustly expressed. However, we observed that the L91P and A212P mutations did result in a modest reduction in protein expression when compared with wild-type or T78A seipin (Fig. 2c). This was not the result of a significant effect of these mutations on mRNA expression levels (Fig. 2d).
Fig. 2.
The pathogenic point mutations T78A, L91P and A212P do not affect seipin protein expression, but L91P and A212P seipin partially mislocalise to the nuclear envelope. (a) HEK293 cells were transfected with empty vector (−), the long form of wild-type seipin with N-terminal triple-FLAG and C-terminal Myc tags (WT) or with identically tagged T78A, L91P or A212P forms. Lysates were separated by SDS-PAGE and immunoblotted for FLAG (α-FLAG) and calnexin (α-Calnexin). (b) C3H10T1/2 cells were transfected with empty vector (−) or with epitope-tagged WT or mutant forms of seipin as in (a). Lysates were separated by SDS-PAGE and immunoblotted for FLAG and calnexin. In (a) and (b) blots are representative of at least three independent experiments. (c) Quantitative analysis of WT, T78A, L91P or A212P forms of seipin protein in C3H10T1/2 cells. FLAG–seipin bands from replicated blots as shown in (b) were normalised to calnexin expression in the same samples and expressed as means ± SEM, n = 3. *p < 0.05 vs WT. (d) C3H10T1/2 cells were transfected with empty vector (m) or with epitope-tagged WT or mutant forms of seipin as in (a). Cells were induced to differentiate for 2 days and expression of mRNA encoding transfected human seipin (hBSCL2) was determined by real-time PCR. Data are normalised to cyclophilin A and expressed as means ± SEM, n = 4. (e) Subconfluent C3H10T1/2 pre-adipocytes were transfected with epitope-tagged WT or mutant forms of seipin and were fixed and immunostained for seipin using anti-Myc antibody or with anti-calnexin antibody to reveal the ER. Individual images are shown in greyscale and merged images show overlay of Myc–seipin (green) and calnexin (red). Scale bars, 10 μm
We and others have previously observed that A212P seipin can display an altered localisation, giving a characteristic ring of fluorescence around the nucleus [13, 20]. As shown in Fig. 2e, we again observed this partial mislocalisation of A212P seipin. A similar effect was observed with L91P seipin. This partial redistribution of L91P and A212P seipin was observed in all cells expressing these constructs whether subconfluent pre-adipocytes (Fig. 2e) or post-confluent cells induced to differentiate for 48 h (electronic supplementary material [ESM] Fig. 1). In contrast, the subcellular distribution of T78A seipin appeared to be identical to that of wild-type seipin.
The T78A, L91P and A212P mutations do not affect the capacity of seipin to bind lipin 1
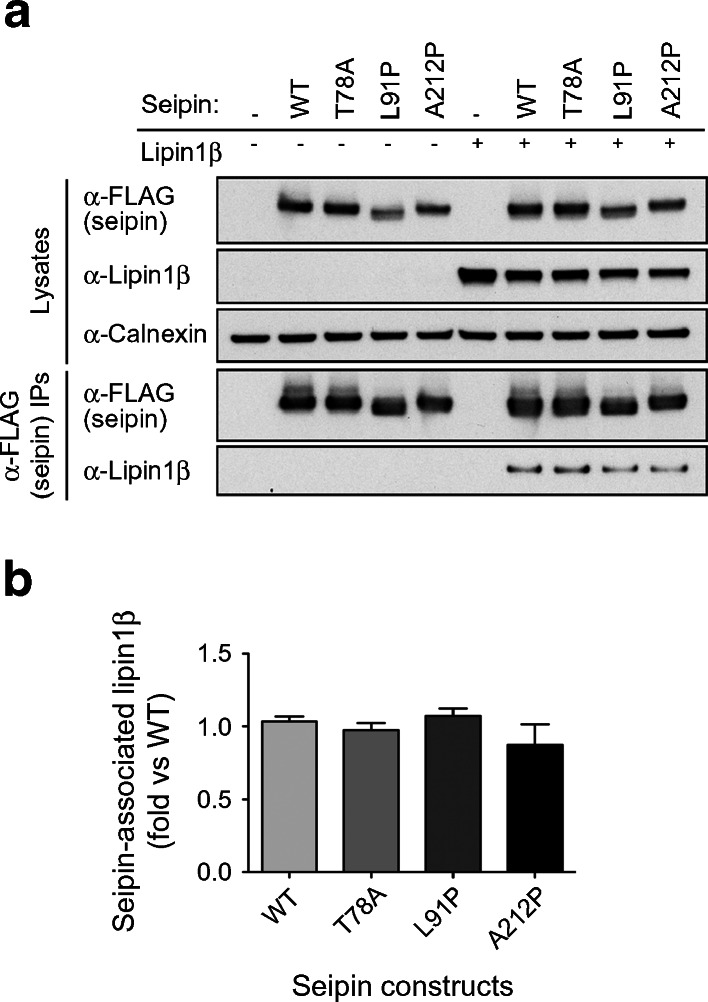
We have recently demonstrated that seipin can bind to the PA phosphatase lipin 1, including endogenous lipin 1 in differentiating cultured adipocytes. This interaction appears to be important for appropriately targeting this pro-adipogenic enzyme to the ER in differentiating pre-adipocytes [17]. We also showed that the premature stop mutants of seipin, E113X, R138X and R275X, appeared unable to bind lipin 1. In addition, the ability of Q391X seipin to bind lipin 1 is significantly reduced [17]. Similar data are shown in ESM Fig. 2. We therefore asked whether the point mutations T78A, L91P and A212P influence lipin 1 binding to seipin. As shown in Fig. 3a, b, we observed no decrease in the ability of seipin to co-immunoprecipitate lipin 1 (quantified data are shown in Fig. 3b). Hence, while the failure to target lipin 1 appropriately offers a potential pathogenic mechanism for the premature stop mutants of seipin, it appears unlikely to explain the failure of adipocyte development in cells expressing the T78A, L91P and A212P mutant forms of seipin.
Fig. 3.
The pathogenic point mutations T78A, L91P and A212P do not affect the capacity of seipin to bind the PA phosphatase lipin 1. (a) HEK293 cells were transfected with empty vector (−), FLAG-tagged long form of wild-type seipin (WT) or with identically tagged T78A, L91P, A212P forms of seipin in the absence or presence of lipin 1β. Cell lysates or anti-FLAG immunoprecipitates were immunoblotted for seipin using anti (α)-FLAG antibodies or lipin 1β using and anti-lipin 1 antibodies, as indicated. Lysates were also probed for calnexin as a loading control. (b) The binding of lipin 1β to WT or mutant forms of seipin was quantified in replicate immunoblots (n = 3), normalised to expression levels and expressed as a fold of that observed with wild-type seipin. Data shown are means ± SEM
L91P and A212P forms of seipin display altered complex formation
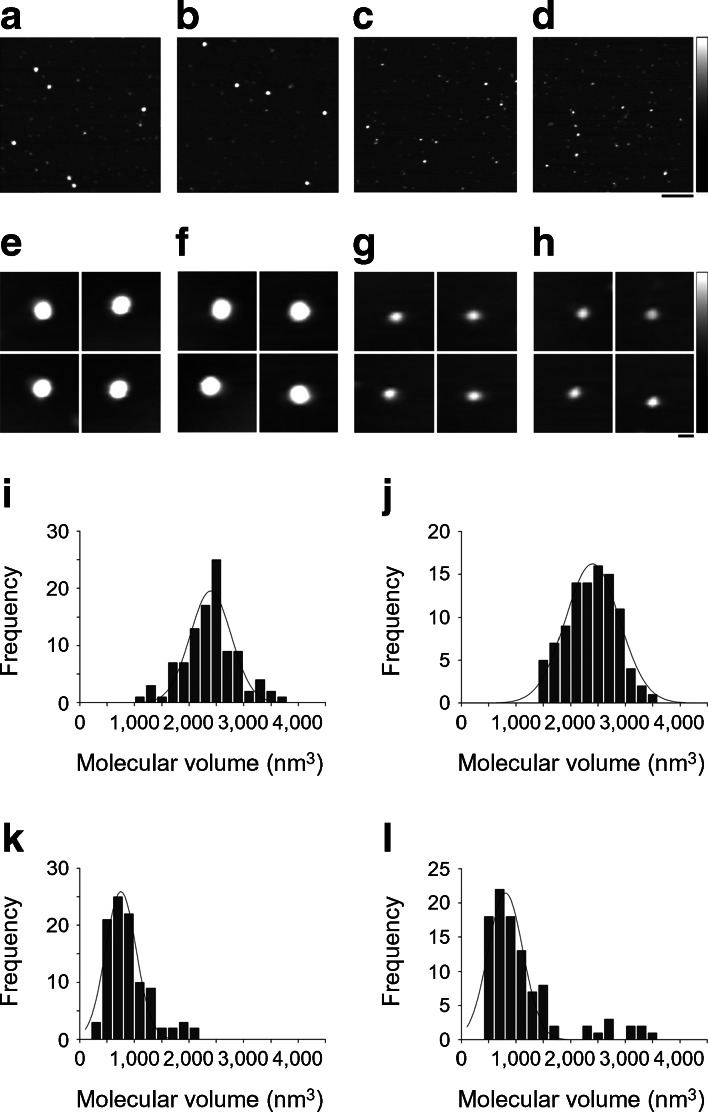
The yeast orthologue of seipin, Fld1p, has been reported to form a homo-oligomer of nine subunits in a circular arrangement [7]. Moreover, analysis of a mutant form of Fld1p carrying the single amino acid substitution G228P, which mimics A212P in the human protein, suggested that this mutation abrogated the ability of seipin to form this oligomeric structure. To date, no studies have examined whether human seipin also forms high-order oligomers or whether pathogenic point mutations affect homo-oligomerisation. To study the architecture of human seipin, we selected AFM. AFM is a scanning probe technique that provides nanometre-scale resolution without the requirement for extensive sample preparation [26]. FLAG-tagged wild-type seipin and the T78A, A212P and L91P mutants were immunopurified and the isolated protein was adsorbed to a mica substrate and imaged using tapping-mode AFM in air. Figure 4a–d shows representative low-magnification images of the four seipin constructs. Wild-type seipin (Fig. 4a) and the T78A mutant (Fig. 4b) appeared as large particles of uniform size. In contrast, the A212P mutant (Fig. 4c) and the L91P mutant (Fig. 4d) appeared as noticeably smaller and less homogenous particles. The galleries of zoomed images of particles shown in Fig. 4e–h confirm the difference in size between the two pairs of constructs; that is, wild-type (Fig. 4e) and T78A (Fig. 4f) vs A212P (Fig. 4g) and L91P (Fig. 4h). Frequency distributions of volumes of the particles, calculated using Equation 1, had peaks at 2,394 ± 36 (mean ± SEM) nm3 (n = 100) for wild-type seipin (Fig. 4i), 2,396 ± 22 nm3 (n = 100) for the T78A mutant (Fig. 4j), 753 ± 20 nm3 (n = 100) for the A212P mutant (Fig. 4k) and 807 ± 33 nm3 (n = 100) for the L91P mutant (Fig. 4l).
Fig. 4.
Wild-type human seipin and T78A mutant seipin form high-molecular-mass oligomers, while L91P seipin and A212P seipin are unable to do so. (a–d) tsA 201 cells were transfected with the long form of wild-type seipin with an N-terminal triple-FLAG tag and a C-terminal Myc tag (a) or with the T78A (b), A212P (c) or L91P (d) mutant forms of this protein. Isolated proteins were subjected to AFM imaging. Scale bar, 200 nm; shade-height scale, 0–5 nm. (e–h) Galleries of zoomed images of wild-type seipin (e) and the T78A (f), A212P (g) and L91P (h) mutants. Scale bar, 20 nm; shade-height scale, 0–5 nm. (i–l) Frequency distributions of molecular volumes of wild-type (i), T78A (j), A212P (k) and L91P (l) seipin. The curves indicate the fitted Gaussian functions
When isolated from transfected tsA 201 cells, the epitope-tagged wild-type seipin and the T78A, A212P and L91P mutants all migrated on polyacrylamide gels at an apparent molecular mass of 80 kDa (data not shown). According to Equation 2, the expected molecular volume for a protein of this size is 152 nm3. It has been shown previously that after solubilisation in Triton X-100, as here, the apparent molecular mass of the seipin complex increases by about 28% because of the presence of bound detergent; this would increase the expected monomer size to 194 nm3. Dividing the observed molecular volume (2,394 nm3 for wild-type seipin) by 194 nm3 gives a value of ∼12, suggesting that wild-type seipin assembles as a dodecamer. According to this analysis, the T78A mutant would also be dodecameric, whereas the A212P and L91P mutants are on average about one-third of this size.
Wild-type seipin and T78A seipin form ordered dodecamers
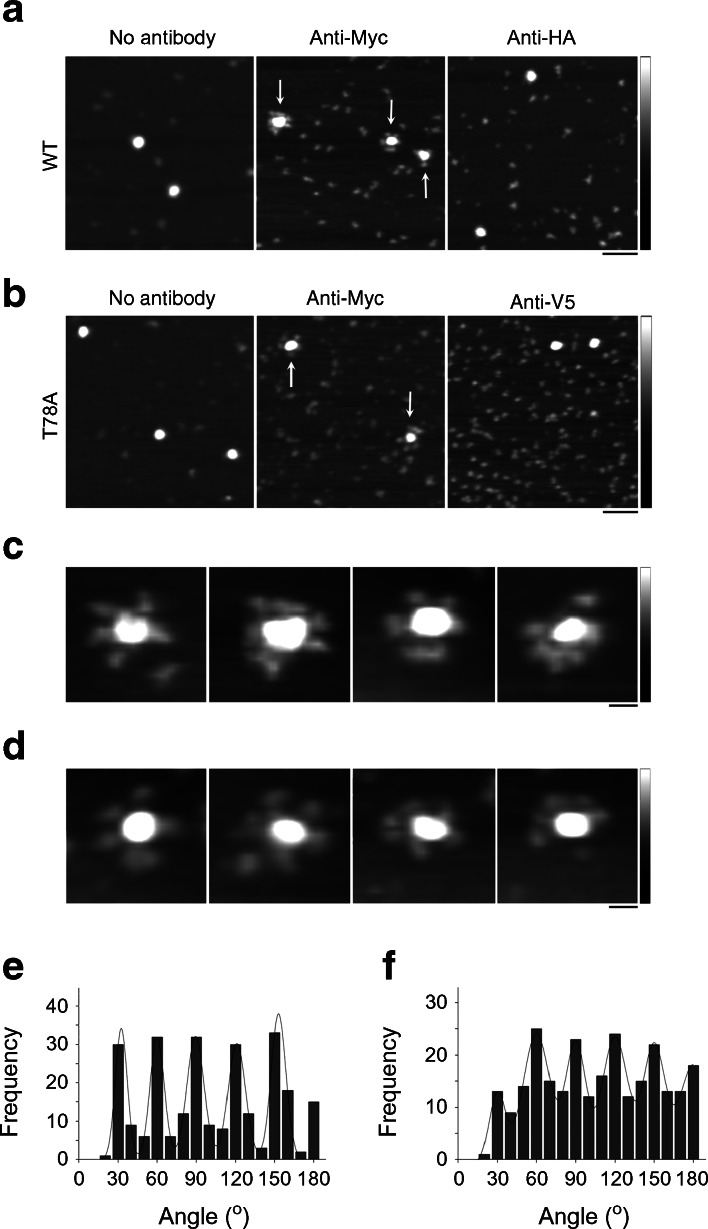
It is well known that measurement of the molecular volumes of proteins by AFM is complicated by factors such as the convolution introduced by the geometry of the scanning tip. Hence, the measured volume is not by itself a reliable indicator of the stoichiometry of the seipin complex. To conclusively establish the stoichiometry of the wild-type and T78A seipin complexes, they were imaged either after incubation with anti-Myc antibodies, which should decorate the C-terminal Myc epitope tag present on each subunit, or anti-HA or anti-V5 antibodies as negative controls. Representative low-magnification AFM images of wild-type seipin are shown in Fig. 5a. In the absence of antibodies, the large particles previously shown in Fig. 4a were again seen (Fig. 5a). After incubation with anti-Myc antibodies, the large particles became decorated by multiple smaller particles (antibodies, indicated by arrows) and took on a rosette-like appearance (Fig. 5a). Many unbound antibodies were also seen on the mica substrate. In contrast, after incubation with anti-HA antibodies, there was very little binding of antibodies to the seipin complexes (Fig. 5a). Quantification of antibody binding revealed that 88% of the seipin complexes were decorated by anti-Myc antibodies (usually multiply), whereas only 18% of the complexes were decorated by anti-HA antibodies (usually singly). A similar antibody decoration profile was seen with the T78A seipin mutant (Fig. 5b). In this case 83% of the seipin particles were decorated by anti-Myc antibodies, compared with 21% with anti-V5 antibodies. Galleries of zoomed images of antibody-decorated wild-type and T78A mutant seipin complexes are shown in Fig. 5c and Fig. 5d, respectively.
Fig. 5.
Both wild-type human seipin and T78A mutant seipin form dodecamers. (a) AFM images of wild-type seipin either alone or after incubation with anti-Myc or anti-HA antibodies. (b) AFM images of T78A mutant seipin either alone or after incubation with either anti-Myc or anti-V5 antibodies. Arrows indicate multiply decorated seipin complexes. Scale bar, 100 nm; shade-height scale, 0–5 nm. (c, d) Gallery of zoomed images showing multiply decorated wild-type (c) or T78A mutant seipin (d). Scale bar, 20 nm; shade-height scale, 0–5 nm. (e, f) Frequency distributions of angles between pairs of antibodies bound to either wild-type (e) or T78A mutant seipin (f). The curves indicate the fitted Gaussian functions
We measured the angles between the anti-Myc antibodies bound to the seipin complexes. This was done by joining the highest point on the central particle (the seipin complex) to the highest points on the peripheral particles (the antibodies) by lines and then determining the angle between the two lines. A frequency distribution of the angles between pairs of antibodies bound to wild-type seipin had several peaks at multiples of 30° (Fig. 5e), consistent with the above suggestion, based on molecular volume, that seipin assembles as a dodecamer. The observed angle profile also suggests that the dodecamer has a circular structure, which becomes decorated around the rim. The repeating 30° separation between bound antibodies excludes a spherical subunit arrangement, which would give a much more confused angle distribution. The same profile of angle peaks was seen with the T78A seipin mutant (Fig. 5f), indicating that it too assembles as a circular dodecamer.
Discussion
Pathogenic mutations in BSCL2, causing severe generalised lipodystrophy, were first reported by Magre and colleagues in 2001 [2]. However, information about the molecular functions of its protein product seipin is only now emerging. In the absence of information regarding its defined functions or structure it has been difficult to uncover how pathogenic mutations in the protein might disrupt the normal function of seipin and cause disease.
We have confirmed that the R275X mutant of seipin appears to be very unstable, in this case using the long translation of the protein bearing the mutation as a single base pair change in the full coding sequence. Once again our data imply that individuals carrying this mutation will be unable to generate even a truncated seipin protein. Our data indicate that this dramatic instability does not occur with any of the other premature stop mutations or missense mutations examined in this study. However, we did observe modest decreases in the expression of the L91P and A212P seipin and acknowledge that such subtle alterations in the stability of these or other mutant seipin proteins might yet prove relevant in vivo. All of the premature stop mutants of seipin examined localise appropriately to the ER membrane in pre-adipocytes. Overall, the current data suggest that the failure to bind lipin 1 appropriately, which we reported in our previous study, offers a plausible pathogenic mechanism for these more stable truncated forms of seipin [17]. In contrast, the missense point mutations of seipin tested here all retain the capacity to bind lipin 1, which we observed for wild-type seipin, suggesting that in itself this is unlikely to explain the inability of these mutants to support adipocyte development in vivo.
Our experiments using AFM reveal that human seipin forms homo-oligomers reminiscent of those reported for the yeast Fld1p. However, instead of the nine subunits seen with the yeast seipin orthologue we find that the human protein assembles into 12-subunit structures. Binns et al postulated that yeast seipin might be structurally involved in the budding or organisation of LDs at the ER [7]. Our work identifying the interaction of seipin with lipin 1 and ongoing studies of other seipin-interacting proteins suggest to us that this oligomeric structure could also be essential for the correct juxtaposition of seipin-associated proteins with each other and/or their substrates. The failure of L91P and A212P seipin to oligomerise evidently could affect either of these potential functions. Hence, the failure to assemble appropriately appears likely to represent a key pathogenic mechanism for L91P and A212P seipin. Fei and colleagues recently demonstrated that A212P seipin does not inhibit triacylglycerol accumulation in oleate-loaded cells as efficiently as wild-type seipin [20]. They also showed that wild-type seipin can co-precipitate other seipin molecules including A212P seipin. These results are consistent with our AFM data showing that human seipin forms oligomers, although it is not clear whether these hybrid complexes of wild-type and A212P seipin form dodecameric structures as does the wild-type seipin alone. If so, this could partly explain why heterozygous carriers of the A212P mutation exhibit no discernible phenotype [2, 4]. A recent study has also reported that A212P might act as a dominant negative inhibitor of adipogenesis if overexpressed [27]. We speculate that this could result from the failure to appropriately form dodecamers when the stoichiometry is shifted to a higher ratio of A212P to wild-type seipin. Evidently AFM offers a means to investigate these possibilities further.
It has been suggested that the G228P mutant of yeast seipin is unstable and that this may result from altered oligomerisation [7]. In our study A212P seipin was expressed robustly although we did observe modestly reduced expression of this and the L91P mutant compared with wild-type seipin [13]. In addition L91P seipin displayed altered mobility on SDS-PAGE. While the reason for this is not clear this could reflect altered glycosylation given the proximity to the known glycosylation site at N88. Overall the data imply that altered oligomerisation could be associated with modestly reduced stability and partially altered localisation for both these mutants.
Finally, it is notable that the T78A seipin protein was indistinguishable from wild-type in any of the aspects of seipin stability, structure or function tested. This observation implies that at least one further molecular mechanism exists that accounts for the inability of individuals carrying this mutation to develop functional adipose tissue. Further information regarding the molecular functions of seipin, perhaps including its interaction with additional binding proteins, may shed light on the specific defect caused by the T78A mutation.
Overall, the results of this study suggest at least four potential mechanisms by which different pathogenic mutations in seipin may lead to a loss of functional protein resulting in failed adipogenesis and ultimately lipodystrophy in affected individuals: (1) decreased protein stability and consequent lack of functional protein (R275X); (2) reduced ability to bind lipin 1 and target this key adipogenic enzyme appropriately to the ER (E113X, R138X, Q391X); (3) failure to oligomerise appropriately and localise exclusively to the ER membrane (L91P and A212P); (4) an as-yet undetermined additional mechanism (T78A). The existence of multiple molecular mechanisms underlying the development of disease due to mutations in only one protein is not unusual. In this regard we note the parallel with partial lipodystrophies caused by mutations in PPARγ, where different mutations can render PPARγ unable to bind DNA appropriately, to become activated by ligands and to associate with co-activators or dissociate from co-repressors [28].
We believe that this study represents the most comprehensive analysis of pathogenic mutations in seipin performed so far and reveals significant new information regarding both the oligomeric structure of wild-type human seipin and the potential molecular mechanisms by which different pathogenic mutations may cause severe lipodystrophy.
Electronic supplementary material
Below is the link to the electronic supplementary material.
The pathogenic point mutations L91P and A212P seipin consistently partially mislocalise to the nuclear envelope in differentiating adipocytes. C3H10T1/2 cells were transfected with empty vector (m), or epitope-tagged wild-type (WT) or T78A, L91P or A212P mutant forms of seipin. Cells were induced to differentiate for 2 days, fixed and immunostained for seipin using anti-Myc antibody (upper panels), or anti-calnexin antibody to reveal the ER (middle panels). Individual images are shown in grayscale, and merged images (lower panels) show overlay of Myc-seipin (green) and calnexin (red). Scale bars, 10 μm. (PDF 1,005 kb)
The pathogenic premature stop mutant forms of seipin display decreased lipin 1 binding capacity. HEK293 cells were transfected with empty vector (-), the long form of wild-type seipin bearing N-terminal triple-FLAG and C-terminal Myc tags (WT), or identically tagged forms of seipin with the point mutations E113X, R138X, R275X or Q391X in the absence or presence of lipin 1β. Cell lysates or anti-FLAG immunoprecipitates were immunoblotted for seipin using anti-FLAG antibodies or lipin 1β using and anti-lipin 1 antibodies, as indicated. Lysates were also probed for calnexin as a loading control. The blot shown is representative of 5 independent experiments. (PDF 1,033 kb)
Acknowledgments
Funding
This work was supported by the Medical Research Council (MRC) (New Investigator Research Grant number GO800203 [to JJR], Program Grant number G09000554 [to SOR]), the Wellcome Trust (grant number 078986/Z/06/Z [to SOR] and grant number 089125/Z/09/Z [to JME]), the Agency for Science, Technology and Research, Singapore (A*STAR) (to MFMS), the MRC Centre for Obesity and Related Metabolic Disorders (MRC-CORD) (grant number GO600717) and the National Institute for Health Research Comprehensive Biomedical Research Centre (grant number CG50826), and a Merit Scholarship from the Islamic Development Bank (to MMUT).
Access to research materials
Where possible, reagents such as novel constructs or cell lines used in this study will be made available by the corresponding author on request. Where reagents originate from other laboratories, details of appropriate contacts will be provided to obtain these materials directly or for consent to be requested so that they may be supplied by us.
Duality of interest
The authors declare that there is no duality of interest associated with the manuscript.
Contribution statement
MFMS, MMUT and RJD performed experiments, analysed data and aided in drafting the manuscript. SOR contributed to data interpretation and manuscript drafting. JME was involved in study design, data interpretation and preparing the manuscript. JJR was responsible for the conception of the study, experimental design, interpretation and preparing the manuscript. All authors approved the submitted version of the manuscript.
Abbreviations
- AFM
Atomic force microscopy
- BSCL
Berardinelli–Seip congenital lipodystrophy
- CGL
Congenital generalised lipodystrophy
- ER
Endoplasmic reticulum
- HA
Haemagglutinin
- LD
Lipid droplet
- PA
Phosphatidic acid
- PPAR
Peroxisome proliferator-activated receptor
Footnotes
M. F. Michelle Sim and M. Mesbah Uddin Talukder contributed equally to this work.
References
- 1.Garg A, Agarwal AK. Lipodystrophies: disorders of adipose tissue biology. Biochim Biophys Acta. 2009;1791:507–513. doi: 10.1016/j.bbalip.2008.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Magre J, Delepine M, Khallouf E, et al. Identification of the gene altered in Berardinelli-Seip congenital lipodystrophy on chromosome 11q13. Nat Genet. 2001;28:365–370. doi: 10.1038/ng585. [DOI] [PubMed] [Google Scholar]
- 3.Miranda DM, Wajchenberg BL, Calsolari MR, et al. Novel mutations of the BSCL2 and AGPAT2 genes in 10 families with Berardinelli-Seip congenital generalized lipodystrophy syndrome. Clin Endocrinol (Oxf) 2009;71:512–517. doi: 10.1111/j.1365-2265.2009.03532.x. [DOI] [PubMed] [Google Scholar]
- 4.Van Maldergem L, Magre J, Khallouf TE, et al. Genotype-phenotype relationships in Berardinelli-Seip congenital lipodystrophy. J Med Genet. 2002;39:722–733. doi: 10.1136/jmg.39.10.722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ito D, Fujisawa T, Iida H, Suzuki N. Characterization of seipin/BSCL2, a protein associated with spastic paraplegia 17. Neurobiol Dis. 2008;31:266–277. doi: 10.1016/j.nbd.2008.05.004. [DOI] [PubMed] [Google Scholar]
- 6.Lundin C, Nordstrom R, Wagner K, et al. Membrane topology of the human seipin protein. FEBS Lett. 2006;580:2281–2284. doi: 10.1016/j.febslet.2006.03.040. [DOI] [PubMed] [Google Scholar]
- 7.Binns D, Lee S, Hilton CL, Jiang QX, Goodman JM. Seipin is a discrete homooligomer. Biochemistry. 2010;49:10747–10755. doi: 10.1021/bi1013003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fei W, Shui G, Gaeta B, et al. Fld1p, a functional homologue of human seipin, regulates the size of lipid droplets in yeast. J Cell Biol. 2008;180:473–482. doi: 10.1083/jcb.200711136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fei W, Shui G, Zhang Y, et al. A role for phosphatidic acid in the formation of "supersized" lipid droplets. PLoS Genet. 2011;7:e1002201. doi: 10.1371/journal.pgen.1002201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Szymanski KM, Binns D, Bartz R, et al. The lipodystrophy protein seipin is found at endoplasmic reticulum lipid droplet junctions and is important for droplet morphology. Proc Natl Acad Sci U S A. 2007;104:20890–20895. doi: 10.1073/pnas.0704154104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wolinski H, Kolb D, Hermann S, Koning RI, Kohlwein SD. A role for seipin in lipid droplet dynamics and inheritance in yeast. J Cell Sci. 2011;124:3894–3904. doi: 10.1242/jcs.091454. [DOI] [PubMed] [Google Scholar]
- 12.Chen W, Yechoor VK, Chang BH, Li MV, March KL, Chan L. The human lipodystrophy gene product Berardinelli-Seip congenital lipodystrophy 2/seipin plays a key role in adipocyte differentiation. Endocrinology. 2009;150:4552–4561. doi: 10.1210/en.2009-0236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Payne VA, Grimsey N, Tuthill A, et al. The human lipodystrophy gene BSCL2/seipin may be essential for normal adipocyte differentiation. Diabetes. 2008;57:2055–2060. doi: 10.2337/db08-0184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen W, Chang B, Saha P, et al. Berardinelli-seip congenital lipodystrophy 2/seipin is a cell-autonomous regulator of lipolysis essential for adipocyte differentiation. Mol Cell Biol. 2012;32:1099–1111. doi: 10.1128/MCB.06465-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peterfy M, Phan J, Xu P, Reue K. Lipodystrophy in the fld mouse results from mutation of a new gene encoding a nuclear protein, lipin. Nat Genet. 2001;27:121–124. doi: 10.1038/83685. [DOI] [PubMed] [Google Scholar]
- 16.Phan J, Reue K. Lipin, a lipodystrophy and obesity gene. Cell Metab. 2005;1:73–83. doi: 10.1016/j.cmet.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 17.Sim MFM, Dennis RJ, Aubry EM, et al. The human lipodystrophy protein seipin is an ER membrane adaptor for the adipogenic PA phosphatase lipin 1. Mol Metab. 2013;2:38–46. doi: 10.1016/j.molmet.2012.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang P, Takeuchi K, Csaki LS, Reue K. Lipin-1 phosphatidic phosphatase activity modulates phosphatidate levels to promote peroxisome proliferator-activated receptor gamma (PPARgamma) gene expression during adipogenesis. J Biol Chem. 2012;287:3485–3494. doi: 10.1074/jbc.M111.296681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang W, Thein S, Guo X, et al. Seipin differentially regulates lipogenesis and adipogenesis through a conserved core sequence and an evolutionarily acquired C-terminus. Biochem J. 2013;452:37–44. doi: 10.1042/BJ20121870. [DOI] [PubMed] [Google Scholar]
- 20.Fei W, Li H, Shui G, et al. Molecular characterization of seipin and its mutants: implications for seipin in triacylglycerol synthesis. J Lipid Res. 2011;52:2136–2147. doi: 10.1194/jlr.M017566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ebihara K, Kusakabe T, Masuzaki H, et al. Gene and phenotype analysis of congenital generalized lipodystrophy in Japanese: a novel homozygous nonsense mutation in seipin gene. J Clin Endocrinol Metab. 2004;89:2360–2364. doi: 10.1210/jc.2003-031211. [DOI] [PubMed] [Google Scholar]
- 22.Grimsey N, Han GS, O'Hara L, Rochford JJ, Carman GM, Siniossoglou S. Temporal and spatial regulation of the phosphatidate phosphatases lipin 1 and 2. J Biol Chem. 2008;283:29166–29174. doi: 10.1074/jbc.M804278200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Payne VA, Au WS, Lowe CE, et al. C/EBP transcription factors regulate SREBP1c gene expression during adipogenesis. Biochem J. 2010;425:215–223. doi: 10.1042/BJ20091112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schneider SW, Lärmer J, Henderson RM, Oberleithner H. Molecular weights of individual proteins correlate with molecular volumes measured by atomic force microscopy. Pflügers Arch. 1998;435:362–367. doi: 10.1007/s004240050524. [DOI] [PubMed] [Google Scholar]
- 25.Neaves KJ, Huppert JL, Henderson RM, Edwardson JM. Direct visualization of G-quadruplexes in DNA using atomic force microscopy. Nucleic Acids Res. 2009;37:6269–6275. doi: 10.1093/nar/gkp679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barrera NP, Edwardson JM. The subunit arrangement and assembly of ionotropic receptors. Trends Neurosci. 2008;31:569–576. doi: 10.1016/j.tins.2008.08.001. [DOI] [PubMed] [Google Scholar]
- 27.Qiu W, Wee K, Takeda K, et al. Suppression of adipogenesis by pathogenic seipin mutant is associated with inflammatory response. PLoS One. 2013;8:e57874. doi: 10.1371/journal.pone.0057874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jeninga EH, Gurnell M, Kalkhoven E. Functional implications of genetic variation in human PPARgamma. Trends Endocrinol Metab. 2009;20:380–387. doi: 10.1016/j.tem.2009.04.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The pathogenic point mutations L91P and A212P seipin consistently partially mislocalise to the nuclear envelope in differentiating adipocytes. C3H10T1/2 cells were transfected with empty vector (m), or epitope-tagged wild-type (WT) or T78A, L91P or A212P mutant forms of seipin. Cells were induced to differentiate for 2 days, fixed and immunostained for seipin using anti-Myc antibody (upper panels), or anti-calnexin antibody to reveal the ER (middle panels). Individual images are shown in grayscale, and merged images (lower panels) show overlay of Myc-seipin (green) and calnexin (red). Scale bars, 10 μm. (PDF 1,005 kb)
The pathogenic premature stop mutant forms of seipin display decreased lipin 1 binding capacity. HEK293 cells were transfected with empty vector (-), the long form of wild-type seipin bearing N-terminal triple-FLAG and C-terminal Myc tags (WT), or identically tagged forms of seipin with the point mutations E113X, R138X, R275X or Q391X in the absence or presence of lipin 1β. Cell lysates or anti-FLAG immunoprecipitates were immunoblotted for seipin using anti-FLAG antibodies or lipin 1β using and anti-lipin 1 antibodies, as indicated. Lysates were also probed for calnexin as a loading control. The blot shown is representative of 5 independent experiments. (PDF 1,033 kb)