Abstract
Cystathionine β-synthase (CBS) is a pyridoxal 5′-phosphate (PLP)-dependent enzyme that catalyzes the condensation of homocysteine with serine to generate cystathionine. Homocystinuria is an autosomal recessive disorder commonly caused by deficiency of CBS activity. Here, we characterized a novel CBS mutation (c.260C>A (p.T87N)) and a previously reported variant (c.700G>A (p.D234N)), found in Venezuelan homocystinuric patients, one nonresponsive and one responsive to vitamin B6. Both mutant proteins were expressed in vitro in prokaryotic and eukaryotic cells, finding lower soluble expression in HEK-293 cells (19% T87N and 23% D234N) compared to wild-type CBS. Residual activities obtained for the mutant proteins were 3.5% T87N and 43% D234N. Gel exclusion chromatography demonstrated a tendency of the T87N mutant to aggregate while the distribution of the D234N mutant was similar to wild-type enzyme. Using immunofluorescence microscopy, an unexpected difference in intracellular localization was observed between the wild-type and mutant proteins. While the T87N mutant exhibited a punctate appearance, the wild-type protein was homogeneously distributed inside the cell. Interestingly, the D234N protein showed both distributions. This study demonstrates that the pathogenic CBS mutations generate unstable proteins that are unable (T87N) or partially unable (D234N) to assemble into a functional enzyme, implying that these mutations might be responsible for the homocystinuria phenotype.
Keywords: Homocystinuria, cystathionine beta-synthase, immunocytochemistry, protein misfolding
1. INTRODUCTION
Cystathionine β-synthase (CBS, EC 4.2.1.22) is a pyridoxal 5′-phosphate (PLP)-dependent enzyme which catalyzes the first step in the transsulfuration pathway, i.e. the condensation of homocysteine and serine to generate cystathionine, the precursor of cysteine. Genetic alterations in the CBS gene are the most common cause of classical homocystinuria (HCU, MIM# 236200), an autosomal recessive disorder characterized by elevated plasma homocysteine levels (usually > 100 μM) and clinical manifestations in the ocular, skeletal, vascular and nervous systems. This disorder has an estimated worldwide frequency of 1:344,000, according to data registered by countries which performed neonatal screening [1]. However, this frequency is expected to be higher since the techniques currently used for homocystinuria screening only detect the most severe cases of the disease [2]. Treatment of homocystinuria is based on methionine intake restriction from the diet and on vitamin B6 (pyridoxine) supplementation, administration of betaine is also recommended in pyridoxine-nonresponsive patients. Two CBS deficiency-associated homocystinuric phenotypes, pyridoxine-responsive and nonresponsive, have been identified based on the decrease in plasma homocysteine levels by vitamin B6 treatment [3].
The active form of the human CBS enzyme, the homotetramer, is formed by 4 subunits of 63 kDa each. Each subunit has a modular organization, an N-terminal catalytic domain that contains the binding sites for the heme group (ligated by C52 and H65), PLP and the substrates; and the C-terminal regulatory domain, which binds S-adenosylmethionine (SAM), an allosteric activator [4, 5]. An activated form of the enzyme is generated upon truncation of the C-terminal regulatory domain. The resulting subunits have a molecular mass of 45 kDa and the truncated enzyme has a four-fold higher turnover number than the full-length enzyme in the absence of SAM [6, 7, 8, 9].
So far, 158 mutations have been described in the human CBS gene (MIM# 613381). Most are missense mutations (http://cbs.lf1.cuni.cz/index.php), many are located in the catalytic domain of the protein and are predicted to impair either cofactor or substrate binding or the formation of the quaternary structure of the protein. Mutations within the C-terminal domain are also capable of affecting the oligomeric state of the enzyme or may impair SAM-dependent regulation [5, 10, 11].
Analysis of mutant CBS proteins has led to some understanding of the biological basis for vitamin B6 responsiveness in homocystinuric patients [12, 13, 14, 15], the role of the heme group in the stability and regulation of the CBS activity [16, 17, 18], the autoinhibitory action of the C-terminal domain and the allosteric activation by SAM [8, 12, 16, 19]. These studies have also provided important insights into the biochemical properties of the enzyme and have contributed to the understanding of genotype-phenotype correlations.
In this study, we have characterized two mutations, c.260C>A (p.T87N) and c.700G>A (p.D234N), found in two Venezuelan homocystinuric patients, one responsive, the other nonresponsive to vitamin B6 treatment. This is the first report of the p.T87N mutation, which was found in compound heterozygosity with the p.G85R mutation on the other allele. The p.T87N mutation is located in exon 2 from the CBS gene, and resulted from a C>A alteration at nucleotide 260 in the cDNA sequence. The p.D234N mutation has been previously described (http://cbs.lf1.cuni.cz/index.php) [20, 21, 22]; however, its effect on the properties of the CBS protein have not been studied. Both mutations are located in the catalytic domain of CBS and affect the oligomeric state and enzyme activity. Furthermore, fluorescence microscopy of transfected HEK-293 cells expressing recombinant wild-type and mutant CBS proteins reveal unexpected differences in intracellular localization.
2. MATERIAL AND METHODS
2.1 Mutation Analyses
Mutations analyzed in this study were found in two homocystinuric patients after evaluating the CBS gene. Genomic DNA was extracted from whole blood and used to analyze the entire coding region and splice junctions of the CBS gene (GenBank AF042836.1) in patient samples, following protocols described by De Lucca et al. [21]. The exon 13 was also analyzed by using the primers 5′-GGAGGGTGAGGTATGAGC-3′ (sense) and 5′-CTGCCT GTAGGTGACTGGGTAC-3′ (antisense). Blood samples were obtained from both parents and two sibs of the Patient 1 to determine inherited pattern of HCU mutations. Genotypic analysis of Patient 2 has been described previously [patient number 3800 in reference 21]. Written informed consent was obtained from all the individuals analyzed in this study.
2.2 Construction of the CBS expression vectors
Human recombinant CBS proteins were expressed using two expression vectors, pGEX4T1/hCBS [6] and pcDNA4A/HisA (Invitrogen, Carlsbad, CA). The pGEX4T1/hCBS vector was used for expressing the recombinant CBS in E. coli as a gluthationine-S-transferase (GST) fusion protein. The commercial pcDNA4A/HisA vector was used for expressing recombinant CBS in HEK-293 cells and contained a poly-histidine (HIS) tag at the N-terminus. The human CBS cDNA was introduced at the KpnI restriction site present in the multiple cloning site in this vector.
The single mutations c.260C>A (p.T87N) and c.700G>A (p.D234N) were introduced into the pGEX4T1/hCBS vector using the QuikChange-II Site-Directed Mutagenesis Kit (Stratagene, Santa Clara, CA). Mutation positions are mentioned according to GenBank sequence AF042836.1 (the A of the start codon is set as nucleotide +1). Sense primers used for site-directed mutagenesis were: 5′-AGAAAATCGGGGACAACCCTATGGTCAGAA-3′ for p.T87N and 5′-CCCCCTGGCTCACTACAACACCACCGCTGATGAG-3′ for p.D234N (nucleotide changes are underlined and shown in bold font). Wild-type CBS cDNA was introduced into the pcDNA4A/HisA vector after amplification from the pHCS3 vector (generously donated by Dr. Kraus, University of Colorado Health Sciences Center, CO) using Pfu Taq Polymerase (Stratagene, Santa Clara, CA) and the following primers: 5′-CGGGTACCCATGCCTTCTGAGACCCCCCA-3′ (sense) and 5′-AGGATCCCCGGGTACCAGCGCTCC-3′ (antisense). Primers included the KpnI restriction site. Mutations p.T87N and p.D234N were also introduced by site-directed mutagenesis. All antisense primers not mentioned before had the primers complementary sense sequences. The presence of the p.T87N and p.D234N mutations and the absence of additional mutations in the entire coding CBS sequence of the constructs were verified by nucleotide sequencing.
2.3 Purification of GST-CBS expressed in E. coli
Wild-type and mutant GST-CBS proteins were purified as described previously [23]. Their heme saturation was evaluated by UV-visible spectroscopy measuring the A280nm/A428nm ratio, where a value of 1 corresponds to 100% heme content as expected for the CBS holoenzyme [24]. Samples were diluted in 50 mM TrisHCl buffer pH 8.6 to ~ 0.35 mg/mL for the wild-type (WT) and the D234N mutant, and 0.5 mg/mL for the T87N protein. Absorption spectra were recorded on a Cary 100 Bio UV-Visible spectrophotometer (Varian). Final protein preparation (10 μg) was loaded onto 10% SDS-PAGE gels according to Laemmli [25]. Gels were either stained with Coomassie blue R-250 or transferred to nitrocellulose membrane in a semidry equipment (Bio-Rad, Hercules, CA) for immunoblot analysis as described by Towbin and coworkers [26]. Chicken anti-CBS polyclonal antibody (1:10,000) and goat anti-chicken monoclonal antibody (Pierce, Rockford, IL) coupled to horseradish peroxidase (1:10,000) were used to detect the recombinant proteins (0.15 μg). Chemiluminescence detection was performed with the ECL Plus Western Blotting Detection System (Amersham, Pittsburgh, PA). Chicken anti-CBS polyclonal antibodies were purified from hen eggs immunized with human CBS following the protocol described by Polson and coworkers [27]. Protein concentration was determined by the Bradford Protein Assay Solution (Bio-Rad, Hercules, CA) using bovine serum albumin as standard.
2.4 Biochemical analyses of purified recombinant CBS proteins
2.4.1 PLP content determination
The PLP contents from wild-type and mutant proteins were determined by hydroxylamine treatment [28], using 3 μM of each protein. A Luminescence Spectrometer LS 50 (Perkin Elmer, Waltham, MA) was used to detect PLP oxime fluorescence (λexc = 353 nm, λem = 446 nm). A standard curve was generated using 0.5-6 μM PLP (Sigma-Aldrich, St. Louis, MO). All assays were carried out in triplicate and the mean ± standard error of the mean (SEM) are reported.
2.4.2 In vitro activity
The enzymatic activities from wild-type and mutant proteins (5 μg) were measured under aerobic conditions as previously described [23]. The enzymatic activity was determined in the absence or presence of the PLP cofactor (250 μM) and SAM (380 μM). Protein concentration was determined before each assay following centrifugation of the samples (5,000 × g) for 5 min at 4 °C. In addition, the final concentration of protein was adjusted using the A428nm/A280nm ratio. All assays were carried out in triplicate and the mean ± standard error of the mean (SEM) are reported.
2.4.3 Oligomeric state
Gel filtration was used to determine the oligomeric state of the purified CBS proteins (80 μg of the wild-type and D234N proteins, and 170 μg of the T87N protein), which were loaded onto a 2×70-cm Sephacryl 200 column in 50 mM Tris-HCl (pH 8), 100 mM KCl, at a flow rate of 2 mL min−1. The column was calibrated using gel filtration standards containing: thyroglobulin (670 kDa), IgG (158 kDa), ovalbumin (44 kDa), myoglobin (17 kDa), and vitamin B12 (1.35 kDa) from Bio-Rad (Hercules, CA).
2.4.4 Thermostability assays
The thermostability from recombinant proteins (wild-type and D234N CBS) was evaluated using an assay described by Evande and coworkers [8]. Briefly, the UV-visible spectrum of the protein (30-100 μg) was recorded at various temperatures (10 to 80 °C). Prior to spectral acquisition, the sample was maintained at the desired temperature for 5 min to achieve thermal equilibration. Then, the A428nm/A280nm values were plotted against temperature to determine the midpoint of thermal denaturation (Tm) after nonlinear regression analysis of the data with a Boltzmann sigmoidal fit. Thermostability was also determined by differential scanning calorimetry (DSC) using a VP-DSC instrument (MicroCal) and 20-30 μM of purified protein dialyzed overnight at 4 °C, against 10 mM HEPES (pH 8). A scan rate of 0.5 °C/min and a temperature range of 10-100 °C were used. The T87N protein had a low A428nm/A280nm value and aggregated easily with an increased in temperature; therefore, it was not possible to perform these assays. All analyses described before were carried out at least three times.
2.5 Expression of recombinant CBS proteins in mammalian cells
Cell culture and transient transfections: HEK-293 cells were maintained at 37 °C with 5% CO2 in DMEM-F12 media supplemented with 10 % fetal calf serum, 1 % glutamax, and 0.05 U.mL−1 streptomycin/50 U.mL−1 penicillin. All reagents were purchased from Gibco (Invitrogen, Carlsbad, CA). Cell cultures were dissociated with trypsin, quantified and plated in T75 flasks at 3.2×106 cells in the above media without antibiotics. The next day, cells were transfected with 15 μg of each vector using Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA) and OptiMEM (Invitrogen, Carlsbad, CA) according to the manufacturer instructions. After 48 h, cells were collected in phosphate buffered saline (137 mM NaCl, 2.7 mM KCl, 4.3 mM Na2HPO4, 1.5 mM KH2PO4), centrifuged at 400 × g for 10 min, and stored at −70°C until use. Cell pellets were resuspended in 200 μL of lysis buffer (250 mM Tris HCl pH 8.6, 150 mM NaCl, 1 mM β-mercaptoethanol, 1% Triton X-100, 0.1 mM PLP, and inhibitors protease cocktail (Sigma-Aldrich, St. Louis, MO)), and centrifuged at 13000 xg for 10 min at 4 °C to obtain the soluble and particulate fractions. These fractions were used for gel electrophoresis and Western blot analyses (5 μg of total protein from soluble fractions and 10 μL from pellet fractions). CBS recombinant proteins were detected by using mouse anti-HIS monoclonal antibody (1:6,000) (Sigma-Aldrich, St. Louis, MO) and the secondary antibody goat anti-mouse monoclonal antibody coupled to horseradish peroxidase (1:10,000) (Pierce, Rockford, IL). Anti-human CBS antibodies obtained from immunized chickens were also used as described in section 2.3. Densitometric analyses were carried out with the QuantityOne software (Bio-Rad, Hercules, CA).
2.5.1 Oligomeric state
Native PAGE was used to assess the CBS quaternary structures expressed in mammalian cells. The proteins (5 μg) were analyzed under non-denaturing conditions in a 4-20 % gradient gel (Bio-Rad, Hercules, CA) over 12 h at 4 °C. Gel was transferred to nitrocellulose membrane as described before. Immunoblotting was used to detect CBS using mouse anti-HIS monoclonal antibody or anti-human CBS antibodies, as previously described. Three independent samples of each protein were analyzed.
2.9 Immunocytochemical detection of CBS proteins in transfected HEK-293 cells
Cells transiently transfected were fixed with 4% paraformaldehyde and permeabilized with 0.25% Triton X-100. Fixed cells were incubated overnight at 4 °C with the primary antibodies. The antibodies used in this study included: mouse anti-polyhistidine (Sigma-Aldrich, St. Louis, MO) diluted 1:6000, rabbit anti-glyceraldehyde 3-phosphate dehydrogenase (GAPDH) (Cell Signal Technologies, Danvers, MA) diluted 1:50, rabbit anti-heat shock protein 70 (Hsp70) (Abcam, Cambridge, MA) and anti-calnexine (Millipore, Billerica, MA) diluted 1:50. The GAPDH, Hsp70 and calnexine proteins were used as cytoplasmic and endoplasmic reticulum markers, respectively. Secondary antibodies were incubated for 1 h at room temperature, protected from light. Goat anti-mouse antibodies conjugated to Alexa 488 or Alexa 594 (Molecular probes, Invitrogen, Carlsbad, CA) and goat anti-rabbit antibodies conjugated to Alexa 594 (Molecular probes, Invitrogen, Carlsbad, CA), diluted at 1:200 and 1:300, respectively, were used. All antibodies were diluted in 2% goat serum (NGS). Nuclei were visualized using DAPI (BD Biosciences, Bedford, MA), Golgi using bodipy FL C5-ceramide (Molecular probes, Invitrogen, Carlsbad, CA) and lysosomes using LysoTracker® Green DND-26 (Molecular probes, Invitrogen, Carlsbad, CA). Coverslips were mounted with mowiol (Calbiochem, Gibbstown, NJ). An Axio Observer Z1 inverted microscope (Carl Zeiss, Thornwood, NY) was used to visualize cells. Pictures were taken with a Photometrics cool snap HQ2 turbo 1394 camera and the RS image version 1.9.2 software (Roper Scientific, Tucson, AZ). Results were obtained from five independent experiments. Statistical analysis was done using one-way analysis of variance with the GraphPad Prism software (version 5.03).
3. RESULTS
3.1 Patients
Patient 1 is a 15 years old Venezuelan homocystinuric patient whose diagnosis was based on clinical manifestations and biochemical profile. This patient had ocular, skeletal, vascular and nervous system alterations, with severe hyperhomocysteinemia (292 μM, normal values (n.v.): <10 μM), and hypermethioninemia (164 μM, n.v.: 5-50 μM) (MD C. L. Domínguez and A. Mahfoud, personal communication). After the analysis of the CBS codifiying region, the p.T87N and the p.G85R mutations were the only alterations found in this patient. The p.T87N variant was confirmed by a second PCR with a MslI restriction enzyme digestion, as described previously in CBS mutation database website by De Lucca (http://cbs.lf1.cuni.cz/index.php).
Blood samples from relatives were obtained to determine the inheritance pattern of the mutations. The p.T87N allele was inherited from the maternal side, while the p.G85R allele came from the paternal side. The patient has two sibs with normal plasma homocysteine levels. However, one of them is heterozygous for the p.T87N mutation, with pectus carinatum and myopia, while the other sibling has no detected mutations but has pectus excavatum. On the other hand, the mother, who is heterozygous for this mutation, has intermediate hyperhomocysteinemia (20 μM) and pectus excavatum.
This patient is of Venezuelan origin until the third generation ancestry with only one foreign great grandfather. She was not compliant with dietary therapy and total plasma homocysteine level was not significantly decreased (169 μM) after one month of treatment with 10 mg of folic acid / 600 mg of vitamin B6 per day, and 600 μg of vitamin B12 every two days (MD C. L. Domínguez, personal communication).
The genotypic and phenotypic analysis of patient 2, who is homozygous for the p.D234N mutation, has been described previously [patient number 3800 in reference 21]. Briefly, this patient was diagnosed at 11 years old, showing hyperhomocysteinemia (143 μM), normal blood methionine levels, skeletal and ocular abnormalities. At the present time, this patient has a total plasma homocysteine level of 50 μM following pyridoxine treatment (MD C. L. Domínguez, personal communication).
3.2 Expression and purification of recombinant CBS proteins
Wild-type and mutant CBS were expressed in E. coli as GST fusion proteins and purified as described by Taoka and cowokers [23]. Recombinant CBS proteins were detected in soluble and particulate fractions from bacterial extracts by immunoblotting using anti-human CBS antibodies, all bands showed similar signal intensity (data not shown). However, after purification, the T87N protein was obtained at ~70% homogeneity as indicated by SDS-PAGE and immunoblotting, while the heme content of this mutant was around 9% of wild-type, as determined by UV-Visible spectroscopy (Supp. Figure 1). Although some degradation bands were observed for this mutant, the main band corresponded to full-length protein (Supp. Figure 1). Omission of partial proteolysis to remove the GST tag did not improve the GST-T87N protein yield. In contrast, the D234N mutant protein was stably expressed in E. coli at roughly equivalent levels as wild-type CBS. The SDS-PAGE and immunoblotting analyses showed around ~90% homogeneity and heme content was similar to wild-type (Supp. Figure 1).
3.3 In vitro biochemical properties of recombinant CBS proteins
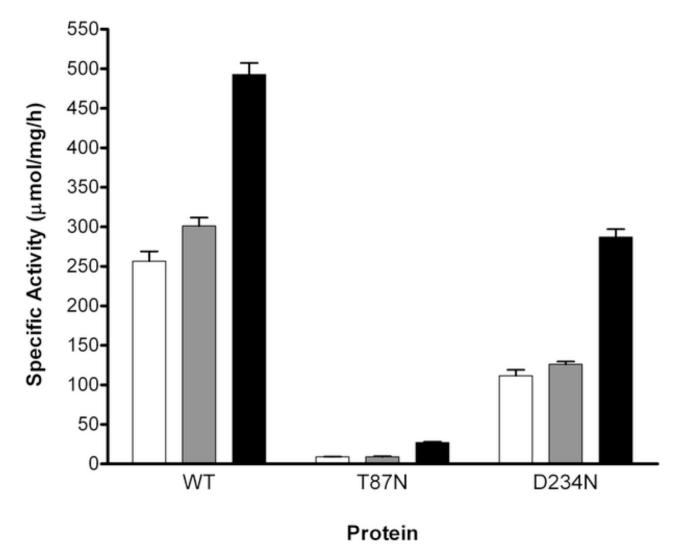
The activity of T87N and D234N mutant proteins was determined and compared to that of wild-type CBS in the absence or presence of PLP (± SAM) (Fig. 1). The activity of T87N CBS was severely affected showing only 3.5% residual activity in the absence of PLP (9.1 ± 0.2 μmol/mg/h versus 257 ± 12 μmol/mg/h for wild-type CBS). The addition of PLP to the reaction mixture did not change its activity. The D234N variant exhibited ~42% residual activity in the absence (112 ± 7 μmol/mg/h) and presence (130 ± 17 μmol/mg/h) of PLP compared to wild-type enzyme (257 ± 12 μmol/mg/h).
Figure 1.
Specific activity from recombinant CBS proteins expressed in E. coli. Proteins was used to evaluate the enzymatic activity as described in Materials and Methods. Activity is indicated as μmol of 14C-cystathionine produced per mg of protein, per hour, at 37 °C. Each bar represents the average ± SEM of at least three independent assays. The enzymatic activity was evaluated in the absence (white bars) or presence of PLP (250 μM) without SAM (grey bars), or in the presence of PLP (250 μM) and SAM (380 μM) (black bars).
The activities of both mutants increased in the presence of SAM, 3-fold for T87N (27 ± 1 μmol/mg/h) and 2½-fold for D234N (287 ± 10 μmol/mg/h), corresponding to 5.5% and 58% wild-type CBS specific activity (493 ± 15 μmol/mg/h), respectively.
Since the p.T87N and p.D234N mutations are located in the catalytic domain of CBS, the PLP content of both proteins expressed in E. coli was evaluated. The PLP content of the T87N and D234N mutants were ~6.5 ± 0.8% and 49 ± 6% respectively, when compared to wild-type.
Since the Venezuelan patient harboring the p.D234N mutation responded positively to pyridoxine treatment, the activity of this mutant was also evaluated following PLP pre-incubation (250 μM). However, the enzymatic activity of the D234N variant did not change with this treatment (111 ± 23 μmol/mg/h).
Size exclusion chromatography was used to evaluate oligomeric states of recombinant proteins. The D234N mutant behaved like wild-type CBS, eluting as a broad peak which included a mixture of oligomeric forms. However, the T87N mutant showed a sharper peak close to void volume, indicating a high tendency of this protein to aggregate [7, 29] (Supp. Fig. 2). Moreover, thermostability assays were not achieved for this mutant because this protein had a low absorbance at 428 nm and easily aggregated with an increase of temperature.
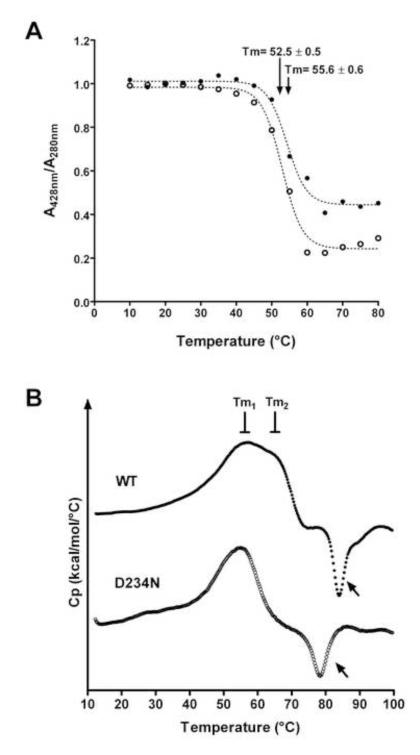
Thermostability assays was used to determine differences between the mutant D234N protein and the wild-type enzyme. When denaturation of the heme binding domain was evaluated (Fig. 2A), the D234N mutant exhibited a lower melting temperature (Tm = 52.5 ± 0.5 °C) than wild-type CBS (55.6 ± 0.6°C). DSC data also indicated a less resistance of the mutant D234N protein towards aggregation/denaturation compare to wild-type enzyme. The wild-type protein showed two transitions in the denaturation trace, at 56 °C (Tm1) and 65 °C (Tm2), whereas for the D234N mutant, only the first transition was observed (56 °C) (Fig. 2B). Additionally, both proteins appeared to aggregate at higher temperatures, since an inverted exothermic trace was seen (Fig. 2B, arrows). However, the inverted trace started at ~70 °C for the D234N protein and at ~80 °C for wild-type CBS. The CBS thermal denaturation was confirmed by native electrophoresis. The CBS samples did not migrate into the gel following DSC analyses (data not shown).
Figure 2.
Evaluation of thermostability for wild-type (•) and D234N (○) CBS proteins. A, The A428 nm/ A280nm ratio obtained for each temperature was plotted and the Tm was estimated from nonlinear regression analysis of the data using a Boltzmann sigmoidal fit (dotted lines). Each symbol represents the mean of three independent assays. B, DSC analysis of protein stability. Original thermograms after baseline subtraction are shown. The traces were vertically displaced on the y-axis to facilitate visualization of both curves. Arrows point out the inverted traces.
3.4 Expression of T87N and D234N mutant proteins in HEK-293 cells
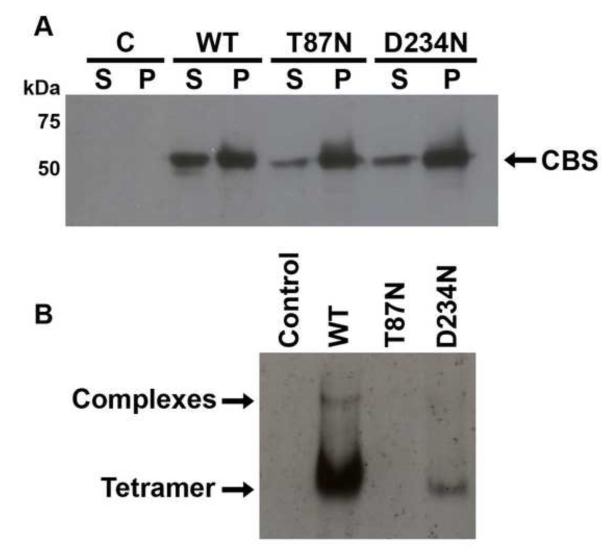
Since skin fibroblasts from patients were not available, the CBS mutants were expressed and characterized in a human cell line, HEK-293. Initially, the amount of soluble protein in cell extracts was evaluated by SDS-PAGE followed by Western blot analysis (Fig. 3A). In all cases, recombinant CBS was detected in both the soluble and particulate fractions from all cell extracts; however, the relative abundance of soluble mutant proteins was significantly lower than wild-type (19% for T87N, 23% for D234N and 46% for wild-type). Although we cannot rule out the possibility that recombinant and endogenous CBS subunits do not co-assemble in HEK-293 cells, we were unable to detect endogenous CBS in these cells.
Figure 3.
Expression of recombinant CBS proteins (WT, T87N and D234N) in transfected HEK-293 extracts. A, Western blot analysis of soluble (S) and pellet (P) fractions from HEK-293 extracts separated by SDS-PAGE. Mouse anti-polyhistidine antibodies were used to detect HIS-tagged CBS. Immunoreactive bands obtained from mutant and wild-type soluble protein fractions were compared by densitometric analyses (see text for values). B, Immunobloting of soluble fractions from cell extracts following separation by native PAGE. Control: cells transfected with the empty pcDNA4A/HisMax vector (negative control). Same results were obtained when blots were stripped and re-probed with anti-human CBS antibodies.
Next, the oligomeric structure of recombinant CBS expressed in HEK-293 cells was evaluated by native PAGE followed by Western blot analysis. CBS tetramers were observed in much lower amounts for the D234N mutant than the wild-type enzyme (Fig. 3B). Immunoreactive protein was not detected with the T87N mutant even when 50 μg of total cell extract was loaded (data not shown). A small proportion of wild-type CBS also formed multimers (Fig. 3B), as reported for human fibroblasts, liver extracts, and transfected CHO cells [2, 17, 30].
3.5 Immunocytochemical analysis of transfected HEK-293 cells
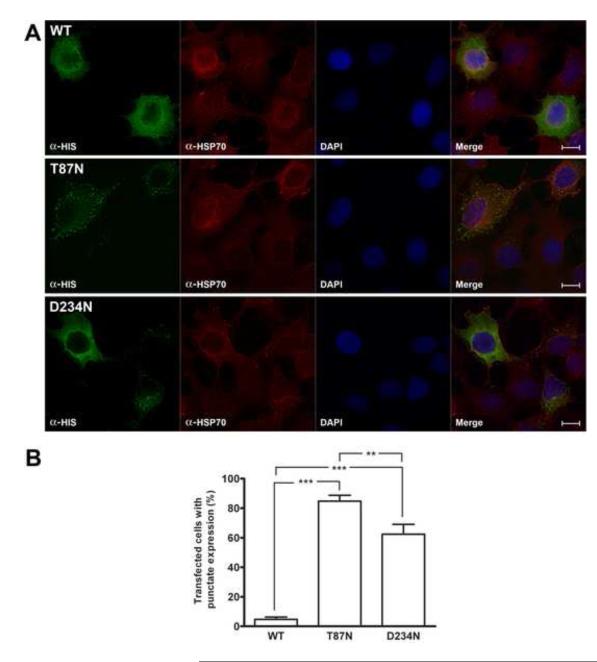
Immunocytochemistry was used to investigate the differences between recombinant wild-type and mutant CBS proteins expressed in HEK-293 cells. Immunofluorescence microscopy of transfected cells from five independent experiments showed that the transfection efficiency was ~ 30 % for all vectors used (data not shown). Surprisingly, the intracellular distribution of the CBS mutants was different from wild-type enzyme, which was homogenously distributed throughout the cell (Fig. 4A, upper panel). The T87N protein exhibited a punctate appearance, which neither co-localized with Hsp70 nor GAPDH, both used as a cytoplasmic markers (Fig. 4A, middle panel, only data from the Hsp70 marker are shown). The D234N mutant exhibited two different distribution patterns (punctuate or homogenous) (Fig. 4A, bottom panel, only data from the Hsp70 marker are shown). Similar results were obtained whether anti-HIS or anti-human CBS antibodies were used for immunocytochemical detection (data not shown). The punctate expression observed for the mutant enzymes (Fig. 4B) suggested retention in a cellular compartment, such as the endoplasmic reticulum, the Golgi apparatus, or the lysosome. To determine which of these compartments might be harboring the mutant CBS proteins, immunocytochemical localization using anti-calnexine (ER protein), bodipy FL C5-ceramide (Golgi tracer) and LysoTracker® Green DND-26 (lysosomes) were performed. Co-localization of the mutant CBS proteins with the ER, Golgi or lysosomal cell compartments were not observed (data not shown).
Figure 4.
Immunofluorescence microscopy from transiently transfected HEK-293 cells. A, Cells expressing recombinant HIS-CBS were detected by using mouse anti-polyhistidine antibody and goat anti-mouse Alexa 594 antibody (green). The cytoplasmic protein Hsp70 was immunodetected with rabbit anti-Hsp70 antibody and goat anti-rabbit Alexa 594 (red). The fourth unlabelled picture in each set is a merge of the other three images. Optical resolution 100X. Scale bar: 5 μm. B, Percentage of transfected cells with punctate expression of recombinant proteins. Results represent the average ± SEM of five independent experiments (***p< 0.001, **p< 0.01).
4. DISCUSSION
In the present study, two mutations (p.T87N and p.D234N) in the CBS protein associated with homocystinuria were characterized. Both mutations were studied in bacterial and mammalian cell expression systems. Our results suggest a possible pathogenic role for these mutations because they affected the oligomeric state, stability and enzymatic activity of the resulting CBS proteins. Patients carrying these mutations had a biochemical phenotype characteristic corresponding to classic homocystinuria. However, the p.T87N (c.260C>A) mutation has only been reported in patient 1 as a compound heterozygous (http://cbs.lf1.cuni.cz/index.php), also harboring the severe p.G85R mutation [14]; therefore, it is difficult to determine the contribution of each mutant allele to patient phenotype.
This patient presents an intermediate phenotype and shows non-responsiveness to vitamin B6 treatment, as stated by Kraus et al. [20] for other homocystinuric patients. Unexpectedly, their relatives carrying the p.T87N mutation had connective tissue abnormalities or hyperhomocysteinemia. Although, heterozygotes for CBS deficiency might have moderate hyperhomocysteinemia after methionine loading and lower enzymatic activity in cultured skin fibroblasts, none connective tissue abnormalities have been reported yet [31, 32]. In addition, fasting levels for Hcy have been shown to be elevated in heterozygous individuals with reduced concentrations of folic acid and vitamin B12 [33]; however, these parameters were not determined in this study. Therefore, we cannot rule out the contribution of some other external or genetic factors (diet, folic acid/vitamin B12 status, and synergistic heterozygosity), which might have contributed to manifestations observed in patient 1 relatives.
The p.T87N mutation affects an evolutionarily conserved residue in the CBS amino acid sequence. It is localized in a connecting loop between the second α-helix and β-sheet at the N-terminal domain. Based on the crystal structure of the truncated CBS dimer, the side chain of T87 was predicted to contribute to stabilization of the hydrophobic dimer interface [34], and interacts with Lys108 and Glu110, which helps to maintain the three dimensional structure of the catalytic core. These residues are highly conserved and form salt bridges with the side chains of two other residues, Glu239 and Arg121, preserving the type II fold from PLP-dependent enzymes [35]. Therefore, the introduction of an asparagine at position 87 might affect the interaction with the adjacent residues, interrupting the structural organization of CBS monomer and generating an unstable protein.
Our results showed that the mutant T87N protein expressed in bacteria has a tendency to form aggregates with very low residual activity, similar to other CBS N-terminal domain mutants expressed in E. coli [14, 28, 36]. Furthermore, results obtained with transfected mammalian cells, such as low amount of soluble T87N CBS enzyme, an absence of correctly assembled protein, and its confinement in what appears to be inclusion bodies, implies that this mutation has an effect on CBS protein folding.
The p.D234N mutation was first described in a heterozygous state in a homocystinuric patient from Puerto Rico with a mild phenotype in which the other allele was not characterized (http://cbs.lf1.cuni.cz/index.php) [20]. This mutation was subsequently found in homozygote state in a Venezuelan homocystinuric patient, who also presented a mild phenotype responsive to pyridoxine treatment [21]. In contrast, El-Said and coworkers [22] described a homozygous patient from Qatar who was nonresponsive to pyridoxine treatment. In our study, the purified D234N protein did not show an increase in enzymatic activity following pre-incubation with PLP, suggesting that the in vivo response to pyridoxine treatment might be complex, potentially involving modifier loci, genetic and epigenetic factors, or differences in the intracellular environment [15, 37].
The D234 residue is highly conserved in mammals, but it is replaced by non polar (Leu y Phe), basic (Arg, Lys, His), acidic (Glu), and uncharged polar (Tyr) residues in other organisms. Lee and coworkers [38] described a different mutation affecting the same codon, c.del700-702GAC (p.del234D), in three Korean patients with an intermediate clinical phenotype. When they expressed the mutation ex vivo in NIH3T3 and COS7 cells, they observed 3% residual enzymatic activity, consistent with the importance of this residue for human CBS.
Results obtained in this study support the negative effect of the p.D234N mutation on cofactor binding and protein stability. The D234N protein showed altered properties related to the active site of the enzyme (changes in kinetic parameters (Supp. Table 1), lower PLP content and activity), even though it was purified from E. coli with a similar yield and quaternary structure than wild-type CBS. However, less soluble and properly assembled subunits were observed when this protein was expressed in HEK-293 cells.
In the CBS structure, D234 is situated on α-helix 6, localized between the hydrophobic heme pocket and the active site containing PLP [34]. This structure (α-helix 6) precedes a group of amino acids, Gln222-Arg224, which are expected to interact with the serine carboxyl group through hydrogen bonds, and contribute to the stability of the aminoacrylate intermediate inside the active site [4, 39, 40]. Moreover, the spatial location of these amino acids is important for accommodating serine in the active site for the enzymatic reaction [41, 42]. As with other missense mutations [43], the substitution of an aspartic acid (negatively charged) for an asparagine (polar uncharged) at the 234 position might disrupt functional amino acid interactions, affecting the spatial arrangement of other residues, like Gln222-Arg224, potentially disrupting the hydrogen bonding network around PLP. These changes could account for the ~2½-fold lower PLP content, a decreased activity and lower Tm obtained for the mutant D234N protein in the thermal denaturation study.
Moreover, our DSC study showed that the p.D234N did not affect the regulatory domain. Pey et al. [44] demonstrated by wild-type enzyme DSC analysis that the thermal denaturation of CBS proteins follow a two independent two-state irreversible denaturation processes, observing two transitions on the thermogram, Tm1 (53 °C) and Tm2 (71 °C). Each peak corresponds to the denaturation of the regulatory domain and the catalytic domain, respectively. The Tm1 of the C-terminal domain obtained for the D234N enzyme did not change respect to the wild-type protein, while the Tm2 for the N-terminal domain of this mutant protein overlapped Tm1, observing a single transition. Pey et al. [44] also observed differences in thermal denaturation profiles between the wild-type and the CBS with missense mutations.
The p.T87N mutation had a major effect on the distribution of CBS since it appeared to be predominantly organized in punctate bodies that did not overlay with the ER, Golgi and lysosomal compartments in HEK-293. The presence of the mutants in punctate or inclusion bodies corresponds to their aggregation behavior observed in vitro, but their nature is presently unknown. Another intriguing possibility is that the punctate structures could represent complexes of CBS with other proteins akin to the purinosome observed for enzymes involved in purine biosynthesis [45]. This aggregation behavior could be responsible for the presence of the T87N protein in the soluble fraction after SDS-PAGE but not native PAGE (Fig. 3), as proposed for the mutant S466L CBS protein [30]. As observed for both mutant proteins, lower amounts of soluble proteins and proper tetramer formation correlated with punctuate intracellular distribution in eukaryotic cells.
One third of CBS mutations predispose the enzyme towards misfolding or misassembly [46], which is commonly observed in other human diseases [47]. Our results suggest that in human cells, the CBS mutants accumulate in intracellular inclusion bodies, perhaps due to their instability. Different approaches have led to improved protein folding and stability of CBS mutants including chemical [48, 49, 50] and molecular chaperones [51], suggesting that homocystinuric patients might benefit from therapies designed to improve the stability of mutant proteins. Recently, the induction of Hsp70 expression in a homocystinuric animal model showed the increase of mutant CBS activity [52].
Further immunocytochemical assays need to be performed to determine if inclusion bodies containing mutant CBS proteins co-localize with proteins from quality control systems such as the proteasome or molecular chaperons. Finally, some of the wild-type CBS protein was found in the cell periphery (data not shown) and might represent the association of recombinant CBS with other proteins in that location. Post-translational modifications of CBS protein that would account for its localization in the proximity of the plasma membrane have not been described yet.
4.1 Conclusions
This study has shown that both mutations influence the folding/assembly of CBS protein, exhibiting a similar punctate distribution in HEK-293 cells. Reduced protein solubility and wrongly sub-unit assembly for mutant variants in eukaryotic cells extracts could not be explained by co-localization with proteosomal or lysosomal markers, which would indicate retention of mutant proteins for its degradation. Therefore, other mechanisms such as association in inclusion bodies, intracellular degradation by cytosolic proteases and/or reduced mRNA translation cannot be ruled out. To our knowledge, this is the first time that immunocytochemical localization has been used to visualize CBS mutant in mammalian cells and reveals differences that might be functionally relevant.
Supplementary Material
HIGHLIGHTS.
Two mutations on CBS gene were expressed finding alterations on enzyme properties
Patients harboring these mutations had different vitamin B6 treatment response
Both mutant proteins showed less residual activities and altered PLP content
Mutant proteins showed different intracellular localization in mammalian cells
ACKNOWLEDGMENTS
We want to thank the patients and their families for their participation in this study. This work was supported by FONACIT N° 2008001053 grant and by the National Institutes of Health (HL58984 to RB). The author thanks C. Castillo, L. García, J. Nuñez, and M. Longart for their help with cell culture and microscopy assays; N. Zerpa and C. Malavé for their collaboration with the production of chicken anti-CBS antibodies; C.L. Domínguez and A. Mahfoud for supplying information about patient treatments; and all personnel from Unidad de Estudio de Errores Innatos del Metabolismo (Fundación IDEA, Venezuela) for technical assistance. We thank Dr Mark Wilson and M. Lakshminarasimhan (Redox Biology Center, University of Nebraska-Lincoln, USA) for their help with the acquisition of DSC data. C. Rizzo for methionine determination from patient samples (Ospedale Pediatrico Bambino Gesù, Italy); and J. Bubis (Universidad Simón Bolívar, Venezuela) for critical reading of the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors declare that they have no conflicts of interest in the research.
6. REFERENCES
- [1].Mudd S, Levy H, Kraus JP. Disorders of transsulfuration. In: Scriver C, Beaudet A, Sly W, Valle D, Childs B, Kinzler K, Vogelstin B, editors. The Metabolic and Molecular Bases of Inherited Disease. McGraw-Hill; New York: 2001. pp. 2007–2056. [Google Scholar]
- [2].Janosík M, Sokolová J, Janošíková B, Krijt J, Klatovská V, Kožich V. Birth Prevalence of Homocystinuria in Central Europe: Frequency and Pathogenicity of Mutation c.1105C>T (p.R369C) in the Cystathionine Beta-Synthase Gene. J Pediatr. 2009;154:431–437. doi: 10.1016/j.jpeds.2008.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Kluijtmans L, Boers G, Kraus JP, Van Den Heuvel L, Cruysberg J, Trijbels F, Blom H. The Molecular Basis of Cystathionine β-Synthase Deficiency in Dutch Patients With Homocystinuria: Effect of CBS Genotype on Biochemical and Clinical Phenotype and on Response to Treatment. Am J Hum Genetic. 1999;65:59–67. doi: 10.1086/302439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Banerjee R, Evande R, Kabil O, Ojha S, Taoka S. Reaction mechanism and regulation of cystathionine beta-synthase. Biochim Biophys Acta. 2003;1647:30–35. doi: 10.1016/s1570-9639(03)00044-x. [DOI] [PubMed] [Google Scholar]
- [5].Miles EW, Kraus JP. Cystathionine beta-synthase: structure, function, regulation, and location of homocystinuria-causing mutations. J Biol Chem. 2004;279:29871–29874. doi: 10.1074/jbc.R400005200. [DOI] [PubMed] [Google Scholar]
- [6].Shan X, Kruger WD. Correction of disease-causing CBS mutations in yeast. Nat Genet. 1998;19:91–93. doi: 10.1038/ng0598-91. [DOI] [PubMed] [Google Scholar]
- [7].Kery V, Poneleit L, Kraus JP. Trypsin Cleavage of Human Cystathionine β-Synthase into an Evolutionarily Conserved Active Core: Structural and Functional Consequences. Arch Biochem Biophys. 1998;355:222–232. doi: 10.1006/abbi.1998.0723. [DOI] [PubMed] [Google Scholar]
- [8].Evande R, Blom H, Boers GH, Banerjee R. Alleviation of intrasteric inhibition by the pathogenic activation domain mutation, D444N, in human cystathionine beta-synthase. Biochemistry. 2002;41:11832–11837. doi: 10.1021/bi026248d. [DOI] [PubMed] [Google Scholar]
- [9].Oliveriusová J, Kery V, Maclean KN, Kraus JP. Deletion mutagenesis of human cystathionine beta-synthase. Impact on activity, oligomeric status, and S-adenosylmethionine regulation. J Biol Chem. 2002;277:48386–48394. doi: 10.1074/jbc.M207087200. [DOI] [PubMed] [Google Scholar]
- [10].Yamanishi M, Kabil O, Sen S, Banerjee R. Structural insights into pathogenic mutations in heme-dependent cystathionine-beta-synthase. J Inorg Biochem. 2006;100:1988–1995. doi: 10.1016/j.jinorgbio.2006.08.020. [DOI] [PubMed] [Google Scholar]
- [11].Koutmos M, Kabil O, Smith JL, Banerjee R. Structural basis for substrate activation and regulation by cystathionine beta-synthase (CBS) domains in cystathionine β-synthase. Proc Natl Acad Sci U S A. 2010;107:20958–20963. doi: 10.1073/pnas.1011448107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Kabil O, Banerjee R. Deletion of the regulatory domain in the pyridoxal phosphate-dependent heme protein cystathionine beta-synthase alleviates the defect observed in a catalytic site mutant. J Biol Chem. 1999;274:31256–31260. doi: 10.1074/jbc.274.44.31256. [DOI] [PubMed] [Google Scholar]
- [13].Tröndle U, Sunder-Plassmann G, Burgmann H, Buchmayer H, Kramer L, Bieglmayer C, Hörl WH, Födinger M. Molecular and clinical characterization of homocystinuria in two Austrian families with cystathionine beta-synthase deficiency. Acta Med Austriaca. 2001;28:145–151. doi: 10.1046/j.1563-2571.2001.01035.x. [DOI] [PubMed] [Google Scholar]
- [14].Maclean KN, Gaustadnes M, Oliveriusova J, Janosík M, Kraus E, Kozich V, Kery V, Skovby F, Rudiger N, Ingerslev J, Stabler S, Allen R, Kraus JP. High homocysteine and thrombosis without connective tissue disorders are associated with a novel class of cystathionine beta-synthase (CBS) mutations. Hum Mutat. 2002;19:641–655. doi: 10.1002/humu.10089. [DOI] [PubMed] [Google Scholar]
- [15].Chen X, Wang L, Fazlieva R, Kruger W. Contrasting behaviors of mutant cystathionine beta-synthase enzymes associated with pyridoxine response. Hum Mutat. 2006;27:474–482. doi: 10.1002/humu.20320. [DOI] [PubMed] [Google Scholar]
- [16].Janosík M, Kery V, Gaustadnes M, Maclean KN, Kraus JP. Regulation of Human Cystathionine β-Synthase y S-Adenosyl-L-methionine: Evidence for Two Catalytically active Conformations Involving an Autoinhibitory Domain in the C-Terminal Region. Biochemistry. 2001a;40:10625–10633. doi: 10.1021/bi010711p. [DOI] [PubMed] [Google Scholar]
- [17].Janosík M, Oliveriusova J, Janosiková B, Sokolová J, Kraus E, Kraus JP, Kozich V. Impaired Heme Binding and Aggregation of Mutant Cystathionine β-Synthase Subunits in Homocystinuria. Am J Hum Genet. 2001b;68:1506–1513. doi: 10.1086/320597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Singh S, Madzelan P, Stasser J, Weeks CL, Becker D, Spiro TG, Penner-Hahn J, Banerjee R. Modulation of the heme electronic structure and cystathionine beta-synthase activity by second coordination sphere ligands: The role of heme ligand switching in redox regulation. J Inorg Biochem. 2009;103:689–697. doi: 10.1016/j.jinorgbio.2009.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Sen S, Yu J, Yamanishi M, Schellhorn D, Banerjee R. Mapping peptides correlated with transmission of intrasteric inhibition and allosteric activation in human cystathionine beta-synthase. Biochemistry. 2005;144:14210–14216. doi: 10.1021/bi051046d. [DOI] [PubMed] [Google Scholar]
- [20].Kraus JP, Janosík M, Kozich V, Mandell R, Shih V, Sperandeo MP, Sebastio G, de Franchis R, Andria G, Kluijtmans LA, Blom H, Boers GH, Gordon RB, Kamoun P, Tsai MY, Kruger WD, Koch HG, Ohura T, Gaustadnes M. Cystathionine beta-synthase mutations in homocystinuria. Hum Mutat. 1999;13:362–375. doi: 10.1002/(SICI)1098-1004(1999)13:5<362::AID-HUMU4>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- [21].De Lucca M, Casique L. Characterization of cystathionine beta-synthase gene mutations in homocystinuric Venezuelan patients: identification of one novel mutation in exon 6. Mol Genet Metab. 2004;81:209–215. doi: 10.1016/j.ymgme.2003.12.003. [DOI] [PubMed] [Google Scholar]
- [22].Elsaid MF, Bener A, Lindner M, Alzyoud M, Shahbek N, Abdelrahman MO, Abdoh G, Bessisso MS, Zschocke J, Hoffmann GF. Are heterozygotes for classical homocystinuria at risk of vitamin B12 and folic acid deficiency? Mol Genet Metab. 2007;92:100–103. doi: 10.1016/j.ymgme.2007.06.010. [DOI] [PubMed] [Google Scholar]
- [23].Taoka S, Ojha S, Shan X, Kruger W, Banerjee R. Evidence for Heme-Mediated Redox Regulation of Human Cystathionine β-Synthase Activity. J Biol Chem. 1998;273:25172–25184. doi: 10.1074/jbc.273.39.25179. [DOI] [PubMed] [Google Scholar]
- [24].Kery V, Bukovska G, Kraus JP. Transsulfuration depends on heme in addition to pyridoxal 5′-phosphate. Cystathionine β-synthase is a heme protein. J Biol Chem. 1994;269:25283–25288. [PubMed] [Google Scholar]
- [25].Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;277:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- [26].Towbin H, Staehelin T, Gordon J. Electrophoresis transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Polson A, Coetzer T, Kruger J, Von Maltzahn E, Van Der Merwe K. Improvements in the isolation of IgY from the yolks of eggs laid by immunized hens. Immunology Invest. 1985;14:323–327. doi: 10.3109/08820138509022667. [DOI] [PubMed] [Google Scholar]
- [28].Ojha S, Wu J, LoBrutto R, Banerjee R. Effects of heme ligand mutations including a pathogenic variant, H65R, on the properties of human cystathionine beta-synthase. Biochemistry. 2002;41:4649–4654. doi: 10.1021/bi011827o. [DOI] [PubMed] [Google Scholar]
- [29].Bukovska G, Kery V, Kraus JP. Expression of human cystathionine beta-synthase in Escherichia coli: purification and characterization. Protein Expr Purif. 1994;5:442–448. doi: 10.1006/prep.1994.1063. [DOI] [PubMed] [Google Scholar]
- [30].Gupta S, Wang L, Hua X, Krijt J, Kozich V, Kruger WD. Cystathionine beta-synthase p.S466L mutation causes hyperhomocysteinemia in mice. Hum Mutat. 2008;29:1048–1054. doi: 10.1002/humu.20773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Tsai MY, Garg U, Key NS, Hanson NQ, Suh A, Schwichtenberg K. Molecular and biochemical approaches in the identification of heterozygotes for homocystinuria. Atherosclerosis. 1996;122:69–77. doi: 10.1016/0021-9150(95)05748-x. [DOI] [PubMed] [Google Scholar]
- [32].Guttormsen AB, Ueland PM, Kruger WD, Kim CE, Ose L, Følling I, Refsum H. Disposition of homocysteine in subjects heterozygous for homocystinuria due to cystathionine beta-synthase deficiency: relationship between genotype and phenotype. Am J Med Genet. 2001;100:204–13. doi: 10.1002/ajmg.1247. [DOI] [PubMed] [Google Scholar]
- [33].El-Said MF, Badii R, Bessisso MS, Shahbek N, El-Ali MG, El-Marikhie M, El-Zyoid M, Salem MS, Bener A, Hoffmann GF, Zschocke J. A common mutation in the CBS gene explains a high incidence of homocystinuria in the Qatari population. Hum Mutat. 2006;27:719. doi: 10.1002/humu.9436. [DOI] [PubMed] [Google Scholar]
- [34].Meier M, Janosík M, Kery V, Kraus JP, Burkhard P. Structure of Human Cystathionine β-Synthase: a unique pyridoxal 5′-phosphate-dependent heme protein. EMBO Journal. 2001;20:3910–3916. doi: 10.1093/emboj/20.15.3910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Taoka S, Lepore BW, Kabil O, Ojha S, Ringe D, Banerjee R. Human cystathionine beta-synthase is a heme sensor protein. Evidence that the redox sensor is heme and not the vicinal cysteines in the CXXC motif seen in the crystal structure of the truncated enzyme. Biochemistry. 2002;41:10454–10461. doi: 10.1021/bi026052d. [DOI] [PubMed] [Google Scholar]
- [36].Sen S, Banerjee R. A pathogenic linked mutation in the catalytic core of human cystathionine beta-synthase disrupts allosteric regulation and allows kinetic characterization of a full-length dimer. Biochemistry. 2007;46:4110–4116. doi: 10.1021/bi602617f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Kruger WD, Wang L, Jhee KH, Singh RH, Elsas LJ., 2nd Cystathionine beta-synthase deficiency in Georgia (USA): correlation of clinical and biochemical phenotype with genotype. Hum Mutat. 2003;22:434–441. doi: 10.1002/humu.10290. [DOI] [PubMed] [Google Scholar]
- [38].Lee SJ, Lee DH, Yoo HW, Koo SK, Park ES, Park JW, Lim HG, Jung SC. Identification and functional analysis of cystathionine beta-synthase gene mutations in patients with homocystinuria. J Hum Genet. 2005;50:648–654. doi: 10.1007/s10038-005-0312-2. [DOI] [PubMed] [Google Scholar]
- [39].Burkhard P, Rao GS, Hohenester E, Schnackerz KD, Cook PF, Jansonius JN. Three-dimensional structure of O-acetylserine sulfhydrylase from Salmonella typhimurium. J Mol Biol. 1998;283:121–133. doi: 10.1006/jmbi.1998.2037. [DOI] [PubMed] [Google Scholar]
- [40].Meier M, Oliveriusova J, Kraus JP, Burkhard P. Structural insights into mutations of cystathionine beta-synthase. Biochim Biophys Acta. 2003;1647:206–213. doi: 10.1016/s1570-9639(03)00048-7. [DOI] [PubMed] [Google Scholar]
- [41].Ozaki S, Nakahara A, Sakaguchi C. Mutagenesis of Gln-142 and Phe-143 of O-acetylserine Sulfhydrylasa. Asian J Biochem. 2009a;4:117–124. [Google Scholar]
- [42].Ozaki S, Sakaguchi C, Nakahara A, Yoshiya M. Mutagenesis Studies of Human Cystathionine β-Synthase: Residues Important for Heme Binding and Substrate Interactions. Protein Pept Lett. 2009b;17:351–355. doi: 10.2174/092986610790780233. [DOI] [PubMed] [Google Scholar]
- [43].Gersting SW, Kemter KF, Staudigl M, Messing DD, Danecka MK, Lagler FB, Sommerhoff CP, Roscher AA, Muntau AC. Loss of function in phenylketonuria is caused by impaired molecular motions and conformational instability. Am J Hum Genet. 2008;83:5–17. doi: 10.1016/j.ajhg.2008.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Pey AL, Majtan T, Sanchez-Ruiz JM, Kraus JP. Human cystathionine beta-synthase (CBS) contains two classes of binding sites for S-adenosyl-L-methionine (SAM): complex regulation of CBS activity and stability by SAM. Biochem J. 2013;449:109–121. doi: 10.1042/BJ20120731. [DOI] [PubMed] [Google Scholar]
- [45].An S, Kumar R, Sheets ED, Benkovic SJ. Reversible compartmentalization of de novo purine biosynthetic complexes in living cells. Science. 2008;320:103–106. doi: 10.1126/science.1152241. [DOI] [PubMed] [Google Scholar]
- [46].Kozich V, Sokolová J, Klatovská V, Krijt J, Janosík M, Jelínek K, Kraus JP. Cystathionine beta-synthase mutations: effect of mutation topology on folding and activity. Hum Mutat. 2010;31:809–819. doi: 10.1002/humu.21273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Gregersen N, Bross P, Vang S, Christensen JH. Protein misfolding and human disease. Annu Rev Genomics Hum Genet. 2006;7:103–124. doi: 10.1146/annurev.genom.7.080505.115737. [DOI] [PubMed] [Google Scholar]
- [48].Singh LR, Chen X, Kozich V, Kruger WD. Chemical chaperone rescue of mutant human cystathionine beta-synthase. Mol Genet Metab. 2007;91:335–342. doi: 10.1016/j.ymgme.2007.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Majtan T, Liu L, Carpenter JF, Kraus JP. Rescue of cystathionine beta-synthase (CBS) mutants with chemical chaperones: purification and characterization of eight CBS mutant enzymes. J Biol Chem. 2010;285:15866–15873. doi: 10.1074/jbc.M110.107722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Kopecká J, Krijt J, Raková K, Kožich V. Restoring assembly and activity of cystathionine b-synthase mutants by ligands and chemical chaperones. J Inherit Metab Dis. 2011;34:39–48. doi: 10.1007/s10545-010-9087-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Singh LR, Kruger WD. Functional rescue of mutant human cystathionine beta-synthase by manipulation of Hsp26 and Hsp70 levels in Saccharomyces cerevisiae. J Biol Chem. 2009;284:4238–4245. doi: 10.1074/jbc.M806387200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Singh LR, Gupta S, Honig NH, Kraus JP, Kruger WD. Activation of mutant enzyme function in vivo by proteasome inhibitors and treatments that induce Hsp70. PLoS Genet. 2010;6:e1000807. doi: 10.1371/journal.pgen.1000807. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.