Abstract
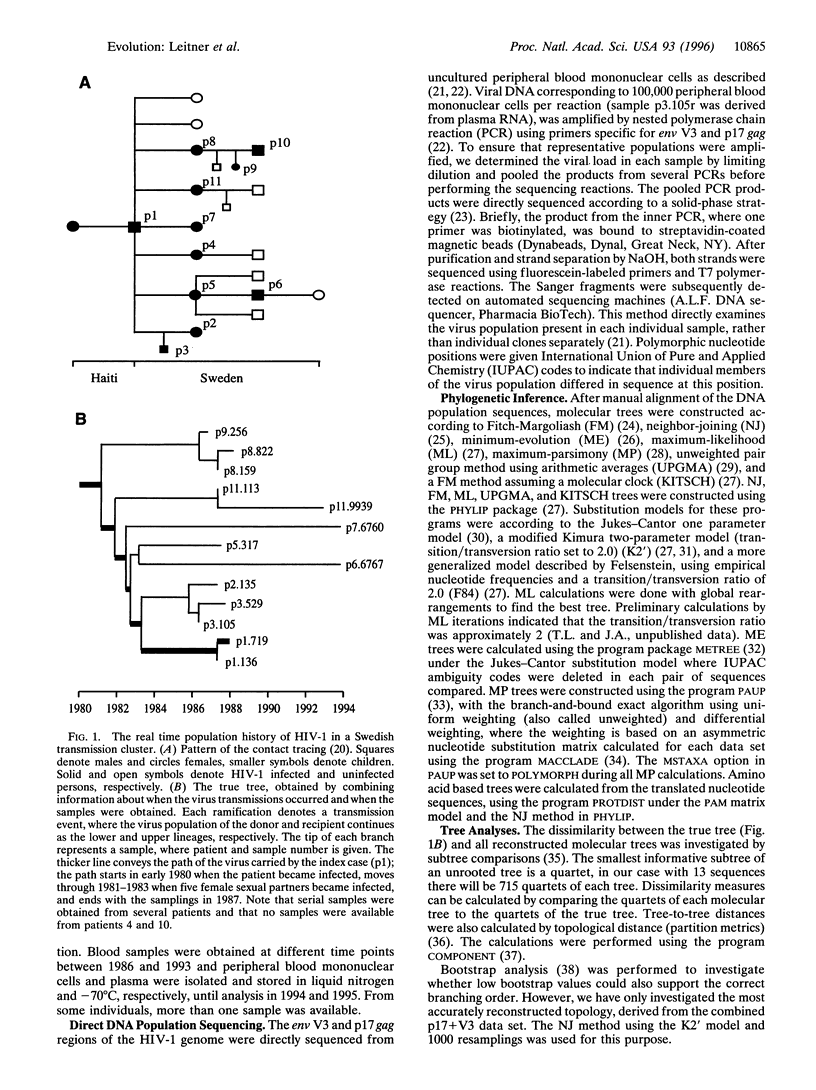
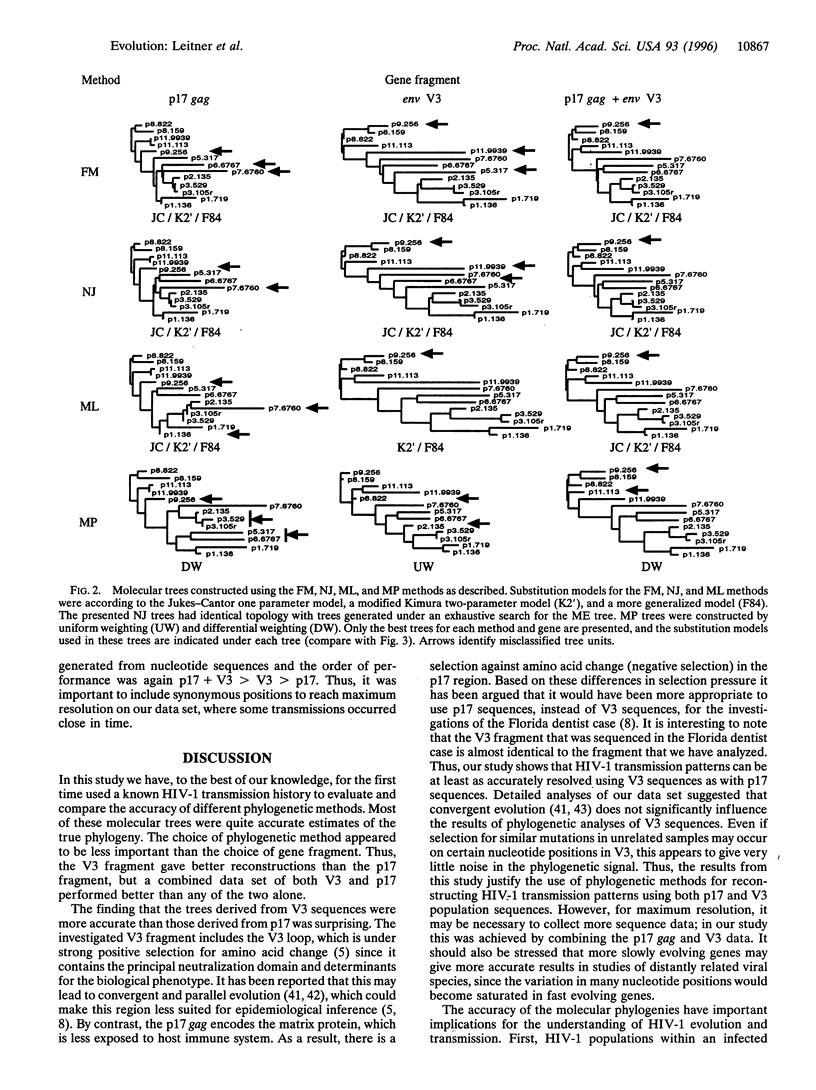
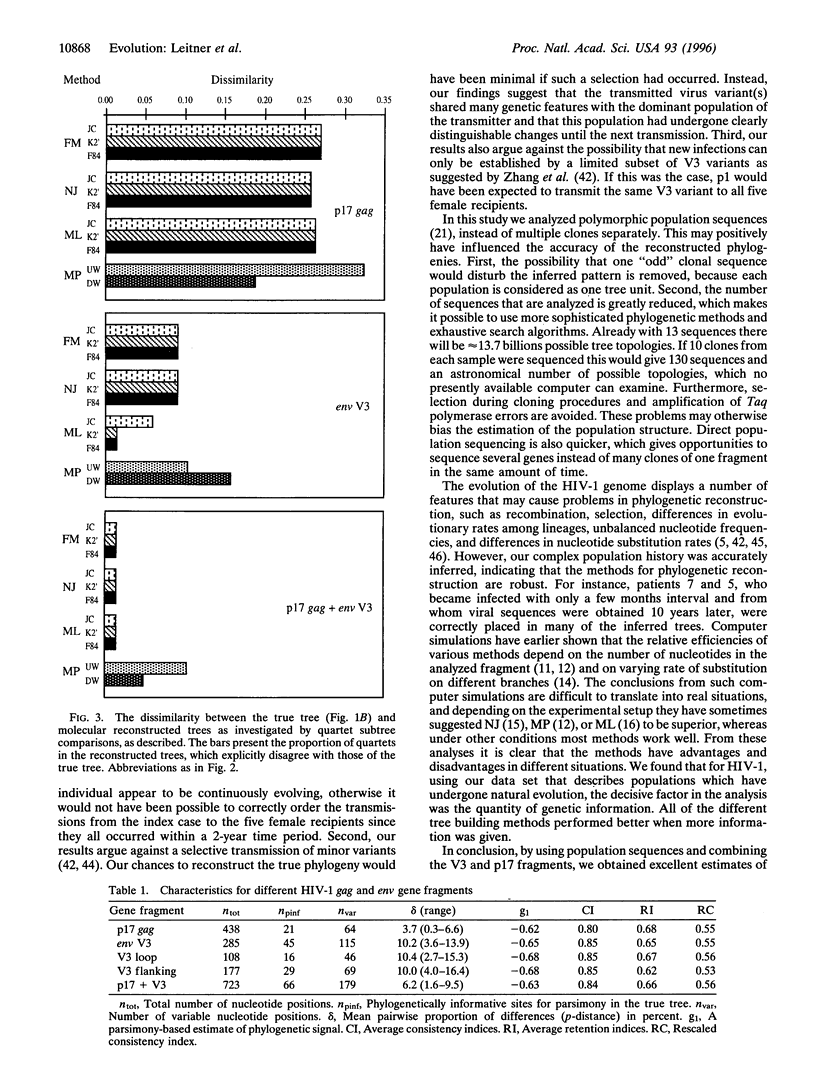
Phylogenetic analyses are increasingly used in attempts to clarify transmission patterns of human immunodeficiency virus type 1 (HIV-1), but there is a continuing discussion about their validity because convergent evolution and transmission of minor HIV variants may obscure epidemiological patterns. Here we have studied a unique HIV-1 transmission cluster consisting of nine infected individuals, for whom the time and direction of each virus transmission was exactly known. Most of the transmissions occurred between 1981 and 1983, and a total of 13 blood samples were obtained approximately 2-12 years later. The p17 gag and env V3 regions of the HIV-1 genome were directly sequenced from uncultured lymphocytes. A true phylogenetic tree was constructed based on the knowledge about when the transmissions had occurred and when the samples were obtained. This complex, known HIV-1 transmission history was compared with reconstructed molecular trees, which were calculated from the DNA sequences by several commonly used phylogenetic inference methods [Fitch-Margoliash, neighbor-joining, minimum-evolution, maximum-likelihood, maximum-parsimony, unweighted pair group method using arithmetic averages (UPGMA), and a Fitch-Margoliash method assuming a molecular clock (KITSCH)]. A majority of the reconstructed trees were good estimates of the true phylogeny; 12 of 13 taxa were correctly positioned in the most accurate trees. The choice of gene fragment was found to be more important than the choice of phylogenetic method and substitution model. However, methods that are sensitive to unequal rates of change performed more poorly (such as UPGMA and KITSCH, which assume a constant molecular clock). The rapidly evolving V3 fragment gave better reconstructions than p17, but a combined data set of both p17 and V3 performed best. The accuracy of the phylogenetic methods justifies their use in HIV-1 research and argues against convergent evolution and selective transmission of certain virus variants.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Albert J., Wahlberg J., Leitner T., Escanilla D., Uhlén M. Analysis of a rape case by direct sequencing of the human immunodeficiency virus type 1 pol and gag genes. J Virol. 1994 Sep;68(9):5918–5924. doi: 10.1128/jvi.68.9.5918-5924.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cann R. L., Stoneking M., Wilson A. C. Mitochondrial DNA and human evolution. Nature. 1987 Jan 1;325(6099):31–36. doi: 10.1038/325031a0. [DOI] [PubMed] [Google Scholar]
- DeBry R. W., Abele L. G., Weiss S. H., Hill M. D., Bouzas M., Lorenzo E., Graebnitz F., Resnick L. Dental HIV transmission? Nature. 1993 Feb 25;361(6414):691–691. doi: 10.1038/361691a0. [DOI] [PubMed] [Google Scholar]
- Doolittle R. F. Convergent evolution: the need to be explicit. Trends Biochem Sci. 1994 Jan;19(1):15–18. doi: 10.1016/0968-0004(94)90167-8. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Phylogenies from molecular sequences: inference and reliability. Annu Rev Genet. 1988;22:521–565. doi: 10.1146/annurev.ge.22.120188.002513. [DOI] [PubMed] [Google Scholar]
- Fitch W. M., Margoliash E. Construction of phylogenetic trees. Science. 1967 Jan 20;155(3760):279–284. doi: 10.1126/science.155.3760.279. [DOI] [PubMed] [Google Scholar]
- Gojobori T., Moriyama E. N., Kimura M. Molecular clock of viral evolution, and the neutral theory. Proc Natl Acad Sci U S A. 1990 Dec;87(24):10015–10018. doi: 10.1073/pnas.87.24.10015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillis D. M., Bull J. J., White M. E., Badgett M. R., Molineux I. J. Experimental phylogenetics: generation of a known phylogeny. Science. 1992 Jan 31;255(5044):589–592. doi: 10.1126/science.1736360. [DOI] [PubMed] [Google Scholar]
- Hillis D. M., Huelsenbeck J. P., Cunningham C. W. Application and accuracy of molecular phylogenies. Science. 1994 Apr 29;264(5159):671–677. doi: 10.1126/science.8171318. [DOI] [PubMed] [Google Scholar]
- Hillis D. M., Huelsenbeck J. P. Signal, noise, and reliability in molecular phylogenetic analyses. J Hered. 1992 May-Jun;83(3):189–195. doi: 10.1093/oxfordjournals.jhered.a111190. [DOI] [PubMed] [Google Scholar]
- Holmes E. C., Brown A. J., Simmonds P. Sequence data as evidence. Nature. 1993 Aug 26;364(6440):766–766. doi: 10.1038/364766b0. [DOI] [PubMed] [Google Scholar]
- Holmes E. C., Zhang L. Q., Robertson P., Cleland A., Harvey E., Simmonds P., Leigh Brown A. J. The molecular epidemiology of human immunodeficiency virus type 1 in Edinburgh. J Infect Dis. 1995 Jan;171(1):45–53. doi: 10.1093/infdis/171.1.45. [DOI] [PubMed] [Google Scholar]
- Holmes E. C., Zhang L. Q., Simmonds P., Rogers A. S., Brown A. J. Molecular investigation of human immunodeficiency virus (HIV) infection in a patient of an HIV-infected surgeon. J Infect Dis. 1993 Jun;167(6):1411–1414. doi: 10.1093/infdis/167.6.1411. [DOI] [PubMed] [Google Scholar]
- Hultman T., Bergh S., Moks T., Uhlén M. Bidirectional solid-phase sequencing of in vitro-amplified plasmid DNA. Biotechniques. 1991 Jan;10(1):84–93. [PubMed] [Google Scholar]
- Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980 Dec;16(2):111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- Leitner T., Escanilla D., Marquina S., Wahlberg J., Broström C., Hansson H. B., Uhlén M., Albert J. Biological and molecular characterization of subtype D, G, and A/D recombinant HIV-1 transmissions in Sweden. Virology. 1995 May 10;209(1):136–146. doi: 10.1006/viro.1995.1237. [DOI] [PubMed] [Google Scholar]
- Leitner T., Halapi E., Scarlatti G., Rossi P., Albert J., Fenyö E. M., Uhlén M. Analysis of heterogeneous viral populations by direct DNA sequencing. Biotechniques. 1993 Jul;15(1):120–127. [PubMed] [Google Scholar]
- Nei M., Takezaki N., Sitnikova T. Assessing molecular phylogenies. Science. 1995 Jan 13;267(5195):253–256. doi: 10.1126/science.7809633. [DOI] [PubMed] [Google Scholar]
- Robertson D. L., Sharp P. M., McCutchan F. E., Hahn B. H. Recombination in HIV-1. Nature. 1995 Mar 9;374(6518):124–126. doi: 10.1038/374124b0. [DOI] [PubMed] [Google Scholar]
- Rzhetsky A., Nei M. Theoretical foundation of the minimum-evolution method of phylogenetic inference. Mol Biol Evol. 1993 Sep;10(5):1073–1095. doi: 10.1093/oxfordjournals.molbev.a040056. [DOI] [PubMed] [Google Scholar]
- Saitou N., Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987 Jul;4(4):406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Saitou N. Property and efficiency of the maximum likelihood method for molecular phylogeny. J Mol Evol. 1988;27(3):261–273. doi: 10.1007/BF02100082. [DOI] [PubMed] [Google Scholar]
- Schrenk F., Bromage T. G., Betzler C. G., Ring U., Juwayeyi Y. M. Oldest Homo and Pliocene biogeography of the Malawi Rift. Nature. 1993 Oct 28;365(6449):833–836. doi: 10.1038/365833a0. [DOI] [PubMed] [Google Scholar]
- Sourdis J., Nei M. Relative efficiencies of the maximum parsimony and distance-matrix methods in obtaining the correct phylogenetic tree. Mol Biol Evol. 1988 May;5(3):298–311. doi: 10.1093/oxfordjournals.molbev.a040497. [DOI] [PubMed] [Google Scholar]
- Strunnikova N., Ray S. C., Livingston R. A., Rubalcaba E., Viscidi R. P. Convergent evolution within the V3 loop domain of human immunodeficiency virus type 1 in association with disease progression. J Virol. 1995 Dec;69(12):7548–7558. doi: 10.1128/jvi.69.12.7548-7558.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vartanian J. P., Meyerhans A., Sala M., Wain-Hobson S. G-->A hypermutation of the human immunodeficiency virus type 1 genome: evidence for dCTP pool imbalance during reverse transcription. Proc Natl Acad Sci U S A. 1994 Apr 12;91(8):3092–3096. doi: 10.1073/pnas.91.8.3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolinsky S. M., Wike C. M., Korber B. T., Hutto C., Parks W. P., Rosenblum L. L., Kunstman K. J., Furtado M. R., Muñoz J. L. Selective transmission of human immunodeficiency virus type-1 variants from mothers to infants. Science. 1992 Feb 28;255(5048):1134–1137. doi: 10.1126/science.1546316. [DOI] [PubMed] [Google Scholar]
- Zhang L. Q., MacKenzie P., Cleland A., Holmes E. C., Brown A. J., Simmonds P. Selection for specific sequences in the external envelope protein of human immunodeficiency virus type 1 upon primary infection. J Virol. 1993 Jun;67(6):3345–3356. doi: 10.1128/jvi.67.6.3345-3356.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]



