Abstract
TNF receptor-associated protein 1 (TRAP1), the main mitochondrial member of the heat shock protein (HSP) 90 family, is induced in most tumor types and is involved in the regulation of proteostasis in the mitochondria of tumor cells through the control of folding and stability of selective proteins, such as Cyclophilin D and Sorcin. Notably, we have recently demonstrated that TRAP1 also interacts with the regulatory protein particle TBP7 in the endoplasmic reticulum (ER), where it is involved in a further extra-mitochondrial quality control of nuclear-encoded mitochondrial proteins through the regulation of their ubiquitination/degradation. Here we show that TRAP1 is involved in the translational control of cancer cells through an attenuation of global protein synthesis, as evidenced by an inverse correlation between TRAP1 expression and ubiquitination/degradation of nascent stress-protective client proteins. This study demonstrates for the first time that TRAP1 is associated with ribosomes and with several translation factors in colon carcinoma cells and, remarkably, is found co-upregulated with some components of the translational apparatus (eIF4A, eIF4E, eEF1A and eEF1G) in human colorectal cancers, with potential new opportunities for therapeutic intervention in humans. Moreover, TRAP1 regulates the rate of protein synthesis through the eIF2α pathway either under basal conditions or under stress, favoring the activation of GCN2 and PERK kinases, with consequent phosphorylation of eIF2α and attenuation of cap-dependent translation. This enhances the synthesis of selective stress-responsive proteins, such as the transcription factor ATF4 and its downstream effectors BiP/Grp78, and the cystine antiporter system xCT, thereby providing protection against ER stress, oxidative damage and nutrient deprivation. Accordingly, TRAP1 silencing sensitizes cells to apoptosis induced by novel antitumoral drugs that inhibit cap-dependent translation, such as ribavirin or 4EGI-1, and reduces the ability of cells to migrate through the pores of transwell filters. These new findings target the TRAP1 network in the development of novel anti-cancer strategies.
Keywords: TRAP1, stress-adaptive response, protein synthesis, protein quality control, co-translational ubiquitination
TNF receptor-associated protein 1 (TRAP1), the only mitochondrial member of the heat shock protein (HSP)90 protein family, is involved in protection from oxidative stress and apoptosis induced by several antitumor agents and other stressors.1 Acute silencing of TRAP1 in tumor cells has been consistently associated with CypD-dependent mitochondrial apoptosis.2 TRAP1-dependent organelle-directed regulation of folding and stability of selective proteins involved in mitochondrial homeostasis, such as Cyclophilin D and Sorcin, is pivotal for the control of tumor cell proteostasis, leading to resistance to apoptosis.3 Remarkably, aberrant deregulation of TRAP1 function has been observed in colorectal4 and prostate carcinomas,5 with potential new opportunities for therapeutic intervention in humans. Evidence suggests that, despite the high homology between all members of the HSP90 chaperone family, TRAP1 has distinct functional properties.6 TRAP1 is involved in endoplasmic reticulum (ER) stress protection7, 8 and some recent findings have reported other sub-cellular localizations of this chaperone.6 In fact, we have recently demonstrated that TRAP1 also localizes in the ER, where it directly interacts with the proteasomal particle TBP7 and controls ubiquitination/degradation levels of specific substrates.9 As a consequence, the expression of two mitochondria-destined proteins, i.e. F1ATPase and the mitochondrial isoform of the calcium-binding protein Sorcin, is decreased in TRAP1 knockdown (KD) cells. Interestingly, the expression of a TRAP1 deletion mutant, which is defective of the mitochondrial targeting sequence and is therefore unable to enter mitochondria, is nonetheless sufficient for rescuing F1ATPase and Sorcin protein levels, thereby demonstrating that TRAP1 exerts an extra-mitochondrial quality control on its client proteins.9 However, the molecular mechanisms responsible for this function still remain to be elucidated. In order to further analyze the roles of TRAP1 in protein quality control, we took advantage of data suggesting that some eukaryotic HSP family members are components of the translational apparatus and function in co-translational processes.10 It is worth noting that the ubiquitin−proteasome system (UPS) itself is involved in the quality control of nascent polypeptides emerging from ribosomes and not yet folded: in fact, it has been estimated that up to 30% of newly synthesized proteins are ubiquitinated and co-translationally degraded by the UPS system,11, 12 while still bound to ribosomes.13 Interestingly, the association between the ribosomal apparatus, the elongation factor eEF1A and the regulatory proteasomal subunits Rpn10 and Rpt114 suggests that close coupling must exist between synthesis, ubiquitination and degradation. Increasing amount of evidence is accumulating on co-translational processes and on the involvement of ribosome-bound chaperones in these events, including the assembly of ribosomes, modulation of translation and targeting of proteins.15 Brandman et al. recently demonstrated that a ribosome-bound quality control complex triggers proteasome-mediated degradation of nascent peptides stalled in the elongation step.16 These data open up a new scenario in which ribosome-associated chaperones act as key regulators of cellular proteostasis through direct or indirect modulation of protein synthesis, folding, assembly and transport. Notably, translation factors are altered in expression or activity in human cancers, and alterations in these proteins and their upstream signaling pathways can have an impact on cancer progression. It has been shown that eIF4E, eIF4G and eIF4A are overexpressed in a wide range of tumors, and drugs designed to inhibit mRNA translation are currently in preclinical and early clinical development.17 Moreover, conditions often found in tumors, such as ER stress, amino acid deprivation and hypoxia, activate eIF2 kinases such as PERK and GCN2, and in turn these kinases phosphorylate eIF2α to inhibit translation.18
Starting from these observations, our study aimed at evaluating the role of TRAP1 in translational control of cancer. Indeed, we demonstrate for the first time that TRAP1 regulation occurs during the translation process through an attenuation of its client protein's synthesis upon several stresses.
Results
TRAP1 involvement in translational control is coupled to ubiquitination/degradation
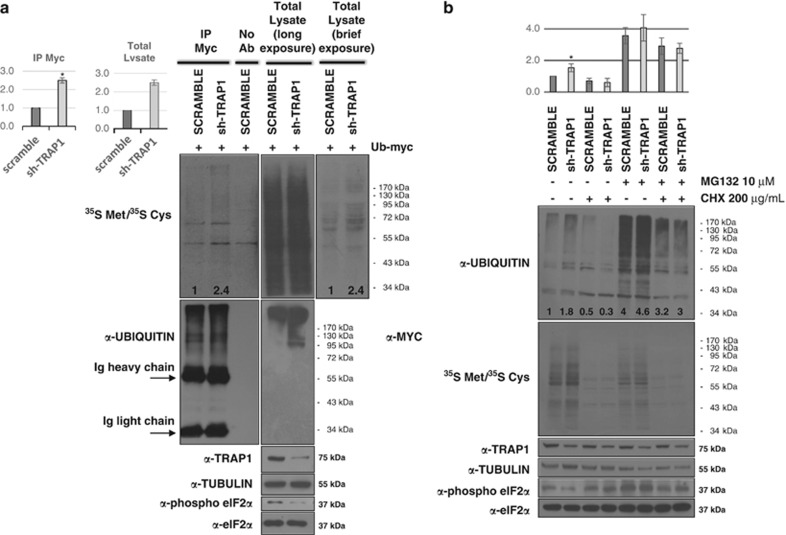
Our previous data demonstrated that TRAP1 silencing strongly increases general protein ubiquitination levels, but this does not correlate with a decreased overall protein stability.9 Taking advantage of previous data12 we hypothesized that the higher ubiquitination (and degradation) level observed in TRAP1 KD HCT116 cells might be co-translational. To achieve this, immunoprecipitates (IP) of Ub conjugates following a brief pulse (1 min) with radiolabeled Met/Cys amino acids were obtained for sh-TRAP1 and scrambled controls. We found that TRAP1-stable interfered colorectal carcinoma (CRC) cells accumulate more than double amounts of ubiquitinated proteins during protein synthesis (Figure 1a, upper panels). As a control, total ubiquitin levels were analyzed both on total lysates and anti-Myc IP upon transfection with Ub-myc expression vector (Figure 1a, middle panels). Surprisingly, control cells incorporate less radioactively labeled amino acids compared with TRAP1 KD cells (total lysates brief exposure), suggesting a role for TRAP1 in the global attenuation of translation. This hypothesis was indeed supported by reduced levels in sh-TRAP1 cells of phospho-eIF2α (Figure 1a, lower panels), which is one of the key regulatory translation factors, whose phosphorylation results in the attenuation of cap-dependent translation while favoring the IRES-dependent one.19
Figure 1.
TRAP1-dependent ubiquitination is due to a co-translational phenomenon. (a) HCT116 cells were transfected with Ubiquitin-myc expression vector. After 48 h, cells were labeled for 1 min with 35S Met/35 S Cys (250 μCi/ml), washed with PBS and lysed in the presence of 200 μg/ml CHX. IP-myc was performed on total lysates and analyzed for autoradiography (20 days) and WB with indicated antibodies. Numbers indicate densitometric intensity of the whole lane, calculated assuming level of control cells equal 1. Graphs represent statistical quantification of densitometric analysis based on two independent replicates, data are expressed as mean±S.D.; *P=0.004. (b) HCT116 cells were treated with CHX (200 μg/ml), MG132 (10 μM) or both and labeled with 35S Met/35S Cys (50 μCi/ml) for 6 h. Total extract were immunoblotted with indicated antibodies. Numbers indicate densitometric intensity of the whole lane, calculated assuming level of control cells (untreated scramble cells) equal 1. Graphs represent statistical quantification of densitometric analysis based on three independent experiments, data are expressed as mean±S.D. from; *P=0.02
To further support the co-translational nature of the high ubiquitin phenotype observed in sh-TRAP1 cells, Met/Cys-labeled-HCT116 cells were treated for 6 h with the protein synthesis inhibitor cycloheximide (CHX), the proteasome inhibitor MG132 or with both of them (Figure 1b). Notably, CHX prevented the stronger ubiquitin phenotype of sh-TRAP1 cells compared with controls, either in the presence or in the absence of MG132. Conversely, in the absence of CHX, Ub levels are higher in TRAP1 KD cells, both in the presence and in the absence of MG132 (Figure 1b, upper panel). The CHX-mediated blockage of protein synthesis reduces global Ub levels and this is per se able to abrogate differences in ubiquitination between sh-TRAP1 and control cells. Accordingly, a time-course experiment performed by treating HCT116 cells with CHX for increasing time intervals shows a progressive decrease in total ubiquitination levels, resulting in complete abolishment of differences between scramble and sh-TRAP1 cells after 6 h (Supplementary Figure S1). Taken together, these results suggest that the high-ubiquitination phenotype of sh-TRAP1 cells is dependent on protein synthesis and that TRAP1 protein quality control is co-translational. However, further studies will be necessary to determine whether this is due to a direct role of TRAP1 in this process or due to an indirect effect of TRAP1 deficiency. Incorporation of radiolabeled aminoacids is shown as a control of CHX treatment (Figure 1b, middle panel). Accordingly, eIF2α phosphorylation decreased in untreated TRAP1 KD cells, and is increased upon CHX treatment both in control and in sh-TRAP1 cells, as a consequence of complete inhibition of protein synthesis.
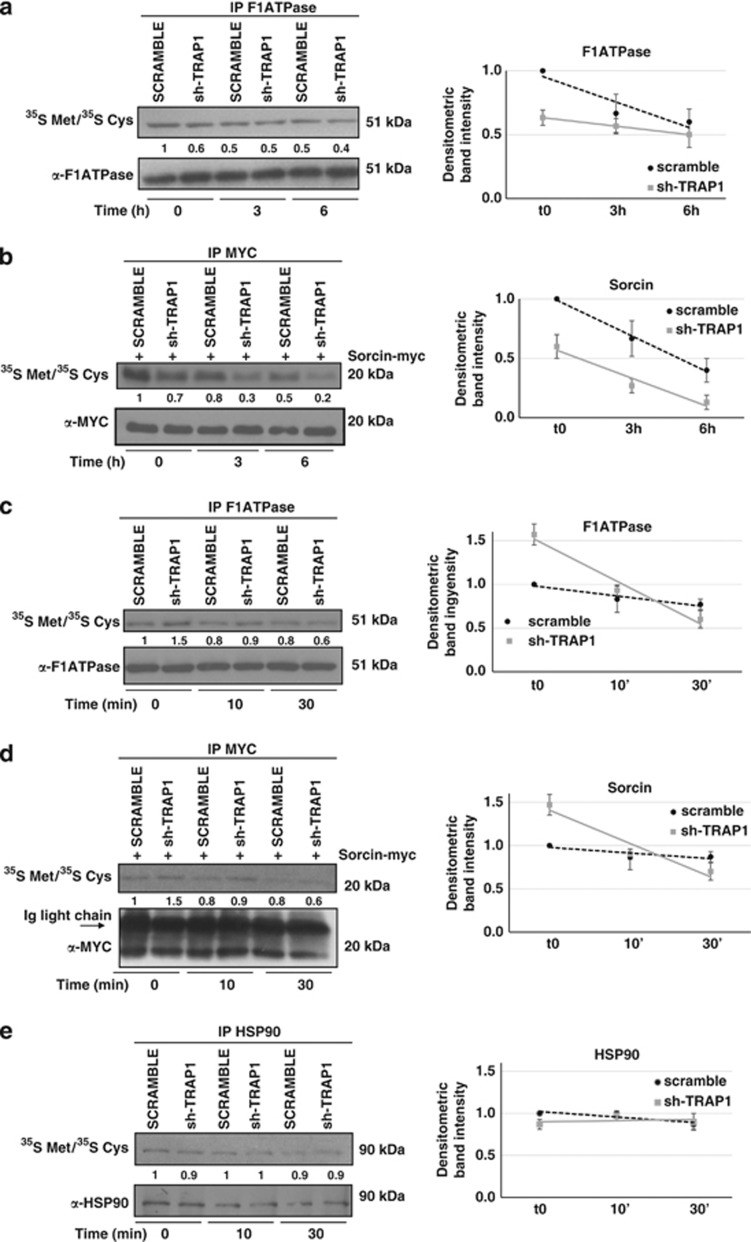
We previously reported that the expression of mitochondrial-destined TRAP1 client proteins (the mitochondrial isoform of Sorcin and the F1ATPase subunit β) was decreased in TRAP1 KD cells as a consequence of their increased ubiquitination.9 In order to analyze the mechanism of this regulation, we measured the stability of both proteins by pulse/chase experiments in TRAP1-interfered HCT116 cells. Consistent with the results obtained on total lysates, Figure 2a and b shows that, despite a difference in total protein amount at time 0, the half-lives of these proteins in TRAP1 KD and control cells are comparable up to 6 h after the pulse. Results of real-time PCR analysis of Sorcin and F1ATPase mRNAs excluded a transcriptional control (data not shown). These data suggest that TRAP1 does not influence long-term stability of client proteins. We then aimed at evaluating whether a selective change in the half-life of Sorcin and F1ATPase could be observed in the early phases of protein synthesis. To achieve this, HCT116 cells were exposed to shorter (30 min) pulses of 35S-labeled Met/Cys followed by short times of chase, to minimize contribution of protein degradation by the proteasome during the pulse: it has been reported that proteins can be degraded by proteasomes within 30 min of their synthesis.20 Indeed, in these experimental conditions, an increased rate of labeling at times 0 in the IPs of both client proteins was observed in TRAP1 KD cells compared with controls (Figure 2c and d); however, they are degraded so fast that, at the steady state, their expression levels are finally lower in sh-TRAP1 clones. Therefore, both synthesis and degradation of specific client proteins are attenuated by TRAP1. These observations suggest that TRAP1-dependent control on Sorcin and F1ATPase is exerted early after synthesis, most likely before their import into the mitochondria. The observed phenomena are selectively directed toward TRAP1 client proteins: in fact, control experiments using the cytosolic HSP90 show that it is not subject to similar mechanisms (Figure 2e).
Figure 2.
Quality control of client proteins is exerted by TRAP1 through coupling of protein synthesis and degradation. (a, b) HCT116 sh-TRAP1 and scramble stable clones were transfected, where indicated (b), with Sorcin-myc expression vector for 24 h, pulse labeled for 1 h as described in Materials and Methods and chased for the indicated times. Immunoprecipatates (IP) were analyzed by autoradiography and WB. (c, d, e) HCT116 sh-TRAP1 and scramble stable clones were transfected, where indicated (d), with Sorcin-myc expression vector for 24 h, pulse labeled for 30 min as described in Materials and Methods and chased for the indicated times. Immunoprecipatates (IP) were analyzed by autoradiography and WB. Numbers indicate densitometric band intensities, each normalized to the respective IP, that have been calculated by assuming protein levels of the control (scramble cells at time 0) equal 1. The data summary (right) shows tendency lines referred to densitometric analysis of all replicates performed. Data are expressed as mean±S.D. from three independent experiments
TRAP1 associates with ribosomes and with members of the translational apparatus
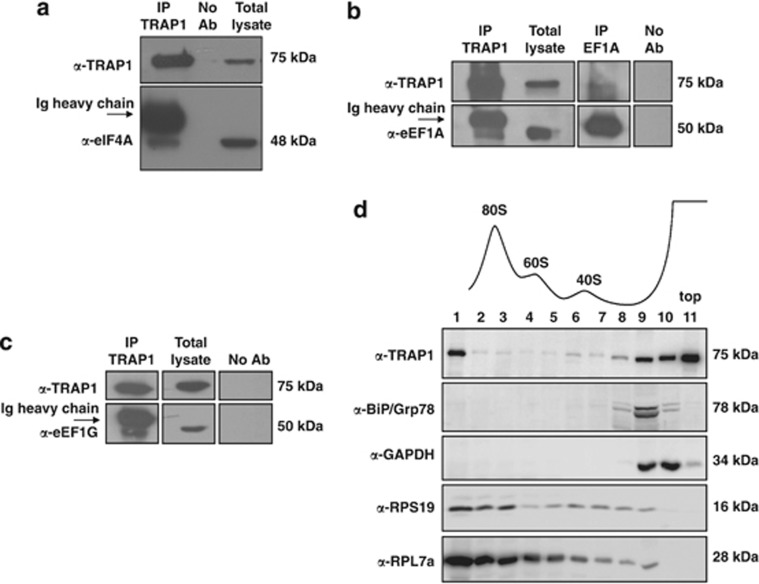
A proteomic analysis of TRAP1 co-IP complexes4, 9 previously allowed us to identify some translational initiation/elongation factors as putative TRAP1 binding proteins. We further confirmed mass spectrum data by co-IP experiments, and observed a selective binding of TRAP1 to factors eIF4A, eEF1A and eEF1G (Figure 3a and c; Supplementary Figure S2A). Accordingly, confocal microscopy analysis showed colocalization of EF1A-GFP fusion protein with TRAP1-cherry, thus supporting their interaction (Supplementary Figure S2B). The relevant finding of TRAP1 interaction with members of the protein synthesis machinery further supported a role of TRAP1 in the regulation of protein synthesis. Indeed, a physical interaction between co-translationally acting chaperones and ribosomes has been widely reported as the primary environment for the correct assembling of nascent polypeptides.15 Therefore, we analyzed whether TRAP1 associates with these organelles, observing that it is present in the ribosomal fractions purified from HCT116 cells (Supplementary Figure S2C). To have further indication on the association of TRAP1 with ribosomes, we separated cytoplasmic extracts from HCT116 cells by ultracentrifugation on sucrose gradients. As shown in Figure 3d, a clear signal of TRAP1 can be observed in the first fraction, indicating that at least part of this protein co-sediments with polysomal complexes, whereas control proteins (BiP/Grp78, glyceraldehyde-3-phosphate dehydrogenase (GAPDH)) are only present in the upper part of the gradient (fractions 9, 10, 11) that includes free cytosolic proteins or light complexes.
Figure 3.
TRAP1 interacts with translation factors and is associated to ribosomes. (a), (b) and (c) Total HCT116 lysates were immunoprecipitated with anti-TRAP1 and anti-eEF1A antibodies and immunoblotted with indicated antibodies. No Ab, total cellular extracts incubated with A/G plus agarose beads without antibody; IP, immunoprecipitation with the corresponding antibodies. (d) Separation of cytoplasmic extracts from HCT116 cells was performed by ultracentrifugation on sucrose gradients as described in Materials and Methods. Absorbance profile as well as the signals of ribosomal protein-specific antibodies indicate the fractions containing ribosomal particles (40S, 60S, 80S) whereas larger polysomes were collected in the first fraction. The upper part of the gradient (fractions 9,10,11) includes free cytosolic proteins or light complexes
TRAP1 regulates the eIF2α pathway
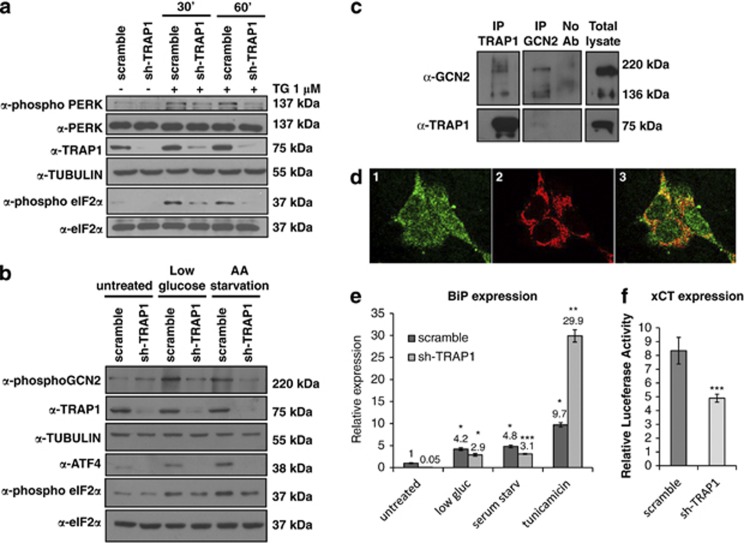
To characterize the signal transduction pathway responsible for the attenuation of protein synthesis by TRAP1, we focused on the eIF2α upstream protein kinases involved in the most common stress adaptive responses of cancer cells.21 Indeed, eIF2α phosphorylation is the first-line response to ER stress and allows the immediate attenuation of global protein synthesis and selective translation of stress-responsive genes, which confers the ability to rapidly respond to changes in extracellular environments. As both PERK and GCN2 kinases are involved in regulation of eIF2α phosphorylation, we further investigated these two pathways in colorectal and breast cancer (data not shown) TRAP1 KD cells. Of note, TRAP1-containing cells exhibit higher activation of PERK (Figure 4a) and, consequently, higher amounts of phospho-eIF2α either before or after ER stress induced by thapsigargin (TG), this being consistent with our previous data on TRAP1 role in resistance to ER stress.9 In parallel experiments, we found higher levels of phospho-GCN2 in TRAP1 scrambled controls than in sh-TRAP1 cells in response to nutrient deprivation (Figure 4b). In addition, a specific interaction between TRAP1 and GCN2 was identified, as confirmed by co-IP experiments (Figure 4c) and confocal microscopy (Figure 4d). This evidence suggests that TRAP1 KD cells are unable to promptly respond to ER stress or nutrient deprivation through PERK/GCN2 activation.
Figure 4.
TRAP1 regulates eIF2α-ATF4 pathway. (a) Scrambled and sh-TRAP1 HCT116 cells were treated with TG (1 μM) for the indicated times. Total lysates were immunoblotted with indicated antibodies. (b) HCT116 cells were starved for 6 h in cysteine/methionine/glutammine-free medium or low glucose (1.5 mg/ml) medium respectively. Total lysates were immunoblotted with the indicated antibodies. (c) TRAP1 and GCN2 co-immunoprecipitate: total HCT116 lysates were immunoprecipitated with anti-TRAP1 and anti-GCN2 antibodies and immunoblotted with indicated antibodies. No Ab, total cellular extracts incubated with A/G plus agarose beads without antibody; IP, immunoprecipitation with the corresponding antibodies. (d) TRAP1/GCN2 colocalization: HCT116 cells were fixed and treated as described in Materials and Methods. Panel 1 shows staining of GCN2 (green); panel 2 shows staining of TRAP1 (red); in Panel 3, a double immunofluorescent staining is shown. (e) Real-time RT-PCR analysis of BiP/Grp78 mRNA expression in scrambled and sh-TRAP1 HCT116 cells exposed to 1 μM TG for 12 h or starved for 48 h in low serum or low glucose medium. All data are expressed as mean±S.D. from two independent experiments; *P≤0.01, **P≤0.001, ***P≤0.0001, n=2. (f) Luciferase assay on xCT promoter constructs transfected in sh-TRAP1 HCT116 cells and scrambled control. All data are expressed as mean±S.D. from two independent experiments; *P≤0.01, **P≤0.001, ***P≤0.0001, n=2
It has been demonstrated that eIF2α phosphorylation induces the preferential translation of a group of stress-responsive mRNAs, including the transcription factor ATF4: accordingly, higher levels of ATF4 were detected in scrambled cells upon amino acid or glucose starvation (Figure 4b). Of note, the re-expression of TRAP1 in sh-TRAP1 cells rescues ATF4 levels (Supplementary Figure S3A), whereas its transient silencing by siRNAs decreases phospho-eIF2α and ATF4 levels (Supplementary Figure S3B).
Preferential activation of eIF2α upregulates ATF4 target genes involved in amino acid synthesis and transport22 as well as in response to oxidative or ER stress, and, among others, xCT, the specific subunit of cystine/glutamate antiporter system,23 and BiP/Grp78, a major ER chaperone essential for protein quality control in the ER.24 Quantitative RT-PCR analysis to assay BiP/Grp78 mRNA levels shows that the expression of this stress-responsive gene is lower in untreated sh-TRAP1 stable transfectants than in control clones; notably, a correlation between TRAP1 and BiP/Grp78 was confirmed in in vivo studies on human breast tumor specimens.25 Remarkably, under stress conditions (i.e., starvation, glucose deprivation, tunicamycin-induced ER stress) the induction of BiP/Grp78 is higher in sh-TRAP1 cells in order to compensate for basal differences in its transcriptional level (Figure 4e). It is worth noting that the results of tunicamycin treatment support our previous data demonstrating the important role of TRAP1 in protection from ER stress.9 Similarly, Luciferase assays using xCT constructs transfected in HCT116 cells confirmed the higher activation of the Luc-reporter gene by the oxidant-responsive xCT promoter in scrambled cells compared with sh-TRAP1 cells (Figure 4f).
Clinical relevance of TRAP1 role in protein synthesis
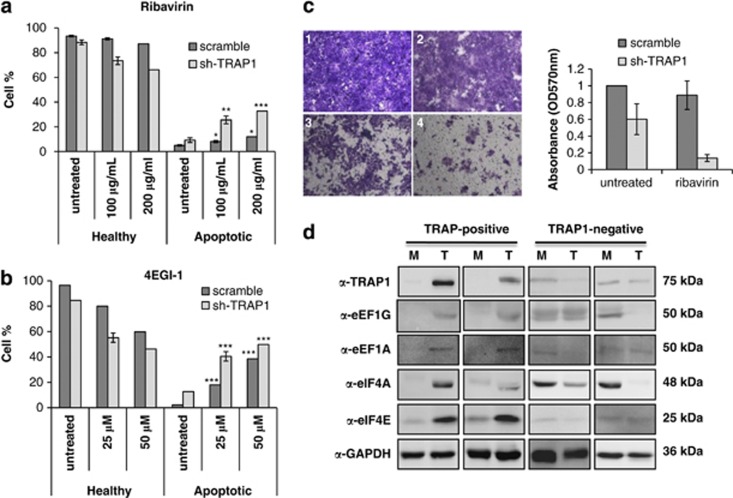
Drugs designed to inhibit mRNA translation are in preclinical development with the aim of becoming anticancer agents.17 Considering the above-demonstrated role of TRAP1 in translational attenuation, we selected two inhibitors of mRNA translation, with the aim of analyzing the response of control and TRAP1 KD cells to these drugs: Ribavirin, an inhibitor of cap-mediated translation,26, 27 and 4EGI-1, a synthetic peptide that binds the translational initiation factor eIF4E and prevents its interaction with eIF4G.28 As shown in Figure 5a and b, a more significant increase in the rate of apoptotic cell death can be observed in sh-TRAP1 stable transfectants. Conversely, cells containing higher TRAP1 levels seem to be less sensitive, especially when it comes to the apoptotic response to Ribavirin. The low sensitivity of TRAP1-containing cells to blockers of cap-dependent mRNA translation is not surprising, considering that an attenuation of protein synthesis is already present in these cells.
Figure 5.
Downregulation of TRAP1 sensitizes HCT116 cells to drugs targeting cap-dependent translation. (a, b) Rates of apoptotic cell death in HCT116 cells treated with Ribavirin (a) (100 or 200 μg/ml) or 4EGI-1 (b) (25 or 50 μM) for 48 h upon stable downregulation of TRAP1. All data are expressed as mean±S.D. from three independent experiments; *P≤0.01, **P≤0.001, ***P≤0.0001, n=3. (c) Representative images (left) of transwell migration assay using 10% FBS as chemoattractant. The data summary (right) shows absorbance of the Crystal violet staining eluted by migrated cells and quantitated by spectrophotometric reading at 570 nm. (d) TRAP1, eIF4A, eIF4E and eEF1G, eEF1A expression in human CRCs. Total cell lysates from four human CRCs (T) and the respective non-infiltrated peritumoral mucosas (M) (2 TRAP1-positive and 2 TRAP1-negative tumors) were separated by SDS-PAGE and immunoblotted using indicated antibodies
It has been proposed that agents interfering with the regulatory mechanism of gene translation (Rapamycin, Ribavirin) could be regarded as leading compounds in the antimetastatic drug development process.29 From these observations and taking into account the fact that a role of TRAP1 in metastatic prostate cancers has been recently demonstrated,5, 30 we analyzed the role of TRAP1 in the multistep process of metastasis by measuring the capability of HCT116 cells to migrate through filters of the Boyden chamber upon TRAP1 interference, and in the presence/absence of mRNA translation inhibitor Ribavirin. As shown in Figure 5c, TRAP1 silencing reduced the ability of CRC cells to migrate. Such a migratory effect was further impaired by Ribavirin, suggesting that the combination of this drug with TRAP1 blockade may efficiently counteract a fundamental step in the metastatic cascade.
Finally, we evaluated whether TRAP1-dependent regulation of the rate of protein synthesis may be relevant in human CRC. To this aim, we analyzed our tissue collection of CRCs, previously characterized for the co-expression of TRAP1 and its client proteins Sorcin, F1ATPase and TBP7.9 We analyzed 10 TRAP1-positive and 10 TRAP1-negative human CRCs for eEF1G, eEF1A, eIF4A and eIF4E expression. Figure 5d reports the immunoblot analysis of these proteins in 4 tumor samples representative of our tumor analysis, whereas their full expression profile in all 20 CRCs is reported in Table 1. Remarkably, the majority of the TRAP1-positive tumors exhibited upregulation of eEF1G (7/10 cases), eEF1A (8/10), eIF4A (5/10 cases) and eIF4E (8/10 cases). In contrast, among 10 tumors with low expression of TRAP1, all exhibited low levels of eIF4A and eIF4E, 9/10 exhibited low expression of eEF1G and 7/10 exhibited low expression of eEF1A. A Chi-square test demonstrated a positive statistical correlation between the expression levels of TRAP1 and those of eEF1G (P=0.02), eIF4A (P=0.039) and eIF4E (P=0.001) and a trend toward a positive correlation between TRAP1 and eEF1A levels (P=0.07).
Table 1. Expression profile of the 4 genes in 20 CRCs.
| eEF1G (P=0.02) | eIF4A (P=0.039) | eIF4E (P=0.001) | eEF1A (P=0.07) | |||||
|---|---|---|---|---|---|---|---|---|
| + | − | + | − | + | − | + | − | |
| TRAP1+ | 7 | 30 | 4 | 6 | 8 | 2 | 8 | 2 |
| TRAP1− | 1 | 9 | 0 | 10 | 0 | 10 | 3 | 7 |
The expression was considered positive when densitometric analysis showed a value higher than 2 compared to surrounding healthy mucosa. For details on case numbers, refer to Supplementary Table 1
Discussion
The best-characterized functions of TRAP1, which is induced in most tumor types, are related to its role in the protection from oxidative stress and mitochondrial cell death6 and in signaling circuitries of mitochondrial integrity and cellular homeostasis.31 However, recent evidence suggests that TRAP1 is involved in the crosstalk between mitochondria and ER and in ER stress protection of tumor cells: in fact, our group for the first time reported a role of TRAP1 in protein quality control due to its interaction with proteasomal components on the outer side of the ER.9
A key finding of the present work, which is a logical continuation and deepening of our previous studies, is the inverse correlation between TRAP1 expression and ubiquitination/degradation of some nascent mitochondrial client proteins. However, further studies are required to provide molecular insights into the mechanism. The regulation of TRAP1 client protein expression in the ER depends on their ubiquitination and does not involve a folding control or changes in the overall protein stability. Several observations argue against a control of folding by TRAP1 in the ER: as TRAP1 client proteins are mitochondrial, they must enter unfolded into the mitochondrial compartment; evidence on the association of ribosomes and mRNA to mitochondria implies that nuclear-encoded mitochondrial proteins can be imported while being translated32 and chaperone linked to protein synthesis provides relevant ‘assistance' to these processes.33, 34 Recent evidence demonstrates that nascent polypeptides often cannot fold stably and are subjected to a robust quality control system that marks them for degradation through co-translational ubiquitination, either during active synthesis, in response to misfolding, or in response to translation stalling.35 Moreover, it has been shown that ribosome-bound molecular chaperones contribute to protect nascent chains from premature ubiquitination and optimize the balance between translation rate and co-translational folding, avoiding aggregation and minimizing co-translational ubiquitination.36 The experimental evidence provided in this article, including the association of TRAP1 with ribosomes and with several translation initiation and elongation factors, suggests that TRAP1 chaperoning activity in the ER involves the regulation of its client protein's synthesis and degradation, most likely at the ER/mitochondria interface. It is worth noting that this novel quality control regulation by ER-associated TRAP1 is additional, non-redundant, complementary and relevant for the antiapoptotic role and for the well-known function of folding regulation by TRAP1 in the mitochondria.2
When the cell faces stress conditions, e.g. starvation or hypoxia, the rate of protein synthesis decreases. In fact, sh-TRAP1 cells show lower levels of phospho-eIF2α in basal conditions and are unable to respond promptly to ER stress and nutrient deprivation by activating PERK and GCN2. Both amino acid and glucose deprivation, stresses found in solid tumors, activate GCN2 to yield an upregulation of ATF4 target genes involved in amino acid synthesis and transport,22 thus making the GCN2-eIF2α-ATF4 pathway critical for maintaining metabolic homeostasis in tumor cells and, in turn, a novel and attractive target for anti-tumor approaches. Consistently, we demonstrated that TRAP1-dependent activation of the GCN2-eIF2α pathway results in a consequent upregulation of ATF4 and induction of its target genes BiP/Grp78 and xCT. This evidence unravels new pathways and protein targets regulated by TRAP1 to yield protection from ER stress, oxidative damage and cell death. Furthermore, BiP/xCT analysis suggests that, under basal conditions, TRAP1 indirectly affects the expression of some genes, especially those involved in the regulation of cellular homeostasis, behaving as a sort of energy saver for the cell. Remarkably, the relevance of these findings is supported by in vivo studies on human breast tumor specimens,25 SAOS-2 osteosarcoma cells, HeLa cervix carcinoma cells and mouse models.36
In our opinion, a relevant result of our study is the evidence that TRAP1 selectively binds members of the translational apparatus (i. e., eIF4A, eEF1A and eEF1G); this observation is herein reported for the first time. Furthermore, TRAP1 is co-expressed with eIF4A, eIF4E, eEF1G and eEF1A in human CRCs, suggesting that a subset of human CRCs upregulates the translational machinery to fulfill the increased demand of protein synthesis upon increase in cell proliferation.17 In this context, the co-upregulation of TRAP1 may, likely, represent a protective mechanism used by cancer cells to induce the synthesis of specific stress-protective proteins.
Targeting specific translation factors that are altered in expression or activity in human cancers offers great promise for the development of a new generation of cancer therapeutics.17 The interaction of TRAP1 with several translation regulatory proteins might have an important impact on their development. Indeed, TRAP1-positive tumor cells exhibit poor responsiveness to mRNA translation inhibitors, whereas the same agents induce higher levels of apoptosis in TRAP1 KD cells. This observation provides useful insights into the mechanism of resistance to these novel agents, which may be poorly active on TRAP1-positive tumors. Furthermore, such evidence is in line with previous observations by our group showing that TRAP1-positive CRC cells are poorly sensitive to chemotherapeutics.37 Altogether this evidence strongly supports the hypothesis that the activation of the TRAP1 pathway represents a major survival mechanism used by cancer cells to escape the activity of several antitumor agents and targets the TRAP1 network in the development of novel anti-cancer drugs to revert resistance to apoptosis.
Finally, our result of ex vivo migration suggests that TRAP1 has a pivotal role in CRC migration, supporting the conclusion that this chaperone is an important element in the multistep process of tumor progression and metastasis dissemination.5, 29 Hence, the inhibition of TRAP1 and TRAP1-interacting translation factors might become a novel approach for successful therapies in human malignancies; in addition, the association of translation inhibitors, which are already in preclinical development, with TRAP1 targeting may efficiently counteract a fundamental step of the metastatic cascade.
Materials and Methods
Cell culture
Human HCT116 colon carcinoma cells were purchased from American Type Culture Collection (ATCC) and cultured in DMEM containing 10% fetal bovine serum and 1.5 mmol/l glutamine. Cell lines are routinely monitored in our laboratory by microscopic morphology check. The authenticity of the cell lines was checked before starting this study, 1 year ago, through standardized techniques by evaluating the presence of a mutation in codon 13 of the Ras proto-oncogene in HCT116 cells, according to ATCC product description. TRAP1-stable interfered cells were obtained as described previously.9
Plasmid generation and transfection procedures
Full-length TRAP1-myc expression vector was obtained as described previously.9 xCT-luc construct was a kind gift from Prof. R Faraonio, University of Naples Federico II, Italy, and was obtained as described previously.38 The eEF1A1-GFP construct was generated using the following primers: forward 5′-ATTAGAATTCTGACTAAGGCCATGGGAAAGGAAAAGACTC-3′ and reverse 5′-ATTAGGATCCAATTTAGCCTTCTGAGCTTTCTGGGC-3′. eEF1A1-6x-his construct, which was a kind gift from A. Lamberti, University of Naples Federico II, was used as a template. PCR-amplified fragments were gel-purified and cloned in-frame into the peGFP-N1 plasmid (Clontech) at the EcoRI and BamHI restriction sites. The TRAP1-cherry construct was generated by cloning a PCR-amplified mCHERRY fragment in-frame into the TRAP1-myc construct at the XhoI and XbaI restriction sites. mCHERRY was amplified using the following primers: forward 5′-ATTACTCGAGGGCGGAGGTGTGAGCAAGG-3′ and reverse 5′-ATTATCTAGACTACTTGTACAGCTC GTCCATGCC-3′. Transient transfection of DNA plasmids was performed with the Polyfect Transfection Reagent (Qiagen). siRNAs of TRAP1 were purchased from Qiagen (cat. no. SI00115150). For control experiments, cells were transfected with a similar amount of scrambled siRNA (Qiagen; cat. no. SI03650318). Transient transfections of siRNAs were performed by using HiPerFect Transfection Reagent (Qiagen) according to the manufacturer's protocol.
WB/Immunoprecipitation analysis
Equal amounts of protein from cell lysates and tumor specimens were subjected to SDS-PAGE and transferred to a PVDF membrane (Millipore). WB analyses were performed as described in.39 Protein immunoprecipitations were carried out on 800 μg of total extracts. Lysates were pre-cleared by incubating with protein A/G-Agarose (Santa Cruz Biotechnologies) for 1 h at 4 °C and then incubated in agitation for 18 h at 4 °C with the antibodies. Subsequently, samples were further incubated for 1 h at 4 °C with fresh beads. Negative control experiments were performed by adding beads only to the cleared lysate. Beads were then collected by centrifugation and washed twice in lysis buffer. Where indicated, protein levels were quantified by densitometric analysis using the software ImageJ.40 The following antibodies from Santa Cruz Biotechnology were used for WB, immunoprecipitation and immunofluorescence analysis: anti-TRAP1 (sc-13557), anti-cMyc (sc-40), anti-Ub (sc-8017), anti-F1ATPase (ATP5B subunit, sc-58619 for IP and sc-C20 for WB), anti-tubulin (sc-8035), anti-HSP90 (sc-1057), anti-eIF2α (sc-133132), anti-CREB2/ATF4 (sc-200), anti-GCN2 (sc-374609 for IP and sc-46338 for IF), anti-GAPDH (sc-69778), anti-pPERK (Thr 981 and sc-32577) and anti-BiP/Grp78 (sc-1051). The following antibodies were also used: anti-phospho eIF2α (p Ser51, #9271), anti-eIF4A (#2013), anti-eIF4E (#2067) and anti-PERK (#3192) from Cell Signaling; anti-phospho GCN2 (p Thr667, #NBP1-4031) and anti-eEF1G (#NB100-223) from Novus Bio; anti-eEF1A (#05-235) from Millipore; and anti-RPS19.41 Rabbit polyclonal anti-RPL3 and rabbit polyclonal anti-L7a antibodies were a kind gift from Prof. G Russo, University of Naples Federico II, Italy.
Ribosome analysis
Preparation of ribosomal fractions was performed as described previously.42 To further separate cytoplasmic extracts, HCT116 was collected by scraping and then resuspended in lysis buffer (10 mM Tris-HCl (pH 7.5), 10 mM NaCl and 10 mM MgCl2, 1 mg/ml 0.5% NP-40, aprotinin, 1 mg/ml leupeptin, 1 mg/ml pepstatinA and 100 mg/ml PMSF). After incubation in ice for 10 min, the extract was centrifuged for 10 min in a microcentrifuge at a maximum speed of 4 °C and the supernatant (cytoplasmic extract) was loaded onto 10–30% linear sucrose gradient containing 30 mM Tris–HCl (pH 7.5), 100 mM NaCl and 10 mM MgCl2. Gradients were centrifuged in a Beckman SW 41 rotor for 4 h at 37000 rpm and then collected in 11 fractions while monitoring the absorbance at 260 nm. The pellet was resuspended in the first fraction (polysomes). All fractions were then precipitated with trichloroacetic acid, resuspended in loading buffer and analyzed by western blot.
Pulse-chase assay
Pulse-chase analysis was performed as described previously.9 Cells were collected at the indicated time points and lysates were separated by 10% SDS-PAGE. Proteins were transferred onto a PVDF membrane and analyzed by autoradiography. The same filters were then probed by WB analysis.
Confocal microscopy
HCT116 cells were fixed with 0.1 M phosphate buffer containing 4% (w/v) paraformaldehyde for 15 min, then blocked and permeabilized with 5% (w/v) BSA, 0.1% (v/v) Triton X-100 and 10% (v/v) FBS in PBS for 20 min at RT and finally stained with primary antibodies and the corresponding secondary TEXAS RED/FITC-conjugated antibodies. Immunofluorescence was analyzed by confocal laser-scanning microscopy using Zeiss 510 LSM (Carl Zeiss Microimaging), equipped with an Argon ionic laser (Carl Zeiss Microimaging) whose wavelength was set to 488 nm, a He–Ne laser whose wavelength was set to 546 nm and with an oil immersion x63/1.4 f objective.
RNA Extraction and real-time RT-PCR analysis
RNA extraction procedures were performed as described in.9 The following primers were used for PCR analysis. BiP/Grp78 – forward 5′-CGTGGATGACCCGTCTGTG-3′, reverse: 5′-CGTCTTTGGTTGCTTGGC-3′ GAPDH – forward 5′-AGGCTGAGAACGGGAAGC-3′, reverse: 5′-CCATGGTGGTGAAGACGC-3′ F1ATPase – forward 5′-GGACTATTGCTATGGATGGTACAG-3′, reverse 5′-CCATGAACTCTGGAGCCTC-3′ Sorcin – forward 5′-GGCCACTCTGCAAGAAGG-3′, reverse 5′-CTATCTGCCCATCCTGTCC-3′. Primers were designed to be intron-spanning. The reaction conditions were 50 °C for 2 min and 95 °C for 2 min, followed by 45 cycles of 15 s at 95 °C, 30 s at 60 °C and 30 s at 72 °C. GAPDH was chosen as the internal control.
Luciferase assay
Luciferase assay was performed with a Dual-Luciferase reporter assay system (Promega) according to the manufacturer's protocol. Firefly luciferase activity was normalized to Renilla internal control luminescence.
Apoptosis and in vitro migration assay
HCT116 cells were treated with Ribavirin (100 or 200 μg/ml) or 4EGI-1 (25 or 50 μM) for 48 h. Apoptosis was evaluated as described in.9 Experiments were performed three times using three replicates for each experimental condition. Migration was assayed using a modified Boyden chamber (Corning Costar) containing a polycarbonate membrane filter (6.5 mm diameter, 8 μm pore size) coated with poly-L-lysine.43 The upper chamber contained cells in DMEM plus 1% FBS in the absence or presence of Ribavirin (100 μg/ml) and the lower chamber contained DMEM plus 10% FBS as a chemoattractant. Cells were incubated for 48 h at 37 °C in 5% CO2. Non-migrated cells were scraped off the upper surface of the membrane with a cotton swab. Migrated cells remaining at the bottom surface were stained with Crystal violet. Staining was eluted with 100 μl of 1% SDS and quantitated by spectrophotometric reading at 570 nm.
Patients
Tumor and normal, non-infiltrated peritumoral mucosa were obtained from 20 patients with CRC during surgical removal of the neoplasm.9 Express written informed consent to use biological specimens for investigational procedures was obtained from all patients.
Statistical analysis
The χ2-Test was used to establish statistical correlation between the expression levels of TRAP1 and those of eIF4A,eIF4E, eEF1A and eEF1G in human CRCs and between apoptotic and gene expression levels in treated versus untreated scramble/sh cells. Statistically significant values (P<0.05) are reported in Table 1. The paired Student T test was used to establish the statistical significance between different levels of apoptosis or gene expression in controls when compared with treated cells or in sh-TRAP1 cells compared with related scramble controls.
All the experiments shown in this article were independently performed three times with similar results, unless otherwise specified.
Acknowledgments
Our special thanks to Professor John Credico for proofreading the manuscript and for suggesting stylistic improvements. This work was supported by the Associazione Italiana per la Ricerca sul Cancro (AIRC) (Grant IG13128), by the Italian Ministry of Health (Grant GR-2010-2310057) to ML and FE and by POR Campania FSE 2007-2013, Project CREME.
Glossary
- TRAP1
TNF receptor-associated protein 1
- HSP
heat shock protein
- KD
knockdown
- UPR
unfolded protein response
- ER
endoplasmic reticulum
- CRC
colorectal carcinoma
- TG
thapsigargin
- Ub
ubiquitin
- UPS
ubiquitin–proteasome system
- shRNA
short-hairpin RNA
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- CHX
cycloheximide
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Disease website (http://www.nature.com/cddis)
Edited by GM Fimia
Supplementary Material
References
- Montesano Gesualdi N, Chirico G, Pirozzi G, Costantino E, Landriscina M, Esposito F. Tumor necrosis factor-associated protein 1 (TRAP-1) protects cells from oxidative stress and apoptosis. Stress. 2007;10:342–350. doi: 10.1080/10253890701314863. [DOI] [PubMed] [Google Scholar]
- Kang BH, Plescia J, Dohi T, Rosa J, Doxsey SJ, Altieri DC. Regulation of tumor cell mitochondrial homeostasis by an organelle-specific Hsp90 chaperone network. Cell. 2007;131:257–270. doi: 10.1016/j.cell.2007.08.028. [DOI] [PubMed] [Google Scholar]
- Plescia J, Salz W, Xia F, Pennati M, Zaffaroni N, Daidone MG, et al. Rational design of shepherdin, a novel anticancer agent. Cancer Cell. 2005;7:457–468. doi: 10.1016/j.ccr.2005.03.035. [DOI] [PubMed] [Google Scholar]
- Landriscina M, Laudiero G, Maddalena F, Amoroso MR, Piscazzi A, Cozzolino F, et al. Mitochondrial chaperone Trap1 and the calcium binding protein Sorcin interact and protect cells against apoptosis induced by antiblastic agents. Cancer Res. 2010;70:6577–6586. doi: 10.1158/0008-5472.CAN-10-1256. [DOI] [PubMed] [Google Scholar]
- Leav I, Plescia J, Goel HL, Li J, Jiang Z, Languino LR, et al. Cytoprotective mitochondrial chaperone TRAP-1 as a novel molecular target in localized and metastatic prostate cancer. Am J Pathol. 2010;176:393–401. doi: 10.2353/ajpath.2010.090521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matassa DS, Amoroso MR, Maddalena F, Landriscina M, Esposito F. New insights into TRAP1 pathway. Am J Cancer Res. 2012;2:235–248. [PMC free article] [PubMed] [Google Scholar]
- Maddalena F, Laudiero G, Piscazzi A, Secondo A, Scorziello A, Lombardi V, et al. Sorcin induces a drug-resistant phenotype in human colorectal cancer by modulating Ca2+ homeostasis. Cancer Res. 2011;71:7659–7669. doi: 10.1158/0008-5472.CAN-11-2172. [DOI] [PubMed] [Google Scholar]
- Takemoto K, Miyata S, Takamura H, Katayama T, Tohyama M. Mitochondrial TRAP1 regulates the unfolded protein response in the endoplasmic reticulum. Neurochem Int. 2011;58:880–887. doi: 10.1016/j.neuint.2011.02.015. [DOI] [PubMed] [Google Scholar]
- Amoroso MR, Matassa DS, Laudiero G, Egorova AV, Polishchuk RS, Maddalena F, et al. TRAP1 and the proteasome regulatory particle TBP7/Rpt3 interact in the endoplasmic reticulum and control cellular ubiquitination of specific mitochondrial proteins. Cell Death Differ. 2012;19:592–604. doi: 10.1038/cdd.2011.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rospert S, Chacinska A. Distinct yet linked: chaperone networks in the eukaryotic cytosol. Genome Biol. 2006;7:208. doi: 10.1186/gb-2006-7-3-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert U, Anton LC, Gibbs J, Norbury CC, Yewdell JW, Bennink JR. Rapid degradation of a large fraction of newly synthesized proteins by proteasomes. Nature. 2000;404:770–774. doi: 10.1038/35008096. [DOI] [PubMed] [Google Scholar]
- Wang F, Durfee LA, Huibregtse JM. A cotranslational ubiquitination pathway for quality control of misfolded proteins. Mol Cell. 2013;50:368–378. doi: 10.1016/j.molcel.2013.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bengtson MH, Joazeiro CA. Role of a ribosome-associated E3 ubiquitin ligase in protein quality control. Nature. 2010;467:470–473. doi: 10.1038/nature09371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang SM, Chen L, Lambertson D, Anand M, Kinzy TG, Madura K. Proteasome-mediated degradation of cotranslationally damaged proteins involves translation elongation factor 1A. Mol Cell Biol. 2005;25:403–413. doi: 10.1128/MCB.25.1.403-413.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preissler S, Deuerling E. Ribosome-associated chaperones as key players in proteostasis. Trends Biochem Sci. 2012;37:274–283. doi: 10.1016/j.tibs.2012.03.002. [DOI] [PubMed] [Google Scholar]
- Brandman O, Stewart-Ornstein J, Wong D, Larson A, Williams CC, Li GW, et al. A ribosome-bound quality control complex triggers degradation of nascent peptides and signals translation stress. Cell. 2012;151:1042–1054. doi: 10.1016/j.cell.2012.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blagden SP, Willis AE. The biological and therapeutic relevance of mRNA translation in cancer. Nat Rev Clin Oncol. 2011;8:280–291. doi: 10.1038/nrclinonc.2011.16. [DOI] [PubMed] [Google Scholar]
- Dang Do AN, Kimball SR, Cavener DR, Jefferson LS. eIF2 α kinases GCN2 and PERK modulate transcription and translation of distinct sets of mRNAs in mouse liver. Physiol Genomics. 2009;38:328–341. doi: 10.1152/physiolgenomics.90396.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spriggs KA, Stoneley M, Bushell M, Willis AE. Re-programming of translation following cell stress allows IRES-mediated translation to predominate. Biol Cell. 2008;100:27–38. doi: 10.1042/BC20070098. [DOI] [PubMed] [Google Scholar]
- Qian SB, Princiotta MF, Bennink JR, Yewdell JW. Characterization of rapidly degraded polypeptides in mammalian cells reveals a novel layer of nascent protein quality control. J Biol Chem. 2006;281:392–400. doi: 10.1074/jbc.M509126200. [DOI] [PubMed] [Google Scholar]
- Moreno-Torres M, Murguía JR. Between Scylla and Charibdis: eIF2alpha kinases as targets for cancer chemotherapy. Clin Transl Oncol. 2011;13:442–445. doi: 10.1007/s12094-011-0680-3. [DOI] [PubMed] [Google Scholar]
- Ye J, Kumanova M, Hart LS, Sloane K, Zhang H, De Panis DN, et al. The GCN2-ATF4 pathway is critical for tumour cell survival and proliferation in response to nutrient deprivation. EMBO J. 2010;29:2082–2096. doi: 10.1038/emboj.2010.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewerenz J, Maher P. Basal levels of eIF2alpha phosphorylation determine cellular antioxidant status by regulating ATF4 and xCT expression. J Biol Chem. 2009;284:1106–1115. doi: 10.1074/jbc.M807325200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen WT, Lee AS. Measurement and modification of the expression level of the chaperone protein and signaling regulator GRP78/BiP in mammalian cells. Methods Enzymol. 2011;490:217–233. doi: 10.1016/B978-0-12-385114-7.00013-1. [DOI] [PubMed] [Google Scholar]
- Maddalena F, Sisinni L, Lettini G, Condelli V, Matassa DS, Piscazzi A, et al. The role of TRAP1 in ER stress protection and protein quality control is relevant for its antiapoptotic activity toward paclitaxel in breast carcinoma cells. Mol Oncol. 2013;7:895–906. doi: 10.1016/j.molonc.2013.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kentsis A, Topisirovic I, Culjkovic B, Shao L, Borden KL. Ribavirin suppresses eIF4E-mediated oncogenic transformation by physical mimicry of the 7-methyl guanosine mRNA cap. Proc Natl Acad Sci USA. 2004;101:18105–18110. doi: 10.1073/pnas.0406927102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kentsis A, Volpon L, Topisirovic I, Soll CE, Culjkovic B, Shao L, et al. Further evidence that ribavirin interacts with eIF4E. RNA. 2005;11:1762–1766. doi: 10.1261/rna.2238705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moerke NJ, Aktas H, Chen H, Cantel S, Reibarkh MY, Fahmy A, et al. Small-molecule inhibition of the interaction between the translation initiation factors eIF4E and eIF4G. Cell. 2007;128:257–267. doi: 10.1016/j.cell.2006.11.046. [DOI] [PubMed] [Google Scholar]
- Jeney A, Kenessey I, Timár F, Oláh J, Pogány G, Babò I, et al. Study of drugs against neoplastic metastasis. Magy Onkol. 2006;50:93–100. [PubMed] [Google Scholar]
- Gao JY, Song BR, Peng JJ, Lu YM. Correlation between mitochondrial TRAP-1 expression and lymph node metastasis in colorectal cancer. World J Gastroenterol. 2012;18:5965–5971. doi: 10.3748/wjg.v18.i41.5965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altieri DC, Stein GS, Lian JB, Languino L. TRAP-1 the mitochondrial Hsp90. Biochim Biophys Acta. 2012;1823:767–773. doi: 10.1016/j.bbamcr.2011.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed AU, Fisher PR. Import of nuclear-encoded mitochondrial proteins: a cotranslational perspective. Int Rev Cell Mol Biol. 2009;273:49–68. doi: 10.1016/S1937-6448(08)01802-9. [DOI] [PubMed] [Google Scholar]
- Albanese V, Yam AY, Baughman J, Parnot C, Frydman J. Systems analyses reveal two chaperone networks with distinct functions in eukaryotic cells. Cell. 2006;124:75–88. doi: 10.1016/j.cell.2005.11.039. [DOI] [PubMed] [Google Scholar]
- Turner GC, Varshavsky A. Detecting and measuring cotranslational protein degradation in vivo. Science. 2000;289:2117–2120. doi: 10.1126/science.289.5487.2117. [DOI] [PubMed] [Google Scholar]
- Duttler S, Pechmann S, Frydman J. Principles of cotranslational ubiquitination and quality control at the ribosome. Mol Cell. 2013;50:379–393. doi: 10.1016/j.molcel.2013.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sciacovelli M, Guzzo G, Morello V, Frezza C, Zheng L, Nannini N, et al. The mitochondrial chaperone TRAP1 promotes neoplastic growth by inhibiting succinate dehydrogenase. Cell Metabolism. 2013;17:988–999. doi: 10.1016/j.cmet.2013.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costantino E, Maddalena F, Calise S, Piscazzi A, Tirino V, Fersini A, et al. TRAP1, a novel mitochondrial chaperone responsible for multi-drug resistance and protection from apoptotis in human colorectal carcinoma cells. Cancer Lett. 2009;279:39–46. doi: 10.1016/j.canlet.2009.01.018. [DOI] [PubMed] [Google Scholar]
- Faraonio R, Vergara P, Di Marzo D, Pierantoni MG, Napolitano M, Russo T, et al. Transcription regulation in NIH3T3 cell clones resistant to diethylmaleate-induced oxidative stress and apoptosis. Antioxid Redox Signal. 2006;8:365–374. doi: 10.1089/ars.2006.8.365. [DOI] [PubMed] [Google Scholar]
- Landriscina M, Fabiano A, Altamura S, Bagalà C, Piscazzi A, Cassano A, et al. Reverse transcriptase inhibitors down-regulate cell proliferation in vitro and in vivo and restore thyrotropin signaling and iodine uptake in human thyroid anaplastic carcinoma. J Clin Endocrinol Metab. 2005;90:5663–5671. doi: 10.1210/jc.2005-0367. [DOI] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nature Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orru S, Aspesi A, Armiraglio M, Caterino M, Loreni F, Ruoppolo M, et al. Analysis of the ribosomal protein S19 interactome. Mol Cell Proteomics. 2007;6:382–393. doi: 10.1074/mcp.M600156-MCP200. [DOI] [PubMed] [Google Scholar]
- Kim TS, Yang CY, Kim HD, Lee JY, Ahn BY, Kim J. Interaction of Hsp90 with ribosomal proteins protects from ubiquitination and proteasome-dependent degradation. Mol Biol Cell. 2006;17:824–833. doi: 10.1091/mbc.E05-08-0713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saadoun S, Papadopoulos MC, Hara-Chikuma M, Verkman AS. Impairment of angiogenesis and cell migration by targeted aquaporin-1 gene disruption. Nature. 2005;434:786–792. doi: 10.1038/nature03460. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.