Abstract
Human-nucleotide-excision repair (NER) deficiency leads to different developmental and segmental progeroid symptoms of which the pathogenesis is only partially understood. To understand the biological impact of accumulating spontaneous DNA damage, we studied the phenotypic consequences of DNA-repair deficiency in Caenorhabditis elegans. We find that DNA damage accumulation does not decrease the adult life span of post-mitotic tissue. Surprisingly, loss of functional ERCC-1/XPF even further extends the life span of long-lived daf-2 mutants, likely through an adaptive activation of stress signaling. Contrariwise, NER deficiency leads to a striking transgenerational decline in replicative capacity and viability of proliferating cells. DNA damage accumulation induces severe, stochastic impairment of development and growth, which is most pronounced in NER mutants that are also impaired in their response to ionizing radiation and inter-strand crosslinks. These results suggest that multiple DNA-repair pathways can protect against replicative decline and indicate that there might be a direct link between the severity of symptoms and the level of DNA-repair deficiency in patients.
Keywords: nucleotide-excision repair, DNA repair, DNA-damage response, DNA damage, aging, C. elegans
DNA damage causes transcription, replication and mitotic stress and induces chromosomal aberrations and mutations that cause cancer.1 Furthermore, it can induce cell death and senescence, leading to functional deterioration, associated with aging. The DNA-damage response (DDR) network, including complementary DNA-repair pathways of nucleotide-excision repair (NER), base-excision repair (BER), mismatch repair (MMR), homologous recombination (HR), non-homologous-end-joining and inter-strand-crosslink repair (ICLR), counteracts genomic erosion.
NER is a conserved and versatile pathway that removes a wide range of DNA-helix-distorting lesions, including those induced by UV light and numerous chemicals.2 The subpathway transcription-coupled NER (TC-NER) removes damage from actively transcribed DNA templates. TC-NER is initiated by lesion-stalled RNA polymerase II and the CSA and CSB proteins. Lesions located anywhere in the genome are repaired by global genome NER (GG-NER), initiated by XPC/hHR23 and UV-DDB complexes. Next, transcription factor TFIIH opens the DNA around the lesion. Together with XPA and RPA, this pre-initiation complex facilitates the orientation of the structure-specific endonucleases ERCC1/XPF and XPG. These incise DNA at the 5′ and 3′ sides of the lesion, respectively, resulting in a single-stranded gap of 25–29 nucleotides that is filled in by DNA synthesis and ligation.
The biological significance of NER is evident from the severe pleiotropic symptoms associated with inherited NER deficiency disorders.3 Mutations in genes affecting GG-NER cause Xeroderma Pigmentosum (XP), characterized by severe photosensitivity and a more than 1000-fold enhanced risk of developing skin cancer. Mutations in ERCC1 and XPF can lead to additional symptoms, such as progressive neurodegeneration, severe developmental failure and segmental progeria.4, 5 Mutations in TC-NER genes, including CSB and XPG, cause different diseases such as Cockayne syndrome (CS), the more severe cerebro-oculo-facio-skeletal syndrome (COFS), a CS/XP combination or UV-sensitive syndrome. CS and COFS are characterized by severe neurodevelopmental defects and segmental progeria. Finally, specific defects in TFIIH can lead to trichothiodystrophy, a severe CS-like disorder.
NER deficiency diseases, including developmental failure and progeroid features, were recapitulated in transgenic mice.1, 3 Progeroid features are also found in other DDR syndromes, which together support the hypothesis that random accumulation of DNA damage contributes to aging.6, 7 However, the molecular mechanisms of DNA-damage-induced aging pathology, the specific contribution of transcription and/or replication stress and the involvement of different DNA lesions and repair pathways remains elusive. Furthermore, mechanisms other than DNA damage accumulation, including defects in transcription regulation, have also been suggested to play a role in the pathogenesis of NER deficiency disorders.8, 9, 10
The nematode Caenorhabditis elegans is well suited as a multicellular model organism to study the in vivo function of NER.11, 12, 13, 14, 15 Whether NER deficiency or accumulating DNA damage is associated with developmental failure and progeroid features in C. elegans is controversial.15 A high dose of UV irradiation arrests development and shortens the life span of C. elegans.16, 17 However, contradictory reports exist as to whether NER-deficient xpa-1 mutant animals have a shortened life span as a consequence of endogenously produced DNA damage.11, 16, 17, 18 Thus, although C. elegans is well-known for its value in identifying genetic pathways that regulate aging,19 it is uncertain whether this animal is suitable for studying the contribution of spontaneously induced DNA damage to aging.
Here, we show that DNA-repair deficiency in C. elegans leads to impaired growth and a progressive, transgenerational decline in replicative capacity reminiscent of proliferative aging, which is directly correlated with the severity of the DNA-repair defect and with a variable transcriptional modulation of growth and stress-response genes.
Results
ERCC-1/XPF-1 deficiency increases lifespan of long-lived mutants
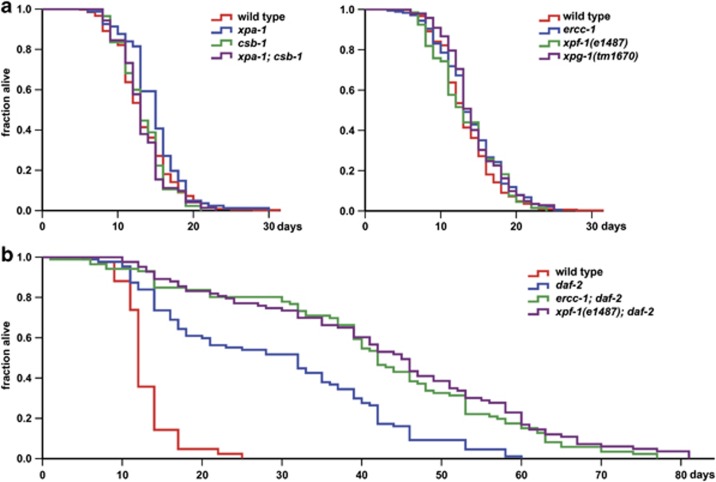
To understand whether DNA-repair deficiency in C. elegans is associated with a shortened life span, we measured the adult life span20 of healthy looking animals with mutated NER genes csb-1, ercc-1, xpa-1, xpf-1, xpg-1 and xpa-1;csb-1 double mutants.21 Strikingly, we did not detect decreased life span in any of the tested mutants (Figure 1a), indicating that NER is not life-span limiting in adult C. elegans.
Figure 1.
NER deficiency does not affect normal life span but loss of ercc-1 and xpf-1 enhances long life span of daf-2 mutants. (a and b) show the fraction of live adult animals with and without mutations in (a) wild type (n=331), xpa-1 (n=81), csb-1 (n=85), xpa-1;csb-1 (n=71), ercc-1 (n=177), xpf-1(e1487; n= 66), xpg-1(tm1670; n= 142). In (b) wild type (n=42), daf-2 (n=87) ercc-1; daf-2 (n= 86), xpf-1; daf-2 (n=83). Mean life span of at least two independent experiments is depicted
The seeming discrepancy compared to mammalian phenotypes may have several explanations. Because of an increased mutation frequency,22 we assume that DNA damage accumulates in some of these mutants. However, accumulation of endogenous DNA damage may not induce functional decline of the mainly post-mitotic tissue in adult worms. Alternatively, as in mammals,6 a compensatory survival response (SR), triggered by DNA damage, may maintain a near-normal life span. Thirdly, the lifespan of C. elegans may be too short to accumulate significant amounts of spontaneous DNA damage. High doses of DNA damage owing to repetitive exposure to UV were previously shown to severely shorten the life span of C. elegans.16 Therefore, we tested whether spontaneous DNA damage accumulation becomes life-span limiting in animals with a longer life span. We generated animals with mutated daf-2 (insulin/Insulin Growth Factor (IGF) receptor), which extends life span by more than twofold,20 and ercc-1 or xpf-1. Surprisingly, ercc-1 and xpf-1 mutations further extend, rather than decrease, the life span of long-lived daf-2 animals (Figure 1b). This intriguing finding suggests that NER deficiency is not life-span limiting in post-mitotic tissue and that a postulated DNA-damage-induced SR even extends life span, together with reduced insulin signaling.
Modulation of growth and stress responses in ercc-1 mutants
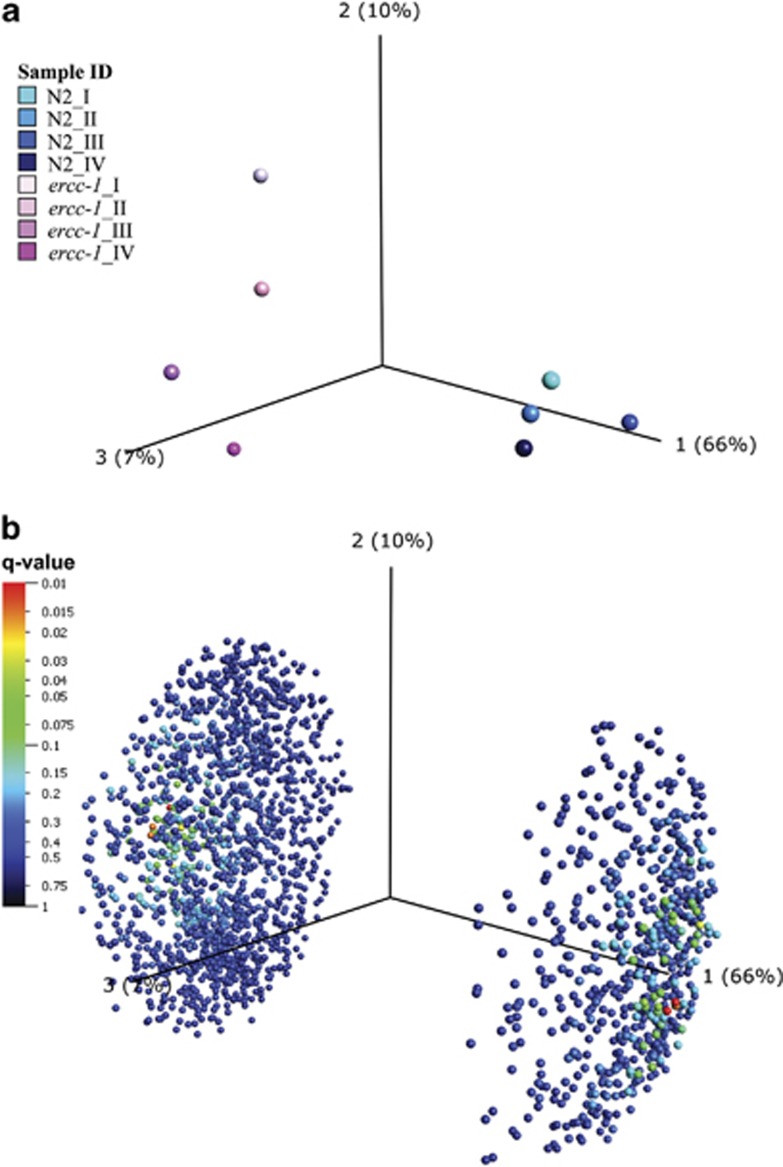
To determine whether SR is indeed present, we analyzed the global gene expression profile of mixed stage ercc-1 mutants. ANOVA analyses revealed that 2139 transcripts were significantly upregulated or downregulated in ercc-1 mutants compared with the wild-type animals (P<0.05; Supplementary Table S1). Principal component analysis (PCA) revealed a clear separation of the ercc-1 mutants from the wild type and, strikingly, a greater biological variance within the group of ercc-1 mutants (Figures 2a and b). Of the 2139 differently regulated transcripts (P<0.05), only 4% showed a more than two-fold change (Supplementary Table S2).
Figure 2.
Factorial map of PCA. PCA on either sample (a) or variables/transcripts (b) between ercc-1 and the wild type (N2) was conducted using the 2139 statistically significant (P<0.05) transcripts found in the ercc-1 mutant group compared with its wild-type counterpart. (a) PCA on the replica samples for ercc-1 and the wild type. The eight samples group together strain-wise. All samples are colored individually (ercc-1 and wild type are colored with different shades of pink and blue, respectively). The PCA indicates that the ercc-1 group has more biological variance than the wild-type control group. (b) PCA on the 2139 statistically significant transcripts between ercc-1 and wild type. The 2139 variables/genes are divided by their variation, found in the data set (Component 1, 2 and 3). The component can thus be interpreted as the variation found within and between the sample groups. For both the samples and the transcript PCA plots, the largest explainable variance is found in component 1, where 66% of the data variation can be explained. This 66% is the difference found between ercc-1 and wild type and thus divides the variables in ercc-1 and wild-type sample groups. Components 2 and 3, with 10 and 7%, respectively, explain the biological variance within each sample group. Here, the variance within the ercc-1 sample group is larger than in the wild-type control sample group, that is, more spread in the direction of components 2 and 3 compared with the wild-type samples. Most variables were found to have the trend upregulated in the ercc-1 group (1530 upregulated, compared with 609 downregulated). For the variable PCA, the transcripts are color-coded based on their Q-value (false discovery rate), where red (Q<0.01) is the lowest Q-value found for a handful of genes. See also Supplementary Tables S1, S3 and Supplementary Figure S1
The differently regulated ercc-1 transcripts were dominated by genes responding to stress, regulating growth and development and genes preferentially expressed in males (Supplementary Tables S1 and S2). Otherwise, the profile reflected an unfocused transcriptome modulation with no overrepresented biological processes (BPs) among the upregulated genes. Interestingly, among the downregulated genes, BP clusters ‘embryonic development ending in birth or egg hatching' and ‘translation' were enriched (Supplementary Figure S1A and Supplementary Table S3), whereas among the upregulated genes, BPs involving development, growth and reproductive process were underrepresented (Supplementary Figure S1B and Supplementary Table S3). Changes observed in ercc-1 mutants could thus be consistent with the idea of a compensatory SR involving suppression of growth and upregulation of stress responses.
As increased stress responses and other transcriptional changes together stimulate longevity in insulin-like-signaling (ILS) mutants,19 we hypothesized that upregulation of stress responses as a result of ercc-1/xpf-1 deficiency might cause additive daf-2 life-span extension (Figure 1b). Interestingly, we noticed a prominent overlap between ercc-1-regulated genes and genes regulated after exposure to silver nanoparticles,23 which are used as an antibacterial agent. A high Pearson correlation (R2=0.73) was observed when genes regulated by more than twofold were compared. This suggests that the PMK-1 p38 MAPK stress-response-signaling pathway, which mediates silver nanoparticle toxicity24 and contributes to increased longevity of daf-2 mutants,25 is activated in ercc-1 mutants. Indeed, we found a strong correlation between genes regulated by PMK-125 and the ercc-1 dataset (R2=0.71 and Fisher exact tests (FET) P<0.0001). This would suggest that NER function is not life-span limiting in post-mitotic tissue, partially on account of an adaptive transcriptome modulation involving activation of p38 MAPK stress-response signaling.
In progeroid NER-deficient mammals, SR involves upregulation of stress responses and suppression of the growth hormone/IGF-1 axis.4, 6 Although our results are reminiscent of such transcriptome modulation, in striking contrast, we observed no enrichment of ILS or BPs involving life-span determination. Transcriptomic comparison of ercc-1 with daf-2,26 age-1 and daf-16 mutants27 also showed no overlapping BPs corresponding to regulation of life span. However, a statistically significant intersection was seen as 20 of the ercc-1-regulated genes (P<0.05) overlap with the 464 daf-2-regulated genes (FET P<0.0001) and 55 genes overlap with the 1254 genes of age-1 and daf-16 mutants (FET P=0.0066). Moreover, comparison of the ercc-1-regulated and daf-16 and age-1 age-related genes revealed a negative correlation (R2=−0.311 and R2=−0.323, respectively). This suggests an overlap between regulated genes in ercc-1 and aging mutants, but in pathways other than ILS.
ercc-1, xpf-1, xpg-1 mutants show growth and developmental defects
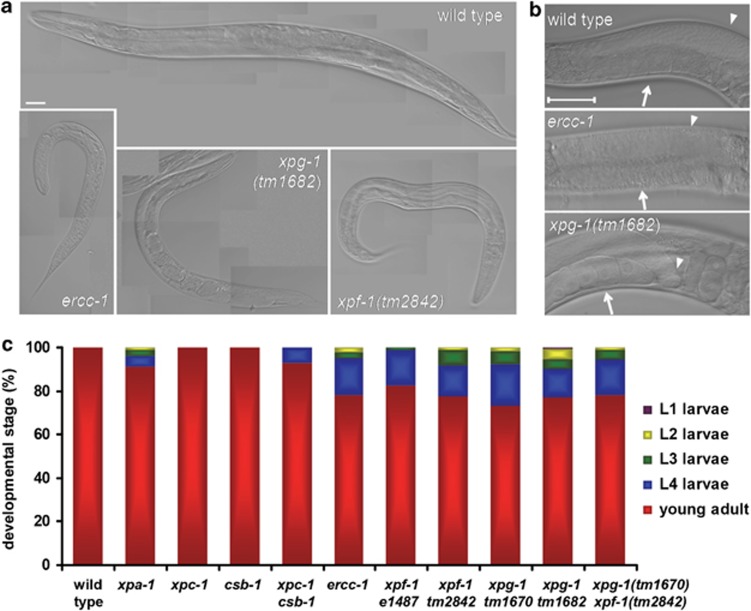
Consistent with the transcriptomic prominence of BPs related to growth and development, we noticed that a fraction of the population of ercc-1, xpf-1 and xpg-1 mutants displayed developmental and growth defects (Figures 3a and b and Table 1). To quantify this, we measured developmental speed of animals laid as eggs over ∼70 h at 20 °C, during which time all wild-type animals reached adulthood (Figure 3c). In contrast, around 20% of ercc-1/xpf-1 mutants failed to reach adulthood in this time period (Figure 3c). Similar defects were observed in another xpf-1 loss-of-function allelic variant (tm2842), which has not been described before but displays UV hypersensitivity that is similar to that of the xpf-1(e1487) mutant12 (Supplementary Figure S2). Furthermore, a comparable developmental delay was observed in two xpg-1 mutant strains (Figure 3c). Growth-retarded animals showed pleiotropic defects (Figures 3a and b). Some animals became adults at a later time point and laid eggs normally, whereas others produced inviable eggs or no eggs at all, or were arrested in development and never reached adulthood. A smaller but noteworthy proportion of xpa-1 animals were also growth-impaired (Figure 3c). This was also observed for xpc-1; csb-1 double mutants, which show a UV sensitivity that is similar to that of the xpa-1 animals, but not in xpc-1 or csb-1 single mutants, which are less UV-sensitive.12 These observations hint at a correlation between the severity of the phenotype and the severity of the DNA-repair deficiency and suggest that DNA-repair-deficient animals are arrested in development or develop slowly if unrepaired, spontaneous DNA damage interferes with genome function.
Figure 3.
ercc-1, xpg-1 and xpg-1 deficiency leads to developmental and growth defects. (a) depicts Nomarski images of a typical young adult wild-type animal and examples of similar age ercc-1, xpg-1 and xpf-1 animals that show growth and developmental defects. Images were taken with the same magnification. Bar in the first image represents 50 μm. (b) shows examples of similar-age normal (wild-type) and morphologically abnormal (ercc-1 and xpg-1) gonad/uterus areas. In the ercc-1 mutant, no healthy oocytes (white arrow) can be discerned in the proximal part of the gonad, whereas in the xpg-1 mutant, oocytes are present but the distal part of the gonad (arrowhead) is mislocalized. The bar in the first image represents 50 μm. (c) depicts the quantification of the growth of different mutants by counting the larval and adult stages that are observed ∼70 h after animals are laid as eggs at 20 °C
Table 1. Brood size of NER mutants.
| Brood size | n | |
|---|---|---|
| Wild type | 291±8 | 11 |
| xpa-1 | 175±19 | 11 |
| xpc-1 | 264±15 | 11 |
| csb-1 | 289±13 | 11 |
| xpc-1 csb-1 | 217±16 | 11 |
| ercc-1 | 151±17 | 17 |
| xpf-1(e1487) | 227±23 | 11 |
| xpf-1(tm2842) | 174±19 | 12 |
| xpg-1(tm1670) | 133±18 | 14 |
| xpg-1(tm1682) | 200±19 | 11 |
| xpg-1(tm1670) xpf-1(tm2842) | 147±11 | 11 |
Shown is the number of eggs laid (brood size).
n denotes the number of animals of which the brood was counted.
ERCC-1, XPF-1 and XPG-1 protect against replicative decline
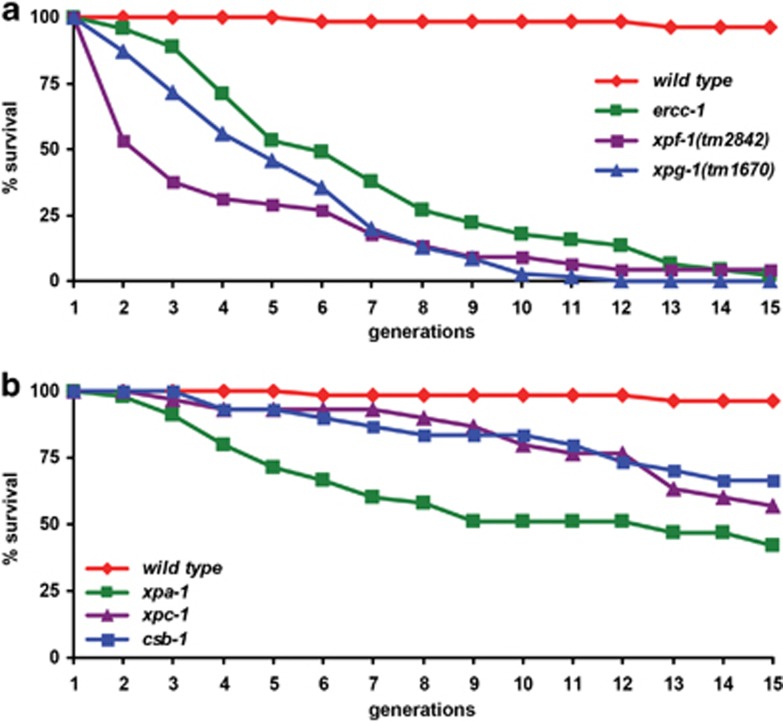
Defects caused by DNA damage accumulation may be more obvious if quantified over a longer period of time than within one generation. Therefore, we tested the transgenerational functional decline of mutant animals by serial passage.28 First, we backcrossed mutants 3–6 times against wild-type animals to exclude the effects of non-specific background mutations. Next, we singled out 15 animals per strain and measured proliferative capacity by quantifying viable progeny over 15 generations. A single animal from the offspring of each of the 15 singles was randomly picked for each generation and transferred to a new plate (Supplementary Figure S3A). We observed a severely reduced proliferative capacity of ercc-1, xpf-1 and xpg-1 animals, compared with wild-type animals (Figure 4a and Supplementary Figure S3B). The xpa-1 animals also displayed a reduction, though less pronounced, of proliferative capacity (Figure 4b). The GG-NER-specific xpc-1 and TC-NER-specific csb-1 strains, which are the mildest NER mutants, showed, correspondingly, the mildest replicative capacity defects. Animals lost viability for different reasons, including sterility, embryonic lethality, (larval) growth arrest and death before reproduction. These results show that the proteins ERCC-1, XPF-1, XPG-1 and, to a lower extent, XPA-1, XPC-1 and CSB-1 protect against stochastic functional decline caused by replicative stress in different tissues.
Figure 4.
ERCC-1, XPF-1, XPG-1 and XPA-1 protect against replicative decline. (a and b) show the survival of successive generations of wild-type and mutant populations if, in each generation, one animal is passaged. Mutant ercc-1, xpf-1 and xpg-1 strains (in A), and to a lower extent, xpa-1 (in B), lose viability in time. The graphs depict cumulative results from multiple experiments at 25 °C. The number of generations (x-axis) is plotted against the surviving percentage. For explanation of the assay and a representative experiment, see Supplementary Figure S3
ERCC-1, XPF-1 and XPG-1 function in multiple repair pathways
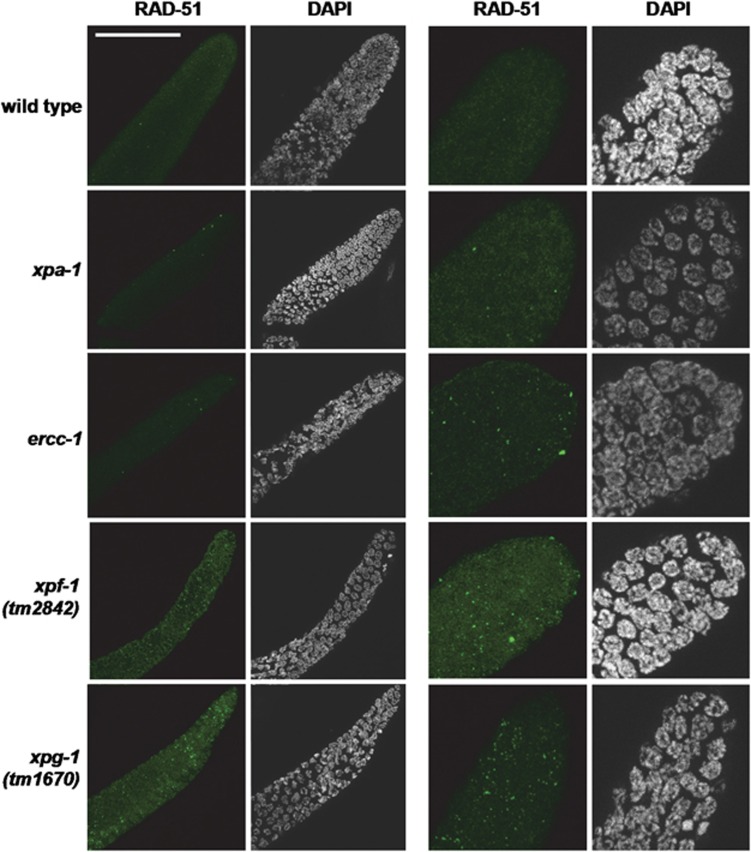
Thus far, we have assumed that DNA damage accumulated in the DNA-repair mutants, which is supported by the increased mutational rate in xpa-1 C. elegans22 and XPA- and ERCC1-deficient mice.29 To test whether DNA damage load is indeed higher in these mutants, we determined RAD-51 foci formation in proliferating germ cells. These foci arise after replication stress from various forms of DNA damage.30 We observed more RAD-51 foci in mitotic germ cells of xpa-1 mutant animals compared with wild-type animals, which was further increased in ercc-1, xpf-1 and xpg-1 mutants (Figure 5). We therefore hypothesize that spontaneously occurring DNA damage does indeed accumulate in these mutants and presume that this leads to the observed phenotypic defects and changes in gene expression.
Figure 5.
xpa-1-, ercc-1-, xpf-1- and xpg-1-deficient animals accumulate RAD-51 foci. RAD-51 foci visualized by the immunofluorescence of gonads of wild-type, xpa-1, ercc-1, xpf-1 and xpg-1 animals. Shown is the distal part of the gonad containing proliferating germ cells prior to meiosis. For each strain, two examples are shown (left and right panels). The panels on the right show a higher magnification of the distal gonad. The bar in the first image represents 50 μm
In xpa-1, ercc-1, xpf-1 and xpg-1 animals, NER is defective,11, 12, 14 suggesting that NER-substrate lesions cause the deleterious phenotype. Nevertheless, the growth, developmental and replicative capacity defects (Figures 3 and 4) and accumulation of RAD-51 foci indicative of DNA damage (Figure 5) are more pronounced in ercc-1, xpf-1 and xpg-1 animals, whereas these animals are not more compromised in NER than are the xpa-1 animals.12 In mammals, orthologs of ERCC-1, XPF-1 and XPG-1 but not of XPA-1 also function in other genome maintenance pathways, such as double-strand break (DSB) repair, ICLR, BER and the response to damage-induced replication problems.31, 32, 33, 34 Also, in C. elegans, there is evidence that ERCC-1 and XPF-1 are involved in DSB and ICL repair.30, 35, 36, 37 Deficiencies in more than one DNA-repair pathway likely result in a higher DNA damage load and therefore stronger defects. We tested whether C. elegans ERCC-1, XPF-1 and XPG-1 are indeed involved in other DNA-repair pathways, as opposed to XPA-1, by measuring survival after DSB, oxidative DNA damage and ICL induction.
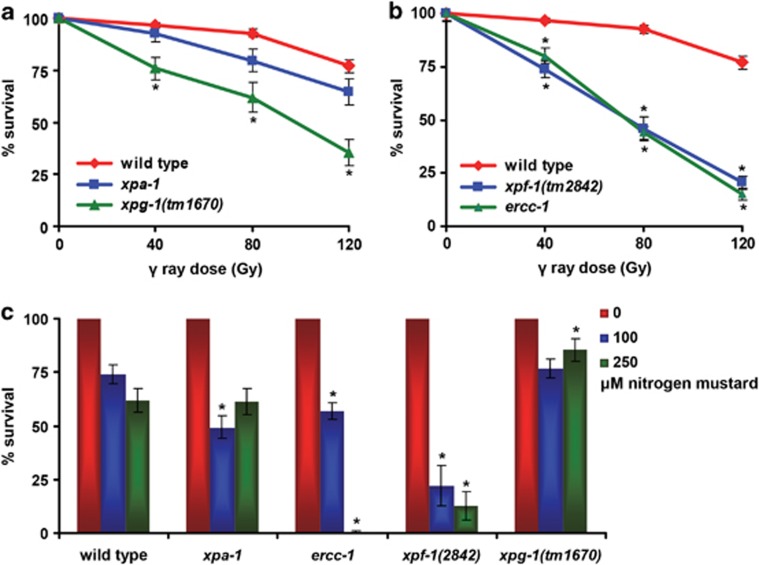
Survival assays showed that ercc-1, xpf-1 and xpg-1 animals were hypersensitive to γ-rays, which mainly induce oxidative DNA damage and strand breaks,38 whereas xpa-1 animals were only marginally sensitive (Figures 6a and b). Additionally, ercc-1 and xpf-1, but not xpa-1 and xpg-1, animals showed hypersensitivity to nitrogen mustard (HN2), which induces monoadducts and ICLs38 (Figure 6c). These results confirm that ERCC-1, XPF-1 and XPG-1 function in different pathways, whereas XPA-1 appears more NER-specific. Importantly, these results likely explain why there is more DNA damage accumulation and consequently a stronger replicative decline in mutants defective in these multifunctional endonucleases, compared with xpa-1.
Figure 6.
DNA damage sensitivities of xpa-1, ercc-1, xpf-1 and xpg-1 mutants. (a and b) show the mean survival of mutant animals following exposure to γ-irradiation. (c) shows the mean survival upon exposure to nitrogen mustard. ercc-1 and xpf-1 animals are strongly hypersensitive to DNA damage induced by UV irradiation (Supplementary Figure S2),12 ionizing radiation and nitrogen mustard. xpg-1 animals are strongly hypersensitive to UV and ionizing radiation, whereas xpa-1 animals are only strongly hypersensitive to UV irradiation.12 Assays were performed at least two times and on independent occasions. Data for xpf-1 and ercc-1 were normalized because untreated mutants already show some embryonic lethality. Error bars denote s.e.m. Asterisks indicate significant differences compared with wild type (P=0.05)
BRC-1 and XPA-1 synergistically maintain transgenerational life span
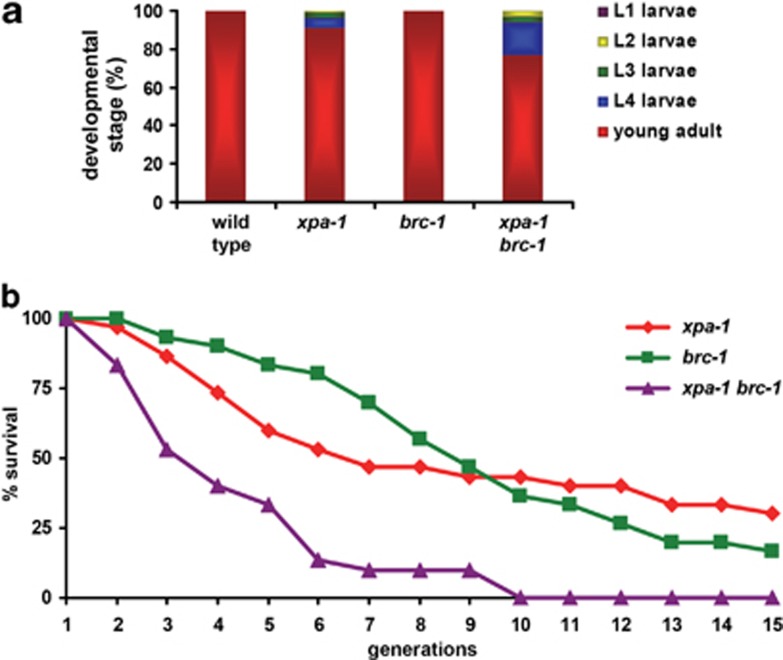
To test the notion that defects in more than one repair pathway lead to stronger developmental and transgenerational replicative failure, we crossed xpa-1 with brc-1 mutants. BRC-1 is orthologous to mammalian BRCA1 and functions in DSB repair through inter-sister meiotic recombination in C. elegans.39 Importantly, brc-1 mutants are hypersensitive to ionizing radiation and ICL-inducing agents, but not to UV irradiation (Supplementary Figure S4)30, 40 and thus resemble ercc-1/xpf-1 mutants except for UV hypersensitivity. Growth and transgenerational replicative capacity were strongly reduced in xpa-1; brc-1 double mutants, compared with their single-mutant counterparts (Figure 7 and Supplementary Figure S5). These results strongly support the hypothesis that the severe defects in ercc-1, xpf-1 and xpg-1 mutants are caused by impairment of multiple DNA-repair pathways, suggesting that these pathways collectively protect against proliferative decline induced by accumulating DNA damage in the population.
Figure 7.
xpa-1; brc-1 double mutants show enhanced developmental and replicative capacity defects. (a) depicts a quantification of the growth of wild-type, xpa-1 and brc-1 single and double mutants, by counting the larval and adult stages that are observed ∼70 h after animals are laid as eggs at 20 °C. (b) Shown is the transgenerational replicative capacity of xpa-1, brc-1 and xpa-1; brc-1 mutants. The cumulative results of two independent experiments at 25 °C are depicted
Discussion
Although C. elegans has proven its power of dissecting the genetic determinants of (post-replicative) aging,19 our analysis shows that this nematode is also well suited for a study of the consequences of DNA-repair deficiency with respect to tissue decline and (replicative) aging. These studies may provide more insight into the etiology of symptoms associated with severe hereditary syndromes caused by DNA-repair defects.
Human aging is associated with a functional decline in both replicating and non-replicating tissues. Replicative decline can be caused by telomere shortening41 (Supplementary Discussion) or other forms of stress, such as accumulation of stochastic DNA damage that physically blocks replication and induces cell cycle checkpoints.42 Direct support of DNA damage contributing to transgenerational functional decline comes from the growth and developmental arrest of germ cells, embryos, larvae and adult animals and reduction in viability that is observed after UV irradiation of C. elegans.11, 12, 16 Here, we show that mere DNA-repair deficiency also impairs replicative capacity, which we hypothesize to be caused by non-repaired endogenously produced DNA lesions. The more pronounced inter-individual variability observed in the phenotype of mutants, compared with wild-type animals, is consistent with the stochastic nature of spontaneous DNA damage, affecting different genes in each individual cell. A statistical representation of this pronounced inter-individual variation, possibly reflecting the stochastic nature of DNA damage, is the larger variance seen in the PCA analysis of ercc-1 mutants (Figures 2a and b).
DNA-damage-induced mutations in essential genes may cause a functional decline, as is observed in MMR mutants.28, 43 We observed typical visible phenotypes, such as dpy and unc, indicative of a mutator phenotype, but at a much lower frequency than the developmental and growth defects. Defects in growth and development were observed in different and independent mutant strains, even after removal of many background mutations by backcrossing against wild-type animals. Furthermore, many defects seemed to occur randomly, as if of a stochastic nature, and defects observed in one generation were not always necessarily transmitted to the next. Therefore, our observations suggest that in addition to mutations, the accumulation of DNA damage itself especially contributes to the decline in growth and fecundity.
DNA damage can directly interfere with progression of transcription and replication. In support of this, UV-induced DNA damage has been shown to impair transcription in C. elegans, leading to degradation of RNA polymerase II and developmental arrest.11 Furthermore, replication fork stalling is suggested by the RAD-51 foci observed in ercc-1, xpf-1 and xpg-1 (Figure 5). However, we do not rule out that other mechanisms could cause developmental defects or changes in gene expression. Active stress signaling by DNA damage detection proteins could be involved, as in transcriptomic reprogramming in xpa-1 mutants.18, 44 Additionally, accumulation of repair intermediates could activate checkpoint signaling,45, 46 although it is unclear whether checkpoint-activating single-stranded gaps will actually be generated in xpa-1, xpf-1 or xpg-1 mutants.47, 48 Furthermore, in replicating mammalian cells and Xenopus egg extracts, XPA was specifically implicated in UV-induced cell-cycle checkpoint activation.49 Unrepaired DNA damage in ercc-1, xpf-1 and xpg-1 mutants could enhance checkpoint signaling via XPA-1, leading to stronger phenotypes, but the similar, albeit less severe, defects in xpa-1 mutants argues against this mechanism. Finally, in mammals, XPC, XPA, XPF and XPG were found to be present at the promoters of active genes,9 and ERCC1, XPF and XPG were specifically implicated in the regulation of hepatic transcription initiation8 and regulation of gene expression,10 suggesting that a defect in transcription could contribute to observed phenotypes. Similar transcription and checkpoint functions have also been described for human BRCA1,50 permitting other interpretations of the brc-1 defects as well. In spite of all these alternatives, there is a striking correlation between severity of symptoms and repair capacity that cannot be readily ignored: xpc-1 and csb-1 mutants have the weakest phenotype and are only defective in a subpathway of NER; xpa-1 and xpc-1; csb-1 double mutants have a defect in NER and an intermediate phenotype; and xpf-1, ercc-1 and xpg-1 mutants have the most severe phenotype and are defective in multiple repair responses.
We and others30, 35, 36, 37 show that ERCC-1/XPF-1 and XPG-1 function in several DNA-repair pathways, as in mammals.31, 32, 33, 34 We hypothesize that these multiple roles cause more severe replicative stress-associated phenotypes, which is supported by the stronger phenotype of the xpa-1;brc-1 double mutant. In mammals, mutations in ERCC1/XPF can cause relatively mild symptoms, including slight photosensitivity and late-onset skin cancer, but can also cause much more extreme symptoms, including progressive neurodegeneration and segmental progeria.4, 5, 51 Similarly, minor mutations in human XPG that impair its NER function lead to XP symptoms, but truncating mutations additionally cause severe CS-like symptoms.52 Some phenotypic differences may be attributed to the different effects of mutations on protein stability, subcellular localization and/or repair functions, leading to differences in the ability to participate in one or more DNA-repair processes.53 This idea is supported by the occurrence of a case of Fanconi Anemia, published upon submission of our work,54 owing to XPF mutations that cause ICLR but not NER deficiency. Our analysis also supports this hypothesis and links involvement in more than one DNA-repair pathway to an enhanced phenotype. The connection between phenotype and level of DNA-repair deficiency further suggests that not only different DNA lesions, including helix-distorting NER substrates, but also ICL and oxidative damage can interfere with cell function and cause aging pathology.44
The lack of life-span differences between wild-type and DNA-repair mutants observed using adult life-span assays20 is in line with earlier studies of xpa-1 animals.11, 16 C. elegans may have developed strategies to preserve rather than discard damaged post-mitotic cells that are not regenerated. This is in line with the finding that DNA-damage-signaling proteins and DDR are downregulated in C. elegans somatic cells.55 Furthermore, negative effects of DNA-repair deficiency might be counteracted by compensatory SRs. This idea is supported by the striking extension of the daf-2 life span by ercc-1/xpf-1 deficiency and the strong correlation between expression profiles associated with ercc-1 and p38 MAPK stress signaling. A similar protective SR elicited by DNA-repair deficiency was observed in TC-NER-deficient mice, which show enhanced survival upon renal-ischemia-reperfusion-induced oxidative stress.56 In C. elegans, loss of germ line stem cells also extends the life span of wild-type and daf-2 mutants.57 As ERCC-1/XPF-1 function in meiotic recombination 36 (unpublished results) and affect embryogenesis (Table 1), they may therefore influence life span by affecting germ line stem cells. This is, however, unlikely because there is no life-span extension in ercc-1 and xpf-1 mutants if daf-2 is unaffected. Furthermore, germ line stem cell numbers in ercc-1 and xpf-1 mutants are not reduced (unpublished results).
Our phenotypic analysis of DNA-repair deficiency in C. elegans provides interesting clues as to the putative causes of some of the symptoms associated with mammalian NER deficiency. Importantly, our results suggest that C. elegans DNA-repair mutants display a DNA-damage-induced decline in growth and fecundity, which can be used to genetically dissect the in vivo mechanisms through which the DDR protects multicellular organisms against genome instability.
Materials and Methods
C. elegans culture
All strains were cultured according to standard methods.58 The alleles used were brc-1(tm1145), csb-1(ok2335), daf-2(e1370), ercc-1(tm2073), mus-81(tm1937), xpa-1(ok698), xpc-1(tm3886), xpf-1/him-9(e1487), xpf-1(tm2842), xpg-1(tm1670) and xpg-1(tm1682). All mutants were backcrossed three to six times every few months. Double mutants were genotyped using PCR.
Life-span assays
Standard life-span assays were performed at 20 °C as previously described.59 Day 1 was defined as the day when animals reached adulthood. Animals were scored every 1 to 3 days. To measure transgenerational replicative capacity, 15 animals per strain were singly seeded and allowed to produce progeny at 25 °C (Supplementary Figure S3A). From each of the 15 progenies, a single animal was randomly picked and transferred to a fresh plate and allowed to produce new offspring. This procedure was repeated for 15 generations. Strains were considered to have lost viability if animals exhibited arrested development, produced no or inviable progeny or died before reproduction. Animals that crawled off the plate were censored.
DNA-repair assays
UV- and ionizing-radiation-survival assays in Figure 6, Supplementary Figure S2A, Supplementary Figures S4A and S4C were performed in the same manner as the previously described ‘germ cell and embryo survival assay'.12 In brief, staged young adults were irradiated by UVB light, emitted by two Philips TL-12 40W tubes (Philips, Eindhoven, The Netherlands), or gamma-rays using a 137Cs source. After a recovery period of 24 h, animals were allowed to lay eggs for 2–3 h on fresh culture plates. The number of eggs laid was determined, and 24 h later, the number of unhatched eggs was determined, from which the survival percentage was calculated. The ‘L1 larvae UV survival' assay in Supplementary Figure S2B and S4D was previously described.12 ICL survival was measured by incubating staged young adult animals for 24 h in M9 buffer, containing nitrogen mustard (mechlorethamine hydrochloride, Sigma-Aldrich Chemie B.V., Zwijndrecht, The Netherlands) and OP50 bacteria. After a 1-h recovery period, animals were allowed to lay eggs for 3–4 h, after which survival of the progeny was scored.
Growth and phenotypic assays
To measure growth, adult animals were allowed to lay eggs for 2 h. After 69 h at 20 °C, the growth of the progeny was scored by counting all the different developmental stages. All experiments were performed multiple times in parallel and at least on two independent occasions. To measure brood size, L4 animals were singly seeded and transferred to a new plate every day. Eggs laid were counted.
Immunofluorescence and microscopy
Images in Figure 3 were acquired using a Zeiss Axio Imager.Z1 (Carl Zeiss B.V., Sliedrecht, The Netherlands). For immunofluorescence on RAD-51, gonads were dissected, freeze-cracked and fixed in 4% paraformaldehyde. Next, gonads were washed, blocked using 0.5% BSA and incubated overnight using a RAD-51 antibody (Novus, Cambridge, UK). The next day, gonads were washed, incubated with a secondary antibody (Alexa-488 conjugated, Invitrogen, Bleiswijk, The Netherlands), washed, DAPI-stained and finally mounted on a glass slide. Images in Figure 5 were acquired using a Zeiss LSM700 confocal microscope.
RNA isolation and microarray processing
Mixed-stage C. elegans ercc-1 and wild-type N2 strains were cultured on E. coli HT115(DE3) pL4440 as a food source for 3–4 days. For each of four biological replicates, worms were collected from 10 to 20 9-cm NGM plates using sucrose floatation. Starved plates were discarded. Packed worm pellets were combined with TRIZOL reagent (Invitrogen) and kept at −80 °C until further processing essentially as previously described.18 Synthesis of double-stranded cDNA, biotin-labeling, purification and fragmentation of aRNA was performed using 3' IVT express kit (Affymetrix, Santa Clara, CA, USA) according to the manufacturer's instructions. Fragmented aRNA samples were hybridized to the Affymetrix GeneChip C. elegans Genome Arrays at NTNU Genomics Resource Center (Trondheim, Norway).
Pre-processing of the raw data (Affymetrix.cel files) was done according to the standard analysis pipeline at the Bioinformatics and Expression Analysis Core Facility at Karolinska Institutet, Huddinge, Sweden (www.bea.ki.se). Briefly, .cel files were imported into Affymetrix Expression Console, pre-processed and normalized using Global Median, following background correction (PM-GCBG) and the summarization method of Plier. No outlier effects were revealed by quality control (QC) plots. The data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus60 and are accessible through GEO Series accession number GSE39145 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc= GSE39145).
Acknowledgments
We thank Ivo van Bostelen, Marcel Tijsterman, Ingrid van der Pluijm and Hanne Kim Skjeldam for support, help with experiments and advice. Some nematode strains used in this work were provided by the Caenorhabditis Genetics Center (funded by NIH National Center for Research Resources), the C. elegans Gene Knockout Consortium and the National Bioresource Project for the nematode. We thank the Association for International Cancer Research (project 08-0084; www.aicr.org.uk), the Netherlands Organization for Scientific Research (NWO; projects 863.08.022 and 912.08.031; www.nwo.nl/), the EU-funded Network of Excellence LifeSpan (FP6 036894; www.lifespannetwork.nl/), the European Research Council (advanced grant 233424; erc.europa.eu), the National Institutes of Health (AG0171242-10; www.nih.gov), the Research Council of Norway FRIBIO (www.forskningsradet.no) and Functional Genomics program, Grant PNRF-143-AI-1/07 from the Polish–Norwegian Research Fund (www.fbn.opi.org.pl) for financial support. Ø.F. was the recipient of a PhD fellowship from EMBIO (University of Oslo). The Affymetrix service was provided by the Norwegian Microarray Consortium (NMC) at the national technology platform, and supported by the functional genomics program (FUGE) of the Research Council of Norway.
Glossary
- BER
base-excision repair
- BP
biological process
- COFS
cerebro-oculo-facio-skeletal syndrome
- CS
cockayne syndrome
- DDR
DNA-damage response
- DSB
double-strand break
- FET
fisher exact tests
- GG-NER
global genome nucleotide excision repair
- HR
homologous recombination
- ICLR
inter-strand crosslink repair
- IGF
insulin growth factor
- ILS
insulin-like signaling
- MMR
mismatch repair
- NER
nucleotide-excision repair
- PCA
principal component analysis
- SR
survival response
- TC-NER
transcription-coupled nucleotide excision repair
- XP
xeroderma pigmentosum
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Edited by B Zhivotovsky
Supplementary Material
References
- Hoeijmakers JH. DNA damage, aging, and cancer. N Engl J Med. 2009;361:1475–1485. doi: 10.1056/NEJMra0804615. [DOI] [PubMed] [Google Scholar]
- Nouspikel TDNA. Repair in mammalian cells: nucleotide excision repair: variations on versatility. Cell Mol Life Sci. 2009;66:994–1009. doi: 10.1007/s00018-009-8737-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diderich K, Alanazi M, Hoeijmakers JH. Premature aging and cancer in nucleotide excision repair-disorders. DNA Repair. 2011;10:772–780. doi: 10.1016/j.dnarep.2011.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niedernhofer LJ, Garinis GA, Raams A, Lalai AS, Robinson AR, Appeldoorn E, et al. A new progeroid syndrome reveals that genotoxic stress suppresses the somatotroph axis. Nature. 2006;444:1038–1043. doi: 10.1038/nature05456. [DOI] [PubMed] [Google Scholar]
- Jaspers NG, Raams A, Silengo MC, Wijgers N, Niedernhofer LJ, Robinson AR, et al. First reported patient with human ERCC1 deficiency has cerebro-oculo-facio-skeletal syndrome with a mild defect in nucleotide excision repair and severe developmental failure. Am J Hum Genet. 2007;80:457–466. doi: 10.1086/512486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garinis GA, van der Horst GT, Vijg J, Hoeijmakers JH. DNA damage and ageing: new-age ideas for an age-old problem. Nat Cell Biol. 2008;10:1241–1247. doi: 10.1038/ncb1108-1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lans H, Hoeijmakers JH. Genome stability, progressive kidney failure and aging. Nat Genet. 2012;44:836–838. doi: 10.1038/ng.2363. [DOI] [PubMed] [Google Scholar]
- Kamileri I, Karakasilioti I, Sideri A, Kosteas T, Tatarakis A, Talianidis I, et al. Defective transcription initiation causes postnatal growth failure in a mouse model of nucleotide excision repair (NER) progeria. Proc Natl Acad Sci USA. 2012;109:2995–3000. doi: 10.1073/pnas.1114941109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le May N, Mota-Fernandes D, Velez-Cruz R, Iltis I, Biard D, Egly JM. NER factors are recruited to active promoters and facilitate chromatin modification for transcription in the absence of exogenous genotoxic attack. Mol Cell. 2010;38:54–66. doi: 10.1016/j.molcel.2010.03.004. [DOI] [PubMed] [Google Scholar]
- Le May N, Fradin D, Iltis I, Bougneres P, Egly JM. XPG and XPF endonucleases trigger chromatin looping and DNA demethylation for accurate expression of activated genes. Mol Cell. 2012;47:622–632. doi: 10.1016/j.molcel.2012.05.050. [DOI] [PubMed] [Google Scholar]
- Astin JW, O'Neil NJ, Kuwabara PE. Nucleotide excision repair and the degradation of RNA pol II by the Caenorhabditis elegans XPA and Rsp5 orthologues, RAD-3 and WWP-1. DNA Repair. 2008;7:267–280. doi: 10.1016/j.dnarep.2007.10.004. [DOI] [PubMed] [Google Scholar]
- Lans H, Marteijn JA, Schumacher B, Hoeijmakers JH, Jansen G, Vermeulen W. Involvement of global genome repair, transcription coupled repair, and chromatin remodeling in UV DNA damage response changes during development. PLoS Genet. 2010;6:e1000941. doi: 10.1371/journal.pgen.1000941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer JN, Boyd WA, Azzam GA, Haugen AC, Freedman JH, Van Houten B. Decline of nucleotide excision repair capacity in aging Caenorhabditis elegans. Genome Biol. 2007;8:R70. doi: 10.1186/gb-2007-8-5-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stergiou L, Doukoumetzidis K, Sendoel A, Hengartner MO. The nucleotide excision repair pathway is required for UV-C-induced apoptosis in Caenorhabditis elegans. Cell Death Differ. 2007;14:1129–1138. doi: 10.1038/sj.cdd.4402115. [DOI] [PubMed] [Google Scholar]
- Lans H, Vermeulen W. Nucleotide excision repair in Caenorhabditis elegans. Mol Biol Int. 2011;2011:542795. doi: 10.4061/2011/542795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd WA, Crocker TL, Rodriguez AM, Leung MC, Lehmann DW, Freedman JH, et al. Nucleotide excision repair genes are expressed at low levels and are not detectably inducible in Caenorhabditis elegans somatic tissues, but their function is required for normal adult life after UVC exposure. Mutat Res. 2010;683:57–67. doi: 10.1016/j.mrfmmm.2009.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyun M, Lee J, Lee K, May A, Bohr VA, Ahn B. Longevity and resistance to stress correlate with DNA repair capacity in Caenorhabditis elegans. Nucleic Acids Res. 2008;36:1380–1389. doi: 10.1093/nar/gkm1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fensgard O, Kassahun H, Bombik I, Rognes T, Lindvall JM, Nilsen H. A two-tiered compensatory response to loss of DNA repair modulates aging and stress response pathways. Aging (Albany, NY) 2010;2:133–159. doi: 10.18632/aging.100127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenyon CJ. The genetics of ageing. Nature. 2010;464:504–512. doi: 10.1038/nature08980. [DOI] [PubMed] [Google Scholar]
- Kenyon C, Chang J, Gensch E, Rudner A, Tabtiang R. A C. elegans mutant that lives twice as long as wild type. Nature. 1993;366:461–464. doi: 10.1038/366461a0. [DOI] [PubMed] [Google Scholar]
- van der Pluijm I, Garinis GA, Brandt RM, Gorgels TG, Wijnhoven SW, Diderich KE, et al. Impaired genome maintenance suppresses the growth hormone--insulin-like growth factor 1 axis in mice with Cockayne syndrome. PLoS Biol. 2007;5:e2. doi: 10.1371/journal.pbio.0050002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denver DR, Feinberg S, Steding C, Durbin M, Lynch M. The relative roles of three DNA repair pathways in preventing Caenorhabditis elegans mutation accumulation. Genetics. 2006;174:57–65. doi: 10.1534/genetics.106.059840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roh JY, Sim SJ, Yi J, Park K, Chung KH, Ryu DY, et al. Ecotoxicity of silver nanoparticles on the soil nematode Caenorhabditis elegans using functional ecotoxicogenomics. Environ Sci Technol. 2009;43:3933–3940. doi: 10.1021/es803477u. [DOI] [PubMed] [Google Scholar]
- Lim D, Roh JY, Eom HJ, Choi JY, Hyun J, Choi J. Oxidative stress-related PMK-1 P38 MAPK activation as a mechanism for toxicity of silver nanoparticles to reproduction in the nematode Caenorhabditis elegans. Environ Toxicol Chem. 2012;31:585–592. doi: 10.1002/etc.1706. [DOI] [PubMed] [Google Scholar]
- Troemel ER, Chu SW, Reinke V, Lee SS, Ausubel FM, Kim DH. p38 MAPK regulates expression of immune response genes and contributes to longevity in C. elegans. PLoS Genet. 2006;2:e183. doi: 10.1371/journal.pgen.0020183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SS, Kennedy S, Tolonen AC, Ruvkun G. DAF-16 target genes that control C. elegans life-span and metabolism. Science. 2003;300:644–647. doi: 10.1126/science.1083614. [DOI] [PubMed] [Google Scholar]
- Budovskaya YV, Wu K, Southworth LK, Jiang M, Tedesco P, Johnson TE, et al. An elt-3/elt-5/elt-6 GATA transcription circuit guides aging in C. elegans. Cell. 2008;134:291–303. doi: 10.1016/j.cell.2008.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degtyareva NP, Greenwell P, Hofmann ER, Hengartner MO, Zhang L, Culotti JG, et al. Caenorhabditis elegans DNA mismatch repair gene msh-2 is required for microsatellite stability and maintenance of genome integrity. Proc Natl Acad Sci USA. 2002;99:2158–2163. doi: 10.1073/pnas.032671599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolle ME, Busuttil RA, Garcia AM, Wijnhoven S, van Drunen E, Niedernhofer LJ, et al. Increased genomic instability is not a prerequisite for shortened lifespan in DNA repair deficient mice. Mutat Res. 2006;596:22–35. doi: 10.1016/j.mrfmmm.2005.11.008. [DOI] [PubMed] [Google Scholar]
- Ward JD, Barber LJ, Petalcorin MI, Yanowitz J, Boulton SJ. Replication blocking lesions present a unique substrate for homologous recombination. EMBO J. 2007;26:3384–3396. doi: 10.1038/sj.emboj.7601766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niedernhofer LJ, Odijk H, Budzowska M, van Drunen E, Maas A, Theil AF, et al. The structure-specific endonuclease Ercc1-Xpf is required to resolve DNA interstrand cross-link-induced double-strand breaks. Mol Cell Biol. 2004;24:5776–5787. doi: 10.1128/MCB.24.13.5776-5787.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Minawi AZ, Saleh-Gohari N, Helleday T. The ERCC1/XPF endonuclease is required for efficient single-strand annealing and gene conversion in mammalian cells. Nucleic Acids Res. 2008;36:1–9. doi: 10.1093/nar/gkm888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trego KS, Chernikova SB, Davalos AR, Perry JJ, Finger LD, Ng C, et al. The DNA repair endonuclease XPG interacts directly and functionally with the WRN helicase defective in Werner syndrome. Cell Cycle. 2011;10:1998–2007. doi: 10.4161/cc.10.12.15878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klungland A, Hoss M, Gunz D, Constantinou A, Clarkson SG, Doetsch PW, et al. Base excision repair of oxidative DNA damage activated by XPG protein. Mol Cell. 1999;3:33–42. doi: 10.1016/s1097-2765(00)80172-0. [DOI] [PubMed] [Google Scholar]
- Pontier DB, Tijsterman M. A robust network of double-strand break repair pathways governs genome integrity during C. elegans development. Curr Biol. 2009;19:1384–1388. doi: 10.1016/j.cub.2009.06.045. [DOI] [PubMed] [Google Scholar]
- Saito TT, Youds JL, Boulton SJ, Colaiacovo MP. Caenorhabditis elegans HIM-18/SLX-4 interacts with SLX-1 and XPF-1 and maintains genomic integrity in the germline by processing recombination intermediates. PLoS Genet. 2009;5:e1000735. doi: 10.1371/journal.pgen.1000735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youds JL, O'Neil NJ, Rose AM. Homologous recombination is required for genome stability in the absence of DOG-1 in Caenorhabditis elegans. Genetics. 2006;173:697–708. doi: 10.1534/genetics.106.056879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedberg EC, Walker GC, Siede W, Wood RD, Schultz RA, Ellenberger T. DNA Repair and Mutagenesis. ASM Press: Washington, DC; 2006. [Google Scholar]
- Adamo A, Montemauri P, Silva N, Ward JD, Boulton SJ, La Volpe A. BRC-1 acts in the inter-sister pathway of meiotic double-strand break repair. EMBO Rep. 2008;9:287–292. doi: 10.1038/sj.embor.7401167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulton SJ, Martin JS, Polanowska J, Hill DE, Gartner A, Vidal M. BRCA1/BARD1 orthologs required for DNA repair in Caenorhabditis elegans. Curr Biol. 2004;14:33–39. doi: 10.1016/j.cub.2003.11.029. [DOI] [PubMed] [Google Scholar]
- Zucchero T, Ahmed S. Genetics of proliferative aging. Exp Gerontol. 2006;41:992–1000. doi: 10.1016/j.exger.2006.06.057. [DOI] [PubMed] [Google Scholar]
- Lombard DB, Chua KF, Mostoslavsky R, Franco S, Gostissa M, Alt FW. DNA repair, genome stability, and aging. Cell. 2005;120:497–512. doi: 10.1016/j.cell.2005.01.028. [DOI] [PubMed] [Google Scholar]
- Tijsterman M, Pothof J, Plasterk RH. Frequent germline mutations and somatic repeat instability in DNA mismatch-repair-deficient Caenorhabditis elegans. Genetics. 2002;161:651–660. doi: 10.1093/genetics/161.2.651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arczewska KD, Tomazella GG, Lindvall JM, Kassahun H, Maglioni S, Torgovnick A, et al. Active transcriptomic and proteomic reprogramming in the C. elegans nucleotide excision repair mutant xpa-1. Nucleic Acids Res. 2013;41:5368–5381. doi: 10.1093/nar/gkt225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marteijn JA, Bekker-Jensen S, Mailand N, Lans H, Schwertman P, Gourdin AM, et al. Nucleotide excision repair-induced H2A ubiquitination is dependent on MDC1 and RNF8 and reveals a universal DNA damage response. J Cell Biol. 2009;186:835–847. doi: 10.1083/jcb.200902150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Driscoll M, Ruiz-Perez VL, Woods CG, Jeggo PA, Goodship JA. A splicing mutation affecting expression of ataxia-telangiectasia and Rad3-related protein (ATR) results in Seckel syndrome. Nat Genet. 2003;33:497–501. doi: 10.1038/ng1129. [DOI] [PubMed] [Google Scholar]
- Wakasugi M, Reardon JT, Sancar A. The non-catalytic function of XPG protein during dual incision in human nucleotide excision repair. J Biol Chem. 1997;272:16030–16034. doi: 10.1074/jbc.272.25.16030. [DOI] [PubMed] [Google Scholar]
- Staresincic L, Fagbemi AF, Enzlin JH, Gourdin AM, Wijgers N, Dunand-Sauthier I, et al. Coordination of dual incision and repair synthesis in human nucleotide excision repair. EMBO J. 2009;28:1111–1120. doi: 10.1038/emboj.2009.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bomgarden RD, Lupardus PJ, Soni DV, Yee MC, Ford JM, Cimprich KA. Opposing effects of the UV lesion repair protein XPA and UV bypass polymerase eta on ATR checkpoint signaling. EMBO J. 2006;25:2605–2614. doi: 10.1038/sj.emboj.7601123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullan PB, Quinn JE, Harkin DP. The role of BRCA1 in transcriptional regulation and cell cycle control. Oncogene. 2006;25:5854–5863. doi: 10.1038/sj.onc.1209872. [DOI] [PubMed] [Google Scholar]
- Gregg SQ, Robinson AR, Niedernhofer LJ. Physiological consequences of defects in ERCC1-XPF DNA repair endonuclease. DNA Repair. 2011;10:781–791. doi: 10.1016/j.dnarep.2011.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharer OD. XPG: its products and biological roles. Adv Exp Med Biol. 2008;637:83–92. doi: 10.1007/978-0-387-09599-8_9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad A, Enzlin JH, Bhagwat NR, Wijgers N, Raams A, Appledoorn E, et al. Mislocalization of XPF-ERCC1 nuclease contributes to reduced DNA repair in XP-F patients. PLoS Genet. 2010;6:e1000871. doi: 10.1371/journal.pgen.1000871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogliolo M, Schuster B, Stoepker C, Derkunt B, Su Y, Raams A, et al. Mutations in ERCC4, encoding the DNA-repair endonuclease XPF, cause Fanconi anemia. Am J Hum Genet. 2013;92:800–806. doi: 10.1016/j.ajhg.2013.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermezovic J, Stergiou L, Hengartner MO, d'Adda di Fagagna F. Differential regulation of DNA damage response activation between somatic and germline cells in Caenorhabditis elegans. Cell Death Differ. 2012;19:1847–1855. doi: 10.1038/cdd.2012.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Susa D, Mitchell JR, Verweij M, van de Ven M, Roest H, van den Engel S, et al. Congenital DNA repair deficiency results in protection against renal ischemia reperfusion injury in mice. Aging Cell. 2009;8:192–200. doi: 10.1111/j.1474-9726.2009.00463.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arantes-Oliveira N, Apfeld J, Dillin A, Kenyon C. Regulation of life-span by germ-line stem cells in Caenorhabditis elegans. Science. 2002;295:502–505. doi: 10.1126/science.1065768. [DOI] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lans H, Jansen G. Multiple sensory G proteins in the olfactory, gustatory and nociceptive neurons modulate longevity in Caenorhabditis elegans. Dev Biol. 2007;303:474–482. doi: 10.1016/j.ydbio.2006.11.028. [DOI] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.