Abstract
Stem cells were characterized by their stemness: self-renewal and pluripotency. Mesenchymal stem cells (MSCs) are a unique type of adult stem cells that have been proven to be involved in tissue repair, immunoloregulation and tumorigenesis. Irradiation is a well-known factor that leads to functional obstacle in stem cells. However, the mechanism of stemness maintenance in human MSCs exposed to irradiation remains unknown. We demonstrated that irradiation could induce reactive oxygen species (ROS) accumulation that resulted in DNA damage and stemness injury in MSCs. Autophagy induced by starvation or rapamycin can reduce ROS accumulation-associated DNA damage and maintain stemness in MSCs. Further, inhibition of autophagy leads to augment of ROS accumulation and DNA damage, which results in the loss of stemness in MSCs. Our results indicate that autophagy may have an important role in protecting stemness of MSCs from irradiation injury.
Keywords: mesenchymal stem cells, autophagy, stemness, reactive oxygen species
Stem cells are defined by their capacity for self-renewal and multipotential differentiation, which include embryonic stem cells and adult stem cells (ASCs). Mesenchymal stem cells (MSCs) are a unique type of ASCs that originate from the mesoderm. They can differentiate into not only mesodermal-lineage cells such as osteoblasts and adipocytes but also ectodermal and endodermal cells.1, 2, 3, 4 MSCs are involved in immunoloregulation, tumorigenesis and tissue repair.
MSCs have an important role in repairing tissue injury. It has been reported that MSCs contribute to several kinds of tissue repair and function recovery by their self-renewal and multidifferentiation potential. Ionizing radiation (IR) is a form of electromagnetic radiation produced by X-ray machines, fluoroscopy, radioactive isotopes, as well as nuclear environmental catastrophe.5 The medical applications of IR have increased in recent decades.6, 7, 8 IR may induce DNA damage, chromosomal aberrations, cell cycle arrest or cell death. Moreover, exposure to irradiation causes cells to generate reactive oxygen species (ROS) and induces single-strand and double-strand DNA breaks.9, 10 An understanding of these radiobiological effects remains a challenge to seek a safe strategy to recover their capacity to repair DNA damage and maintain their stemness in MSCs exposed to irradiation.
Autophagy is characterized by the formation of a double-membrane vesicle, called autophagosome, which engulfs cytoplasmic components and delivers them to lysosomes for degradation.11 Autophagy is a conserved proteolytic mechanism that degrades cytoplasmic material, including cell organelles, and is important in maintaining intracellular homeostasis and keeping the cell healthy, which can be activated as adaptive response to adverse environmental conditions, such as deprivation of nutrients, hypoxia and different types of therapeutic stress.12, 13, 14, 15, 16, 17 Autophagy is an important event in many different kinds of cells,18 including erythrocytes,19 lymphocytes20 and monocytes.21 Mortensen et al.22 reported that autophagic mechanisms were activated in hematopoietic stem cells (HSCs). HSCs lacking autophagy-related gene ATG7, an essential autophagy protein, were unable to survive in vivo. These results suggest that autophagy is important for the stemness maintenance and differentiation in HSCs. As a consequence, we investigated the role of autophagy in preserving self-renewal and multidifferentiation potential in MSCs. Interestingly, we found that autophagy induced by nutrient deprivation or rapamycin can reverse injury in MSCs exposed to irradiation. We hypothesized that induction of autophagy could protect MSCs against irradiation and maintain their multipotential status. The purpose of this study is to investigate the role of autophagy in maintenance of stemness in MSCs during exposure to IR and the potential mechanisms.
Results
Irradiation impairs the stemness of MSCs
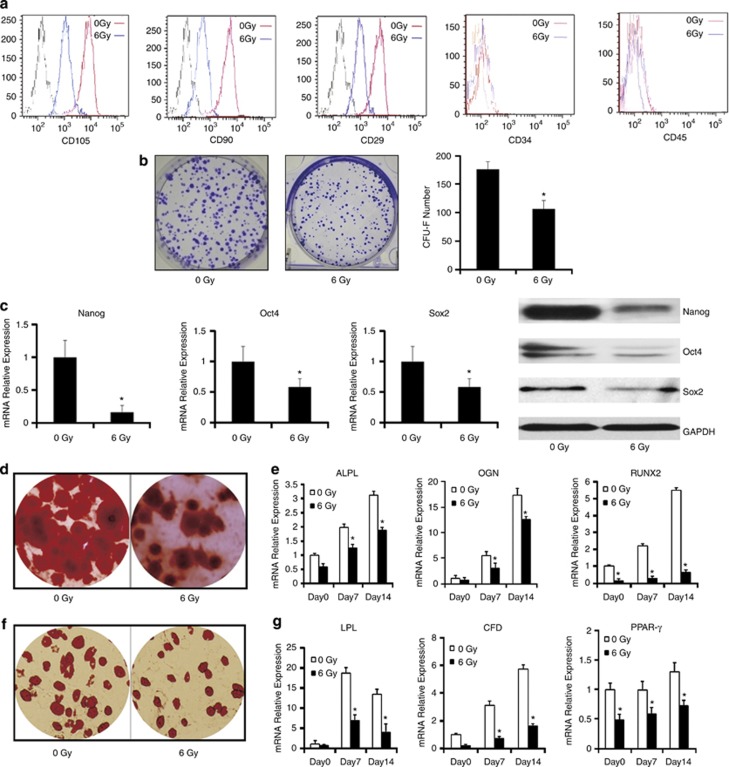
As shown in Figure 1a, MSCs were positive for specific surface markers such as CD105, CD90 and CD29, but negative for the hematopoietic cell-surface markers like CD34 and CD45. The expression of CD105, CD90 and CD29 in irradiated MSCs appeared to be less than that in control group.
Figure 1.
The effect of irradiation injury on stemness of MSCs in vitro. MSCs were irradiated at 6 Gy and collected at 6 h after irradiation. (a) MSCs surface markers in different groups were detected by flow cytometry. (b) CFU-F assays. The number of colonies was determined after 14 days of culture. (c) Real-time PCR and western blotting were employed to examine the expression of stemness markers Nanog, Oct4 and Sox2 of MSCs exposed to irradiation compared with control groups. (d) Osteogenic differentiation of MSCs was detected by Alizarin Red stain. (e) The quantitative expression of osteogenic marker genes ALPL, OGN and RUNX2 were measured by real-time PCR at 0, 7 and 14 days. (f) Adipogenic differentiation of MSCs was detected by Oil red-O. (g) The quantitative expression of adipogenesis marker genes LPL, CFD and PPAR-γ were measured by real-time PCR at 0, 7 and 14 days. The data presented are from three replicates as mean±S.E. *P<0.05
A series of the irradiation doses, from 1 to 10 Gy, were employed to determine a proper dose that has an impact on the stemness without inhibiting the proliferation in MSCs. We found that the colony formation of MSCs was effectively inhibited by 6 Gy dose irradiation while the cell viability of MSCs was not (Supplementary Figure 1). Then, apoptosis of MSCs at 0, 6, 24 and 48 h after 6 Gy irradiation was examined with PI-Annexin V-FITC assay. The results demonstrated that there was no significant effect of 6 Gy dose irradiation on apoptosis of MSCs at early stage. (Supplementary Figure 2).
Efficiency of self-renewal was assessed by the rate of colony formation in clony-forming unit-fibroblast (CFU-F) assay. Both CFU-F numbers and the mean colony size were significantly smaller in irradiated MSCs than that in control group (Figure 1b). The calculated efficiency for CFU-F varied significantly between cell cultures obtained from two groups. MSCs and irradiated MSCs possessed the CFU-F efficiency of 35.2 (±2.69) and 20.6% (±3.06%), respectively.
We were particularly interested in understanding whether pluripotency genes were also affected by irradiation. Importantly, well-known embryonic transcription factors such as Nanog, Oct4 and Sox2, which were expressed in hESCs and were critical for maintaining pluripotency and self-renewal, were also expressed in MSCs. Real-time PCR and western blotting were performed to quantify the expression of Nanog, Oct4 and Sox2 in MSCs (Figure 1c). The expression of Nanog, Oct4 and Sox2 in irradiated MSCs decreased significantly compared with the control group.
To examine the effect of irradiation on MSCs osteogenesis, MSCs were cultured with osteogenic induction medium after exposed to irradiation. MSCs were collected for 21 days, and Alizarin Red S staining was used to examine the mineralization status of the cells at the end of culture. With osteogenic induction, irradiated MSCs exhibited a less calcium deposition than that of control (Figure 1d). Real-time PCR was employed to detect the mRNA levels of ALPL, OGN and RUNX2, the osteogenic-related markers and transcription factor gene in MSCs with osteogenic induction for 0, 7 and 14 days. The ALPL, OGN and RUNX2 mRNA expression levels in both groups were gradually elevated at day 14. During the 14-day period of osteogenic induction, irradiated MSCs showed a relative lower level of ALPL and OGN compared with the control groups. Similarly, the mRNA expression level of RUNX2 markedly decreased in the irradiated MSCs group compared with control group at day 14 (Figure 1e).
The effect of irradiation on MSCs adipogenesis was also investigated. Irradiated MSCs were cultured in the adipogenic medium. After 21 days of adipogenic induction, irradiated MSCs showed remarkably reduced Oil red-O+ staining compared with control (Figure 1f). The mRNA expression of adipogenic-related markers and transcription factor LPL, CFD and PPAR-γ in the irradiated MSCs were assessed at 0, 7 and 14 days of adipogenic differentiation as well. In the irradiated MSCs group, the mRNA expression levels of LPL and CFD were significantly suppressed, whereas PPAR-γ showed slight decrease in mRNA expression of the irradiated MSCs group (Figure 1g). All the data implied that irradiation injured the self-renewal and multidifferentiation potential of MSCs.
Starvation/rapamycin reduce the injury of MSCs induced by irradiation
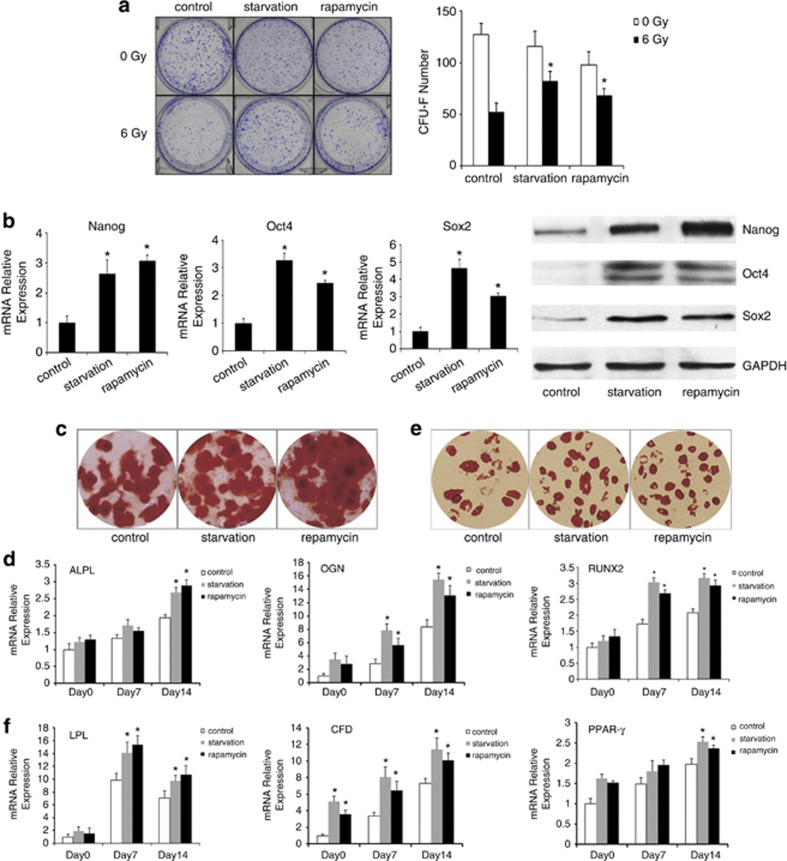
Irradiated MSCs were pretreated with starvation or rapamycin to induce autophagy. As shown in Figure 2a, the calculated efficiency for CFU-F of irradiated MSCs was lower than those of starvation- or rapamycin-pretreated group. Irradiated MSCs showed CFU-F efficiency of 10.4% (±1.72%), irradiated MSCs pretreated with starvation or rapamycin showed CFU-F efficiency of 16.4% (±1.84%) and 13.6% (±1.34%). The expression of pluripotent transcription factors Nanog, Oct4 and Sox2 were upregulated when irradiated MSCs were pretreated with starvation or rapamycin (Figure 2b).
Figure 2.
MSCs pretreated with starvation or rapamycin maintained stemness after irradiation. (a) CFU-F assays. The number of colonies was determined after 14 days of culture. (b) The expression of stemness markers Nanog, Oct4 and Sox2 of irradiated MSCs pretreated with starvation or rapamycin measured by real-time PCR and western blotting. (c) Osteogenic differentiation of irradiated MSCs pretreated with starvation or rapamycin was detected by Alizarin Red stain. (d) The quantitative expression of osteogenic marker genes ALPL, OGN and RUNX2 were measured by real-time PCR at 0, 7 and 14 days. (e) Adipogenic differentiation of irradiated MSCs pretreated with starvation or rapamycin was detected by Oil red-O. (f) The quantitative expression of adipogenesis marker genes LPL, CFD and PPAR-γ were measured by real-time PCR at 0, 7 and 14 days. The data presented are from three replicates as mean ±S.E. *P<0.05
The actual amount of calcium deposition and the mRNA expression of lineage-specific-related markers and transcription factor for osteocytes ALPL, OGN and RUNX2 were increased in the irradiated MSCs pretreated with starvation (Figures 2c and d). The induced adipocytes were increased and the mRNA expression of adipogenic markers LPL, CFD and PPAR-γ also increased in the irradiated MSCs pretreated with starvation compared with control group (Figures 2e and f). Similar results could be observed when MSCs were pretreated with rapamycin. These observations indicated that irradiated MSCs pretreated with starvation or rapamycin possess a high capacity of expansion and multilineage differentiation in vitro than those of irradiated MSCs.
Autophagy is induced by starvation or rapamycin in irradiated MSCs
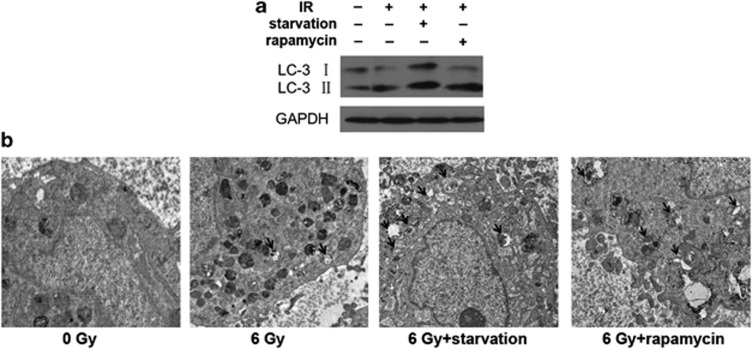
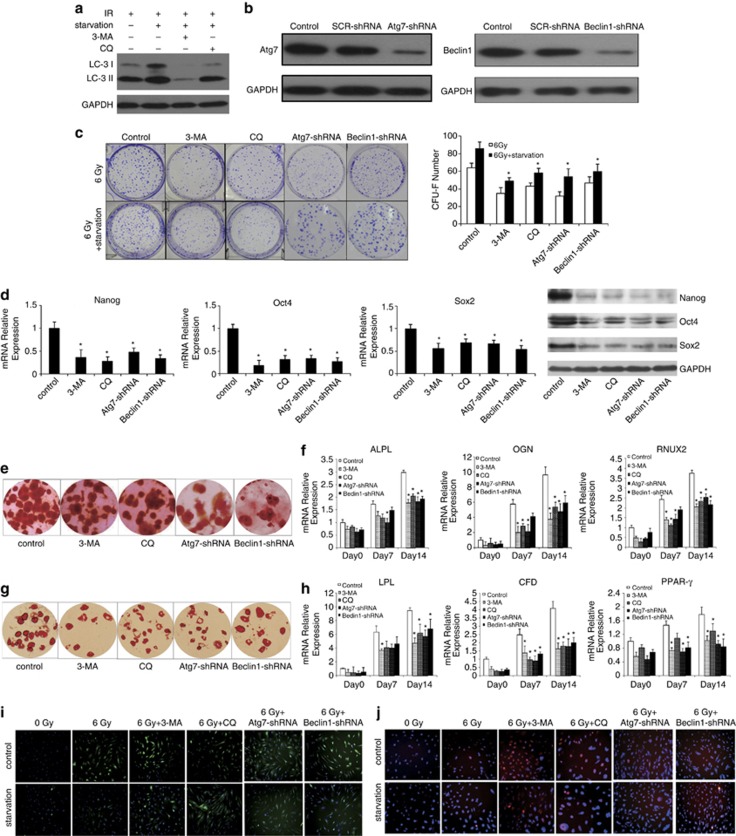
Subsequently, we investigated the autophagy in irradiated MSCs pretreated with starvation or rapamycin, a well-described inducer of autophagy. Microtubule-associated protein light chain 3 (LC3) expression is the most commonly used marker for autophagosome formation. Autophagy induction leading to LC3 is cleaved to produce LC3-I, which is localized on the membrane of autophagosomes. LC3-II is a lipidated form of LC3-I. We examined the expression of LC3-I (18 kDa) and LC3-II (16 kDa) in MSCs after irradiation by western blotting. The level of LC3-II increased slightly in irradiated MSCs and in rapamycin-pretreated groups. Meanwhile, the amount of LC3-II increased significantly in the irradiated MSCs pretreated with starvation than that in the control group (Figure 3a). Electron microscopic analysis was employed to observe autophagsome formation. The results showed the presence of characteristic double-membrane organelles in irradiated MSCs pretreated with starvation or rapamycin (Figure 3b). All of these results suggested that starvation or rapamycin induces autophagy in irradiated MSCs.
Figure 3.
Examination of autophagy in MSCs pretreated with starvation or rapamycin exposed to irradiation. (a) Total protein extracts were analyzed by western blotting with antibody against LC3. GAPDH expression was used as control. (b) Electron micrographs exhibited numerous vacuoles with cytoplasmic content in irradiated MSCs pretreated with starvation or rapamycin; most of them were clearly identified as being autophagosomes. Scale bar: 2 μm
We examined the p53 expression and activation in irradiated MSCs and found that the level of p53 activation increased in irradiated MSCs compared with control group. We also detected the level of p53 activation in irradiated MSCs pretreated with starvation. However, we observed that there was no significant difference of p53 activation between irradiated MSCs pretreated with starvation and irradiated MSCs (Supplementary Figure 3).
Autophagy contributes to maintain stemness of irradiated MSCs by ROS deletion
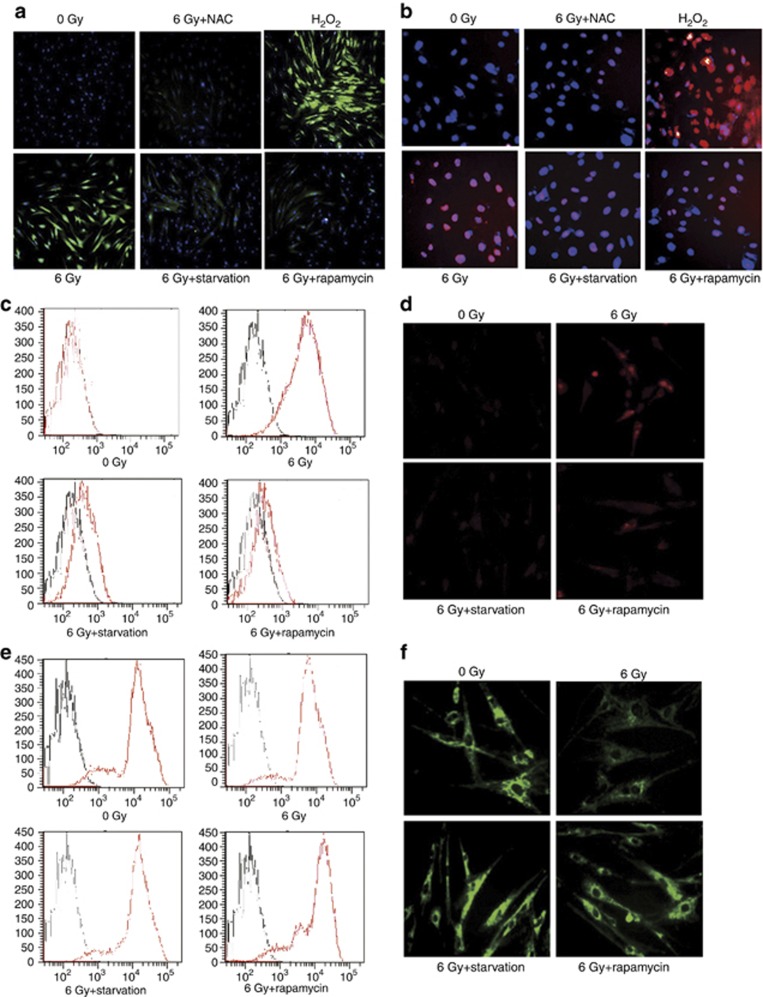
We next explored the underlying mechanism that autophagy may protect MSCs against irradiation injury. Irradiation induces ROS generation that leads to DNA damage. However, autophagy has been shown to be regulated by ROS.23, 24, 25 The loss of autophagy can induce ROS accumulation and DNA damage.26 It is possible that the elevated autophagy in irradiated MSCs may also serve as an adaptation to prevent the accumulation of ROS; hence, we detected the intracellular ROS level by using fluorescent probe dichlorofluorescin diacetate (DCF-DA). Irradiation increased the intracellular ROS level in MSCs. However, the intracellular ROS level decreased in MSCs pretreated with starvation or rapamycin, suggesting that autophagy contributes to minimize the accumulation of ROS (Figure 4a). The increase of ROS induced by irradiation was completely reversed by incubation with the antioxidant N-acetylcysteine (NAC, 2 mM). In contrast, MSCs that treated with H2O2 (100 μM) showed an increased ROS accumulation (Figure 4a). The excessive ROS would result in numerous forms of DNA damage, including DNA double-strand breaks (DSBs). γ-H2A.X has a key role in recruitment of repair proteins to the sites of DNA DSBs.27 γ-H2A.X in MSCs was examined by immunofluorescence to investigate whether DSBs were induced in MSCs exposed to irradiation. As shown in Figure 4b, the intensity of γ-H2A.X was increased in irradiated MSCs, which indicated that more DNA damage occurred in MSCs when exposed to irradiation. However, γ-H2A.X levels decreased in irradiated MSCs pretreated with starvation or rapamycin. Further, treatment with NAC could abrogate the effect of oxidative DNA damage on MSCs (Figure 4b), confirming that irradiation-induced DNA damage is mediated by ROS generation.
Figure 4.
Autophagy decreases ROS generation and alleviates DNA damage in irradiated MSCs. (a) Irradiated MSCs pretreated with starvation or rapamycin were stained with DCF-DA to determine ROS levels measured by immunofluorescence. Cell nucleus was stained with Hoechst 33258. (b) MSCs were stained with γ-H2A.X antibody to determine DNA damage. Cell nucleus was stained with DAPI. Images were captured with fluorescence microscope, magnification × 100. (c and d) Mitochondrial ROS staining with MitoSOX was measured by FACS and immunofluorescence. (e and f) MMP staining with rhodamine 123 was measured by FACS and immunofluorescence
Although ROS can be generated through non-mitochondrial mechanisms, mitochondria is the main intracellular source of ROS in most of tissues. Mitochondrial membrane potential (MMP) reflects the functional state of mitochondria; maintenance of membrane potential is an essential property of mitochondria. In addition, accumulated ROS resulted in MMP depolarization and mitochondria damage. Therefore, we assessed mitochondria ROS with MitoSOX Red (Invitrogen, Carlsbad, CA, USA; Figures 4c and d) and MMP with Rohdamin123 (Figures 4e and f) in MSCs measured by FACS and immunofluorescence. The mitochondrial ROS level in irradiated MSCs alone was higher than that in irradiated MSCs pretreated with starvation or rapamycin. MMP went depolarized in irradiated MSCs but maintained in irradiated MSCs pretreated with starvation or rapamycin. These results indicate that autophagy protects MSCs from irradiation injury by decreasing ROS and reducing DNA damage.
The protective effect from autophagy on the stemness of MSCs was suppressed by autophagy inhibitor
Autophagy was suppressed by chemical inhibitors to detect the role of autophagy in maintaining stemness of irradiated MSCs. The most extensively used autophagy inhibitor is 3-methyladenine (3-MA), which blocks the formation of autophagosomes by inhibiting the class III phosphatidylinositol 3-kinase.28 Chloroquine (CQ) is another autophagy inhibitor, which inhibits autophagy at a later step in the pathway, that caused an additional accumulation of LC3-II. The addition of 3-MA significantly reduced the expression of LC3-II, and CQ accumulated LC3-II expression in irradiated MSCs pretreated with starvation (Figure 5a). After autophagy inhibition with 3-MA and CQ, irradiated MSCs pretreated with starvation demonstrated lower CFU-F efficiency than control (Figure 5c). The expressions of Nanog, Oct4 and Sox2 were downregulated by autophagy inhibitor 3-MA and CQ in irradiated MSCs pretreated with starvation (Figure 5d). When autophagy in irradiated MSCs pretreated with starvation was deleted by autophagy inhibitor, the potential of osteogenesis and adipogenesis differentiation in MSCs decreased significantly (Figures 5e and g). Meanwhile, we also observed downregulated expression of mRNA associated with osteoblastic and adipocytic differentiation in irradiated MSCs pretreated with starvation when autophagy was inhibited (Figures 5f and h). We also inhibited autophagy in irradiated MSCs pretreated with starvation by short hairpin RNA (shRNA)-ATG7 or shRNA-Beclin1. As shown in Figures 5b–h, the expression of ATG7 and Beclin1 was significantly inhibited and the two shRNAs, respectively, diminished the protection of autophagy on MSCs stemness.
Figure 5.
Inhibition autophagy results in stemness injury and ROS increase in irradiated MSCs. (a) LC3 expression was detected by western blotting assay in irradiated MSCs pretreated with starvation by autophagy inhibitor 3-MA or CQ. (b) Irradiated MSCs pretreated with starvation were transfected with shRNAs to knockdown the autophagy-associated genes ATG7 and Beclin1. The bottom panel is a GAPDH-loading control. (c) CFU-F assays. The number of colonies was determined after 14 days of culture. (d) The expression of stemness markers Nanog, Oct4 and Sox2 of irradiated MSCs pretreated with starvation by inhibiting autophagy were measured by real-time PCR and western blotting. (e) Osteogenic differentiation of irradiated MSCs pretreated with starvation by autophagy inhibitors was stained with Alizarin Red S. (f) The expression of osteogenic markers ALPL, OGN and RUNX2 were measured by real-time PCR. (g) Adipogenic differentiation of irradiated MSCs pretreated with starvation by autophagy inhibitors were stained with Oil red-O. (h) The expression of adipogenic markers LPL, CFD and PPAR-γ were measured by real-time PCR. (i) Irradiated MSCs pretreated with starvation by autophagy inhibitors stained with DCF-DA to detect ROS level were measured by immunofluorescence. Cell nucleus was stained with Hoechst 33258. (j) Irradiated MSCs pretreated with starvation by autophagy inhibitors were stained with γ-H2A.X antibody to determine DNA damage. Cell nucleus was stained with DAPI. The data presented are from three replicates as mean±S.D. *P<0.05
Inhibition of autophagy in irradiated MSCs pretreated with starvation by autophagy inhibitor resulted in an increase in total ROS levels compared with control group (Figure 5i). Strikingly, the inhibition of autophagy resulted in an increase in DNA damage, as reflected by the greater number of γ-H2A.X foci in irradiated MSCs pretreated with starvation (Figure 5j). These data indicated that autophagy served as decreasing ROS levels and reducing DNA damage in MSCs.
Discussion
In this study, we demonstrated that autophagy had a critical role in stemness maintenance of MSCs from IR injury. We found that stemness of MSCs was impaired by irradiation, whereas this process could be reversed by autophagy induction. Further, we showed that autophagy decreased ROS accumulation, relieved DNA damage and maintained stemness in MSCs when MSCs were exposed to IR.
Stem cells were characterized by their stemness: self-renewal and pluripotency. Self-renewal was described as the proliferation of undifferentiated stem cells without lineage commitment,29 whereas pluripotency refers to the ability of stem cells differentiating into a variety of cell lineages.30 MSCs, one type of human ASCs, were identified by their capacity to adhere to plastic and generate colony-forming unit-fibroblasts (CFU-F) in culture, as well as by their potential to differentiate into multiple lineages. Stem cells need to protect their genome from damage to maintain their pool and self-renewal capacity.31 We revealed that induction of autophagy by starvation or rapamycin protected MSCs from injury induced by irradiation and maintained their stemness. When MSCs are exposed to irradiation, the self-renewal capacity and maintenance of pluripotency in culture is attenuated. Self-renewal capacity of MSCs was routinely detected by CFU-F assays. MSCs exhibited higher CFU-F efficiency, whereas irradiated MSCs possessed lower CFU-F efficiency. Autophagy inhibition could further decrease the CFU-F efficiency of irradiated MSCs. Indeed, MSCs have been reported to express a large number of genes including embryonic transcription factors Nanog, Oct4 and Sox2, which regulated the stemness of MSCs.32, 33, 34, 35 In our study, the loss of pluripotency was confirmed by significant downregulation of Nanog, Oct4 and Sox2 expression in MSCs during exposure to irradiation, which, however, was recovered by inducing autophagy. Our result demonstrated that the osteogenic and adipogenic differentiation potential of MSCs were damaged by irradiation, which was consistent with the previous study.36
Autophagy is important in maintaining intracellular homeostasis. On one hand, autophagy eliminates toxic and damaged cellular components. On the other hand, this process delivers new precursors for synthesis of macromolecules. It has been recently shown that autophagy is required for the maintenance of HSCs.22, 37 The authors showed that ATG7−/− hematopoietic stem and progenitor cells significantly accumulated more aberrant mitochondria, elevated mitochondrial superoxide levels, DNA damage and apoptosis. They also demonstrated that those cells failed to form secondary colonies in vitro. The importance of autophagy as an enhancer of ‘stemness' has been further strengthened by a study showing that autophagy is required for self-renewal and differentiation of adult human stem cells. Salemi et al.38 had shown that under conditions of autophagy blockade, epidermal stem cells, dermal stem cells and HSC underwent a loss of their self-renewal and differentiation capacities. Our results showed that the basal activity of autophagy in MSCs was very low. However, administration of rapamycin or serum-free starvation was sufficient to induce autophagy in vitro. Our data suggest that autophagy has an adaptive system to help MSCs overcoming irradiation stress. When cells are exposed to IR, decomposition reactions occur and a variety of ROS are generated.39 Various defense mechanisms have been developed to protect cells against oxidative stress, such as upregulation of antioxidants, removal of specific proteins by the ubiquitin–proteasome system and removal of damaged proteins and organelles by autophagy.40, 41 When ROS is at high cytotoxic level, the possible role of autophagy is restraining oxidative damage from reaching cytotoxic levels as well as maintaining energy homeostasis. Jang et al.42 have reported that a low oxygen niche in the bone marrow limits ROS production, resulting in better protection and higher self-renewal potential for HSCs. An increase of ROS level in irradiated MSCs has been shown in Figure 4. We also found an increasing frequency in DNA damage in irradiated MSCs. Therefore, we assume that increasing ROS levels induced by irradiation leads to DNA damage, thus, destroys the stemness and multipotential of MSCs.
Most ROS was generated in cells by the mitochondrial respiratory chain.43 ROS production by mitochondria can act as signaling molecules regulating a variety of cellular functions, including cell survival or senescence.44, 45 Genetically engineered mouse models have provided abundant evidence for the important role of autophagy in mitochondrial integrity and ROS control. For example, AMPK- and ULK1- (Atg1 homolog) knockout liver exhibits p62 accumulation and defective mitophagy.46 Downstream of Atg1, FIP200(Atg17 homolog) knockout livers exhibit significantly increase in the mitochondrial mass and ROS.37 Ubiquitinated proteins and ROS accumulate in Atg5- and Atg7-knockout mice or cells.47, 48, 49, 50 Autophagy is well known in mitochondrial quality control by degrading damaged mitochondria.51 The reduction in MMP could reflect an accumulation of damaged mitochondria. Hence, we assessed the mitochondrial ROS and MMP by FACS and immunofluorescence. The results demonstrated induction of autophagy in irradiated MSCs reduced mitochondrial ROS generation and maintained MMP. These data suggest that autophagy may degrade defective mitochondria, prevent excessive ROS generation that could induce DNA damage and protect MSCs from irradiation injury.
We provide evidence that irradiation impair the stemness of MSCs by inducing ROS accumulation. Autophagy can delete ROS and reduce DNA damage in irradiated MSCs. These results indicate that autophagy serves as a protective mechanism for MSCs stemness when they are exposed to irradiation. Autophagy-inducing agents may be used as candidates for protecting stem cells from irradiation injury.
Materials and Methods
Culture of human MSCs
Human MSCs from umbilical cord (a gift from Institute of Health Sciences and Shanghai Institute of Immunology, Chinese Academy of Sciences, Shanghai, China;)52 were cultured in Dulbecco's modified Eagle's medium low glucose (Gibco, Los Angeles, CA, USA) with 10% fetal bovine serum (FBS, Gibco) and used for experiments.
Irradiation procedure
MSCs were irradiated with ELEKTA Synergy Linear Accelerator (Cravoley, UK) at 6 Gy (a dose rate of 350 cGy/min) over an appropriate field size in the Irradiation Centre of the Eastern Hepatobiliary Surgery Hospital. Control cells were removed from the incubator at the same time without radiation exposure.
We replaced the culture medium with serum and antibiotic-free culture medium 6 h before irradiation. CQ (10 μM) or 3-MA (2 mM) was added into culture medium 2 h before irradiation. In another group, 2 h before irradiation, we added rapamycin (200 nM). After irradiation, the medium was removed and changed to standard culture medium. Cells were incubated at atmospheric conditions of 5% CO2.
Flow cytometry
Six hours after irradiation, surface markers for differently treated MSCs were quantified by flow cytometry using anti-CD105-FITC, anti-CD90-FITC, anti-CD29-FITC, anti-CD34-PE and anti-CD45-PE (Biolegend, San Diego, CA, USA) antibodies. Briefly, 1 × 105 cells were washed twice with phosphate-buffered saline (PBS), resuspended in 100 μl of PBS containing monoclonal antibodies and incubated for 30 min at 4 °C. The cells were then washed twice and resuspended in 300 μl of PBS. Fluorescence analysis was performed with a flow cytometer (FACS Aria, BD, Franklin Lakes, NJ, USA).
Cell counting Kit-8
The measurement of viable cell mass was performed with Cell Counting Kit-8 (Dojindo, Kumamoto, Japan), and living cells were counted with WST-8. Cells (5 × 103 cells per well) were first seeded in 96-well plates for irrdiation. After 72 h, 10 μl solution from Cell Counting Kit-8 was added to each well. These plates were continuously incubated for 2 h in a humidified CO2 incubator at 37 °C. Finally, the absorbance of sample taken from each well was measured on a microplate reader (Synergy HT, Bio-Tek, Vinooski, VT, USA) at 450 nm.
Cell apoptosis assay
For cell apoptosis analysis, ∼1 × 106 cells were collected. PI-Annexin V-FITC assay was used to measure apoptotic cells by flow cytometry according to the manufacturer's instructions (Nanjing Keygen Biotech, Nanjing, China). Briefly, cells were collected by trypsinization, washed with ice-cold PBS twice and resuspended in 300 μl 1 × binding buffer containing 5 μl Annexin V and 5 μl PI for 30 min at room temperature in the dark. After incubation, at least 10 000 cells were measured on a BD FACS Aria flow cytometer.
Colony formation assay
Colony-forming unit-fibroblast (CFU-F) assay was performed to assess the capacity and efficiency for self-renewal. MSCs were seeded at a density of 50 cells/cm2 in triplicates. After 14 days, cells were washed with PBS, fixed in methanol and stained with 2% crystal violet for 10 min. Excess stain was removed with PBS. Stained colonies made up of more than 50 cells were scored as CFU and were counted.
MSCs differentiation assays
Differentiation of MSCs into adipogenic and osteogenic lineages were verified following irradiation. Cells were seeded in 24-well culture plates (2 × 104 cells per well). Six hours after irradiation, MSCs were cultured with specialized media according to the desired differentiation.
Osteogenic differentiation
MSCs were treated with osteogenic culture medium to induce osteogenic differentiation (Cyagen, Santa Clara, CA, USA) for 21 days, with medium being changed every 3 days. Osteogenic medium consisted of 10% FBS, 100 U/ml penicillin, 100 μg/ml streptomycin, 0.2 mM ascorbate, 10 mM β-glycerolphosphate and 0.1 μM dexamethasone. At the end of osteogenic induction, cells were washed with PBS and fixed in 4% formaldehyde, and then stained with Alizarin Red S (Sigma-Aldrich, St. Louis, MO, USA) to access osteogenic differentiation.
Adipogenic differentiation
For the first 3 days, MSCs were cultivated in adipogenic differentiation medium A (induction medium,) with 10% FBS, 100 U/ml penicillin, 100 μg/ml streptomycin, 1 μM dexamethasone, 100 μM indomethacin, 0.5 mM methyisobutylxanthine and 10 μM insulin. Then the cells were cultured in differentiation medium B (maintenance medium) with 10% FBS, 100 U/ml penicillin, 100 μg/ml streptomycin and 5 μg/ml insulin for 24 h, and then the medium was changed to differentiation medium A. After three cycles of induction/maintenance, cells were cultivated in adipogenic differentiation medium B for additional 7 days by replacing the medium every 3 days (all supplements from Cyagen). For evaluation of adipocytes, cells were fixed in 4% formaldehyde, washed with PBS and stained with 0.2% Oil red-O Solution (Sigma, St. Louis, MO, USA) to detect lipid droplets within the cells.
Real-time PCR
Total RNA was isolated by using Trizol reagent (Invitrogen) according to the manufacturer's specifications. cDNA was reverse-transcribed using the RevertAid RT-PCR system (Fermentas, Pittsburgh, PA, USA). Real-time PCR was performed by mixing cDNA with primers and Maxima SYBR Green qPCR Master Mix (Applied Biosystems, Carlsbad, CA, USA). Real-time PCR was performed using a Stratagene Mx3000P Real-time PCR System with supplied software (Applied Biosystems), according to the manufacturer's instructions. RNA expression levels were compared after normalization with GAPDH. The primer sequences used in this study are listed in Table 1.
Table 1. List of primers used for real-time PCR (GAPDH was used as an internal control).
| Gene product | Primers product |
|---|---|
| ALPL | Fw: 5′-TGCTCTGCGCAGGATTGGAACA-3′ |
| Rv: 5′-AGGCAGGTGCCAATGGCCAGTA-3′ | |
| RUNX2 | Fw: 5′-ACAGAACCACAAGTGCGGTGCA-3′ |
| Rv: 5′-TGGTGTCACTGTGCTGAAGAGGC-3′ | |
| OGN | Fw: 5′-TGCCTTGATAGGAGGAAAACA-3′ |
| Rv: 5′-GATCCCCAAAAGCATTTAAGG-3′ | |
| LPL | Fw: 5′-TTACCCAGTGTCCGCGGGCT-3′ |
| Rv: 5′-AGACGACTCGGGGCTTCTGCA-3′ | |
| PPAR-γ | Fw: 5′-CGCCGTGGCCGCAGATTTGAA-3′ |
| Rv: 5′-AGAGATC CACGGAGCTGATCCCA-3′ | |
| CFD | Fw: 5′-TCTACACCCGCGTGGCGAGCTATGC-3′ |
| Rv: 5′-GACTTCATTGCTCGGGACTTTGTTG-3′ | |
| NANOG | Fw: 5′-CATTCTCCTGCCTCAGCCTC-3′ |
| Rv: 5′-CCCGTCTCTACTAAAAATACAAAA-3′ | |
| OCT4 | Fw: 5′-CAGCCTCTTGAGTAGCTGGG-3′ |
| Rv: 5′-GTCAGGAGTTCGAGACCAGC-3′ | |
| SOX2 | Fw: 5′-GGCAGCTACAGCATGATGCAGGAGC-3′ |
| Rv: 5′-CTGGTCATGGAGTTGTACTGCAGG-3′ | |
| GAPDH | Fw: 5′-AGAAGGCTGGGGCTCATTTG-3′ |
| Rv: 5′-AGGGGCCATCCACAGTCTTC-3′ |
ROS levels and MMP detection
Cellular peroxide levels were assessed by staining with 5 μM DCF-DA (Sigma) for 30 min at 37 °C, cell nucleus was stained with Hoechst 33258 (1 μM, Sigma) for 20 min and analyzed by using a fluorescence microscope. For detection and quantification of mitochondria-derived ROS, MSCs were loaded with a fluorogenic probe MitoSOX Red (2.5 μM, Invitrogen) for 10 min. Flow cytometer was performed to measure MitoSOX Red fluorescence in MSCs. For simultaneous measurement of MMP, MSCs were stained with rhodamine 123 (5 μg/ml, Sigma) in the cultures for 30 minutes at 37 °C. MSCs were observed under fluorescence microscope and quantified by flow cytometer.
Staining for γ-H2A.X
For evaluating the effect of irradiation on DNA damage, we assayed the expression of γ-H2A.X in MSCs. MSCs were washed with PBS before fixation in 4% (v/v) paraformaldehyde at 6 h after irradiation. Cells were permeabilized with 0.1% Triton X-100 in TBS, and blocked using TBS containing 1% BSA and 10% goat serum for 1 h at room temperature. Cells were then stained using an anti-γH2AX rabbit monoclonal antibody (1 : 200, Cell Signaling, Boston, MA, USA) overnight at 4 °C. Thereafter, the cells were washed and incubated with goat anti-rabbit IgG antibodies (1 : 400, Invitrogen) for 1 h at 4 °C. Cell nucleus was subsequently stained with DAPI. After washing and being mounted, the cells were examined under a fluorescence microscope (Olympus, Tokyo, Japan).
Western blotting analysis
MSCs were washed in cold-buffered PBS and lysed in RIPA buffer with 1 mM PMSF on ice. Cell lysates were centrifuged (12 000 r.p.m., 10 min) at 4 °C, and the protein supernatant was transferred into new tubes. The concentration of the protein samples was determined with BCA Protein Assay Kit (Pierce, Rockford, IL, USA). A 20-μg sample of the total protein was resolved using 12% SDS-PAGE and transferred onto PVDF membranes. The membranes were blocked in Tris-buffered saline containing Tween 20 with 5% nonfat milk at room temperature for 2 h. Primary antibodies to detect LC3 (1 : 1000, Novus Biologicals, Inc, Littleton, CO, USA), ATG7 (1 : 2000, Novus Biologicals, Inc) and Beclin1 (1 : 2000, Novus Biologicals, Inc) were incubated overnight with the membranes at 4 °C. Membranes were incubated with horseradish peroxidase-conjugated anti-rabbit secondary antibodies (1 : 8000, Dako, Carpinteria, CA, USA), and proteins were detected by enhanced chemiluminescence (Beyotime, Shanghai, China). GAPDH was used as the internal control to normalize the loading materials.
Electron microscopy
MSCs were fixed in a solution of 2.5% glutaraldehyde acid in 0.1 M PBS buffer (pH 7.4) for 2 h at room temperature, then incubated in 1% osmium tetroxide in 0.1 M PBS buffer (pH 7.4) for 2 h, dehydrated in solutions of ethanol and acetone, embedded in Araldite (Basel, Switzerland) and finally solidified. Fifty nanometer sections were cut on a LKB-I ultramicrotome and picked up on copper grids, post-stained with uranyl acetate and lead citrate, and examined in a Philips CM-120 transmission electron microscopy (Eindhoven, The Netherlands).
Short hairpin RNA
shRNA candidate target sequences to Atg7 is 5′-CCAAGGTCAAAGGACGAAGAT-3′ and to Beclin1 is 5′-GCAGATGAGGAAGATCGCCTT-3′. The oligonucleotides encoding the shRNA sequence were inserted into the GFP express vector pGCL-GFP (Shanghai GeneChem, Shanghai, China). SCR-shRNA was used as a negative RNAi control. The recombinant virus was packaged using Lentivector Expression Systems (Shanghai GeneChem). Irradiated MSCs preteated with starvation were infected. Then the cells were observed under fluorescence microscope and collected at 48 h after transfection.
Statistical analysis
All experiments were performed at least in triplicate. All data are presented as mean±S.D. of the replicates. Statistical analysis was performed by Student's t-test (two-tailed) using the Microsoft Excel Analysis Tool Pak (Microsoft, Redmond, WA, USA). Value of P<0.05 was considered statistically significant.
Acknowledgments
This project was supported by Key Basic Research Project of China (Grant number 2012CBA01303, 2011CB966200, 2010CB945600, 2011CB965100); Key project of National Natural Science Foundation of China (Grant number 81030041); National Natural Science Foundation of China (Grant number 31171321, 81101622); Special Funds for National key Sci-Tech Sepcial Project of China (Grant number 2012ZX10002-016, 2012ZX10002011-011); Shanghai Science and Technology Committee (Grant number 11ZR1449500, 12ZR1439800, 11nm0504700); Shanghai Municipal Health Bureau (Grant number XYQ2011044, 20114004) and Science Fund for Creative Research Groups, NSFC, China (Grant number 81221061).
Glossary
- ASCs
adult stem cells
- ATG
autophagy-related homolog
- CFU-F
clony-forming unit-fibroblast
- CQ
chloroquine
- FBS
fetal bovine serum
- HSC
hematopoietic stem cell
- IR
ionizing radiation
- LC3
microtubule-associated protein-1 light chain 3
- 3-MA
3-methyladenine
- MMP
mitochondrial membrane potential
- MSCs
mesenchymal stem cells
- NAC
N-acetylcysteine
- ROS
reactive oxygen species
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Disease website (http://www.nature.com/cddis)
Edited by A Finazzi-Agró
Supplementary Material
References
- Prockop DJ. Marrow stromal cells as stem cells for nonhematopoietic tissues. Science. 1997;276:71–74. doi: 10.1126/science.276.5309.71. [DOI] [PubMed] [Google Scholar]
- Granero-Molto F, Weis JA, Longobardi L, Spagnoli A. Role of mesenchymal stem cells in regenerative medicine: application to bone and cartilage repair. Expert Opin Biol Ther. 2008;8:255–268. doi: 10.1517/14712598.8.3.255. [DOI] [PubMed] [Google Scholar]
- Salem HK, Thiemermann C. Mesenchymal stromal cells: current understanding and clinical status. Stem Cells. 2010;28:585–596. doi: 10.1002/stem.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dezawa M, Ishikawa H, Itokazu Y, Yoshihara T, Hoshino M, Takeda S, et al. Bone marrow stromal cells generate muscle cells and repair muscle degeneration. Science. 2005;309:314–317. doi: 10.1126/science.1110364. [DOI] [PubMed] [Google Scholar]
- Wilson KD, Sun N, Huang M, Zhang WY, Lee AS, Li Z, et al. Effects of ionizing radiation on self-renewal and pluripotency of human embryonic stem cells. Cancer Res. 2010;70:5539–5548. doi: 10.1158/0008-5472.CAN-09-4238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer J, Czito B, Yin FF, Willett C. Advanced radiation therapy technologies in the treatment of rectal and anal cancer: intensity-modulated photon therapy and proton therapy. Clin Colorectal Cancer. 2007;6:348–356. doi: 10.3816/CCC.2007.n.003. [DOI] [PubMed] [Google Scholar]
- Schulz-Ertner D. The clinical experience with particle therapy in adults. Cancer J. 2009;15:306–311. doi: 10.1097/PPO.0b013e3181b01922. [DOI] [PubMed] [Google Scholar]
- Pepek JM, Willett CG, Czito BG. Radiation therapy advances for treatment of anal cancer. J Natl Compr Canc Netw. 2010;8:123–129. doi: 10.6004/jnccn.2010.0008. [DOI] [PubMed] [Google Scholar]
- Wang Y, Liu L, Pazhanisamy SK, Li H, Meng A, Zhou D. Total body irradiation causes residual bone marrow injury by induction of persistent oxidative stress in murine hematopoietic stem cells. Free Radic Biol Med. 2010;48:348–356. doi: 10.1016/j.freeradbiomed.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadet J, Delatour T, Douki T, Gasparutto D, Pouget JP, Ravanat JL, et al. Hydroxyl radicals and DNA base damage. Mutat Res. 1999;424:9–21. doi: 10.1016/s0027-5107(99)00004-4. [DOI] [PubMed] [Google Scholar]
- Klionsky DJ. Autophagy: from phenomenology to molecular understanding in less than a decade. Nat Rev Mol Cell Biol. 2007;8:931–937. doi: 10.1038/nrm2245. [DOI] [PubMed] [Google Scholar]
- Klionsky DJ, Emr SD. Autophagy as a regulated pathway of cellular degradation. Science. 2000;290:1717–1721. doi: 10.1126/science.290.5497.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinet W, De Meyer GR, Andries L, Herman AG, Kockx MM. In situ detection of starvation-induced autophagy. J Histochem Cytochem. 2006;54:85–96. doi: 10.1369/jhc.5A6743.2005. [DOI] [PubMed] [Google Scholar]
- Kondo Y, Kanzawa T, Sawaya R, Kondo S. The role of autophagy in cancer development and response to therapy. Nat Rev Cancer. 2005;5:726–734. doi: 10.1038/nrc1692. [DOI] [PubMed] [Google Scholar]
- Rouschop KM, Ramaekers CH, Schaaf MB, Keulers TG, Savelkouls KG, Lambin P, et al. Autophagy is required during cycling hypoxia to lower production of reactive oxygen species. Radiother Oncol. 2009;92:411–416. doi: 10.1016/j.radonc.2009.06.029. [DOI] [PubMed] [Google Scholar]
- Pursiheimo JP, Rantanen K, Heikkinen PT, Johansen T, Jaakkola PM. Hypoxia-activated autophagy accelerates degradation of SQSTM1/p62. Oncogene. 2009;28:334–344. doi: 10.1038/onc.2008.392. [DOI] [PubMed] [Google Scholar]
- Papandreou I, Lim AL, Laderoute K, Denko NC. Hypoxia signals autophagy in tumor cells via AMPK activity, independent of HIF-1, BNIP3, and BNIP3L. Cell Death Differ. 2008;15:1572–1581. doi: 10.1038/cdd.2008.84. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Levine B. Autophagy in mammalian development and differentiation. Nat Cell Biol. 2010;12:823–830. doi: 10.1038/ncb0910-823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandoval H, Thiagarajan P, Dasgupta SK, Schumacher A, Prchal JT, Chen M, et al. Essential role for Nix in autophagic maturation of erythroid cells. Nature. 2008;454:232–235. doi: 10.1038/nature07006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pua HH, Guo J, Komatsu M, He YW. Autophagy is essential for mitochondrial clearance in mature T lymphocytes. J Immunol. 2009;182:4046–4055. doi: 10.4049/jimmunol.0801143. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Morgan MJ, Chen K, Choksi S, Liu ZG. Induction of autophagy is essential for monocyte-macrophage differentiation. Blood. 2012;119:2895–2905. doi: 10.1182/blood-2011-08-372383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortensen M, Soilleux EJ, Djordjevic G, Tripp R, Lutteropp M, Sadighi-Akha E, et al. The autophagy protein Atg7 is essential for hematopoietic stem cell maintenance. J Exp Med. 2011;208:455–467. doi: 10.1084/jem.20101145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, McMillan-Ward E, Kong J, Israels SJ, Gibson SB. Mitochondrial electron-transport-chain inhibitors of complexes I and II induce autophagic cell death mediated by reactive oxygen species. J Cell Sci. 2007;120 (Pt 23:4155–4166. doi: 10.1242/jcs.011163. [DOI] [PubMed] [Google Scholar]
- Scherz-Shouval R, Shvets E, Fass E, Shorer H, Gil L, Elazar Z. Reactive oxygen species are essential for autophagy and specifically regulate the activity of Atg4. EMBO J. 2007;26:1749–1760. doi: 10.1038/sj.emboj.7601623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewaele M, Maes H, Agostinis P. ROS-mediated mechanisms of autophagy stimulation and their relevance in cancer therapy. Autophagy. 2010;6:838–854. doi: 10.4161/auto.6.7.12113. [DOI] [PubMed] [Google Scholar]
- Mathew R, Karp CM, Beaudoin B, Vuong N, Chen G, Chen HY, et al. Autophagy suppresses tumorigenesis through elimination of p62. Cell. 2009;137:1062–1075. doi: 10.1016/j.cell.2009.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem. 1998;273:5858–5868. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- Petiot A, Ogier-Denis E, Blommaart EF, Meijer AJ, Codogno P. Distinct classes of phosphatidylinositol 3′-kinases are involved in signaling pathways that control macroautophagy in HT-29 cells. J Biol Chem. 2000;275:992–998. doi: 10.1074/jbc.275.2.992. [DOI] [PubMed] [Google Scholar]
- Orford KW, Scadden DT. Deconstructing stem cell self-renewal: genetic insights into cell-cycle regulation. Nat Rev Genet. 2008;9:115–128. doi: 10.1038/nrg2269. [DOI] [PubMed] [Google Scholar]
- Zhang H, Wang ZZ. Mechanisms that mediate stem cell self-renewal and differentiation. J Cell Biochem. 2008;103:709–718. doi: 10.1002/jcb.21460. [DOI] [PubMed] [Google Scholar]
- Naka K, Hirao A. Maintenance of genomic integrity in hematopoietic stem cells. Int J Hematol. 2011;93:434–439. doi: 10.1007/s12185-011-0793-z. [DOI] [PubMed] [Google Scholar]
- Riekstina U, Cakstina I, Parfejevs V, Hoogduijn M, Jankovskis G, Muiznieks I, et al. Embryonic stem cell marker expression pattern in human mesenchymal stem cells derived from bone marrow, adipose tissue, heart and dermis. Stem Cell Rev. 2009;5:378–386. doi: 10.1007/s12015-009-9094-9. [DOI] [PubMed] [Google Scholar]
- Greco SJ, Liu K, Rameshwar P. Functional similarities among genes regulated by OCT4 in human mesenchymal and embryonic stem cells. Stem Cells. 2007;25:3143–3154. doi: 10.1634/stemcells.2007-0351. [DOI] [PubMed] [Google Scholar]
- Go MJ, Takenaka C, Ohgushi H. Forced expression of Sox2 or Nanog in human bone marrow derived mesenchymal stem cells maintains their expansion and differentiation capabilities. Exp Cell Res. 2008;314:1147–1154. doi: 10.1016/j.yexcr.2007.11.021. [DOI] [PubMed] [Google Scholar]
- Pierantozzi E, Gava B, Manini I, Roviello F, Marotta G, Chiavarelli M, et al. Pluripotency regulators in human mesenchymal stem cells: expression of NANOG but not of OCT-4 and SOX-2. Stem Cells Dev. 2011;20:915–923. doi: 10.1089/scd.2010.0353. [DOI] [PubMed] [Google Scholar]
- Li J, Kwong DL, Chan GC. The effects of various irradiation doses on the growth and differentiation of marrow-derived human mesenchymal stromal cells. Pediatr Transplant. 2007;11:379–387. doi: 10.1111/j.1399-3046.2006.00663.x. [DOI] [PubMed] [Google Scholar]
- Liu F, Lee JY, Wei H, Tanabe O, Engel JD, Morrison SJ, et al. FIP200 is required for the cell-autonomous maintenance of fetal hematopoietic stem cells. Blood. 2010;116:4806–4814. doi: 10.1182/blood-2010-06-288589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salemi S, Yousefi S, Constantinescu MA, Fey MF, Simon HU. Autophagy is required for self-renewal and differentiation of adult human stem cells. Cell Res. 2012;22:432–435. doi: 10.1038/cr.2011.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewing D, Jones SR. Superoxide removal and radiation protection in bacteria. Arch Biochem Biophys. 1987;254:53–62. doi: 10.1016/0003-9861(87)90080-4. [DOI] [PubMed] [Google Scholar]
- Grune T, Merker K, Sandig G, Davies KJ. Selective degradation of oxidatively modified protein substrates by the proteasome. Biochem Biophys Res Commun. 2003;305:709–718. doi: 10.1016/s0006-291x(03)00809-x. [DOI] [PubMed] [Google Scholar]
- Kiffin R, Bandyopadhyay U, Cuervo AM. Oxidative stress and autophagy. Antioxid Redox Signal. 2006;8:152–162. doi: 10.1089/ars.2006.8.152. [DOI] [PubMed] [Google Scholar]
- Jang YY, Sharkis SJ. A low level of reactive oxygen species selects for primitive hematopoietic stem cells that may reside in the low-oxygenic niche. Blood. 2007;110:3056–3063. doi: 10.1182/blood-2007-05-087759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poyton RO, Ball KA, Castello PR. Mitochondrial generation of free radicals and hypoxic signaling. Trends Endocrinol Metab. 2009;20:332–340. doi: 10.1016/j.tem.2009.04.001. [DOI] [PubMed] [Google Scholar]
- Zhang DX, Gutterman DD. Mitochondrial reactive oxygen species-mediated signaling in endothelial cells. Am J Physiol Heart Circ Physiol. 2007;292:H2023–H2031. doi: 10.1152/ajpheart.01283.2006. [DOI] [PubMed] [Google Scholar]
- Balaban RS, Nemoto S, Finkel T. Mitochondria, oxidants, and aging. Cell. 2005;120:483–495. doi: 10.1016/j.cell.2005.02.001. [DOI] [PubMed] [Google Scholar]
- Egan DF, Shackelford DB, Mihaylova MM, Gelino S, Kohnz RA, Mair W, et al. Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science. 2011;331:456–461. doi: 10.1126/science.1196371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R, et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–889. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- Komatsu M, Waguri S, Chiba T, Murata S, Iwata J, Tanida I, et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441:880–884. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- Wu JJ, Quijano C, Chen E, Liu H, Cao L, Fergusson MM, et al. Mitochondrial dysfunction and oxidative stress mediate the physiological impairment induced by the disruption of autophagy. Aging. 2009;1:425–437. doi: 10.18632/aging.100038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takamura A, Komatsu M, Hara T, Sakamoto A, Kishi C, Waguri S, et al. Autophagy-deficient mice develop multiple liver tumors. Genes Dev. 2011;25:795–800. doi: 10.1101/gad.2016211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priault M, Salin B, Schaeffer J, Vallette FM, di Rago JP, Martinou JC. Impairing the bioenergetic status and the biogenesis of mitochondria triggers mitophagy in yeast. Cell Death Differ. 2005;12:1613–1621. doi: 10.1038/sj.cdd.4401697. [DOI] [PubMed] [Google Scholar]
- Ren G, Zhang L, Zhao X, Xu G, Zhang Y, Roberts AI, et al. Mesenchymal stem cell-mediated immunosuppression occurs via concerted action of chemokines and nitric oxide. Cell Stem Cell. 2008;2:141–150. doi: 10.1016/j.stem.2007.11.014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.