Abstract
LIM homeobox domain 6 (LHX6) is a putative transcriptional regulator that controls the differentiation and development of neural and lymphoid cells. However, the function of LHX6 in cancer development remains largely unclear. Recently, we found that LHX6 is hypermethylated in lung cancer. In this study, we analysed its epigenetic regulation, biological functions, and related molecular mechanisms in lung cancer. Methylation status was evaluated by methylation-specific PCR and bisulfite genomic sequencing. LHX6 mRNA levels were measured in relation to the methylation status. The effects of LHX6 expression on tumourigenesis were studied in vitro and in vivo. LHX6 was readily expressed in normal lung tissues without methylation, but was downregulated or silenced in lung cancer cell lines and tissues with hypermethylation status. Treatment of lung cancer cells with the demethylating agent 5-aza-2′-deoxycytidine restored LHX6 expression. Moreover, LHX6 hypermethylation was detected in 56% (52/93) of primary lung cancers compared with none (0/20) of the tested normal lung tissues. In lung cancer cell lines 95D and H358, forced expression of LHX6 suppressed cell viability, colony formation, and migration, induced apoptosis and G1/S arrest, and inhibited their tumorigenicity in nude mice. On the other hand, knockdown of LHX6 expression by RNA interference increased cell proliferation and inhibited apoptosis and cell cycle arrest. These effects were associated with upregulation of p21 and p53, and downregulation of Bcl-2, cyclinD1, c-myc, CD44, and MMP7. In conclusion, our results suggest that LHX6 is a putative tumour suppressor gene with epigenetic silencing in lung cancer.
Keywords: LHX6, DNA methylation, tumour suppressor, lung cancer, epigenetics
Lung cancer is one of the most common malignancies and the leading cause of cancer-related death worldwide.1 Although traditional cytology and imaging technology have important roles in the diagnosis of lung cancer, 67% of newly diagnosed lung cancer patients are still detected at an advanced stage.2 Effective early diagnosis and targeted therapies to reduce mortality would benefit from a better understanding of the key molecular changes in normal cells, which lead to precancerous lesions and malignant tumour cells.
Lung carcinogenesis is a multi-phase process involving the accumulation of multiple genetic and epigenetic changes.3, 4, 5, 6 Alterations in normal DNA methylation patterns are fundamental to the tumorigenic process that contributes to all of the typical hallmarks of a cancer cell.7, 8 Hypermethylation of tumour suppressor genes is associated with epigenetically mediated gene silencing. Recently, an increasing number of genes inactivated by CpG island hypermethylation have been recognised as important components involved in multistep cancer development in humans.9, 10, 11, 12, 13 Characterisation of novel functional genes associated with promoter methylation may help us to understand the molecular mechanisms of alterations in tumour-suppressive pathways involved in lung carcinogenesis, and find better potential targets for the diagnosis and treatment of lung cancer.
The human LIM homeobox domain 6 (LHX6) gene is located on 9q33.2 and encodes a LIM homeodomain transcription factor involved in embryogenesis, more specifically in head development.14 LHX6 activity is required for normal tangential and radial migration of GABAergic interneurons and specification of neuronal subtypes in the neocortex and hippocampus.15 LHX6 and LHX8 co-ordinately induce neuronal expression of Sonic hedgehog that controls the generation of interneuron progenitors.16 In addition, LHX6 hypermethylation acts as a sensitive methylation marker in head and neck carcinomas and has an important role in early diagnosis of cervical cancer.17, 18, 19 Recently, we identified LHX6 hypermethylation in lung cancer. However, the role of LHX6 in lung cancer remains unclear. In the present study, we investigated the epigenetic regulation and biological function of LHX6 and its molecular basis in lung cancer.
Results
LHX6 is downregulated or inactivated in lung cancer tissues and cell lines
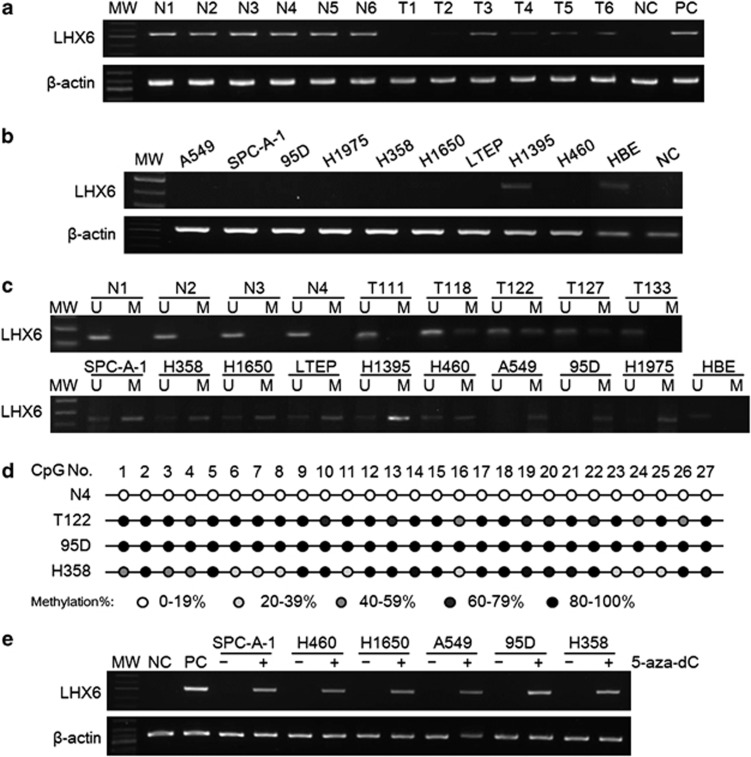
First, we performed reverse transcription-polymerase chain reaction (RT-PCR) to detect LHX6 mRNA expression in 10 primary lung cancer tissues and their corresponding adjacent non-tumour tissues. As shown in Figure 1a, LHX6 expression was downregulated in lung cancer tissues compared with that in the adjacent normal tissues. Next, we examined LHX6 expression in nine lung cancer cell lines and the normal human bronchial epithelial (HBE) cell line by RT-PCR and quantitative RT-PCR. As shown in Figure 1b and Supplementary Figure S1, LHX6 expression was reduced or silenced in eight (90%) cell lines, but was readily detected in H1395 cells and the normal cell line HBE. These results suggest aberrant gene silencing of LHX6 in lung cancer.
Figure 1.
Epigenetic silencing of LHX6 in lung cancer tissues and cell lines. (a) Representative results of LHX6 mRNA expression in tissues. LHX6 mRNA expression was significantly decreased in primary lung cancer tissues compared with that in the corresponding normal tissues as detected by RT-PCR. (b) mRNA expression of LHX6 in lung cancer cell lines was determined by semi-quantitative RT-PCR with β-actin as the internal control. LHX6 was weakly or not expressed in all lung cancer cell lines except for H1395. (c) Representative MSP results of the LHX6 methylation status in human tissues and cell lines. LHX6 was unmethylated in human normal lung tissues, but frequently hypermethylated in human lung cancer tissues and cell lines. MW, molecular weight; M, methylated product; U, unmethylated product; N, normal tissue; T, lung cancer tissue. (d) BGS analysis confirmed the methylation status of LHX6 in human tissues and cell lines. LHX6 was fully methylated in 95D cells, partially methylated in H358 and T122 cells, and unmethylated in normal tissue. (e) LHX6 mRNA expression was restored in lung cancer cell lines after treatment with the demethylation agent 5-aza-dC. ‘–' indicates DMSO control; and ‘+' indicates 5-aza-dC
LHX6 hypermethylation is associated with transcriptional silencing
To clarify whether expression of the LHX6 gene was regulated by DNA hypermethylation, we first analysed the methylation status of LHX6 in human cell lines by methylation-specific polymerase chain reaction (MSP). MSP region information is shown in Supplementary Figure S2a. The reliability of the MSP results was verified by DNA sequencing (Supplementary Figure S2b). As shown in Figure 1c, we found that all lung cancer cell lines in our study showed hypermethylation, whereas HBE cells exhibited an unmethylated status. However, the methylation pattern was heterogeneous in the CpG island region. Fully methylated LHX6 was detected in cell lines A549, 95D, and H1975, whereas LHX6 was partly methylated in SPC-A-1, H358, H1650, LTEP, H1395, and H460 cell lines, which showed downregulation of LHX6 expression. The methylation status was inversely correlated with the LHX6 expression level in all lung cancer cell lines and HBE cells except in H1395 cells. These results suggest that LHX6 hypermethylation is associated with transcriptional downregulation or silencing.
Next, we detected the LHX6 methylation status in 93 cases of human primary lung cancer and 20 cases of normal lung tissue. Using MSP, we found LHX6 hypermethylation in 52 (including 43 samples with partial methylation and 9 samples with fully methylation) out of 93 (56%) lung cancer samples and no LHX6 hypermethylation (0/20) in normal lung tissues (representative results are shown in Figure 1c). Next, we analysed the association of the LHX6 methylation status and clinicopathological characteristics in 62 patients with lung cancer. However, as shown in Supplementary Table S1, there was no correlation between LHX6 methylation and clinicopathological features such as age, gender, histological type, pathological stage, or differentiation status.
We performed bisulfite genomic sequencing (BGS) to provide a detailed map of the DNA methylation pattern within the CpG island region of the LHX6 gene. The size of the PCR product was 324 bp and included the transcriptional start site (–99 to +225 bp; Supplementary Figure S2a). Representative results are shown in Figure 1d. In 27 CpG sites of this region, LHX6 was fully hypermethylated in 95D cells. In H358 cells and lung cancer tissue T122, LHX6 was partially methylated. As expected, LHX6 was unmethylated in HBE cells expressing LHX6. These results are in agreement with those of MSP, and suggest that DNA methylation may be responsible for loss of LHX6 expression.
5-aza-2′-deoxycytidine restores LHX6 expression
To further test the hypothesis that loss of LHX6 expression is caused by hypermethylation of the LHX6 gene, lung cancer cell lines were treated with the demethylating agent 5-aza-2′-deoxycytidine (5-aza-dC). Representative results of semi-quantitative RT-PCR and quantitative RT-PCR analyses are illustrated in Figure 1e and Supplementary Figure S3, respectively. As expected, lung cancer cell lines treated with 5-aza-dC, which initially showed high levels of LHX6 methylation, were induced to express LHX6. Quantitative RT-PCR analyses showed that the ratio of LHX6 to β-actin transcripts was increased several fold in the 5-aza-dC-treated group compared with that in the control group. These results strongly indicated that hypermethylation is responsible for defective expression of LHX6 in lung cancer.
LHX6 inhibits cell growth in vitro and in vivo
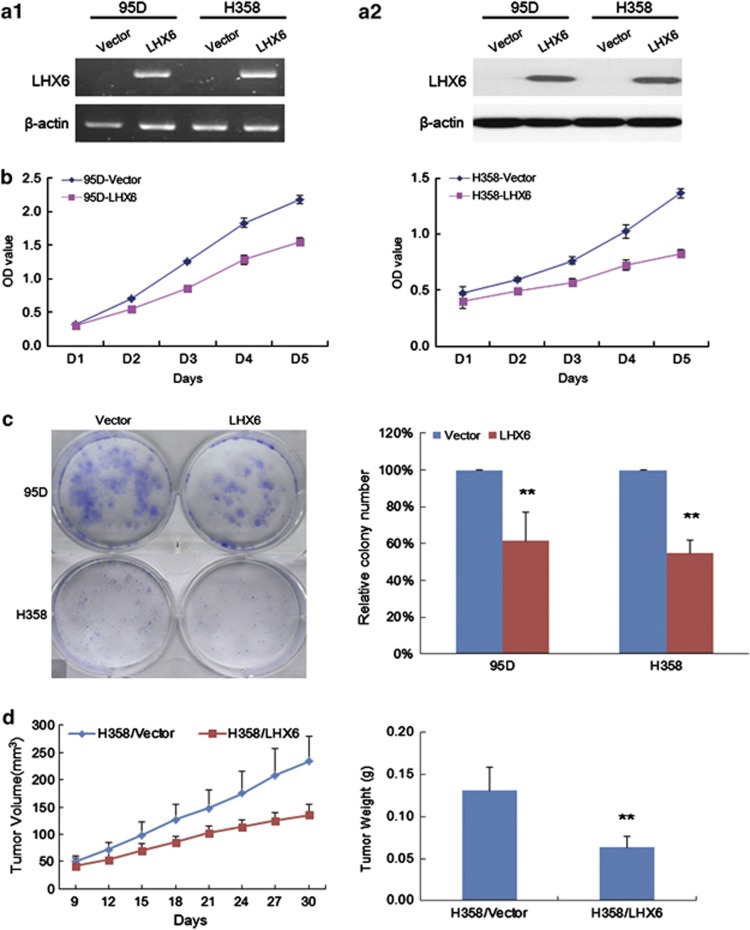
To evaluate the functional significance of LHX6 in lung cancer, we examined growth suppression by overexpression of LHX6 in 95D and H358 cells that showed loss of LHX6 expression and CpG island hypermethylation. The transfection efficiency is shown in Supplementary Figure S4. Re-expression of LHX6 in these cell lines was evidenced by RT-PCR and western blot analyses (Figure 2a). The results of the cell proliferation assay indicated that ectopic expression of LHX6 significantly inhibited the growth of 95D and H358 cells compared with that in the control groups (Figure 2b). To confirm the inhibitory effect of LHX6 on the growth of lung cancer cell lines, we used a colony formation assay using 95D and H358 cells. Compared with control vector-transfected cells without LHX6 expression, we found a greatly reduced number of colonies formed by LHX6-transfected 95D and H358 cells (P<0.01; Figure 2c). These data further demonstrated that LHX6 inhibits lung cancer cell proliferation.
Figure 2.
LHX6 inhibits cell growth. (a) Re-expression of LHX6 mRNA and protein in transfected cell lines 95D and H358 was confirmed by RT-PCR (a1) and western blotting (a2), respectively. (b) LHX6 significantly inhibited the proliferation of 95D and H358 cells as shown by CCK-8. These data are shown as the mean±S.D. of three independent experiments. (c) Colony formation assays were performed to confirm the effect of LHX6 on cell growth. Quantitative analysis of colony numbers is shown in the right panel. Compared with empty vector transfectants, a significant reduction of colony numbers was observed for LHX6 transfectants. (d) Inhibition of tumour growth by LHX6 expression in vivo. Growth curves of tumours in nude mice showed that LHX6 transfectants grew slower than empty vector transfectants. **P<0.01
We confirmed the effect of LHX6 on cell proliferation by examining LHX6 overexpression in a tumourigenesis assay using nude mice. Tumour growth of H358 cells overexpressing LHX6 was significantly reduced compared with that of control cells (Figure 2d). The mean tumour weight was significantly less in nude mice injected with LHX6-overexpressing H358 cells compared with that in mice injected with control vector-transfected cells (P<0.01; Figure 2d). These results suggest that LHX6 is a tumour suppressor gene that negatively regulates tumour growth both in vitro and in vivo.
LHX6 induces apoptosis and cell cycle arrest
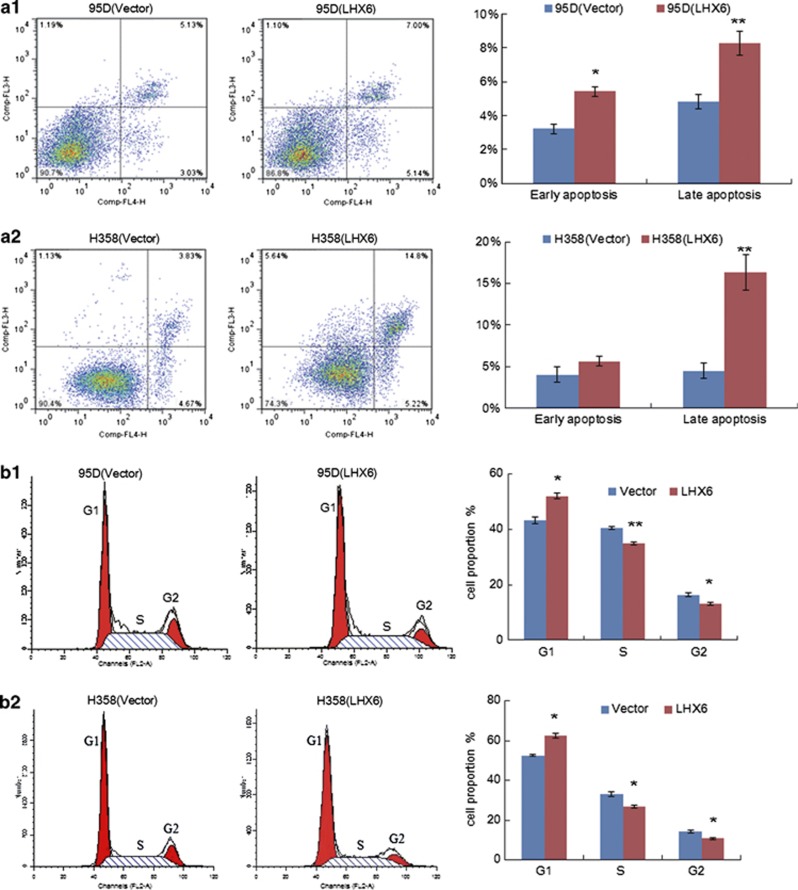
We next determined whether inhibition of tumour cell growth by LHX6 is related to apoptosis and cell cycle arrest. First, we performed an apoptosis assay by flow cytometry (Figure 3a). We found that ectopic expression of LHX6 caused a significant increase in early apoptosis and late apoptosis compared with that of control vector-transfected 95D (Figure 3a(1)) and H358 (Figure 3a(2)) cells. In addition, morphological observation and Hoechst 33258 staining were performed to definitively establish the effect of LHX6 expression on apoptosis (Supplementary Figure S5).
Figure 3.
Effect of LHX6 expression on apoptosis and the cell cycle. (a) Flow cytometric assays with Annexin V-APC and 7-AAD double staining of 95D (a1) and H358 (a2) cells. Left panel shows representative dots plots and right panels show the quantitative analysis. (b) Cell cycle analysis by flow cytometry. The distribution of empty or LHX6 expression vector-transfected 95D (b1) and H358 (b2) cells in cell cycle phases. The percentages are represented by bar graphs. Each experiment was repeated three times. *P<0.05; **P<0.01
Next, the effect of LHX6 expression on the cell cycle was evaluated by flow cytometry (Figure 3b). Among 95D cells without exogenous LHX6 expression, the percentages of cells in G1, S, and G2 phases were 43.26±1.10, 40.39±0.44, and 16.35±0.66%, respectively. In contrast, among 95D cells with re-expression of LHX6, the percentages of cells in G1, S, and G2 phases were 51.96±1.00, 34.86±0.52, and 13.18±0.48%, respectively (Figure 3b(1)). This trend was also found in H358 cells. The percentages of H358 cells in G1, S, and G2 phases were 52.71±0.47, 32.95±1.22, and 14.35±0.74%, respectively. In contrast, among H358 cells with re-expression of LHX6, the percentages of cells in G1, S, and G2 phases were 62.48±1.19, 26.70±0.76, and 10.83±0.43%, respectively (Figure 3b(2)). These results demonstrate a reduction in the percentages of cells in S and G2 phases, and retention of 95D and H358 cells with restoration of LHX6 expression in G1 phase (P<0.05).
LHX6 expression induces cell migration
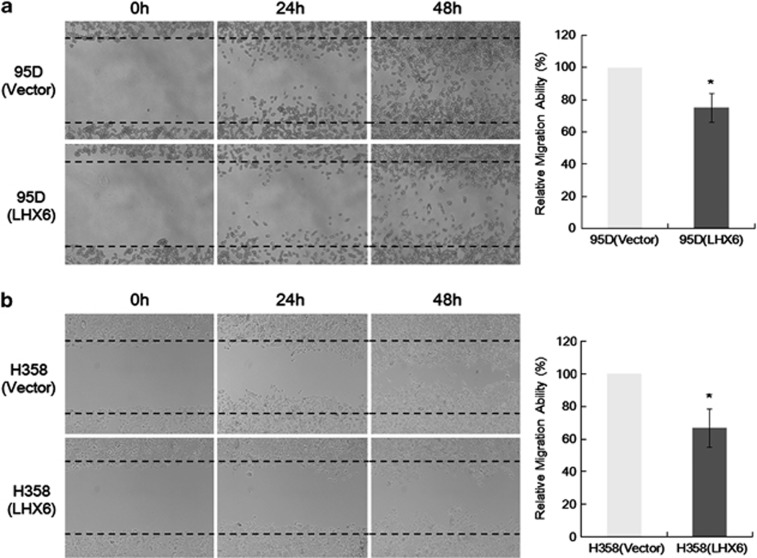
The effect of LHX6 expression on lung cancer cell motility was investigated by a wound healing assay. LHX6 transfectants spread along wound edges much slower than that of control vector transfectants (Figure 4). Quantitative analyses at 48 h confirmed a significant reduction in wound closure by LHX6-transfected 95D (Figure 4a) and H358 (Figure 4b) cells compared with that of empty vector-transfected control cells.
Figure 4.
Cell motility as determined by a wound healing assay. The ability of LHX6 transfectants to spread along wound edges in 95D (a) and H358 (b) cells was significantly reduced compared with that of control cells. Quantification of the migrated cells is shown in the right panel. All data represent the mean±S.D. of three independent experiments. *P<0.05
Knockdown of LHX6 promotes cell proliferation
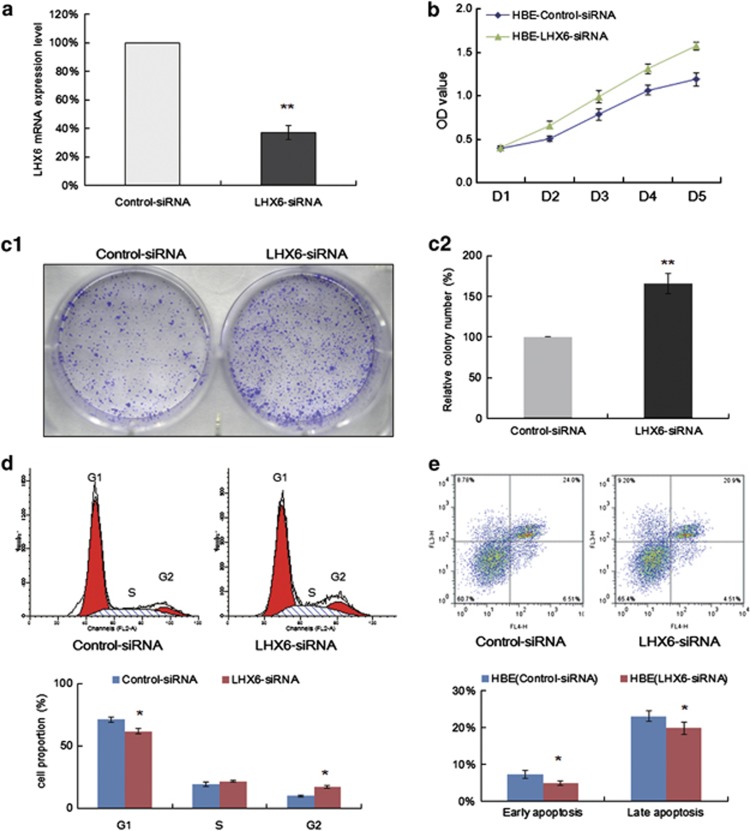
To confirm the inhibitory effect of LHX6 expression on cell growth, we knocked down LHX6 expression in the LHX6-expressing cell line HBE by siRNA. Real-time quantitative RT-PCR showed that LHX6 expression was significantly decreased by LHX6 siRNA (Figure 5a). We found that knockdown of LHX6 promoted the growth of HBE cells by a CCK-8 cell viability assay (Figure 5b). Accordingly, knockdown of LHX6 significantly increased colony formation by more than 60% compared with that of cells transfected with control siRNA (Figure 5c). These results further indicated that LHX6 acts as a potential tumour suppressor gene in lung cancer.
Figure 5.
Effect of LHX6 knockdown on cell growth. (a) LHX6 mRNA-targeting siRNA and a non-targeting control siRNA were transfected into LHX6-expressing HBE cells. The efficiency of LHX6 knockdown was examined by real-time RT-PCR. (b) siRNA-mediated knockdown of LHX6 induced cell proliferation as shown by CCK-8. (c) Colony formation assays confirmed the effect of LHX6 expression on cell growth. Colony numbers were increased by knockdown of LHX6. Colonies were photographed under a phase-contrast microscope (left, c1). Average numbers of colonies are represented by the bar graph (right, c2). Each experiment was repeated three times. (d) Cell cycle analysis by flow cytometry. The distribution of siRNA negative control- and LHX6 siRNA-transfected HBE cells in cell cycle phases. The percentages are represented by the bar graph. Each experiment was repeated three times. (e) The effect of LHX6 knockdown on apoptosis was analysed by flow cytometry. *P<0.05; **P<0.01
Next, we further examined the effect of LHX6 knockdown on cell cycle regulation. Among HBE cells transfected with negative control siRNA, 70.92±2.37% were in G1 phase, 19.18±1.76% were in S phase, and 9.91±0.61% were in G2 phase. In contrast, among LHX6 siRNA-transfected HBE cells, 61.65±1.99% were in G1 phase, 21.57±0.85% were in S phase, and 16.79±1.14% were in G2 phase (P<0.05) (Figure 5d). Thus, LHX6-targeted siRNA decreased the number of cells in G1 phase, whereas significantly increasing the number of cells in G2 phase, suggesting that loss of LHX6 expression increases cellular proliferation.
Finally, we examined the effect of LHX6 knockdown on apoptosis by flow cytometry. Our results showed that both early apoptotic cells (7.26±1.05% versus 4.96±0.63%) and late apoptotic cells (23.00±1.41% versus 19.75±1.63%) among LHX6 siRNA-transfected HBE cells decreased significantly compared with those among control siRNA-transfected HBE cells (P<0.05) (Figure 5e).
Molecular targets of LHX6 in cell lines
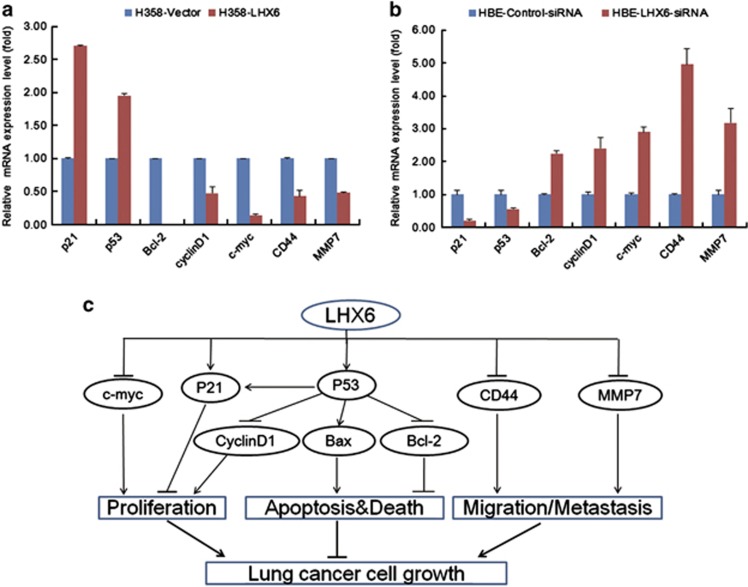
To gain insights into the molecular mechanisms of the tumour-suppressive effect of LHX6, LHX6-modulated downstream target genes were characterised by quantitative RT-PCR analysis of H358 cells stably transfected with LHX6. As shown in Figure 6a, LHX6 increased expression of the pro-apoptotic gene p53 and downregulated the anti-apoptotic gene Bcl-2. LHX6 also enhanced expression of p21, a cell cycle regulator. LHX6 exerted an anti-proliferative effect by downregulating expression of cell proliferation genes cyclinD1 and c-myc, which are both important G1 phase regulators. Moreover, inhibition of cell migration by LHX6 resulted from downregulation of CD44 and MMP7 expression. On the other hand, loss of LHX6 expression had the opposite effect on regulation of these genes (Figure 6b). The results suggest that LHX6 acts as tumour suppressor by regulating the expression of important genes involved in proliferation, apoptosis, and migration pathways (Figure 6c).
Figure 6.
Effect of LHX6 on its downstream genes in cancer pathways. (a) Ectopic expression of LHX6 enhanced the expression of p21 and p53, and decreased the expression of Bcl-2, cyclinD1, c-myc, CD44, and MMP7. (b) Expression of p21 and p53 decreased, whereas the expression of Bcl-2, cyclinD1, c-myc, CD44, and MMP7 increased in HBE cells with knockdown of LHX6. (c) Schematic diagram for the mechanisms of LHX6 as a tumour suppressor
Discussion
Several lines of evidence support the hypothesis that epigenetic changes by DNA methylation have an important role in lung carcinogenesis.20, 21, 22, 23 Recently, epigenetic silencing of many novel genes that function as putative tumour suppressor genes has been reported to contribute to human lung cancer.9, 10, 11, 21, 23 In this study, we identified a typical CpG island located in the transcription region of the LHX6 gene, which showed a hypermethylation status in lung cancer. To our knowledge, this is the first study of epigenetic regulation of LHX6 expression and its function in lung cancer.
In this study, we found that LHX6 hypermethylation is frequent and cancer specific in lung cancer. Our results suggest that LHX6 methylation is associated with tumorigenesis. In accordance with our study, methylation levels of the LHX6 promoter are also strongly associated with cervical carcinogenesis and cancer development.18, 19 This epigenetic alteration of the LHX6 gene begins at a relatively early stage, suggesting its potential as a biomarker for early diagnosis and prevention of head and neck carcinomas, and cervical cancer.17, 18
We also found that the reduction of LHX6 expression was related to CpG island methylation of LHX6. Furthermore, a high dose of 5-aza-dC stimulated LHX6 re-expression in most of the lung cancer cell lines, suggesting that CpG island methylation is the predominant regulatory mechanism of LHX6 inactivation in lung cancer. However, compared with 5-aza-dC alone, exposure to various concentrations of 5-aza-dC with trichostatin A increases LHX6 expression more significantly in cervical cell lines.18, 19 Thus, other epigenetic modifications such as histone modifications may be involved in silencing LHX6 expression in lung cancer. Accordingly, studies on the human LHX3 gene have shown that cell-specific expression of two different transcripts of LHX3 is driven by epigenetic and genetic regulatory systems.24 Considering that LHX3 and LHX6 belong to the same family, it is plausible to assume that human LHX6 expression may be regulated in a similar manner. In the future, the mechanism of LHX6 regulation requires further study.
LHX6 is a putative transcriptional regulator that controls the differentiation and development of neural and lymphoid cells, particularly in the central nervous system.14, 15 In addition to its established role in muscle development, little is known about the function of LHX6, especially in cancer. Recently, several other LHX genes have also been implicated in tumourigenesis.25, 26, 27, 28, 29 LHX4 may act as a tumour suppressor by downregulation of alpha-fetoprotein expression in hepatocarcinogenesis.25 Inactivation of LHX8 expression reduces the transcription of pro-apoptotic genes bax, caspase-2, and caspase-3 in mouse oocytes.26 Epigenetic alteration of the LHX9 gene is involved in glioma cell invasiveness and migration.27 Hypermethylation-mediated reduction of LMX1A expression inhibits tumourigenesis, epithelial-to-mesenchymal transition, and stem-like properties of the epithelia in gastric and ovarian cancers.28, 29 In addition, several novel epigenetically silenced genes have been found to suppress cell growth in numerous types of cancer.30, 31, 32, 33, 34, 35 These studies have suggested that LHX6 may have a potential tumour suppressor role in lung cancer.25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35
Thus, the tumour suppressor role of LHX6 in lung cancer was investigated both in vitro and in vivo. Ectopic expression of LHX6 in the silenced 95D and H358 cell lines showed a significant growth suppressive effect by inhibition of cell proliferation and colony formation. A decrease of tumour growth by LHX6 was confirmed by the reduction of tumourigenesis in nude mice. On the other hand, siRNA-mediated knockdown of LHX6 in HBE cells resulted in significantly increased cell proliferation. These results indicate that LHX6 functions as a tumour suppressor in lung carcinogenesis. Similar to our findings, overexpression of LHX6 in cervical cancer cells causes a reduction of cell proliferation and suppresses the tumorigenic phenotype, further demonstrating the growth suppressive effects of LHX6 expression and suggesting that it may be a novel tumour suppressor gene in cervical cancer.19
LHX6 is a LIM homeobox transcription factor expressed during embryogenesis. However, the molecular mechanisms that regulate LHX6 transcriptional activity are unknown. Previously, LHX6 and an interaction with PITX2 were found to regulate cell proliferation in the cervical loop and promote cell differentiation in the anterior region of the incisor.36 Recently, Zhang et al.37 demonstrated that LHX6 directly interacts with PITX2 to inhibit PITX2 transcriptional activity and activation of multiple promoters. PITX2 directly interacts with β-catenin to synergistically regulate LEF-1 expression, which regulates the activity of several growth control genes.38, 39 These findings demonstrate new molecular mechanisms of LHX6 for normal craniofacial and tooth development. In our study, we also found that LHX6 decreased the expression of Wnt signalling target genes cyclinD1, c-myc, CD44, and MMP7. Our results suggest that LHX6 may be an important negative regulator of the WNT signalling pathway that is involved in many biological processes including embryogenesis and carcinogenesis.40 However, further definition and investigation of LHX6-regulated genes are critical to understand the pathways through which LHX6 exerts its tumour-suppressive activity.
In conclusion, we have identified LHX6 as a novel functional tumour suppressor gene that is silenced by CpG island methylation in lung cancer. LHX6 contributes to the suppression of tumourigenesis by decreasing cell proliferation and inducing apoptosis and cell cycle arrest by regulating several important signalling pathways. LHX6 methylation may serve as a potential epigenetic biomarker for early diagnosis of patients with lung cancer.
Materials and Methods
Cell lines and tissue samples
Lung cancer cell lines (A549, SPC-A-1, 95D, H1975, H358, H1650, LTEP, H1395, and H460) and an immortalised HBE cell line were obtained from the American Type Culture Collection (Manassas, VA, USA) and Cell Biology Institute of Chinese Academy of Science (Shanghai, China). All cell lines were cultured in RPMI-1640 medium (Gibco BRL, Rockville, MD, USA) supplemented with 10% foetal bovine serum (Gibco BRL) and incubated in a humidified atmosphere with 5% CO2 at 37 °C.
A total of 93 primary lung cancer tissues and 20 normal lung tissues were obtained during biopsy before any therapeutic intervention at the Affiliated Xi'nan Hospital of the Third Military Medical University. The tumours were classified according to the WHO histological typing of lung tumours, and staged following the TNM classification of malignant tumours defined by the International Union against Cancer. All experiments and procedures were approved by the Clinical Research Ethics Committee of the Third Military Medical University.
RNA extraction, RT-PCR, and real-time quantitative RT-PCR analyses
Total RNA was extracted from cells and tissues with Trizol (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's protocol. cDNA was synthesised from 1 μg total RNA using a PrimeScript RT reagent Kit with gDNA Eraser (Takara, Otsu, Japan). RT-PCR and real-time quantitative RT-PCR analyses were performed as described previously with the housekeeping gene β-actin used as an internal control.41, 42, 43 Primer sequences are listed in Supplementary Table S2.
DNA extraction, MSP, and BGS
Genomic DNA from cell lines and tissues was isolated by a DNA extraction kit (Promega, Madison, WI, USA) and chemically modified using an EZ DNA Methylation-Gold Kit (Zymo Research, Orange, CA, USA) according to the manufacturer's instructions. Primer pairs used for MSP and BGS are listed in Supplementary Table S2. For MSP, the protocol was carried out as reported previously.44, 45, 46 For BGS, the PCR products were purified and inserted into a pGEM-T easy vector (Promega). Ten colonies were chosen randomly for sequencing.
Demethylation agent 5-aza-dC treatment
Demethylation was performed as reported previously.41 Briefly, lung cancer cells were seeded at a density of 1 × 106 cells per ml in 10-cm dishes. After 24 h of culture, cells were treated with 10 μM 5-aza-dC (Sigma, St. Louis, MO, USA) for 3 days. Control cells were treated in parallel with DMSO for each time point. The medium was replaced every day. Cells were harvested and mRNA expression of LHX6 was analysed by RT-PCR and quantitative RT-PCR.
Construction of the LHX6 overexpression vector
cDNA corresponding to the full-length human LHX6 gene was verified by sequencing and subcloned into the mammalian expression vector pIRES2-EGFP (Invitrogen). Lung cancer cell lines were transfected using X-treme Gene HP DNA transfection reagent (Roche, Mannheim, Germany) according to the manufacturer's instructions. Clones with stable transfection of LHX6 or empty vector were established with G418 (Invitrogen) selection for 2–3 weeks. Gene and protein expression of LHX6 was confirmed by RT-PCR and western blot analyses.
Knockdown of LHX6 by siRNA
Four siRNA sequences and a negative control sequence of the siRNA annealing sequence were designed, synthesised, and then subcloned into pcDNA6.2 GW/EmGFP siRNA vectors (Invitrogen). The HBE cell line with LHX6 expression was transfected with the vectors carrying LHX6 or negative control siRNAs. Knockdown efficiency was evaluated at 48 h after transfection by real-time quantitative RT-PCR. The siRNA with the highest knockdown efficiency was used for functional studies. Stably transfected cells with LHX6 knockdown were selected with 0.6 mg/ml Blasticidin S HCl (Invitrogen) for colony formation assays.
Cell viability assay
Briefly, 8 × 103 cells per well were seeded in 96-well plates and transiently transfected with pIRES2-EGFP-LHX6, LHX6 siRNA, or control vectors. After 1–5 days of transfection, cell viability was evaluated by Cell Counting Kit-8 (CCK-8; Dojindo, Kumamoto, Japan) according to the manufacturer's instructions. Data were expressed as the optical density measured at a wavelength of 450 nm. Experiments were carried out in triplicate.
Colony formation assay
For the overexpression assay, 95D and H358 cells (1 × 105 cells/well) were seeded in 24-well plates and transfected with pIRES2-EGFP-LHX6 or empty vectors. For the knockdown assay, HBE cells were transfected with LHX6 siRNA or control vectors. At 2 days post transfection, cells were passaged at a 1:20 ratio into six-well plates containing medium with G418 or Blasticidin S HCl. After culturing for 14–21 days, cells were fixed with 4% paraformaldehyde and then stained with crystal violet. Colonies with more than 50 cells per colony were counted. All experiments were conducted three times in triplicates.
Hoechst 33258 staining
Hoechst 33258 staining was carried out at 48 h post transfection. The cells were fixed with 4% paraformaldehyde for 15 min, washed with PBS, and then stained with Hoechst 33258 (Beyotime, Jiangsu, China) for 5 min. After two washes with PBS, morphological changes of apoptotic nuclei were observed under a fluorescence microscope with excitation and emission at 346 and 460 nm, respectively.
Flow cytometry
Cells with overexpression or knockdown of LHX6 were fixed in 70% ethanol and stained with 15 μg/ml propidium iodide at 4 °C in the dark. The cells were then analysed by a FACSCalibur (BD Biosciences, Franklin Lakes, NJ, USA). Cell cycle profiles were determined using ModFitLT software (Becton Dickinson, San Diego, CA, USA). Apoptosis was determined by dual staining with an Annexin V-APC and 7-AAD Apoptosis Detection kit (Keygen, Nanjing, China) according to the manufacturer's instructions, and then analysed by FlowJo software (TreeStar, San Carlos, CA, USA).
Wound healing assay
Cell migration was assessed by a scratch wound assay. Briefly, 95D and H358 cells (5 × 105 cells/well) stably transfected with pIRES2-EGFP-LHX6 or empty vector were cultured in six-well plates. At 90% confluence, three scratch wounds across each well were made using a P-200 pipette tip. At 0, 24 and 48 h of culture, images were taken of the wound closure areas. The experiment was performed in triplicate wells and repeated three times.
In vivo tumorigenicity
H358 cells (5 × 106 cells in 0.2 ml PBS) stably transfected with pIRES2-EGFP-LHX6 or empty vector were injected subcutaneously into the dorsal flank of 4-week-old female Balb/c nude mice (n=4 per group). Tumour volume (mm3) was estimated by measuring the longest (a) and shortest (b) diameter of the tumour and calculated as ab2/2. All experimental procedures were approved by the Animal Ethics Committee of the Third Military Medical University.
Western blot analysis
Total protein was extracted from cell lines, and the protein concentration was measured by the Bradford method (Bio-Rad Laboratories, Hercules, CA, USA). Protein samples (40 μg) were separated by 10% SDS-PAGE and then transferred to an equilibrated polyvinylidene difluoride membrane (Amersham Biosciences, Buckinghamshire, UK). After incubation at 4 °C overnight with a primary antibody against LHX6 (Santa Cruz Biotechnology, Santa Cruz, CA, USA) and then reacting with the secondary antibody, the proteins were detected by enhanced chemiluminescence (Amersham Corporation, Arlington Heights, IL, USA).
Statistical analysis
Statistical analyses were performed with SPSS 13.0 software (SPSS, Chicago, IL, USA). Data were expressed as the mean±S.D. Results were evaluated using the t-test, Fisher's exact test, and Mann–Whitney U-test. A P-value<0.05 was considered significant.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (81202238, 81030052, 81071695), Natural Science Foundation Project of CQ CSTC of China (cstc2011jjA10098), and the Third Military Medical University (2011XQN07). We thank YJ Li for technical assistance. The current study does not have any commercial connection with any other people or organisation.
Glossary
- 5-aza-dC
5-aza-2′-deoxycytidine
- LHX6
LIM Homeobox domain 6
- BGS
bisulfite genomic sequencing
- MSP
methylation-specific polymerase chain reaction
- RT-PCR
reverse transcription-polymerase chain reaction
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Disease website (http://www.nature.com/cddis)
Edited by A Stephanou
Supplementary Material
References
- Siegel R, Naishadham D, Jemal A. Cancer statistics. CA Cancer J Clin. 2013;2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- Mascaux C, Peled N, Garg K, Kato Y, Wynes MW, Hirsch FR. Early detection and screening of lung cancer. Expert Rev Mol Diagn. 2010;10:799–815. doi: 10.1586/erm.10.60. [DOI] [PubMed] [Google Scholar]
- Breuer RH, Pasic A, Smit EF, van Vliet E, Vonk Noordegraaf A, Risse EJ, et al. The natural course of preneoplastic lesions in bronchial epithelium. Clin Cancer Res. 2005;11:537–543. [PubMed] [Google Scholar]
- Thorgeirsson TE, Geller F, Sulem P, Rafnar T, Wiste A, Magnusson KP, et al. A variant associated with nicotine dependence, lung cancer and peripheral arterial disease. Nature. 2008;452:638–642. doi: 10.1038/nature06846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteller M. Cancer epigenomics: DNA methylomes and histone-modification maps. Nat Rev Genet. 2007;8:286–298. doi: 10.1038/nrg2005. [DOI] [PubMed] [Google Scholar]
- Risch A, Plass C. Lung cancer epigenetics and genetics. Int J Cancer. 2008;123:1–7. doi: 10.1002/ijc.23605. [DOI] [PubMed] [Google Scholar]
- Feil R, Fraga MF. Epigenetics and the environment: emerging patterns and implications. Nat Rev Genet. 2012;13:97–109. doi: 10.1038/nrg3142. [DOI] [PubMed] [Google Scholar]
- Rodríguez-Paredes M, Esteller M. Cancer epigenetics reaches mainstream oncology. Nat Med. 2011;17:330–339. doi: 10.1038/nm.2305. [DOI] [PubMed] [Google Scholar]
- Cui T, Chen Y, Yang L, Knösel T, Huber O, Pacyna-Gengelbach M, et al. The p53 target gene desmocollin 3 acts as a novel tumor suppressor through inhibiting EGFR/ERK pathway in human lung cancer. Carcinogenesis. 2012;33:2326–2333. doi: 10.1093/carcin/bgs273. [DOI] [PubMed] [Google Scholar]
- Jia Y, Yang Y, Brock MV, Zhan Q, Herman JG, Guo M. Epigenetic regulation of DACT2, A key component of the Wnt signaling pathway in human lung cancer. J Pathol. 2013;230:194–204. doi: 10.1002/path.4073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Chen Y, Cui T, Knösel T, Zhang Q, Albring KF, et al. Desmoplakin acts as a tumor suppressor by inhibition of the Wnt/β-catenin signaling pathway in human lung cancer. Carcinogenesis. 2012;33:1863–1870. doi: 10.1093/carcin/bgs226. [DOI] [PubMed] [Google Scholar]
- Liu W, Li X, Chu ES, Go MY, Xu L, Zhao G, et al. Paired box gene 5 is a novel tumor suppressor in hepatocellular carcinoma through interaction with p53 signaling pathway. Hepatology. 2011;53:843–853. doi: 10.1002/hep.24124. [DOI] [PubMed] [Google Scholar]
- Xu L, Li X, Chu ES, Zhao G, Go MY, Tao Q, et al. Epigenetic inactivation of BCL6B, a novel functional tumour suppressor for gastric cancer, is associated with poor survival of gastric cancer. Gut. 2012;61:977–985. doi: 10.1136/gutjnl-2011-300411. [DOI] [PubMed] [Google Scholar]
- Grigoriou M, Tucker AS, Sharpe PT, Pachnis V. Expression and regulation of Lhx6 and Lhx7, a novel subfamily of LIM homeodomain encoding genes, suggests a role in mammalian head development. Development. 1998;125:2063–2074. doi: 10.1242/dev.125.11.2063. [DOI] [PubMed] [Google Scholar]
- Liodis P, Denaxa M, Grigoriou M, Akufo-Addo C, Yanagawa Y, Pachnis V. Lhx6 activity is required for the normal migration and specification of cortical interneuron subtypes. J Neurosci. 2007;27:3078–3089. doi: 10.1523/JNEUROSCI.3055-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flandin P, Zhao Y, Vogt D, Jeong J, Long J, Potter G, et al. Lhx6 and Lhx8 coordinately induce neuronal expression of Shh that controls the generation of interneuron progenitors. Neuron. 2011;70:939–950. doi: 10.1016/j.neuron.2011.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estécio MR, Youssef EM, Rahal P, Fukuyama EE, Góis-Filho JF, Maniglia JV, et al. LHX6 is a sensitive methylation marker in head and neck carcinomas. Oncogene. 2006;25:5018–5026. doi: 10.1038/sj.onc.1209509. [DOI] [PubMed] [Google Scholar]
- Jung S, Jeong D, Kim J, Yi L, Koo K, Lee J, et al. The role of hLHX6-HMR as a methylation biomarker for early diagnosis of cervical cancer. Oncol Rep. 2010;23:1675–1682. doi: 10.3892/or_00000811. [DOI] [PubMed] [Google Scholar]
- Jung S, Jeong D, Kim J, Yi L, Koo K, Lee J, et al. Epigenetic regulation of the potential tumor suppressor gene, hLHX6.1, in human cervical cancer. Int J Oncol. 2011;38:859–869. doi: 10.3892/ijo.2011.904. [DOI] [PubMed] [Google Scholar]
- Esteller M. Epigenetics in cancer. N Engl J Med. 2008;358:1148–1159. doi: 10.1056/NEJMra072067. [DOI] [PubMed] [Google Scholar]
- Okamoto J, Hirata T, Chen Z, Zhou HM, Mikami I, Li H, et al. EMX2 is epigenetically silenced and suppresses growth in human lung cancer. Oncogene. 2010;29:5969–5975. doi: 10.1038/onc.2010.330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, An Q, Li L, Zhang D, Huang J, Feng X, et al. Hypermethylation of p16INK4a in Chinese lung cancer patients: biological and clinical implications. Carcinogenesis. 2003;24:1897–1901. doi: 10.1093/carcin/bgg169. [DOI] [PubMed] [Google Scholar]
- Tessema M, Klinge DM, Yingling CM, Do K, Van Neste L, Belinsky SA. Re-expression of CXCL14, a common target for epigenetic silencing in lung cancer, induces tumor necrosis. Oncogene. 2010;29:5159–5170. doi: 10.1038/onc.2010.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaden BC, MIII Garcia, Smith TP, Rhodes SJ. Two promoters mediate transcription from the human LHX3 gene: involvement of nuclear factor I and specificity protein 1. Endocrinology. 2006;147:324–337. doi: 10.1210/en.2005-0970. [DOI] [PubMed] [Google Scholar]
- Hung TM, Hu RH, Ho CM, Chiu YL, Lee JL, Jeng YM, et al. Downregulation of alpha-fetoprotein expression by LHX4: a critical role in hepatocarcinogenesis. Carcinogenesis. 2011;32:1815–1823. doi: 10.1093/carcin/bgr219. [DOI] [PubMed] [Google Scholar]
- Choi Y, Ballow DJ, Xin Y, Rajkovic A. Lim homeobox gene, lhx8, is essential for mouse oocyte differentiation and survival. Biol Reprod. 2008;79:442–449. doi: 10.1095/biolreprod.108.069393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vladimirova V, Mikeska T, Waha A, Soerensen N, Xu J, Reynolds PC, et al. Aberrant methylation and reduced expression of LHX9 in malignant gliomas of childhood. Neoplasia. 2009;11:700–711. doi: 10.1593/neo.09406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong W, Feng L, Xie Y, Zhang H, Wu Y. Hypermethylation-mediated reduction of LMX1A expression in gastric cancer. Cancer Sci. 2011;102:361–366. doi: 10.1111/j.1349-7006.2010.01804.x. [DOI] [PubMed] [Google Scholar]
- Chao TK, Yo YT, Liao YP, Wang YC, Su PH, Huang TS, et al. LIM-homeobox transcription factor 1, alpha (LMX1A) inhibits tumourigenesis, epithelial-mesenchymal transition and stem-like properties of epithelial ovarian cancer. Gynecol Oncol. 2013;128:475–482. doi: 10.1016/j.ygyno.2012.12.018. [DOI] [PubMed] [Google Scholar]
- Melotte V, Lentjes MH, van den Bosch SM, Hellebrekers DM, de Hoon JP, Wouters KA, et al. N-Myc downstream-regulated gene 4 (NDRG4): a candidate tumor suppressor gene and potential biomarker for colorectal cancer. J Natl Cancer Inst. 2009;101:916–927. doi: 10.1093/jnci/djp131. [DOI] [PubMed] [Google Scholar]
- Yu J, Ma X, Cheung KF, Li X, Tian L, Wang S, et al. Epigenetic inactivation of T-box transcription factor 5, a novel tumor suppressor gene, is associated with colon cancer. Oncogene. 2010;29:6464–6474. doi: 10.1038/onc.2010.370. [DOI] [PubMed] [Google Scholar]
- Das PM, Thor AD, Edgerton SM, Barry SK, Chen DF, Jones FE. Reactivation of epigenetically silenced HER4/ERBB4 results in apoptosis of breast tumor cells. Oncogene. 2010;29:5214–5219. doi: 10.1038/onc.2010.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du W, Wang S, Zhou Q, Li X, Chu J, Chang Z, et al. ADAMTS9 is a functional tumor suppressor through inhibiting AKT/mTOR pathway and associated with poor survival in gastric cancer. Oncogene. 2013;32:3319–3328. doi: 10.1038/onc.2012.359. [DOI] [PubMed] [Google Scholar]
- Cheung KF, Lam CN, Wu K, Ng EK, Chong WW, Cheng AS, et al. Characterization of the gene structure, functional significance, and clinical application of RNF180, a novel gene in gastric cancer. Cancer. 2012;118:947–959. doi: 10.1002/cncr.26189. [DOI] [PubMed] [Google Scholar]
- Xu X, Liu RF, Wan BB, Xing WM, Huang J, Han ZG. Expression of a novel gene FAM43B repressing cell proliferation is regulated by DNA methylation in hepatocellular carcinoma cell lines. Mol Cell Biochem. 2011;354:11–20. doi: 10.1007/s11010-011-0800-y. [DOI] [PubMed] [Google Scholar]
- Kioussi C, Briata P, Baek SH, Rose DW, Hamblet NS, Herman T, et al. Identification of a Wnt/β-Catenin→Pitx2 pathway mediating cell-type-specific proliferation during development. Cell. 2011;111:673–685. doi: 10.1016/s0092-8674(02)01084-x. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Gutierrez D, Li X, Bidlack F, Cao H, Wang J, et al. The LIM Homeodomain Transcription Factor LHX6: a transcriptional repressor that interacts with Pituitary homeobox 2 (PITX2) to regulate odontogenesis. J Biol Chem. 2013;288:2485–2500. doi: 10.1074/jbc.M112.402933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vadlamudi U, Espinoza HM, Ganga M, Martin DM, Liu X, Engelhardt JF, et al. PITX2, beta-catenin and LEF-1 interact to synergistically regulate the LEF-1 promoter. J Cell Sci. 2000;118:1129–1137. doi: 10.1242/jcs.01706. [DOI] [PubMed] [Google Scholar]
- Baek SH, Kioussi C, Briata P, Wang D, Nguyen HD, Ohgi KA, et al. Regulated subset of G1 growth-control genes in response to derepression by the Wnt pathway. Proc Natl Acad Sci USA. 2003;100:3245–3250. doi: 10.1073/pnas.0330217100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polakis P. Wnt signaling and cancer. Genes Dev. 2000;14:1837–1851. [PubMed] [Google Scholar]
- Liu WB, Han F, Jiang X, Yang LJ, Li YH, Liu Y, et al. ANKRD18A as a novel epigenetic regulation gene in lung cancer. Biochem Biophys Res Commun. 2012;429:180–185. doi: 10.1016/j.bbrc.2012.10.116. [DOI] [PubMed] [Google Scholar]
- Liu WB, Liu JY, Ao L, Zhou ZY, Zhou YH, Cui ZH, et al. Dynamic changes in DNA methylation during multistep rat lung carcinogenesis induced by 3-methylcholanthrene and diethylnitrosamine. Toxicol Lett. 2009;189:5–13. doi: 10.1016/j.toxlet.2009.04.022. [DOI] [PubMed] [Google Scholar]
- Liu WB, Liu JY, Ao L, Zhou ZY, Zhou YH, Cui ZH, et al. Epigenetic silencing of cell cycle regulatory genes during 3-methylcholanthrene and diethylnitrosamine induced multistep rat lung cancer. Mol Carcinog. 2010;49:556–565. doi: 10.1002/mc.20621. [DOI] [PubMed] [Google Scholar]
- Liu WB, Ao L, Zhou ZY, Cui ZH, Zhou YH, Yuan XY, et al. CpG island hypermethylation of multiple tumor suppressor genes associated with loss of their protein expression during rat lung carcinogenesis induced by 3-methylcholanthrene and diethylnitrosamine. Biochem Biophys Res Commun. 2010;402:507–514. doi: 10.1016/j.bbrc.2010.10.061. [DOI] [PubMed] [Google Scholar]
- Liu WB, Ao L, Cui ZH, Zhou ZY, Zhou YH, Yuan XY, et al. Molecular analysis of DNA repair gene methylation and protein expression during chemical-induced rat lung carcinogenesis. Biochem Biophys Res Commun. 2011;408:596–601. doi: 10.1016/j.bbrc.2011.04.067. [DOI] [PubMed] [Google Scholar]
- Liu WB, Cui ZH, Ao L, Zhou ZY, Zhou YH, Yuan XY, et al. Aberrant methylation accounts for cell adhesion-related gene silencing during 3-methylcholanthrene and diethylnitrosamine induced multistep rat lung carcinogenesis associated with overexpression of DNA methyltransferases 1 and 3a. Toxicol Appl Pharmacol. 2011;251:70–78. doi: 10.1016/j.taap.2010.12.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.