Abstract
Somatic cells can be converted into induced pluripotent stem cells (iPSCs) by forced expression of various combinations of transcription factors, but the molecular mechanisms of reprogramming are poorly understood. Specifically, evidence that the reprogramming process can take many distinct routes only begins to emerge. It is definitively established that p53 deficiency greatly enhances reprogramming, revealing p53's barrier function for induced pluripotency, but the role of its homologs p63 and p73 are unknown. Here we report that in stark contrast to p53, p73 has no role in reprogramming. However, p63 is an enabling (rather than a barrier) factor for Oct4, Sox2 and Klf4 (OSK) and Oct4 and Sox2 (OS), but not for Oct4 and Klf4 (OK) reprogramming of mouse embryonic fibroblasts. Specifically, p63 is essential during reprogramming for maximum efficiency, albeit not for the ability to reprogram per se, and is dispensable for maintaining stability and pluripotency of established iPSC colonies. ΔNp63, but not TAp63, is the principal isoform involved. Loss of p63 can affect reprogramming via several mechanisms such as reduced expression of mesenchymal–epithelial transition and pluripotency genes, hypoproliferation and loss of the most reprogrammable cell populations. During OSK and OS reprogramming, different mechanisms seem to be critical, such as regulation of epithelial and pluripotency genes in OSK reprogramming versus regulation of proliferation in OS reprogramming. Finally, our data reveal three different routes of reprogramming by OSK, OS or OK, based on their differential p63 requirements for iPSC efficiency and pluripotency marker expression. This supports the concept that many distinct routes of reprogramming exist.
Keywords: p63, ΔNp63, reprogramming, pluripotency, iPSCs
Reprogramming somatic cells into induced pluripotent stem cells (iPSCs) provides a powerful tool for the generation of autologous patient-specific cell therapies and for studying disease-specific stem cells.1 Mouse and human iPSCs are typically produced via forced expression of various combinations of transcription factors, most often Oct4, Sox2, Klf4 and c-Myc (OSKM) or without c-Myc (OSK).2, 3, 4, 5 Moreover, reprogramming by only two factors can be achieved, especially on less differentiated cells.6, 7, 8, 9, 10
For mechanistic studies of reprogramming, mouse embryo fibroblasts (MEFs) are the most commonly used source of somatic cells.10, 11, 12, 13, 14, 15 These studies showed that reprogramming is largely a stochastic process that relies on multiple independent epigenetic events.12, 16, 17 Thus, individual cells convert into iPSCs with different latencies described by a Gaussian distribution.10, 16, 17 Indeed, individual cells undergoing reprogramming show considerable variations in their gene expression patterns12 and can concomitantly express mixed lineage-specific genes, indicating disrupted homeostasis.10 Nonetheless, several studies also provided evidence for some deterministic steps, such as accelerated cell division and decreased cell size in all MEFs that become reprogrammed,13 and mesenchymal–epithelial transition (MET).11, 14, 15 Interestingly, both the stochastic process and MET are amenable to regulation, and one common factor capable of doing so is p53.
p53 is a tumor-suppressor mutated in over half of human cancers.18 It maintains genomic stability and induces cell cycle arrest, apoptosis or replicative senescence via transcription-dependent and -independent pathways.19, 20 In addition, several studies have established that p53 is a potent barrier of reprogramming, which is not surprising given the mechanistic overlap between reprogramming and tumorigenesis.13, 21, 22, 23, 24, 25, 26, 27, 28, 29 Thus, p53 deficiency by genetic knockout (KO) or chemical knockdown greatly enhances reprogramming efficiency, whereas p53 stabilization reduces it. p53 serves as reprogramming barrier by several different mechanisms. First, the superior reprogramming of p53-deficient cells is due to their enhanced proliferation.13, 28 Second, p53 prevents reprogramming of cells that carry various types of DNA damage by activating apoptosis.22 Finally, p53 can block the MET transcriptional program by inhibiting Klf4.29
The p53 homologs p63 and p73 are major regulators of epithelial and neural development, respectively. p63 KO mice lack skin and epithelial appendages including teeth, hair follicles and mammary epithelium and so on, and have truncated or missing limbs due to apical ectodermal ridge defects.30, 31 p73 KO mice exhibit defective embryonic and adult neurogenesis associated with hydrocephalus, cortical thinning and hippocampal dysgenesis.32, 33, 34 Both p63 and p73 exist in multiple N- and C-terminal isoforms. Thus, full-length TAp63/TAp73, which carry the N-terminal transcriptional activation (TA) domain, can induce cell cycle arrest and apoptosis similar to p53, whereas the N-terminally truncated ΔNp63/ΔNp73 often act in a dominant-negative manner by inhibiting full-length family members, including p53.35, 36, 37 p63 and p73 have been implicated in oncogenesis,35, 36, 37 but their role in somatic reprogramming has barely been investigated.34, 38 Here we show for the first time that ΔNp63 is a potent positive regulator of reprogramming. Moreover, we show that different three- and two-factor reprogramming pathways have different p63 dependencies, providing evidence that reprogramming can proceed via multiple distinct routes.
Results
p63 is essential for reprogramming by OSK and Oct4 and Sox2 (OS), but not by Oct4 and Klf4 (OK) factor combinations
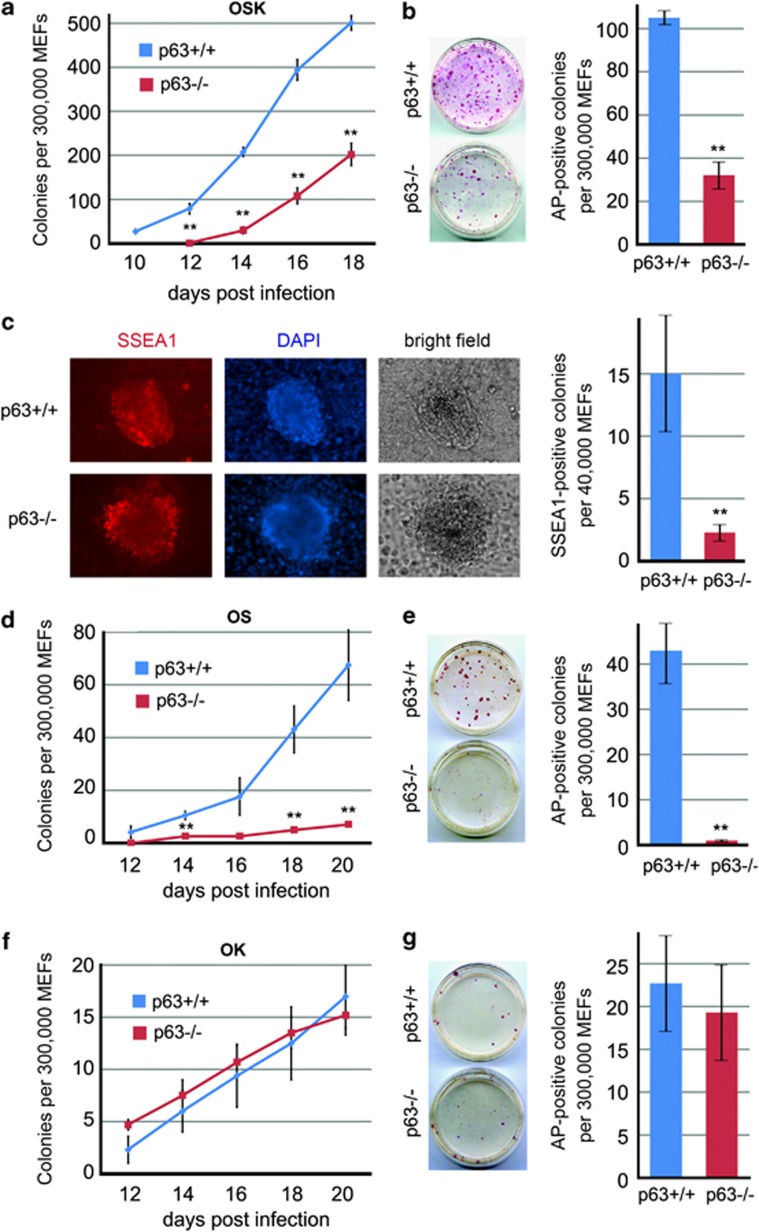
To investigate a possible role of the p53 homologs p63 and p73 in reprogramming, we transduced wild-type (WT) and corresponding null MEFs from littermate embryos with OSK. Consistent with previous results,21, 22, 23, 24, 25, 26, 27, 28, 29 p53−/− MEFs were reprogrammed with much higher efficiency compared with WT MEFs, while p73 deficiency did not affect reprogramming or pluripotency of established iPSC colonies, as we recently reported34 (Supplementary Figures 1A, C–F). In striking contrast, reprogramming efficiency of p63−/− MEFs was markedly reduced compared with WT controls (Figure 1a, Supplementary Figure 1B). Thus, we set to investigate the precise role of p63 in reprogramming.
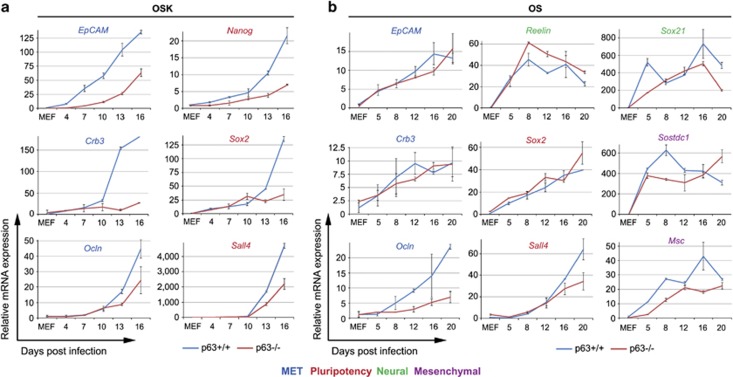
Figure 1.
p63 is essential for OSK and OS, but not for OK reprogramming. Littermate WT and p63−/− MEFs grown and treated identically were transduced with the following combinations of transcription factors: OSK (a–c), OS (d and e) or OK (f and g). Representative littermate pairs of 12 OSK, 4 OS and 4 OK reprogramming experiments are shown. (a, d and f) Emerging iPSC-like colonies in triplicate plates were counted at indicated time points (d.p.i.) and mean±S.D. was plotted. (b, e and g) At the last time point of reprogramming (i.e., before cell culture became overgrown or very few new colonies were formed), plates were fixed and stained for the pluripotency marker AP. Representative images (left) and mean±S.D. from triplicates (right) are shown. (c) WT and p63−/− MEFs were OSK transduced in 24-well plates and immunostained for the pluripotency marker SSEA1. DAPI was used to visualize nuclei. Representative images (left) and mean±S.D. from triplicates (right) are shown. Note that SSEA1 staining could not be done on OS and OK-transduced MEFs in 24-well plates because of much lower reprogramming efficiencies with two factors.10 **P<0.01
First, we followed the kinetics of OSK-induced conversion of WT and p63−/− MEFs into iPSC-like colonies, as defined by morphological criteria (small cells forming highly compact round masses with shiny sharp borders). The iPSC nature of these colonies was confirmed by alkaline phosphatase (AP) and stage-specific embryonic antigen 1 (SSEA1) staining, teratoma assays and so on (see below). We found that reprogramming efficiency of p63−/− MEFs was significantly decreased (Figure 1a) and, comparing nine different MEFs (from five independent pregnancies), was on average threefold lower than in WT controls (0.066±0.061% versus 0.154±0.108%, P=0.02). In three additional embryo pairs, reprogramming efficiency of p63−/− MEFs was even more drastically reduced, that is, 10-, 25- and 160-fold lower than in WT controls. Furthermore, when WT and p63−/− colonies were stained for pluripotency markers AP and SSEA1, the reduction in reprogramming efficiency of p63−/− MEFs was confirmed and averaged 2.5-fold and 9.1-fold, respectively, in three independent embryo pairs (Figures 1b and c).
We recently showed that MEFs can be successfully reprogrammed into iPSCs using OS and OK two-factor combinations, albeit with lower efficiencies.10 Thus, we tested if p63 deficiency would also affect two-factor reprogramming. Strikingly, in five experiments, p63−/− MEFs were strongly defective in OS reprogramming, but appeared completely normal in OK reprogramming (Figures 1d–g). These differences were not because of technical failures because: (i) WT and p63−/− MEFs expressed similar levels of reprogramming factors at the mRNA and protein level (Figure 3e, Supplementary Figures 2A–C); (ii) WT and p63−/− MEFs had similar transduction efficiencies, confirmed by parallel green fluorescent protein (GFP) transductions followed by fluorescence-activated cell sorting (FACS) analysis (90.4±6.2% versus 88.5±4.7% cells, n=8 and 10, P=0.45); (iii) in multiple individual OSK, OS and OK-derived WT and p63−/− colonies we confirmed integration of the introduced factors, while excluding contamination by unintended ones (Supplementary Figure 3). Altogether, these data indicate that p63 is essential for reprogramming with OSK and OS, but is dispensable for OK reprogramming. This strongly suggests that reprogramming with OK likely undertakes a different molecular route than reprogramming initiated by OSK and OS.
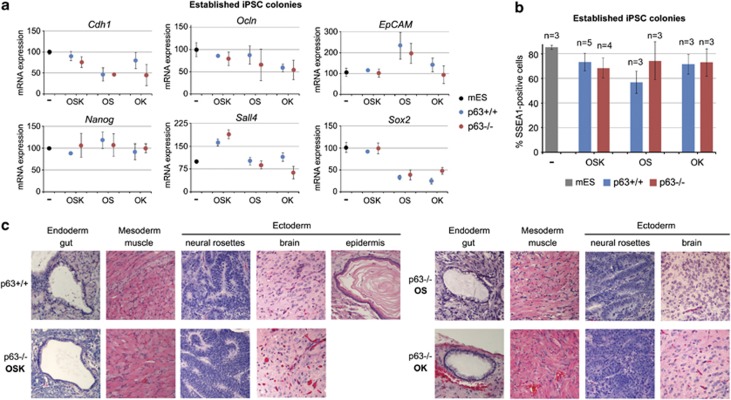
p63 is dispensable for stability and pluripotency of established iPSC colonies
To determine whether p63 is essential during the reprogramming process or for maintenance of established iPSC colonies in OSK and OS reprogramming, we first propagated individual primary p63−/− iPSC colonies. This was readily achievable and no growth defects or loss of undifferentiated morphology was observed. Furthermore, p63−/− iPSC colonies maintained their undifferentiated phenotype after passaging, confirmed by expression of MET and pluripotency markers. Thus, WT and corresponding p63−/− OSK, OS and OK-derived iPSC colonies expressed similar levels of Cdh1, Occludin, EpCAM, Nanog, Sall4 and Sox2 by qRT-PCR and SSEA1 by FACS analysis (Figures 2a and b). Moreover, exogenous reprogramming factors were silenced in both WT and p63−/− iPSC colonies, as expected (Supplementary Figure 2D).
Figure 2.
p63 is dispensable for maintenance and pluripotency of established iPSC colonies. (a) Individual OSK, OS and OK-derived WT and p63−/− iPSC colonies were propagated, harvested at passage 2 and analyzed by qRT-PCR for expression of MET (top) and pluripotency genes (bottom). Expression was normalized to HPRT and set to 100 for untreated mouse ES cell line W4 (mES), which served as a control. Mean±S.D. from duplicates of one clone (mES and OSK) or duplicates of two clones (OS and OK) are shown. (b) Individual OSK, OS and OK-derived WT and p63−/− iPSC colonies were propagated (passages 1–3) and analyzed by FACS for percent of SSEA1-positive cells. Mean±S.D. is shown, n, number of clones analyzed. (c) Teratoma assay. Individual OSK, OS and OK-derived WT and p63−/− iPSCs (passages 3–4) were subcutaneously injected into nude mice. Teratomas were dissected, histologically processed and hematoxylin and eosin stained. Representative teratomas derived from two OSK, two OS and one OK p63−/− iPSCs are shown
We next asked if p63 deficiency affects pluripotency, that is, the ability of iPSCs to produce three germ layers by teratoma assays. We found that OSK, OS and OK-derived p63−/− iPSCs produced derivatives of all three germ layers (Figure 2c). The most common tissues included neuroectoderm and mature brain tissue (ectoderm), smooth and striated muscle (mesoderm) and gastrointestinal glands (endoderm). In addition, p63−/− iPSCs produced bronchial epithelium, intestinal crypts, exocrine pancreas (endoderm), adipocytes and cartilage (mesoderm) (Supplementary Figure 4). However, a prominent difference from WT iPSCs was that none of seven p63−/− iPSCs produced with OSK, OS and OK formed squamous epithelial ‘pearls', consistent with the phenotype of p63 KO mice that completely lack epidermis and other squamous epithelia.31 Taken together, these data indicate that p63 is largely dispensable for pluripotency and maintenance of iPSCs once they are established, but points to its essential role during the reprogramming process.
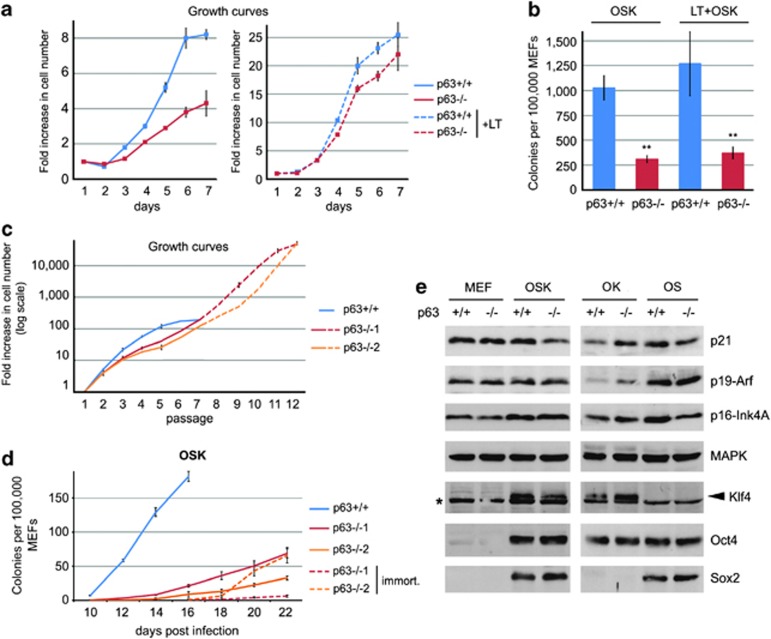
The p63−/− OSK reprogramming defect is not because of decreased cell proliferation or upregulation of reprogramming barriers
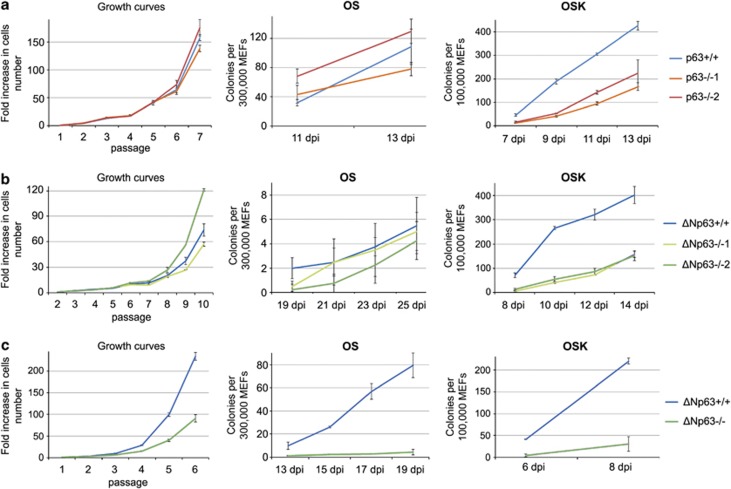
It was shown that the efficiency of reprogramming depends on normal cell proliferation.13, 28 Consistent with the reported hypoproliferation of p63−/− keratinocytes,39 we observed that p63−/− MEFs proliferate slower than their WT counterparts (Figures 3a and c). Thus, we tested whether improving proliferation of p63−/− MEFs would restore their OSK reprogramming efficiency. To this end, we transduced p63−/− and WT MEFs with SV40 large T antigen (LT), which enhanced and essentially equalized their proliferation (Figure 3a). Importantly, however, LT failed to improve OSK reprogramming of p63−/− MEFs (Figure 3b). Second, we made use of two independent p63−/− MEF cultures that spontaneously immortalized in vitro (Figure 3c). Despite excellent proliferation, their reprogramming efficiency was still significantly below that of WT MEFs and comparable to parental p63−/− MEFs efficiency (Figure 3d). Moreover, no enhanced senescence or apoptosis was detected in p63−/− MEFs during OSK, OS and OK reprogramming (Supplementary Figures 5A–C). Altogether, these data establish that the reduced OSK reprogramming efficiency of p63−/− MEFs is not caused by hypoproliferation.
Figure 3.
The reprogramming defect of p63−/− MEFs is not because of decreased proliferation or upregulation of reprogramming barriers. (a and b) WT and p63−/− MEFs were transduced with SV40 LT antigen (LT), which essentially equalizes their proliferation rate, but not their OSK reprogramming. Growth curves (a) and OSK reprogramming efficiency at 14 d.p.i. (b) of parental and LT-transduced WT and p63−/− MEFs is shown. Mean±S.D. of triplicates. **P<0.01. One of two independent experiments with similar results is shown. (c and d) Two p63−/− MEFs (p63−/−1 and p63−/−2) spontaneously immortalized during prolonged in vitro passaging, which improved their proliferation rate, but not OSK reprogramming. Growth curves (c) and OSK reprogramming (d), mean±S.D. of triplicates. Dashed lines correspond to immortalized MEFs. (e) Immunoblot analysis of the indicated proteins in non-transduced WT and p63−/− MEFs and in OSK, OS and OK-transduced WT and p63−/− MEFs at 5 d.p.i. MAPK serves as loading control. Note that Klf4, Oct4 and Sox2 antibodies detect both the endogenous and exogenous proteins. Arrowhead shows the specific Klf4 band right above a nonspecific band (*). The immunoblot of different OSK-transduced MEFs was repeated three times with similar results
We previously reported that MEFs lacking the p63 homolog p73 exhibit a compensatory upregulation of p53, an established reprogramming barrier.21, 22, 23, 24, 25, 26, 27, 28, 29 Thus, we asked whether p53 is also activated in p63−/− MEFs. However, neither p53 protein nor its sensitive targets p21 and miR34a/b/c, also known reprogramming barriers,40, 41, 42, 43 are upregulated in p63−/− MEFs before and during OSK, OS and OK reprogramming (Figure 3e, Supplementary Figures 5D and E). Similarly, p53 shRNAs do not rescue their reprogramming efficiency (Figure 5g). Moreover, the tumor suppressors p19-Arf and p16-Ink4A, which are known reprogramming barriers in mouse and human fibroblasts,23, 24 were shown to partially mediate skin and limb defects of p63−/− mice.39 However, p19 and p16 are again unchanged in p63−/− MEFs before and during reprogramming (Figure 3e). Thus, the defective OSK and OS reprogramming of p63−/− MEFs cannot be explained by upregulation of likely reprogramming barriers in this context.
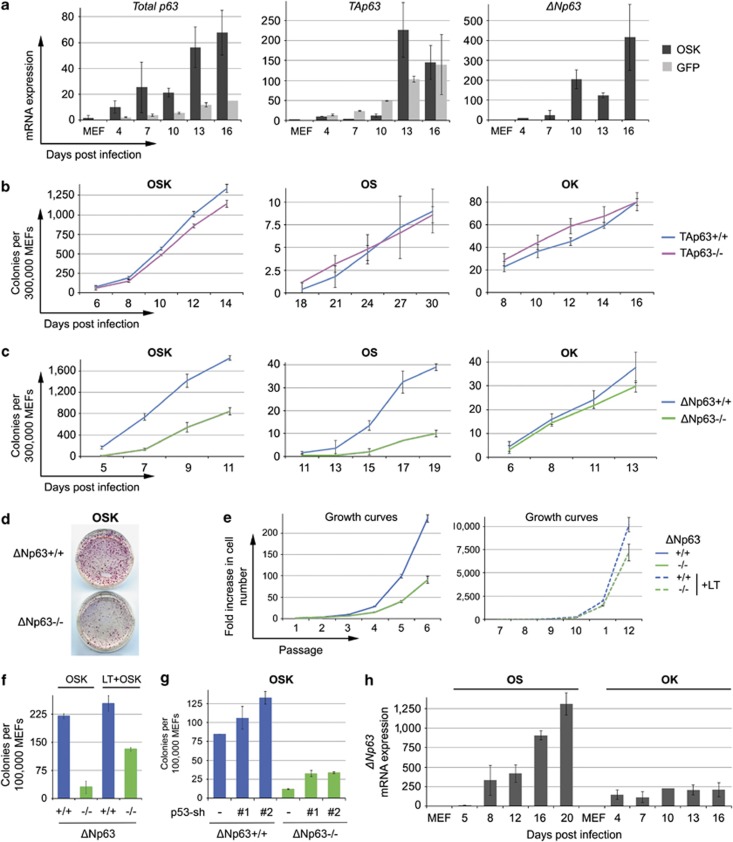
Figure 5.
ΔNp63, but not TAp63 isoform is involved in OSK and OS reprogramming. (a) WT MEFs transduced with OSK or control GFP viruses were harvested at indicated time points (d.p.i.). Expression of total p63, TAp63 and ΔNp63 mRNAs was analyzed by qRT-PCR. All gene expression levels were normalized to HPRT and set to 10 for OSK-transduced MEFs at 4 d.p.i. One of three independent experiments with similar results is shown. Mean±S.D. (b–d) Littermate WT and TAp63−/− MEFs (b) or WT and ΔNp63−/− MEFs (c and d) were transduced with OSK, OS or OK. Emerging iPSC-like colonies in triplicate plates were counted at indicated time points (d.p.i.) and mean±S.D. was plotted. A representative experiment of three TAp63−/− reprogramming experiments (from three different TAp63−/− embryos) and four ΔNp63−/− reprogramming experiments (from four different ΔNp63−/− embryos) are shown. (d) A representative AP staining of OSK-transduced ΔNp63−/− and WT MEFs. (e, f) WT and ΔNp63−/− MEFs were transduced with SV40 LT before OSK reprogramming. LT essentially equalized proliferation (e), but not OSK reprogramming efficiency (f, scored at 9 d.p.i.). Mean±S.D. of triplicates. (g) p53 knockdown by two different p53 shRNAs does not rescue OSK reprogramming of ΔNp63−/− MEFs (efficiency scored at 10 d.p.i.). Mean±S.D. (h) qRT-PCR of ΔNp63 mRNA expression during OS and OK reprogramming of WT MEFs. Expression levels were normalized to HPRT and set to 10 for OS-transduced MEFs at 5 d.p.i. Mean±S.D. The specificity of p63, TAp63 and ΔNp63 qRT-PCR reactions was confirmed by sequencing of the corresponding PCR products
Reduction of MET and pluripotency genes in reprogramming of p63−/− MEFs
As we narrowed the requirement for p63 in OSK and OS reprogramming to stages before fully established iPSC colonies and their maintenance, we next asked if transcriptional programs are changed in p63−/− MEFs. It was proposed that MET is an early and necessary step in MEF reprogramming, followed by activation of core pluripotency genes.11, 14 Indeed, we found that expression of all analyzed MET and pluripotency genes was significantly reduced during OSK reprogramming of p63−/− MEFs, including EpCAM, Ocln, Crb3, Nanog, Sall4 and Sox2 (Figure 4a, Supplementary Figure 6A).
Figure 4.
p63 is essential for MET and pluripotency genes expression in OSK reprogramming, but not OS reprogramming. WT and p63−/− MEFs undergoing reprogramming with OSK (a) or OS (b) were harvested at indicated time points (d.p.i.). Expression of indicated genes was analyzed by qRT-PCR, normalized to HPRT and set to 1 for non-transduced MEFs in each graph. Gene names are color-coded as follows: MET genes in blue, pluripotency genes in red, OS-specific neural progenitor markers in green and OS-specific mesenchymal progenitor markers in purple. Note that Nanog was not induced by OS (data not shown). Mean±S.D.
Furthermore, we previously showed that MET and pluripotency genes are also activated during two-factor reprogramming, albeit at much lower levels.10 Thus, we tested their expression in OS and OK reprogramming. As predicted from normal OK reprogramming, marker expression was not reduced in p63−/− MEFs (Supplementary Figure 6B). However, in OS reprogramming expression of Ocln (MET) and Sall4 (pluripotency) was reduced in p63−/− MEFs, whereas all other markers were normal (Figure 4b). Moreover, we previously reported that lineage-specific genes are differentially upregulated during OS and OK reprogramming of WT MEFs.10 Thus, we tested if OS-specific genes are affected in p63−/− MEFs. We found that only one of four genes (Musculin) was reduced in p63−/− MEFs, whereas the others were normal (Reelin, Sox21 and Sostdc1; Figure 4b). Altogether, these data suggest that while p63 is critical for both efficient OSK and OS reprogramming, the underlying molecular mechanisms of p63 requirement are different. In OSK reprogramming, p63 appears to be a master regulator of the transcriptional network engaging MET and pluripotency genes. In contrast, the role of p63 in OS reprogramming does not seem to involve regulation of the majority of MET, pluripotency and lineage-specific genes, but rather may be related to proliferation (see below).
The lack of ΔNp63 is responsible for OSK and OS reprogramming defects
The two major p63 isoforms, TAp63 and ΔNp63, are transcribed from alternative promoters and have different functions.44 Specifically, ΔNp63 is highly expressed in the basal layer of epidermis, largely drives keratinocyte differentiation in vivo and is proposed to be responsible for maintenance of epidermal progenitor/stem cells.45, 46, 47, 48 Similarly, TAp63 is expressed in hair follicle bulge cells and dermal papillae cells (niches for epidermal and dermal stem cells, respectively) and is implicated in the maintenance of adult dermal and epidermal precursors.49 Thus, both TAp63 and ΔNp63 could in principle be involved in reprogramming, given the stem cell-like nature of iPSCs. To test this, we first analyzed expression of TAp63 and ΔNp63 mRNAs during OSK reprogramming of WT MEFs by qRT-PCR. We found that expression of total p63 as well as TAp63 and ΔNp63 (but not p53 or p73) gradually increases during reprogramming (Figure 5a, Supplementary Figure 6C). However, induction of TAp63 is nonspecific, as it also occurs in control GFP-transduced MEFs. In contrast, ΔNp63 is strongly upregulated by OSK but not by GFP viruses. These data strongly suggest that ΔNp63, but not TAp63, regulates reprogramming.
To directly test the hypothesis that ΔNp63 is essential for reprogramming, we made use of isoform-specific TAp63−/− and ΔNp63−/− MEFs.48, 49 As predicted from mRNA expression patterns, TAp63−/− MEFs were reprogrammed normally by OSK, OS and OK (Figure 5b). In contrast, ΔNp63−/− MEFs were strongly deficient in OSK and OS, but not in OK reprogramming, completely reproducing the phenotype of global p63−/− MEFs (Figures 5c and d). Furthermore, ΔNp63−/− MEFs were hypoproliferative, whereas TAp63−/− MEFs were not (Figure 5e and data not shown). Moreover, ectopic LT restored proliferation of ΔNp63−/− MEFs, but did not restore their OSK reprogramming, and neither did shRNA-mediated knockdown of p53 (Figures 5e–g).
Of note, ΔNp63 mRNA is much more strongly induced in OS than in OK reprogramming of WT MEFs, both in amplitude and in kinetics (Figure 5h), providing a mechanistic explanation for why p63 is critical for OS, but not for OK reprogramming. Altogether, these data unequivocally establish that the lack of ΔNp63 isoform is responsible for the reduced OSK and OS reprogramming efficiency of global p63−/− MEFs.
Despite several attempts, we could not rescue OSK reprogramming of p63−/− MEFs by ectopic ΔNp63 expression (which was confirmed). This is most likely because of the fact that it is virtually impossible to mimic in vitro the proper timing and levels of ΔNp63 expression observed in vivo, both known to be crucial for physiologic activities of transcription factors in general.
Hypoproliferation may be responsible for defective OS reprogramming of p63−/− MEFs
On occasion, some p63−/− and ΔNp63−/− MEFs showed normal OS reprogramming (two out of six global p63−/− and one out of four ΔNp63−/− MEF cultures from certain embryos). When we analyzed growth curves of these cultures, we found that their proliferation was largely normal (Figures 6a and b). However, OSK reprogramming of these same MEFs was still defective (Figures 6a and b right panels). On the other hand, all p63−/− and ΔNp63−/− MEFs with defective OS reprogramming were also hypoproliferative (Figure 6c and data not shown). These correlations strongly suggest that during OS reprogramming, ΔNp63 is critical mostly to maintain cell proliferation.
Figure 6.
Reprogramming efficiency with OS but not OSK correlates with proliferation in p63−/− and ΔNp63−/− MEFs. p63−/− (a) or ΔNp63−/− (b and c) MEFs that proliferate normally (a and b) or are proliferation-defective (c) were reprogrammed with OS and OSK as indicated, side-by-side with their WT littermate controls. The number of iPSC-like colonies at indicated time points (d.p.i.) was plotted. Concomitant growth curve analysis shows a strong correlation of proliferative capacity with OS reprogramming, but not with OSK reprogramming. Mean±S.D. from triplicates
ΔNp63 is essential for maintenance of the most reprogrammable MEF populations
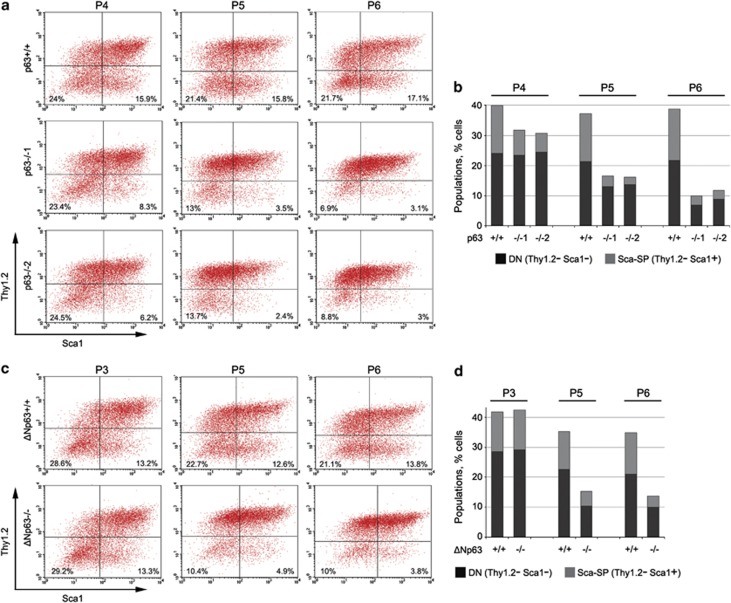
We recently showed that WT MEFs are heterogeneous and can be FACS-sorted into distinct sub-populations based on the expression of two cell-surface molecules, Sca1 (labeling mesenchymal and epithelial progenitors) and Thy1.2 (labeling more differentiated lipo- and myo-fibroblasts), so that Thy1.2-negative populations (Sca1 single-positive, SP; and Thy1.2/Sca1 double-negative, DN) had greatly increased OSK and especially OS and OK reprogramming efficiencies.10 Thus, we tested the abundance of these most reprogrammable populations in p63−/− and ΔNp63−/− MEFs. We found that both global p63−/− and ΔNp63−/− MEFs progressively lose Sca1-SP and DN populations after passage 3 (Figure 7). Of note, we routinely use passages 2–3 MEFs, suggesting that this loss of the most reprogrammable fractions in p63−/− and ΔNp63−/− MEFs occurs early during reprogramming and likely contributes to the low efficiency.
Figure 7.
ΔNp63 is critical for maintenance of the most reprogrammable MEF populations. (a and c) Live MEFs from two p63−/− and one ΔNp63−/− embryos and corresponding WT littermate controls were analyzed by FACS for Thy1.2 and Sca1 cell surface expression at the indicated passages (P3-P6). (b and d) The percentage of double-negative (DN, Thy1.2-negative/Sca1-negative) and Sca1-SP cells was quantified and plotted over time
Interestingly, the rescue of cell proliferation by LT also restored the Sca1-SP and DN populations in ΔNp63−/− MEFs (Supplementary Figures 7A and B). These exact MEFs also had the strongest (albeit incomplete) rescue of OSK reprogramming (Figure 5f). In contrast, LT did not restore Sca1-SP and DN fractions nor did it restore OSK reprogramming in global p63−/− MEFs (Supplementary Figures 7C–E). This correlation suggests that progressive loss of highly reprogrammable Sca1-SP and DN populations is an additional mechanism underlying the reprogramming defects of p63−/− and ΔNp63−/− MEFs.
Discussion
Since 2008, a plethora of studies has identified the tumor-suppressor p53 as a strong reprogramming barrier. However, whether and how p53 homologs p63 and p73 can regulate reprogramming was unknown. Using a loss-of-function approach, we show here and in a previous report34 that p73−/− MEFs reprogram normally and are fully pluripotent, indicating that p73 has no role in reprogramming. On the other hand, forced ΔNp73 overexpression was found to significantly improve speed and efficiency of reprogramming of human fibroblasts.39 As ΔNp73 is a well-known dominant-negative p53 family member,32, 50 its positive effect on reprogramming is most likely mediated by antagonizing p53 activity. This, however, was not directly tested.
In contrast to ΔNp73 overexpression or loss of p53, we show here that p63 is a promoter rather than a barrier of reprogramming. The major conclusions from our study are: (1) p63 is essential for OSK and OS, but not for OK reprogramming of MEFs, (2) p63 is essential during reprogramming for maximum efficiency, albeit not for the ability to reprogram per se, and is dispensable for maintaining stability and pluripotency of established iPSC colonies, (3) ΔNp63, but not TAp63, is the principal isoform involved in reprogramming, (4) during OSK and OS reprogramming, different p63-mediated mechanisms are critical, that is the regulation of MET and pluripotency genes in OSK reprogramming versus regulation of proliferation in OS reprogramming, (5) p63/ΔNp63 deficiency also causes loss of the most reprogrammable MEF sub-populations. Finally, our data suggest that different combinations of transcription factors initiate distinct routes of reprogramming, which supports the stochastic model of reprogramming. Altogether, (i) the differential effect of OSK/OS versus OK factors; (ii) the complete reproducibility of the p63−/− effects by isoform-specific ΔNp63−/− MEFs; and (iii) the high reproducibility of results in multiple independent experiments with MEFs derived from many independent pregnancies, implies a specific enabling role of ΔNp63 in reprogramming.
The finding that p63 regulates OSK and OS reprogramming by different mechanisms is intriguing. The most logical explanation is that these different factor combinations induce iPSCs via distinct molecular routes/intermittent cell types, each having a differential requirement for p63. Indeed, all tested MET and pluripotency genes were reduced in p63−/− cells in OSK reprogramming, but only a few were reduced in OS reprogramming. This is consistent with the observation that even WT MEFs activate epithelial genes much weaker in OS versus OSK reprogramming, and instead activate neural and mesenchymal genes.10 It is likely that these neural and mesenchymal precursors do not require p63 for maintenance of their cell identity. In contrast, OSK reprogramming upregulates epithelial genes and therefore requires p63 as a known epithelial master regulator.39, 44, 45, 46, 47, 48 Of note, we found that p63/ΔNp63−/− MEFs are also strongly impaired in OSKM reprogramming, a factor combination also known to induce MET11, 14 (Supplementary Figure 8). It remains to be identified which specific p63 transcriptional targets are involved in MET and reprogramming in general, and how they differ from those that mediate p63 function in epithelial development. Also, it will be interesting to see whether p63 also regulates reprogramming by other commonly used factor combinations, for example, those containing LIN28 and Nanog.
In contrast to OSK, OS reprogramming is sensitive to hypoproliferation of p63−/− MEFs. This is not surprising, given the essential role of cell proliferation in reprogramming13, 28 and the known regulation of proliferation by p63.31, 45 The reason why OSK reprogramming cannot be rescued by improving proliferation is likely due to defective downstream events such as MET and pluripotency gene activation.
In stark contrast to OSK and OS, OK reprogramming of p63−/− MEFs is normal. A number of reasons may explain this. First, as the principal isoform, ΔNp63, is upregulated much stronger during OSK and OS than during OK reprogramming, it suggests that the OK combination does not invoke molecular pathways/intermittent cell types that absolutely require ΔNp63. Second, similarly to OS, the MET program is only weakly activated in OK reprogramming.10 Third, although OS and OSK infections visibly enhance proliferation, OK infections generally do not, independent of the genotype (data not shown). Thus, the reduced proliferation capacity of p63−/− MEFs is irrelevant in OK reprogramming. Finally, p63/ΔNp63−/− MEFs progressively lose the most reprogrammable cell populations. Interestingly, we noticed that OK and OS reprogramming of sorted WT MEFs differs in that the OK efficiency is highest in the DN population, whereas the OS efficiency is highest in the Sca1-SP population (AN, unpublished data). As the Sca1-SP population (best reprogrammable by OS) is lost faster in p63−/− MEFs than the DN population (best reprogrammable by OK), this difference likely contributes to the defective OS (but not OK) reprogramming.
There is a known antagonistic interplay between p63 and Klf4, another master regulator of the epidermis51, 52 and a reprogramming factor. In skin, ΔNp63 and Klf4 are expressed in opposite compartments in the basal and superficial layers of the epidermis, respectively, likely because p63 directly binds to the Klf4 promoter and represses it.53, 54 Thus, p63−/− MEFs might in fact be expected to have increased levels and/or activity of endogenous Klf4, and as a result enhanced OK reprogramming efficiency because of additive effects of endogenous and exogenous Klf4. Indeed, we found that two out of four MET markers were upregulated in p63−/− MEFs during OK reprogramming. Also, the OK reprogramming efficiency of p63/ΔNp63−/− MEFs was in fact above the WT controls in two out of five experiments (data not shown). However, this positive effect might be counterbalanced by the loss of the most reprogrammable MEF populations, resulting in an overall unchanged reprogramming efficiency.
Materials and Methods
Mice and cell culture
This study was approved by the Institutional Animal Care and Use Committee at Stony Brook University. Heterozygous p63+/−,31 ΔNp63+/−,48 TAp63+/−,49 p53+/−55 and p73+/−32 mice were intercrossed within the genotype to obtain littermate KO and corresponding WT control embryos. MEFs were derived from E13.5 embryos using standard procedures and cultured in DMEM supplemented with 10% FBS, 1 × penicillin/streptomycin and 1 × antibiotic/antimycotic (Gibco, Grand Island, NY, USA) on 0.1% gelatin-coated tissue culture plates (Millipore, Billerica, MA, USA). iPSCs were maintained in iPSC medium consisting of DMEM/F12 supplemented with 10% KO serum replacement (KSR, Invitrogen, Grand Island, NY, USA), 1 × nonessential amino acids, 1 × L-glutamine, 1 × penicillin/streptomycin, 55 μM β-mercaptoethanol (Invitrogen) and 1000 μ/ml mouse leukemia inhibitory factor (LIF, Millipore or eBioscience, San Diego, CA, USA).
Generation of iPSCs
iPSCs were generated, as described elsewhere.56 Briefly, human Klf4, Oct4 and Sox2 in the retroviral vector Rebna were transfected into packaging Phoenix E cells using Lipofectamine reagent (Invitrogen; either two or three at once at equal amounts or separately, in which case viral supernatants were mixed just before use). After 24 h, transfected Phoenix E cells were selected with 2 μg/ml puromycin for 2–3 days and then switched to MEF media. After 12 h, viral supernatants were collected approximately every 12 h until cultures became overgrown, followed by 1 : 6–1 : 10 splitting and another round of puromycin selection. Passages 2–3 MEFs from littermate embryos were plated in 6 cm plates at 250 000 cells per plate for OSK reprogramming or 350 000 cells per plate for OS and OK reprogramming (for OSK reprogramming, 150 000 MEFs in six-well plates were also often used) and infected with fresh or once frozen 45 μm-filtered viral supernatants five times approximately every 12 h (LT-expressing MEFs were plated at 150 000 per 6 cm plate). At 24 h after the last infection, cells were switched to iPSC medium. Stable undifferentiated iPSC colonies were manually picked at 14–21 days post infection (d.p.i.) and expanded on feeder cells (irradiated mouse embryo fibroblasts, GlobalStem, Rockville, MD, USA).
AP and SSEA1 staining
AP staining was routinely done at the last time point of each experiment (i.e., before cell cultures became overgrown or when very few new colonies appeared), by fixing the culture with 4% PFA for 7 min at room temperature, followed by three PBS washes (including one overnight wash at 4 °C) and staining with Alkaline Phosphatase Detection Kit (Millipore) according to the manufacturer's instructions. For SSEA1 immunostaining, iPSCs induced with OSK from 40 000 MEFs in 24-well plates were fixed with 4% paraformaldehyde for 30 min at room temperature, washed twice with PBS and blocked/permeabilized in PBS with 5% heat-inactivated horse serum, 1% BSA and 0.2% Triton-X-100 for 20 min at room temperature, followed by overnight incubation at 4 °C with mouse PE-conjugated SSEA1 antibody (eBioscience, 1 : 100) in the same blocking/permeabilization solution. Fluorescence images were acquired at 10 × magnification using an Axiovert 200 M microscope (Zeiss, Thornwood, NY, USA) and AxioVision Software (Zeiss).
Quantitative real-time PCR
Total RNA was extracted using Trizol reagent (Invitrogen), and cDNA was prepared with Super Script II reverse transcriptase (Invitrogen). Real-time PCR was performed using the QuantiTect SYBR Green PCR kit (Qiagen, Valencia, CA, USA) on an Opticon 2 instrument (MJ Research Inc., Watertown, MA, USA) and analyzed with Opticon Monitor software. Samples were analyzed in duplicates and normalized to HPRT. The primers for Cdh1 (Tm 82.8 °C), EpCAM (Tm 78.9 °C), HPRT (Tm 75.6 °C), Nanog (Tm 78.6 °C), Oct4 (Tm 81.8 °C), Sall4 (Tm 82.4 °C) and Sox2 (Tm 85.8 °C)10 and the primers for p63 (Tm 85.2 °C), TAp63 (Tm 81.4 °C) and ΔNp63 (Tm 85.2 °C)57 were previously described; melting temperatures of the PCR products are given in parentheses. Information about additional primers is provided in Supplementary Table 1. All the primers were mouse specific and were used at the following PCR conditions: 94 °C 1 min, 60 °C 1 min, 72 °C 1.5 min (42 cycles), except for Sox2 (30 s at each temperature). Note that for qRT-PCR analysis during reprogramming (Figure 4, Supplementary Figure 6B), transduced MEFs were grown in iPSC medium without LIF, except when otherwise noted (Supplementary Figure 6A), which prevented growth of actual iPSC colonies. Although reprogramming in the presence of LIF represents the normal process, it is avoided in RT-PCR reactions, because it leads to artificial amplification of the genes specifically expressed in iPSC colonies, because of their superior proliferation comparing with the rest of the cells.13
FACS analysis and teratoma assay
For FACS analysis, iPSC colonies were harvested by Accutase treatment (Sigma, St. Louis, MO, USA) and depleted of feeder cells by a 30-min incubation on gelatin-coated plates, while MEFs were harvested by 0.05% Trypsin. The following conjugated antibodies were used for FACS analysis of live cells: PE-SSEA1, APC-Sca1 (both from eBioscience) and PE-Thy1.2 (BD Biosciences, San Jose, CA, USA). Analysis was performed on FACS Calibur instrument (Becton-Dickinson, Franklin Lakes, NJ, USA) using CellQuest software (BD Biosciences). For teratoma assay, iPSCs were resuspended in PBS/Matrigel 3 : 1 (Invitrogen) and subcutaneously injected into the flanks of CD1 athymic nude mice (Harlan, Frederick, MD, USA) at 2 × 106 iPSCs in 100 μl per site. When tumors reached 1 cm3 in size (at 4–7 weeks), they were harvested and processed for histology.
Generation of stable SV40 LT-expressing MEFs and p53 shRNAs
pBABE-LT vector (Addgene, Cambridge, MA, USA) was transfected into packaging Phoenix E cells using Lipofectamine. After 12 h, 400 000 MEFs (passages 2–3) from littermate embryos were plated in 6 cm plates and infected with fresh 45 μm-filtered viral supernatant five times approximately every 12 h, followed by incubation in MEF medium for 4 days and 2–3 rounds of puromycin selection of stably transfected MEFs afterward. p53 shRNA #1 was from Addgene (cat. no. 19751) and #2 was previously described.58
Immunoblot and growth curve analyses
Immunoblot analysis was carried out by standard methods using the following primary antibodies: p21 (F-5, Santa Cruz, Dallas, TX, USA), p19-Arf (ab80-100, Abcam, Cambridge, MA, USA), p16-Ink4A (M-156, Santa Cruz), MAPK (1B3B9, Millipore), Klf4 (ab34814, Abcam), Oct4 (ab19857, Abcam) and Sox2 (AB5603, Millipore). For growth curve analysis, 300 000 MEFs were plated in six-well plates and counted/re-plated every 3 days.
Acknowledgments
This work was funded by NCI grant CA93853 (to UMM), NYSTEM grant N08T-040 (to UMM), a NIDDK T32 Fellowship (to EMA) and the Stem Cell Facility grant NYSTEM #C026716.
Glossary
- AP
alkaline phosphatase
- DN
Thy1.2/Sca1 double-negative
- d.p.i.
days post infection
- FACS
fluorescence-activated cell sorting
- GFP
green fluorescent protein
- iPSCs
induced pluripotent stem cells
- KO
knockout
- LT
large T antigen
- MEFs
mouse embryo fibroblasts
- MET
mesenchymal–epithelial transition
- OK
Oct4 and Klf4
- OS
Oct4 and Sox2
- OSKM
Oct4, Sox2, Klf4 and c-Myc
- SP
Sca1 single-positive
- TA
transcription activation
- WT
wild-type
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Edited by G Melino
Supplementary Material
References
- Stadtfeld M, Hochedlinger K. Induced pluripotency: history, mechanisms, and applications. Genes Dev. 2010;24:2239–2263. doi: 10.1101/gad.1963910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Wernig M, Meissner A, Cassady JP, Jaenisch R. c-Myc is dispensable for direct reprogramming of mouse fibroblasts. Cell Stem Cell. 2008;2:10–12. doi: 10.1016/j.stem.2007.12.001. [DOI] [PubMed] [Google Scholar]
- Nakagawa M, Koyanagi M, Tanabe K, Takahashi K, Ichisaka T, Aoi T, et al. Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts. Nat Biotech. 2008;26:101–106. doi: 10.1038/nbt1374. [DOI] [PubMed] [Google Scholar]
- Martinez-Fernandez A, Nelson TJ, Ikeda Y, Terzic A. c-MYC independent nuclear reprogramming favors cardiogenic potential of induced pluripotent stem cells. J Cardiovasc Transl Res. 2010;3:13–23. doi: 10.1007/s12265-009-9150-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JB, Zaehres H, Wu G, Gentile L, Ko K, Sebastiano V, et al. Pluripotent stem cells induced from adult neural stem cells by reprogramming with two factors. Nature. 2008;454:646–650. doi: 10.1038/nature07061. [DOI] [PubMed] [Google Scholar]
- Kim JB, Greber B, Araúzo-Bravo MJ, Meyer J, Park KI, Zaehres H, et al. Direct reprogramming of human neural stem cells by OCT4. Nature. 2009;461:649–643. doi: 10.1038/nature08436. [DOI] [PubMed] [Google Scholar]
- Kim JB, Sebastiano V, Wu G, Araúzo-Bravo MJ, Sasse P, Gentile L, et al. Oct4-induced pluripotency in adult neural stem cells. Cell. 2009;136:411–419. doi: 10.1016/j.cell.2009.01.023. [DOI] [PubMed] [Google Scholar]
- Hester ME, Song S, Miranda CJ, Eagle A, Schwartz PH, Kaspar BK. Two factor reprogramming of human neural stem cells into pluripotency. PLoS One. 2009;4:e7044. doi: 10.1371/journal.pone.0007044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemajerova A, Kim SY, Petrenko O, Moll UM. Two-factor reprogramming of somatic cells to pluripotent stem cells reveals partial functional redundancy of Sox2 and Klf4. Cell Death Differ. 2012;19:1268–1276. doi: 10.1038/cdd.2012.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samavarchi-Tehrani P, Golipour A, David L, Sung HK, Beyer TA, Datti A, et al. Functional genomics reveals a BMP-driven mesenchymal-to-epithelial transition in the initiation of somatic cell reprogramming. Cell Stem Cell. 2010;7:64–77. doi: 10.1016/j.stem.2010.04.015. [DOI] [PubMed] [Google Scholar]
- Buganim Y, Faddah DA, Cheng AW, Itskovich E, Markoulaki S, Ganz K, et al. Single-cell expression analyses during cellular reprogramming reveal an early stochastic and a late hierarchic phase. Cell. 2012;150:1209–1222. doi: 10.1016/j.cell.2012.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith ZD, Nachman I, Regev A, Meissner A. Dynamic single-cell imaging of direct reprogramming reveals an early specifying event. Nat Biotechnol. 2010;28:521–526. doi: 10.1038/nbt.1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Liang J, Ni S, Zhou T, Qing X, Li H, et al. A mesenchymal-to-epithelial transition initiates and is required for the nuclear reprogramming of mouse fibroblasts. Cell Stem Cell. 2010;7:51–63. doi: 10.1016/j.stem.2010.04.014. [DOI] [PubMed] [Google Scholar]
- Chen J, Liu J, Yang J, Chen Y, Chen J, Ni S, et al. BMPs functionally replace Klf4 and support efficient reprogramming of mouse fibroblasts by Oct4 alone. Cell Res. 2011;21:205–212. doi: 10.1038/cr.2010.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna J, Carey BW, Jaenisch R. Reprogramming of somatic cell identity. Cold Spring Harb Symp Quant Biol. 2008;73:147–155. doi: 10.1101/sqb.2008.73.025. [DOI] [PubMed] [Google Scholar]
- Hanna JH, Saha K, Jaenisch R. Pluripotency and cellular reprogramming: facts, hypotheses, unresolved issues. Cell. 2010;143:508–525. doi: 10.1016/j.cell.2010.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu-Monette ZY, Medeiros LJ, Li Y, Orlowski RZ, Andreeff M, Bueso-Ramos CE, et al. Dysfunction of the TP53 tumor suppressor gene in lymphoid malignancies. Blood. 2012;119:3668–3683. doi: 10.1182/blood-2011-11-366062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talos F, Moll UM. Role of the p53 family in stabilizing the genome and preventing polyploidization. Adv Exp Med Biol. 2010;676:73–91. doi: 10.1007/978-1-4419-6199-0_5. [DOI] [PubMed] [Google Scholar]
- Vaseva AV, Moll UM. The mitochondrial p53 pathway. Biochim Biophys Acta. 2009;1787:414–420. doi: 10.1016/j.bbabio.2008.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Yin X, Qin H, Zhu F, Liu H, Yang W, et al. Two supporting factors greatly improve the efficiency of human iPSC generation. Cell Stem Cell. 2008;3:475–479. doi: 10.1016/j.stem.2008.10.002. [DOI] [PubMed] [Google Scholar]
- Marión RM, Strati K, Li H, Murga M, Blanco R, Ortega S, et al. A p53-mediated DNA damage response limits reprogramming to ensure iPS cell genomic integrity. Nature. 2009;460:1149–1153. doi: 10.1038/nature08287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utikal J, Polo JM, Stadtfeld M, Maherali N, Kulalert W, Walsh RM, et al. Immortalization eliminates a roadblock during cellular reprogramming into iPS cells. Nature. 2009;460:1145–1148. doi: 10.1038/nature08285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Collado M, Villasante A, Strati K, Ortega S, Cañamero M, et al. The Ink4/Arf locus is a barrier for iPS cell reprogramming. Nature. 2009;460:1136–1139. doi: 10.1038/nature08290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamura T, Suzuki J, Wang YV, Menendez S, Morera LB, Raya A, et al. Linking the p53 tumour suppressor pathway to somatic cell reprogramming. Nature. 2009;460:1140–1144. doi: 10.1038/nature08311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong H, Takahashi K, Ichisaka T, Aoi T, Kanagawa O, Nakagawa M, et al. Suppression of induced pluripotent stem cell generation by the p53-p21 pathway. Nature. 2009;460:1132–1135. doi: 10.1038/nature08235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye D, Wang G, Liu Y, Huang W, Wu M, Zhu S, et al. MiR-138 promotes induced pluripotent stem cell generation through the regulation of the p53 signaling. Stem Cells. 2012;30:1645–1654. doi: 10.1002/stem.1149. [DOI] [PubMed] [Google Scholar]
- Hanna J, Saha K, Pando B, van Zon J, Lengner CJ, Creyghton MP, et al. Direct cell reprogramming is a stochastic process amenable to acceleration. Nature. 2009;462:595–601. doi: 10.1038/nature08592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosh R, Assia-Alroy Y, Molchadsky A, Bornstein C, Dekel E, Madar S, et al. p53 counteracts reprogramming by inhibiting mesenchymal-to-epithelial transition. Cell Death Differ. 2013;20:312–320. doi: 10.1038/cdd.2012.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills AA, Zheng B, Wang XJ, Vogel H, Roop DR, Bradley A. p63 is a p53 homologue required for limb and epidermal morphogenesis. Nature. 1999;398:708–713. doi: 10.1038/19531. [DOI] [PubMed] [Google Scholar]
- Yang A, Schweitzer R, Sun D, Kaghad M, Walker N, Bronson RT, et al. p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature. 1999;398:714–718. doi: 10.1038/19539. [DOI] [PubMed] [Google Scholar]
- Yang A, Walker N, Bronson R, Kaghad M, Oosterwegel M, Bonnin J, et al. p73-deficient mice have neurological, pheromonal and inflammatory defects but lack spontaneous tumours. Nature. 2000;404:99–103. doi: 10.1038/35003607. [DOI] [PubMed] [Google Scholar]
- Talos F, Abraham A, Vaseva AV, Holembowski L, Tsirka SE, Scheel A, et al. p73 is an essential regulator of neural stem cell maintenance in embryonal and adult CNS neurogenesis. Cell Death Differ. 2010;17:1816–1829. doi: 10.1038/cdd.2010.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandrova EM, Talos F, Moll UM. p73 is dispensable for commitment to neural stem cell fate, but is essential for neural stem cell maintenance and for blocking premature differentiation. Cell Death Diff. 2013;20:368. doi: 10.1038/cdd.2012.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandrova EM, Moll UM. Role of p53 family members p73 and p63 in human hematological malignancies. Leuk Lymphoma. 2012;53:2116–2129. doi: 10.3109/10428194.2012.684348. [DOI] [PubMed] [Google Scholar]
- Rufini A, Agostini M, Grespi F, Tomasini R, Sayan BS, Niklison-Chirou MV, et al. p73 in cancer. Genes Cancer. 2011;2:491–502. doi: 10.1177/1947601911408890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melino G. p63 is a suppressor of tumorigenesis and metastasis interacting with mutant p53. Cell Death Diff. 2011;18:1487–1499. doi: 10.1038/cdd.2011.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Cheng Z, Yang Z, Zheng J, Lin T. DNp73 improves generation efficiency of human induced pluripotent stem cells. BMC Cell Biol. 2012;13:9. doi: 10.1186/1471-2121-13-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su X, Cho MS, Gi YJ, Ayanga BA, Sherr CJ, Flores ER. Rescue of key features of the p63-null epithelial phenotype by inactivation of Ink4a and Arf. EMBO J. 2009;28:1904–1915. doi: 10.1038/emboj.2009.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L, He X, Lim LP, de Stanchina E, Xuan Z, Liang Y, et al. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447:1130–1134. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi YJ, Lin CP, Ho JJ, He X, Okada N, Bu P, et al. miR-34 miRNAs provide a barrier for somatic cell reprogramming. Nat Cell Biol. 2011;13:1353–1360. doi: 10.1038/ncb2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonini D, Russo MT, De Rosa L, Gorrese M, Del Vecchio L, Missero C. Transcriptional repression of miR-34 family contributes to p63-mediated cell cycle progression in epidermal cells. J Invest Dermatol. 2010;130:1249–1257. doi: 10.1038/jid.2009.438. [DOI] [PubMed] [Google Scholar]
- Lake BB, Fink J, Klemetsaune L, Fu X, Jeffers JR, Zambetti GP, et al. Context-dependent enhancement of induced pluripotent stem cell reprogramming by silencing Puma. Stem Cells. 2012;30:888–897. doi: 10.1002/stem.1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Candi E, Dinsdale D, Rufini A, Salomoni P, Knight RA, Mueller M, et al. TAp63 and DeltaNp63 in cancer and epidermal development. Cell Cycle. 2007;6:274–285. doi: 10.4161/cc.6.3.3797. [DOI] [PubMed] [Google Scholar]
- Truong AB, Kretz M, Ridky TW, Kimmel R, Khavari PA. p63 regulates proliferation and differentiation of developmentally mature keratinocytes. Genes Dev. 2006;20:3185–3197. doi: 10.1101/gad.1463206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Candi E, Rufini A, Terrinoni A, Dinsdale D, Ranalli M, Paradisi A, et al. Differential roles of p63 isoforms in epidermal development: selective genetic complementation in p63 null mice. Cell Death Differ. 2006;13:1037–1047. doi: 10.1038/sj.cdd.4401926. [DOI] [PubMed] [Google Scholar]
- Koster MI, Dai D, Marinari B, Sano Y, Costanzo A, Karin M, et al. p63 induces key target genes required for epidermal morphogenesis. Proc Natl Acad Sci USA. 2007;104:3255–3260. doi: 10.1073/pnas.0611376104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano RA, Smalley K, Magraw C, Serna VA, Kurita T, Raghavan S, et al. ΔNp63 knockout mice reveal its indispensable role as a master regulator of epithelial development and differentiation. Development. 2012;139:772–782. doi: 10.1242/dev.071191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su X, Paris M, Gi YJ, Tsai KY, Cho MS, Lin YL, et al. TAp63 prevents premature aging by promoting adult stem cell maintenance. Cell Stem Cell. 2009;5:64–75. doi: 10.1016/j.stem.2009.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grob TJ, Novak U, Maisse C, Barcaroli D, Lüthi AU, Pirnia F, et al. Human delta Np73 regulates a dominant negative feedback loop for TAp73 and p53. Cell Death Differ. 2001;8:1213–1223. doi: 10.1038/sj.cdd.4400962. [DOI] [PubMed] [Google Scholar]
- Jaubert J, Cheng J, Segre JA. Ectopic expression of kruppel like factor 4 (Klf4) accelerates formation of the epidermal permeability barrier. Development. 2003;130:2767–2777. doi: 10.1242/dev.00477. [DOI] [PubMed] [Google Scholar]
- Segre JA, Bauer C, Fuchs E. Klf4 is a transcription factor required for establishing the barrier function of the skin. Nat Genet. 1999;22:356–360. doi: 10.1038/11926. [DOI] [PubMed] [Google Scholar]
- Dai X, Segre JA. Transcriptional control of epidermal specification and differentiation. Curr Opin Genet Dev. 2004;14:485–491. doi: 10.1016/j.gde.2004.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordani N, Pozzi S, Martynova E, Fanoni D, Borrelli S, Alotto D, et al. Mutant p53 subverts p63 control over KLF4 expression in keratinocytes. Oncogene. 2011;30:922–932. doi: 10.1038/onc.2010.474. [DOI] [PubMed] [Google Scholar]
- Jacks T, Remington L, Williams BO, Schmitt EM, Halachmi S, Bronson RT, et al. Tumor spectrum analysis in p53-mutant mice. Curr Biol. 1994;4:1–7. doi: 10.1016/s0960-9822(00)00002-6. [DOI] [PubMed] [Google Scholar]
- Alexandrova EM, Moll UM. Generation of p53-deficient induced pluripotent stem cells from mouse embryo fibroblasts. Methods Mol Biol. 2013;962:157–164. doi: 10.1007/978-1-62703-236-0_13. [DOI] [PubMed] [Google Scholar]
- Wolff S, Talos F, Palacios G, Beyer U, Dobbelstein M, Moll UM. The alpha/beta carboxy-terminal domains of p63 are required for skin and limb development. New insights from the Brdm2 mouse which is not a complete p63 knockout but expresses p63 gamma-like proteins. Cell Death Differ. 2009;16:1108–1117. doi: 10.1038/cdd.2009.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickins RA, Hemann MT, Zilfou JT, Simpson DR, Ibarra I, Hannon GJ, et al. Probing tumor phenotypes using stable and regulated synthetic microRNA precursors. Nat Genet. 2005;37:1289–1295. doi: 10.1038/ng1651. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.