Abstract
Human papillomavirus (HPV) infection, most commonly genotype 16 of the alpha-9 family, is implicated in the etiology of a subset of oropharyngeal squamous cell carcinomas (OPSC) worldwide. Data are scarce regarding OPSC in South Africans, and three prior studies suggest no significant etiologic role for HPV. We aimed to investigate for evidence of HPV etiology in OPSCs from black South Africans by polymerase chain reaction (PCR) methodologies with determination of HPV subtype by sequencing, in situ hybridization (ISH), and p16INK4a immunohistochemistry (IHC), as a surrogate marker for an HPV-driven tumor. It was hypothesized that HPV-driven tumors would be positive by PCR plus IHC and/or ISH whereas OPSCs with HPV background infections (HPV-passenger) would be positive by PCR alone. Formalin-fixed, paraffin-embedded tissues from 51 OPSCs collected between 2005 and 2010 from 41 patients were analyzed for HPV by GP5+6+ PCR (targeting the HPV L1 region), pU-1M/pU-2R PCR (targeting the HPV E6/E7 region) and HPV-31 specific PCR (targeting the E5 region), chromogenic ISH, and p16INK4a IHC. All cases positive by PCR were subject to sequencing to determine HPV genotype. The patient mean age was 58.0 years and 88 % were male. Of the 51 evaluable tumors, 48 (94.1 %) were positive for HPV DNA by PCR: 25 (49.1 %) met criteria for an HPV-driven tumor, 23 (45.1 %) for HPV-passenger, and 3 (5.9 %) were HPV-unrelated. Sequencing of the PCR-positive cases revealed the following genotypes: combined HPV-16 and 31 (41.7 %), HPV-31 (25.0 %), HPV-16 (22.9 %), combined HPV-16 and 18 (6.3 %), and a single case each of HPV 18 and HPV 33. Studies via ISH were negative in all cases. In accordance with worldwide trends but contrary to prior South African data, HPV likely plays an etiologic role in a significant subset (at least 49.1 %) of OPSC in black South Africans. We found that the alpha-9 HPV family, particularly HPV-16 and 31 either in combination or separately, to predominate in our sample tumors. The use of multiple PCR primers increased sensitivity of viral detection, and a HPV-31 specific primer confirmed the presence of this genotype in many samples. Further studies including HPV E6/E7 mRNA assays are needed to better elucidate the pathogenic role of HPV in black South African OPSCs.
Keywords: Human papillomavirus, HPV-16, HPV-31, South Africa, Oropharyngeal squamous cell carcinoma
Introduction
Every year there are an estimated 633,000 new cases of head and neck cancer worldwide with human papillomavirus (HPV) infection implicated as an important etiologic agent in a subset of cases: predominantly HPV-16 [genus alpha-papillomavirus, species 9 (also includes types 31, 33, 35, 52, 58) and HPV-18 (genus alpha-papillomavirus, species 7 (also includes 39, 45, 59, 68)] [1]. The link between HPV infection and squamous cell carcinoma is strong for oropharyngeal squamous cell carcinomas (OPSC), with oropharynx defined as the area including the posterior one-third of the tongue, palatine and pharyngeal tonsils, bounded inferiorly by the epiglottis and superiorly by the soft palate [1, 2]. Identification of HPV-related tumors is important due to different molecular profiles, treatment, and prognosis compared to a classic OPSC thought to arise from the ‘chemical carcinogenesis’ pathway wherein tumorigenesis is associated with tobacco, betel nut, and alcohol use and is seen predominantly in elderly patients [3–11]. A proposed combined pathway (wherein both a chemical insult and HPV infection are implicated in tumorigenesis) has unclear clinical significance [12–14]. While older age is significantly associated with increased prevalence of the non-HPV OPSC pathway (with some authors suggesting a breakpoint of 60 years to favor the non-HPV pathway), both HPV-related and HPV-unrelated tumors can be found in all age groups [4, 14, 15]. Histologic features suggesting an HPV-driven tumor can be seen on routine hematoxylin and eosin (H & E) tissue sections and are well-described in the literature as a non-keratinizing squamous cell carcinoma demonstrating pushing borders and areas of comedo-type necrosis, with cytologic features including indistinct cell membranes and small nucleoli [2, 15, 16]. With an improved prognosis compared with a non-HPV related tumor, there is strong interest in determining the best strategies for diagnosing a suspected HPV-driven OPSC.
Given the acceptance of HPV as an etiology for OPSC worldwide, we noted that this disease had not been documented in the South African population in three identified relevant prior studies of oral squamous cell carcinomas. Van Rensburg et al. [17] in 1995 evaluated 66 patients (unspecified race) with oral squamous cell carcinomas of unspecified sites via in situ hybridization (ISH) for HPV subtypes 6, 11, 16, and 18 and immunohistochemical stain (IHC) for the viral L1 capsid. This study yielded negative results except for one case with ISH positivity in normal epithelium adjacent to tumor. Van Rensburg et al. [18] in 1996 analyzed 146 oral squamous cell carcinomas from black South Africans via polymerase chain reaction (PCR) for HPV subtypes 6, 11, 16, and 18 with Southern blot confirmation of positive results and calculated an HPV prevalence of 1.4 % (one case each of HPV-11 and HPV-16). Most recently, Boy et al. [19] in 2006 studied 59 cases of oral squamous cell carcinoma with a pan-HPV ISH and PCR for HPV 16 and 18, and found seven (11.9 %) positive cases (all HPV-18) via PCR, with uniformly negative results by ISH. The combined results of these three studies suggest that HPV is likely not a significant factor in the development of oral squamous cell carcinoma in South Africans. Other data from South Africa indirectly link oral cancers to HPV: epidemiologic studies revealed a strong correlation between cervical squamous cell carcinoma (with well-established link to HPV) and oral squamous cell carcinoma in black, “mixed-race”, and white South Africans [20]. In this present study we limit the investigation to assessing for HPV in OPSC; this is in comparison to the three prior studies wherein the oral site was unspecified in the two Van Rensburg studies, while Boy et al. specifically include both oropharyngeal and oral cavity lesions. This present study also limits the patient samples to black South Africans as a measure to limit confounding variables.
While the majority of the current literature strongly supports a link between HPV and oral squamous cell carcinoma worldwide, [13, 21–26] prevalence of HPV-linked tumors does vary by geographic location with some areas (North America, Japan, and Australia) demonstrating a strong link while others (China and Africa) showing low or no association [24, 26]. Data regarding African patients are limited compared to data captured regarding North American and European patients: for example, one of the largest meta-analyses (60 studies totaling 5,046 samples using PCR for HPV identification in head and neck squamous cell carcinomas) only identified a single African study [26]. Outside of Africa, recent analysis comparing Caucasian American versus African American patients reveals that in high stage OPSC, HPV is more common in the tumors of the Caucasians [27]. Comparison of disease prevalence in different studies is limited by factors including differing diagnostic methodologies and anatomic sites (some studies limited analysis to oropharynx, while others include oral cavity or any head and neck site) [24, 25, 28]. Regarding anatomic site of disease, the author of one large review found that 51 % of tonsillar carcinomas were HPV positive [29], while another study including all oropharyngeal sites yielded an overall prevalence of 25.9 % [26], supporting evidence that HPV-related disease prevalence varies depending on site. Additionally, studies have shown that HPV prevalence estimates are highly dependent on the HPV testing methods applied and their analytical sensitivity for HPV [30–33].
The IHC stain p16INK4a is used in research and in the clinic as evidence to support an HPV etiology for a head and neck or anogenital squamous cell carcinoma. Two key HPV oncogenes (E6 and E7), which respectively inactivate p53 and retinoblastoma (Rb) tumor suppressor functions, are factors contributing to a loss of normal cell cycle control. An HPV-driven tumor will, by functional inactivation of Rb, characteristically overexpress p16INK4a; conversely, a non-HPV related tumor will usually, but not always, have absent or weak stain with p16INK4a [3, 9, 10, 34–36]. Use of p16INK4a as a proxy for HPV infection also allows for evaluation of the morphologic features and exclusion of areas that may be p16INK4a positive without meeting diagnostic histologic criteria for squamous cell carcinoma. With much of the basic research based in the cervical carcinoma literature, it is uncertain whether or not the relationship of HPV infection to p16INK4a expression in head and neck tumors directly parallels that found in cervical tumors; p16INK4a expression might be impacted by non-HPV related factors such as smoking or alcohol abuse, or further mediated by genetic or epigenetic events.
In the present study we evaluate OPSCs using four methodologies: three different PCR methodologies to assay for presence of HPV, sequencing to determine HPV genotype, chromogenic in situ hybridization (CISH) for HPV, and p16INK4a IHC. A combination of the above techniques allows for sensitive detection of HPV DNA via PCR as well as localization of HPV effect via either CISH or IHC. Debate regarding the best methodology for diagnosing an HPV-driven squamous cell carcinoma is ongoing [5, 13, 32, 36]. One study on paraffin-embedded tissue (using loss of heterozygosity profile and mRNA expression of E6 on frozen material as a criterion standard) found that the combination of PCR for HPV and p16INK4a IHC yielded nearly 100 % sensitivity and 100 % sensitivity for the presence of HPV with associated molecular changes [37]. Termine et al. [24] in a meta-analysis suggest that adequate evaluation demands at least a sensitive marker for HPV (for example, PCR) in combination with a localization marker such as ISH to show that the associated changes are in lesional tissue. The molecular marker p16INK4a in multiple studies has validated to be a useful surrogate for HPV infection, as well as localization marker, and further as an independent prognostic indicator of a treatment-responsive tumor when the stain is positive [5, 9, 13, 22, 37–39]. Positivity via CISH, particularly detection of a punctate staining pattern, is a useful specific test via demonstration of HPV viral integration into the DNA of the host cell; however, CISH is less sensitive and may yield false negative results in cases of low viral copy numbers [36]. By using PCR, CISH, and p16INK4a IHC we aim to divide the OPSCs into three groups: first, HPV as driver of tumor (HPV positive by PCR plus IHC and/or ISH); second, HPV as a passenger within/adjacent to tumor (HPV positive by PCR negative by IHC and/or ISH), third, HPV negative tumor [40, 41].
Materials and Methods
All research was performed under the auspices of the Institutional Review Board of the University of Vermont and the research protocols of the University of the Witwatersrand. We identified 55 cases of OPSC from black South Africans diagnosed between 2005 and 2010. Formalin-fixed paraffin-embedded (FFPE) tissue blocks for each case were provided with gender, race, age, and location of tumor provided (age specified for all but one patient). All further studies were performed at the University of Vermont.
DNA Extraction
Ten 5 μm sections of FFPE tissues were cut into 1.5 mL tubes. As a precaution against specimen cross-contamination, after each FFPE block the microtome blade and mounting was wiped with xylene substitute [Hemo-De® (Fisher Scientific, Pittsburgh, PA)], ethanol and then DNA Away™ (Fisher Scientific, Pittsburgh, PA) to removes traces of DNA or DNase. Tissues were dewaxed by xylene washes followed by ethanol rinses, and then air dried. DNA was extracted from the specimens using a proteinase K and column purification method according to supplier instructions (Genomic DNA Clean & Concentrator™, Zymo Research Corp., Irvine, CA).
HPV Genotyping
Polymerase chain reaction procedures were performed in a PCR workstation (UVP, LLC, Upland, CA) enabling the inactivation of any contaminating DNA by UV irradiation of PCR pipettes and tubes racks, as well as water, buffer, and other reagents. Additionally, the procedure included the substitution of dTTP with dUTP and the incorporation of a uracil N-glycosylase (HK™-UNG Thermolabile Uracil N-Glycosylase, Epicentre Biotechnologies, Madison, WI) pre-PCR incubation step for the elimination of any contaminating HPV amplicons from prior rounds of PCR [42]. All PCR reactions included ~100 ng of DNA extract. Samples were confirmed as amenable to PCR by amplification of a β-globin fragment ~209 base pairs in size using PC03/5 primers [PCO3: 5′-ACACAACTGTGTTCACTAGC-3′; PCO5: 5′-GAAACCCAAGAGTCTTCTCTG-3′] [42]. Screening for HPV in the specimens was performed by three separate PCR assays: firstly, by GP5+/6+ PCR screening for at least 37 different HPV types using primers that target the L1 region; secondly (further to emerging data in the course of the study showing the frequent detection of HPV-31) pU1-M/pU-2R PCR was performed targeting the E6/E7 region with detection of HPV types 16, 18, 31, 33, 52 and 58 and thirdly, PCR using HPV-31 specific primers targeting the E5 region to confirm HPV-31 results [43, 44]. PCR method details are shown in Table 1.
Table 1.
PCR Primer Sequences
| PCR Assay | Primer Sequences (5′–3′) | Region | Amplicon size | Ref. |
|---|---|---|---|---|
| GP5+/6+ | Fwd (GP5+): TTTGTTACTGTGGTAGATACTAC Rev (GP6+): GAAAAATAAACTGTAAATCATATTC |
L1 | ~140 bp | [44] |
| pU1-M/pU-2R | Fwd (pU-1M): TGTCAAAAACCGTTGTGTCC Rev (pU-2R): GAGCTGTCGCTTAATTGCTC |
E6/E7 | ~230–270 bp | [43] |
| HPV-31 | Fwd (Pr1): CTACAGTAAGCATTGTGCTATGC Rev (Pr2): ACGTAATGGAGAGGTTGCAATAACCC |
E5 | ~153 bp | [33] |
BLAST search confirmed 100 % homology with HPV-31, 39 % (Pr1) and 27 % (Pr2) homology for HPV-16 with no 3′-end matches, and 48 % (Pr1) and 31 % (Pr2) homology for HPV-18 with no 3′-end matches
Polymerase chain reaction controls included HPV plasmid DNA [including HPV-31 (American Type Culture Collection, Manassas, VA, ATCC® #65446)], DNA extracts from CaSki and SiHa cells (ATCC® #s CRL-1550™ and HTB-35™, both HPV-16 positive); HeLa cells (ATCC® # CCL-2™, HPV-18 positive); and C-33 A cells (ATCC® # HTB-31™; HPV negative) and also control reactions set up with no added DNA. Presence of an amplicon of expected size following 2.0 % agarose gel electrophoresis was taken as evidence of HPV positivity for each assay (Fig. 1). PCR positive specimens were purified using a DNA Clean & Concentrator™ column (Zymo Research Corp., Irvine, CA). Cycle sequencing was performed on the purified DNA by the Vermont Cancer Center DNA Analysis Core Facility staff on an Applied Biosystems® 3013 Genetic Analyzer (ABI, Foster City, CA). Sequencing data were assessed for HPV genotype by NCBI BLAST search, with an example of the output via all three PCR methodologies shown in Fig. 2.
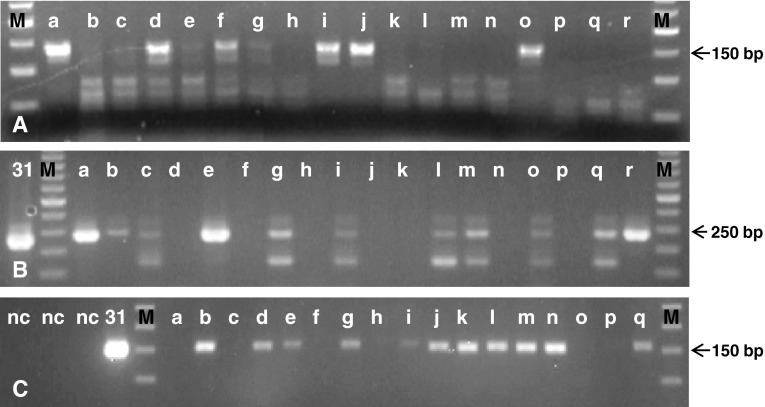
Fig. 1.
Representative agarose gel electrophoresis of PCR amplicons. a GP5+/6+ PCR demonstrating ~140 base pair amplification from the HPV L1 open reading frame (ORF); amplicons of predicted size apparent in lanes a, d, f, g, i, j, and o. b pU1-M/pU-2R PCR demonstrating ~240 base pair amplification from the HPV E6/E7 ORF; amplicons of predicted size apparent in lanes a, b, c, e, g, i, l, m, o, q, r. c HPV-31 primer specific PCR demonstrating ~155 base pair amplification from the HPV E5 ORF; amplicons of predicted size apparent in lanes b, d, e, g, i, j, k, l, m, n, q. Confirmation of amplicon identity as HPV sequences by cycle sequencing (Fig. 2). M—50 base pair ladder; lower case letters: samples; 31—HPV-31 plasmid control; nc—negative controls
Fig. 2.
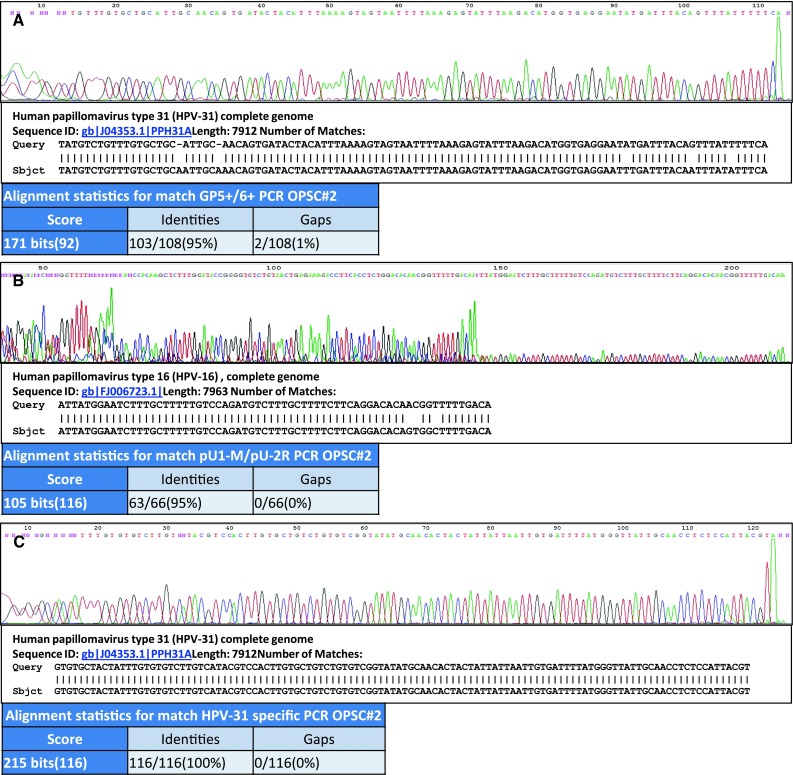
Sequencing data from an oropharyngeal tumor from one patient [Lab Identifier #2 (Table 2)] demonstrating the detection of HPV-31 or HPV-16 dependent upon the PCR assay employed. a GP5+/6+ PCR electropherogram data together with NCBI BLAST search alignment showing 95 % homology with HPV-31 genome bases 6,577–6,684 with results suggestive of a possible HPV-31 sub-type. Homology with HPV-16 is 67/77 (87 %) of the shown sequence. b pU1-M/pU-2R PCR electropherogram data together with NCBI BLAST search alignment showing 95 % homology with HPV-16 genome bases 419–484 and suggestive of a possible HPV-16 sub-type. Homology with HPV-31 is 54/65 (83 %) of the sequence shown. [Note: Competing bases at the 5′- end of the sequence compromised full sequence BLAST analysis; this is most likely due to the shorter co-amplicons visible in Fig. 1b.] c HPV-31 specific PCR electropherogram data together with NCBI BLAST search alignment showing 100 % homology with HPV-31 genome bases 3,874–3,989. Homology with HPV-16 is 75/112 (67 %) of the sequence shown. ‘Query’ shows the sequence obtained from the patient sample; ‘Sbject’ shows the closest identity match reference sequence supplied by NCBI BLAST search
Chromogenic In Situ Hybridization
Chromogenic in situ hybridization utilizing tyramide amplification was performed as previously described [45, 46]. Samples were hybridized via a biotinylated Wide Spectrum HPV DNA Probe Cocktail detecting HPVs 6, 11, 16, 18, 31, 33, 35, 45, 51, and 52 (Dako North America Inc., Carpinteria, CA). Hybridization events were demonstrated via a GenPoint™ Catalyzed Signal Amplification system according to supplier instructions (Dako North America Inc., Carpinteria, CA) using 3′-amino-9-ethylcarbazole (AEC) chromagen. Signal patterns were defined as ‘diffuse’ when nuclei were completely stained (episomal HPV), ‘punctate’ if distinct dot-like intra-nuclear signal (integrated HPV), mixed ‘diffuse and punctate’, or negative [47]. Cervical carcinoma samples were used as positive controls, with omission of HPV probe from the hybridization mix as the negative control.
Immunohistochemistry
p16INK4a IHC was performed using clone G175-405, BD (Parmingen™, San Diego, CA). Sections of FFPE tissue 5 μm thick were dewaxed, incubated in 10 mM sodium citrate pH 6.0 at 98 °C for 20 min, and then cooled for a further 20 min. After buffer rinses, tissues were immersed in 3 % hydrogen peroxide in methanol for 20 min to block endogenous peroxidase. After further buffer washes, slides were immersed for 20 min in serum-free protein block (Dako North America Inc., Carpinteria, CA). Following additional buffer washes, the tissues were incubated at room temperature for 30 min with p16INK4a antibodies, diluted 1:50. Positivity was demonstrated with application of the EnVision™ polymer system (Dako North America Inc., Carpinteria, CA) and staining with 3,3′-diaminobenzidene (DAB) chromagen. Positive controls included cervical lesions known to overexpress p16INK4a. Negative controls were performed using a murine HPV-associated IgG antibody directed against a non-mammalian protein (Dako North America Inc., Carpinteria, CA). Staining for p16INK4a was positive in the tumor if either: moderate cytoplasmic or nuclear staining in >50 % of tumor, or areas of strong non-focal nuclear and cytoplasmic staining, as corresponds to criteria used by others in assessment of HPV status in OPSC [5, 13, 15, 16].
Results
The findings of this study are summarized in Table 2. Following GP5+/6+ PCR, 41 (80.4 %) of samples tested HPV positive on the basis of agarose gel data; subsequent amplicon genotyping by sequencing revealed HPV types 16 [n = 5 (9.8 %)], 18 [n = 4 (7.8 %)], and 31 [n = 32 (62.8 %)]. With an unexpectedly high percentage of specimens testing HPV-31 positive, all specimens were screened with pU1-M/pU2-R primers: 37 specimens tested HPV positive; including HPV types 16 [n = 34 (66.7 %)], 31 [n = 2 (3.9 %)], 33 [n = 1 (2.0 %)]. Seven specimens that were HPV negative by GP5+/6+ PCR were positive by pU1-M/pU2-R PCR and eleven samples positive by GP5+/6+ PCR were negative by pU1-M/pU2-R PCR; three specimens were HPV negative by both assays. HPV-31 specific PCR was then performed on specimens detected as HPV-31 positive by either of the general PCR assays. All specimens [with DNA sample remaining (n = 29)] that originally tested as positive for HPV-31 by L1 targeted PCR showed a band of predicted size for HPV-31 after E5 region PCR, by gel electrophoresis. HPV-31 identity was confirmed by sequencing for 14 (48.3 %) samples; for 15 (51.7 %) samples there was a sequencing reaction failure (the study had insufficient resources to repeat these assays).
Table 2.
Data summary of p16INK4a immunohistochemical and HPV type sequencing results
| Lab identifier | p16INK4a IHC | PCR: GP5+/6+ | PCR: pU-1M/pU-2R | PCR: HPV-31 specific | Combined HPV genotypes |
|---|---|---|---|---|---|
| 1 | Negative | 31 | 16 | 31 | 16, 31 |
| 2 | Positive | 31 | 16 | 31 | 16, 31 |
| 3 | Positive | 31 | Negative | 31 | 31 |
| 41 | Positive | Negative | 16 | Not done | 16 |
| 52 | Positive | 31 | 16 | a | 16, 31 |
| 6 | Negative | 31 | Negative | 31 | 31 |
| 7 | Negative | 31 | Negative | a | 31 |
| 8 | Negative | Negative | 16 | Not done | 16 |
| 9 | Positive | 16 | 16 | Not done | 16 |
| 10 | Negative | 16 | Negative | Not done | 16 |
| 113 | Negative | 18 | 16 | Not done | 16, 18 |
| 12 | Positive | Negative | 16 | Not done | 16 |
| 13 | Positive | Negative | 16 | Not done | 16 |
| 14 | Negative | 31 | 16 | a | 16, 31 |
| 154 | Positive | 31 | 16 | 31 | 16, 31 |
| 164 | Negative | Negative | 16 | Not done | 16 |
| 17 | Positive | 31 | Negative | a | 31 |
| 18 | Positive | 31 | 16 | a | 16, 31 |
| 19 | Positive | 18 | 16 | Not done | 16, 18 |
| 20 | Negative | Negative | Negative | Not done | Negative |
| 215 | Negative | 31 | 16 | 31 | 16, 31 |
| 22 | Positive | 31 | 31 | a | 31 |
| 23 | Negative | 31 | 16 | a | 16, 31 |
| 24 | Negative | Negative | Negative | Not done | Negative |
| 25 | Positive | 16 | 16 | Not done | 16 |
| 26 | Positive | 31 | Negative | 31 | 31 |
| 276 | Positive | 31 | Negative | 31 | 31 |
| 288 | Positive | 16 | 16 | Not done | 16 |
| 29 | Negative | 31 | 16 | 31 | 16, 31 |
| 30 | Positive | 18 | Negative | Not done | 18 |
| 318 | Negative | Negative | 33 | Not done | 33 |
| 32 | Negative | 31 | 16 | 31 | 16, 31 |
| 33 | Positive | 31 | 16 | a | 16, 31 |
| 34 | Negative | 31 | 16 | 31 | 16,31 |
| 35 | Negative | 31 | 31 | a | 31 |
| 36 | Negative | 31 | 16 | a | 16, 31 |
| 37 | Positive | 31 | Negative | a | 31 |
| 38 | Positive | 31 | 16 | a | 16, 31 |
| 395 | Negative | 31 | 16 | a | 16, 31 |
| 40 | Positive | 31 | 16 | 31 | 16, 31 |
| 416 | Positive | Negative | Negative | Not done | Negative |
| 42 | Positive | Negative | 16 | Not done | 16 |
| 43 | Positive | 18 | 16 | Not done | 16, 18 |
| 442 | Positive | 31 | 16 | a | 16, 31 |
| 457 | Negative | 31 | 16 | a | 16, 31 |
| 463 | Negative | 31 | Negative | 31 | 31 |
| 471 | Negative | 31 | Negative | b | 31 |
| 487 | Negative | 31 | 16 | b | 16, 31 |
| 497 | Negative | 31 | 16 | 31 | 16, 31 |
| 507 | Negative | 31 | 16 | b | 16, 31 |
| 51 | Positive | 16 | 16 | Not done | 16 |
| Total positive | 26 (51.0 %) | 41 (80.4 %) | 37 (72.6 %) | – | 48 (94.1 %) |
aBand seen of appropriate size detected on gel electrophoresis but with sequencing reaction failure
bInsufficient genetic material remaining for re-PCR analysis
Superscripts indicate tumors from same patient
p16INK4a staining was scored as positive in 26/51 (51.0 %) specimens; 23/49 (46.9 %) HPV PCR positive specimens were negative by p16INK4a IHC; 25/49 (51.0 %) HPV positive specimens were positive by p16INK4a IHC; one-third (33.3 %) of specimens were HPV negative by GP5+/6+ or pU-1M/pU-2R PCR and positive by p16INK4a IHC. HPV CISH staining was negative in all cases.
The breakdown of samples by classification and genotype is shown in Table 3. In the HPV as driver category, the most common HPV identified was an HPV-16/HPV-31 co-infection tied with HPV-16 alone (8 cases of each). Evaluation of the HPV as passenger category yielded similar results with 23.5 % of the total cases demonstrating an HPV-16/HPV-31 co-infection, comprising more than half of the HPV as passenger cases.
Table 3.
Class assignment and associated HPV genotype
| Tumor classification (n = 51) | Number (% of total) | HPV genotype | Number (% of total) |
|---|---|---|---|
| HPV as driver (HPV by PCR and p16INK4a positive) | 25 (49.1) | ||
| HPV-16 and HPV-31 | 8 (15.7) | ||
| HPV-16 | 8 (15.7) | ||
| HPV-31 | 6 (11.8) | ||
| HPV-16 and HPV-18 | 2 (3.9) | ||
| HPV-18 | 1 (2.0) | ||
| HPV as passenger (HPV by PCR positive and p16INK4a negative) | 23 (45.1) | ||
| HPV-16 and HPV-31 | 12 (23.5) | ||
| HPV-31 | 6 (11.8) | ||
| HPV-16 | 3 (5.9) | ||
| HPV-33 | 1 (2.0) | ||
| HPV-16 and HPV-18 | 1 (2.0) | ||
| HPV-negative (HPV by PCR negative and p16INK4a positive or negative) | 3 (5.9) | – | 3 (5.9) |
The initial study included 55 samples analyzed by PCR with sequencing, CISH for HPV, and p16INK4a IHC. Examples of negative and positive IHC staining for p16INK4a are shown in Fig. 3, along with an example of an H&E stained section of an HPV-driven tumor showing classic histologic features. Four cases were excluded for the following reasons: one pediatric patient with respiratory papillomatosis, two tumor blocks were inadequate for all necessary studies, and one duplicated tumor (separately accessioned tissue from the same patient and listed site, and with same HPV subtypes identified) yielding 51 tissue samples from 41 patients. We allowed multiple tumors per patient to remain in our analysis in cases where the tumors were either from different listed sites, collected at different times, or yielded different HPV genotype results by sequencing. For example, one patient had two tumors removed from the oropharynx 4 years apart, and one typed as HPV-16/HPV-31, and the other as HPV-16/HPV-18. Ages were provided for all except one patient and ranged from 36 to 91 (mean 57.5). The subjects were predominantly (36 of 41) male (demographic data in Table 4).
Fig. 3.
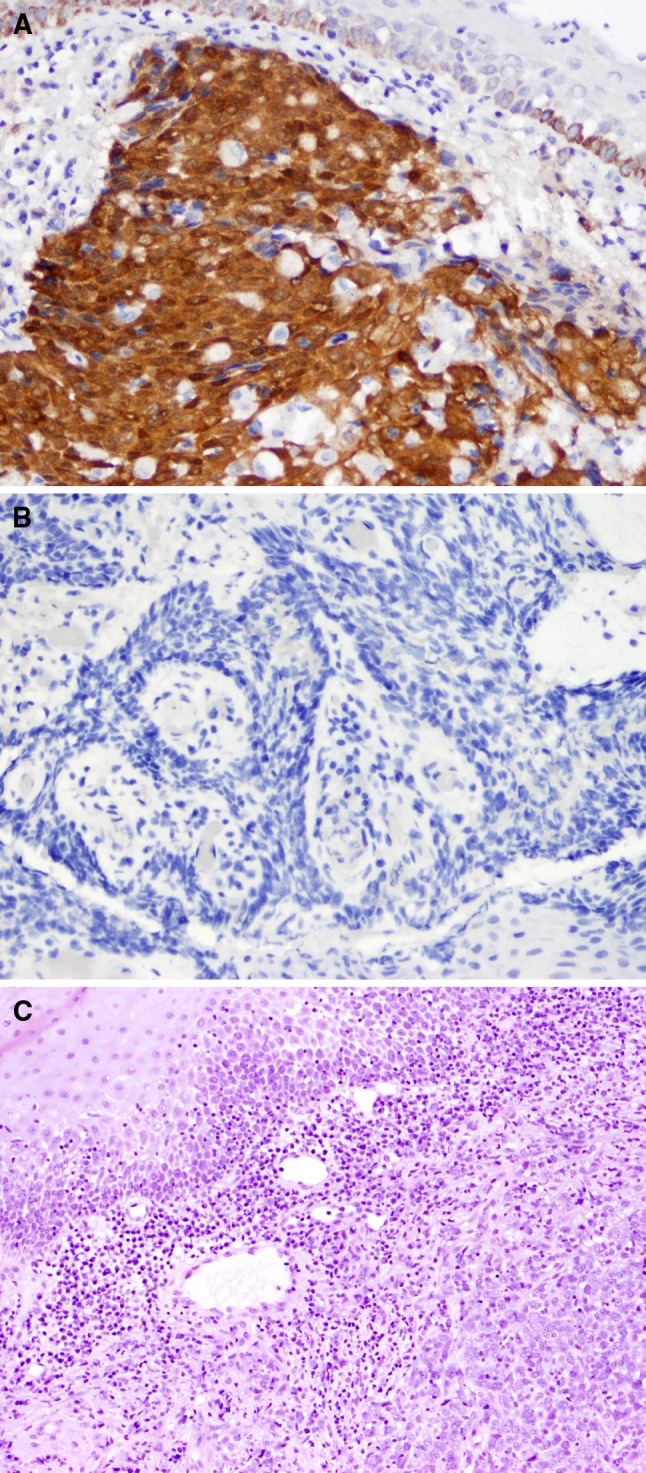
a p16INK4a positive OPSC (A) (40×); a p16INK4a negative OPSC b (40×). The histologic features (H & E stain) of a HPV-driven (p16INK4a positive and HPV positive) OPSC demonstrating non-keratinizing squamous cell carcinoma at lower right, surrounded by chronic inflammation with representative normal squamous epithelium seen at upper left c (20×)
Table 4.
Patient demographics and tumor sites
| Patient characteristics (n = 41) | Number (percent) | |
|---|---|---|
| Gender | Male | 36 (87.8) |
| Female | 5 (12.2) | |
| Age | Mean: 57.5 Range: 36–91 Standard deviation: 11.8 |
41 |
| Tumor site (n = 51) | Base of tongue or base of tongue/tonsil | 14 (27.5) |
| Tonsil | 14 (27.5) | |
| Oropharynx (unspecified) | 23 (45.1) |
One patient age not provided
Discussion
There have been few prior studies of OPSC and HPV in the South African population. Worldwide prevalence studies have shown that HPV genotype 16 is most commonly identified in OPSC associated with HPV. A review by Syrjänen found that of 432 patients with tonsillar OPSC, HPV-16 was the most common genotype identified (84 %), with 4.6 % HPV-33, and 2.8 % HPV-31 [29]. A larger meta-analysis of 5046 patients found 35.6 % of OPSCs positive for HPV; however, in this study only one of the 60 included papers studied an African population (Sudanese) [26]. Termine et al’s meta-analysis of 62 studies of head and neck and oral squamous cell carcinomas included three African studies (the South African papers previously discussed) as the only representations of the African continent [24]. Another meta-analysis of HPV-associated head and neck carcinoma studies limited itself to focus on HPV-16 [25]. More recent data from France assessed 534 patients and found that HPV-16 was the most common isolate with a prevalence of 89.7 % in HPV- positive OPSC [48]. While many large studies suggest that HPV-16 is the most commonly identified HPV genotype in both oral and oropharyngeal squamous cell carcinomas, our data suggest that in South African OPSCs, HPV-16 may be often present in a co-infection with its alpha-9 family relative, HPV-31. As HPV-31 was not expected to be commonly isolated, an HPV-31 specific primer set was used to confirm HPV-31 presence: for example, in nine of the 30 samples identified as HPV-31 by GP5+6+ and HPV-16 by pU-1M/pU-2R, the HPV-31 specific primer was confirmatory. The variability in HPV genotype identified according to primer set used may be due to different affinities inherent in the primer design.
These results were also unexpected in the context of what is known of the dominant HPV genotypes in South African cervical carcinoma. In South Africa, HPV genotypes 16 and 18 are most commonly isolated from cervical carcinoma (correlating well with worldwide data), and comprise together 62.8 % of cases. HPV-31 is found in 4.3 % of South African cervical carcinomas, corresponding to the worldwide reported rate of 3.6 % [49]. The most common HPV genotypes isolated in cervical cancer specimens (World Health Organization data) as compared to the most common HPV genotypes in this study from the HPV- driven group is shown in Table 5.
Table 5.
Most common HPV genotypes isolated from South African OPSCs compared to HPV genotypes from South African cervical carcinomas (adapted from WHO data) [49]
| South Africa OPSCs (current study) | South Africa cervical carcinoma (WHO data) |
|---|---|
| HPV-16 and HPV-31 (32.0 %) | HPV-16 (52.1 %) |
| HPV-16 (32.0 %) | HPV-18 (10.7 %) |
| HPV-31 (24.0 %) | HPV-33 (9.1 %) |
| HPV-16 and HPV-18 (8.0 %) | HPV-31 (4.2 %) |
| HPV-18 (4.0 %) | HPV-45 (3.3 %) |
By using multiple PCR assays, we illustrated the inter-assay discrepancies in terms of positivity and HPV-genotype, as has been previously described and discussed in the cervical cancer literature leading some to advocate the use of multiple primer sets to enhance sensitivity. (30–33) Meta-analyses and reviews of head and neck SCC generally accept multiple PCR platforms for determination of HPV status as there is no criterion standard [24–26]. Our laboratory most often uses the GP5+6+ primer set as it amplifies a short PCR product highly suited for use on FFPE DNA extracts. While we had used the pU-1M/pU-2R product before, we had no prior experience with the HPV-31 specific primers. By using multiple primers, we limited false negative results by PCR due to particular known weaknesses of the primers; for example, the HPV PCR assay for L1 sequences might have resulted in false negative data if the L1 region was abrogated by HPV integration into the cell genome [50]. HPV types 16, 31, 35, 33, 52, 58 and 67 (alpha-9 group) are part of same HPV species group, and recent full genome sequencing data indicates that HPV 16 and 31 are part of a consistent clade with a recent common ancestor [51, 52]. The alpha-9 group is separated into different HPV genotypes via a convention wherein a separate type is defined when there is at least 10 % different in the L1 sequence region [52]. While the genotypes are related, they should be separately identifiable by sequencing. It is possible that different PCR primer assays yield different results due to preferential annealing events during PCR that, in combination with cycle sequencing used for identifying HPV types, results in detection of a single rather than a dual genotype in cases where there is co-infection. While it is interesting to determine which type of HPV is associated with the tumor, all types identified are high risk. With a marketed prophylactic HPV vaccine in South Africa, determination of predominant genotypes may be of future public health benefit in tracking possible disease prevention in head and neck sites.
The three identified prior studies in South African patients did not show any significant evidence of HPV-related oral squamous cell carcinoma. These results may reflect limitations of testing methodologies or a low incidence of HPV-related disease during the time periods examined. In all three studies the authors focused on oral squamous cell carcinoma with Boy et al. specifically including some oropharyngeal sites; analysis of these sites may yield lower HPV positivity as data suggest that HPV-related tumors are most consistently associated with the oropharynx [24, 25, 48]. The sensitive methodologies chosen focused on HPV-16 and HPV-18 without assay coverage for other high risk genotypes, including 31, 33, and 45, while the pan-HPV assays were lower sensitivity methodologies (see Table 6).
Table 6.
Methodologies of prior OPSC HPV South African studies
In the present study we focused on black South Africans for two reasons: the prevalence of OPSC disease is high in this population, and few studies have examined the HPV-related contribution to the disease. In South Africa, oropharyngeal cancer rates are not separately reported, but oral cancer rates are higher in the black population compared to Caucasian: 10.68 % of all cancers in black males compared to 2.55 % of all cancers in white males, with corresponding rates of 2.35 % of all cancers in black females compared to 1.06 % of all cancers in white [53, 54]. Risk factors (classically tobacco and alcohol) for the chemical carcinogenesis pathway are also prevalent which complicates data analysis. Tobacco use is particularly common: data from 2000 indicate that 36.6 % of Caucasian and 22.7 % of black South Africans use tobacco [55]. There are design limitations to this study that limit the generalizability including the limited geographic area sampled (the Witwatersrand region of South Africa), the under-representation of female patients, and absence of data regarding patients’ tobacco usage.
As HPV-related OPSCs have a better prognosis and less risk of relapse [6–8, 15, 34] and longer survival [8], determination of HPV positivity has clinical importance. We demonstrated evidence of HPV etiology in at least 49.1 % of OPSC (HPV as driver) and possible HPV involvement in an additional 45.1 % (HPV as passenger). The patients with HPV driven tumors should have a favorable prognosis, and may need less aggressive therapy. This simplified model may help stratify patients, but the results must be interpreted with the understanding that multiple genetic insults are needed for carcinogenesis as well as the recognition that risky behaviors tend to cluster together [14].
Regarding the CISH results, all cases in this series were negative; however, we did have two positive ISH results (albeit showing positive cells in a minority of tumor cells) among the other OPSC from the same cohort that were not considered in this study (one case omitted as patient had a history of respiratory papillomatosis with known HPV 11, and one case from a white, not black, South African OPSC). With the understanding that the CISH was (partially) working in some cases in this series, but not as many as we would expect given the PCR and IHC results, we attempted to re-optimize the CISH (e.g. by inclusion of a wider range of digestion times); however, HPV signal remained undetectable. In previous studies, our lab has demonstrated HPV by CISH in a high percentage of cervical and head and neck tumors from the USA, India, and Brazil, as well as in an ongoing study of ~150 local (Vermont) oropharyngeal tumors [45–47]. The absence of HPV CISH positive black South African OPSCs might be a consequence of tissue fixation conditions: possibly, protease pre-treatment conditions that are a critical part of the assay, either under-revealed target DNA or too readily resulted in excessive erosion of HPV sequences in the tissues; either eventuality would result in loss of CISH positive signals.
The lack of detection of HPV by CISH partially undermines the assignment of HPV-driver status. While we had hoped to correlate the results of CISH with PCR, we instead used only the PCR data to define an HPV positive tumor. This is a less robust test than CISH as PCR is highly sensitive but has poor specificity since adjacent uninvolved epithelium may contain HPV virus, and hence a positive result may indicate that HPV was present in the tissue block but not necessarily in the tumor. While demonstration of positive p16INK4a results by IHC in lesional tissue supports an HPV-driven tumor diagnosis, it is not specific for an HPV-driven tumor as p16INK4a expression/repression could occur for reasons of genomic abrogation (such as loss of heterozygosity) or epigenetic events such as promoter silencing by hypermethylation. Further, downstream events can diminish the effect of p16INK4a status and subsequently call into question the utility of p16INK4a IHC as prognostic marker. Our binary definition of p16INK4a tumors (including assessment of strength of nuclear and cytoplasmic staining) while used in many other published studies is imperfect and there is ongoing research suggesting that the location of staining in head and neck tumors in general should be critically revisited as it may impact prognosis [56].
For future studies, micro-dissection techniques may help address the problem of uncertainty regarding HPV viral location when using PCR methodology on FFPE tissue. In Boy et al’s [19] prior study of OPSC in South Africa using PCR and ISH the HPV positivity was ascribed to be most likely either commensal infection or contaminant; in the present study we use p16INK4a as supportive evidence that the HPV virus detected is likely tumorigenic. This study is composed predominantly of male patients and is geographically limited to the Witwatersrand area. A larger study enriched with female patients and drawing from other regions of South Africa will be helpful in the further elucidation of OPSCs in this population.
Conclusions
In accordance with worldwide trends, HPV appears to play a role in the pathogenesis of OPSC in black South Africans. HPV is likely the driver of tumorigenesis in at least 49.1 % of the cases examined in this study. A more robust estimate of HPV-driver status might be gained by HPV E6/E7 mRNA expression analysis possibly by a highly sensitive pan-high-risk HPV E6/E7 RNA in situ hybridization assay [57]. In concordance with the results of several meta-analyses primarily of Western developed nations, alpha-9 HPV viruses are pre-dominant; interestingly and uniquely, our data suggest HPV-31 may be especially significant in the etiology of OPSCs from black South Africans along with HPV-16, the most well-studied genotype associated with OPSC in non-African populations. With these preliminary data, we caution against using PCR or other diagnostic modality restricted to a limited number of HPV genotypes when assessing tumors from Africa for HPV status. Further studies, possibly including alternative in situ hybridization methodologies such as HPV RNA targets to help confirm ‘driver’ status, are needed to corroborate our data as presented here.
Acknowledgments
Vermont Cancer Center DNA Analysis Core Facility
References
- 1.Ferlay J, Shin HR, Bray F, et al. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127(12):2893–2917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 2.Slootweg PJ, Eveson JW. Tumours of the oral cavity and oropharynx. In: Barnes L, Eveson JW, Reichart P, editors. Pathology and genetics of head and neck tumours. Lyon: IARC Press; 2005. pp. 165–210. [Google Scholar]
- 3.Braakhuis BJM, Leemans CR, Brakenhoff RH. A genetic progression model of oral cancer: current evidence and clinical implications. J Oral Pathol Med. 2004;33:317–322. doi: 10.1111/j.1600-0714.2004.00225.x. [DOI] [PubMed] [Google Scholar]
- 4.Leemans CR, Braakhuis BJM, Brakenhoff RH. The molecular biology of head and neck cancer. Nat Rev Cancer. 2011;11:9–22. doi: 10.1038/nrc2982. [DOI] [PubMed] [Google Scholar]
- 5.Lewis JS, Thorstad WL, Chernock RD, et al. p16 positive oropharyngeal squamous cell carcinoma: an entity with a favorable prognosis regardless of tumor HPV status. Am J Surg Path. 2010;34(8):1088–1096. doi: 10.1097/PAS.0b013e3181e84652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fakhry C, Westra WH, Li S, et al. Improved survival of patients with human papillomavirus-positive head and neck squamous cell carcinoma in a prospective clinical trial. J Natl Cancer Inst. 2008;100(4):261–269. doi: 10.1093/jnci/djn011. [DOI] [PubMed] [Google Scholar]
- 7.Ragin CC, Taioli E. Survival of squamous cell carcinoma of the head and neck in relation to human papillomavirus infection: review and meta-analysis. Int J Cancer. 2007;121(8):1813–1820. doi: 10.1002/ijc.22851. [DOI] [PubMed] [Google Scholar]
- 8.Ang KK, Harris J, Wheeler R, et al. Human papillomavirus and survival of patients with oropharyngeal cancer. N Engl J Med. 2010;363(1):24–35. doi: 10.1056/NEJMoa0912217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kumar B, Cordell KG, Lee JS, et al. EGFR, p16, HPV Titer, Bcl-xL and p53, sex, and smoking as indicators of response to therapy and survival in oropharyngeal cancer. J Clin Oncol. 2008;26(19):3128–3137. doi: 10.1200/JCO.2007.12.7662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Braakhuis BJ, Snijders PJ, Keune WJ, et al. Genetic patterns in head and neck cancers that contain or lack transcriptionally active human papillomavirus. J Natl Cancer Inst. 2004;96(13):998–1006. doi: 10.1093/jnci/djh183. [DOI] [PubMed] [Google Scholar]
- 11.Chen SF, Yu FS, Chang YC, et al. Role of human papillomavirus infection in carcinogenesis of oral squamous cell carcinoma with evidences of prognostic association. J Oral Pathol Med. 2012;41(1):9–15. doi: 10.1111/j.1600-0714.2011.01046.x. [DOI] [PubMed] [Google Scholar]
- 12.Maxwell JH, Kumar B, Feng FY, et al. Tobacco use in human papillomavirus-positive advanced oropharynx cancer patients related to increased risk of distant metastases and tumor recurrence. Clin Cancer Res. 2010;16(4):1226–1235. doi: 10.1158/1078-0432.CCR-09-2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weinberger PM, Yu Z, Haffty BG, et al. Molecular classification identifies a subset of human papillomavirus–associated oropharyngeal cancers with favorable prognosis. J Clin Oncol. 2006;24(5):736–747. doi: 10.1200/JCO.2004.00.3335. [DOI] [PubMed] [Google Scholar]
- 14.Smith EM, Rubenstein LM, Haugen TH, et al. Tobacco and alcohol use increases the risk of both HPV-associated and HPV-independent head and neck cancers. Cancer Causes Control. 2010;21(9):1369–1378. doi: 10.1007/s10552-010-9564-z. [DOI] [PubMed] [Google Scholar]
- 15.Chernock RD, El-Mofty SK, Thorstad WL, et al. HPV-related nonkeratinizing squamous cell carcinoma of the oropharynx: utility of microscopic features in predicting patient outcomes. Head Neck Pathol. 2009;3:186–194. doi: 10.1007/s12105-009-0126-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.El-Mofty SK, Patil S. Human papillomavirus (HPV)-related oropharyngeal nonkeratinizing squamous cell carcinoma: characterization of a distinct phenotype. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 2006;101:339–345. doi: 10.1016/j.tripleo.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 17.Van Rensburg EJ, van Heerden WF, Venter EH, et al. Detection of human papillomavirus DNA with in situ hybridisation in oral squamous carcinoma in a rural black population. S Afr Med J. 1995;85(9):894–896. [PubMed] [Google Scholar]
- 18.Van Rensburg EJ, Engelbrecht S, Van Heerden WF, et al. Human papillomavirus DNA in oral squamous cell carcinomas from an African population sample. Anticancer Res. 1996;16(2):969–973. [PubMed] [Google Scholar]
- 19.Boy S, Van Rensburg EJ, Engelbrecht S, et al. HPV detection in primary intra-oral squamous cell carcinomas–commensal, aetiological agent or contamination? J Oral Pathol Med. 2006;35(2):86–90. doi: 10.1111/j.1600-0714.2006.00385.x. [DOI] [PubMed] [Google Scholar]
- 20.Postma TC, Van Heerden WF. Is the human papillomavirus a mutual aetiological agent in oral and cervical squamous cell carcinoma? Anticancer Res. 2003;23(4):3509–3512. [PubMed] [Google Scholar]
- 21.Tran N, Rose BR, O’Brien CJ. Role of human papillomavirus in the etiology of head and neck cancer. Head Neck. 2007;29(1):64–70. doi: 10.1002/hed.20460. [DOI] [PubMed] [Google Scholar]
- 22.Syrjänen S. Human papillomavirus (HPV) in head and neck cancer. J Clin Virol. 2005;32(Suppl 1):S59–S66. doi: 10.1016/j.jcv.2004.11.017. [DOI] [PubMed] [Google Scholar]
- 23.Fischer CA, Kampmann M, Zlobec I, et al. p16 expression in oropharyngeal cancer: its impact on staging and prognosis compared with the conventional clinical staging parameters. Ann Oncol. 2010;10:1961–1966. doi: 10.1093/annonc/mdq210. [DOI] [PubMed] [Google Scholar]
- 24.Termine N, Panzarella V, Falaschini S, et al. HPV in oral squamous cell carcinoma vs head and neck squamous cell carcinoma biopsies: a meta-analysis (1988–2007) Ann Oncol. 2008;19(10):1681–1690. doi: 10.1093/annonc/mdn372. [DOI] [PubMed] [Google Scholar]
- 25.Hobbs CG, Sterne JA, Bailey M, et al. Human papillomavirus and head and neck cancer: a systematic review and meta-analysis. Clin Otolaryngol. 2006;31(4):259–266. doi: 10.1111/j.1749-4486.2006.01246.x. [DOI] [PubMed] [Google Scholar]
- 26.Kreimer AR, Clifford GM, Boyle P, et al. Human papillomavirus types in head and neck squamous cell carcinomas worldwide: a systematic review. Cancer Epidemiol Biomarkers Prev. 2005;14(5):467–475. doi: 10.1158/1055-9965.EPI-04-0551. [DOI] [PubMed] [Google Scholar]
- 27.Chernock RD, Zhang Q, El-Mofty SK, et al. Human papillomavirus-related squamous cell carcinoma of the oropharynx: a comparative study in whites and African Americans. Arch Otolaryngol Head Neck Surg. 2011;137(2):163–169. doi: 10.1001/archoto.2010.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sturgis EM, Cinciripini PM. Trends in head and neck cancer incidence in relation to smoking prevalence: an emerging epidemic of human papillomavirus-associated cancers? Cancer. 2007;110(7):1429–1435. doi: 10.1002/cncr.22963. [DOI] [PubMed] [Google Scholar]
- 29.Syrjänen S. HPV infections and tonsillar carcinoma. J Clin Path. 2004;57:449–455. doi: 10.1136/jcp.2003.008656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Evans MF, Adamson CS, Schned LM, et al. HPV is detectable in virtually all abnormal cervical cytology samples after reinvestigation of HPV negatives with multiple alternative PCR tests. Diagn Mol Pathol. 2010;19(3):144–150. doi: 10.1097/PDM.0b013e3181c1482c. [DOI] [PubMed] [Google Scholar]
- 31.Chan PK, Cheung TH, Tam AO, et al. Biases in human papillomavirus genotype prevalence assessment associated with commonly used consensus primers. Int J Cancer. 2006;118(1):243–245. doi: 10.1002/ijc.21299. [DOI] [PubMed] [Google Scholar]
- 32.Chan PK, Picconi MA, Cheung TH, et al. Laboratory and clinical aspects of human papillomavirus testing. Crit Rev Clin Lab Sci. 2012;49(4):117–136. doi: 10.3109/10408363.2012.707174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Karlsen F, Kalantari M, Jenkins A, et al. Use of multiple PCR primer sets for optimal detection of human papillomavirus. J Clin Microbiol. 1996;34(9):2095–2100. doi: 10.1128/jcm.34.9.2095-2100.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Klussmann JP, Gültekin E, Weissenborn SJ, et al. Expression of p16 protein identifies a distinct entity of tonsillar carcinomas associated with human papillomavirus. Am J Pathol. 2003;162(3):747–753. doi: 10.1016/S0002-9440(10)63871-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gillison ML, Koch WM, Capone RB, et al. Evidence for a causal association between human papillomavirus and a subset of head and neck cancers. J Natl Cancer Inst. 2000;92(9):709–720. doi: 10.1093/jnci/92.9.709. [DOI] [PubMed] [Google Scholar]
- 36.Braakhuis BJM, Brakenhoff RH, Meijer CJ, et al. Human papillomavirus in head and neck cancer: the need for a standardized assay to assess the full clinical importance. Eur J Cancer. 2009;45:2935–2939. doi: 10.1016/j.ejca.2009.08.018. [DOI] [PubMed] [Google Scholar]
- 37.Smeets SJ, Hesselink AT, Speel EJ, et al. A novel algorithm for reliable detection of human papillomavirus in paraffin embedded head and neck cancer specimen. Int J Cancer. 2007;121(11):2465–2472. doi: 10.1002/ijc.22980. [DOI] [PubMed] [Google Scholar]
- 38. Singhi AD, Westra WH. Comparison of human papillomavirus in situ hybridization and p16 immunohistochemistry in the detection of human papillomavirus-associated head and neck cancer based on a prospective clinical experience. Cancer. 2010;116(9):2166–73. [DOI] [PubMed]
- 39.Romagosa C, Simonetti S, López-Vicente L, et al. p16(Ink4a) overexpression in cancer: a tumor suppressor gene associated with senescence and high-grade tumors. Oncogene. 2011;30(18):2087–2097. doi: 10.1038/onc.2010.614. [DOI] [PubMed] [Google Scholar]
- 40.Evans MF, Matthews A, Kandil D, et al. Discrimination of ‘driver’ and ‘passenger’ HPV in tonsillar carcinomas by the polymerase chain reaction, chromogenic in situ hybridization, and p16INK4a immunohistochemistry. Head Neck Pathol. 2011;5(4):344–348. doi: 10.1007/s12105-011-0282-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Matsukura T, Sugase M. Pitfalls in the epidemiologic classification of human papillomavirus types associated with cervical cancer using polymerase chain reaction: driver and passenger. Int J Gynecol Cancer. 2008;18:1042–1050. doi: 10.1111/j.1525-1438.2007.01157.x. [DOI] [PubMed] [Google Scholar]
- 42.de Roda Husman AM, Snijders PJ, Stel HV, et al. Processing of long-stored archival cervical smears for human papillomavirus detection by the polymerase chain reaction. Br J Cancer. 1995;72:412–417. doi: 10.1038/bjc.1995.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fujinaga Y, Shimada M, Okazawa K, et al. Simultaneous detection and typing of genital human papillomavirus DNA using the polymerase chain reaction. J Gen Virol. 1991;72(Pt 5):1039–1044. doi: 10.1099/0022-1317-72-5-1039. [DOI] [PubMed] [Google Scholar]
- 44.Evans MF, Adamson CS, Simmons-Arnold L, et al. Touchdown General Primer (GP5+/GP6+) PCR and optimized sample DNA concentration support the sensitive detection of human papillomavirus. BMC Clin Pathol. 2005;5:10. doi: 10.1186/1472-6890-5-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Evans MF, Mount SL, Beatty BG, et al. Biotinyl-tyramide-based in situ hybridization signal patterns distinguish human papillomavirus type and grade of cervical intraepithelial neoplasia. Mod Pathol. 2002;15:1339–1347. doi: 10.1038/modpathol.3880698. [DOI] [PubMed] [Google Scholar]
- 46.Evans MF, Aliesky HA, Cooper K. Optimization of biotinyl tyramide- based in situ hybridization for sensitive background-free applications on formalin-fixed, paraffin-embedded tissue specimens. BMC Clin Pathol. 2003;3(1):2. doi: 10.1186/1472-6890-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cooper K, Herrington CS, Stickland JE, et al. Episomal and integrated human papillomavirus in cervical neoplasia shown by non-isotopic in situ hybridisation. J Clin Pathol. 1991;44:990–996. doi: 10.1136/jcp.44.12.990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.St Guily JL, Jacquard AC, Prétet JL, et al. Human papillomavirus genotype distribution in oropharynx and oral cavity cancer in France-The EDiTH VI study. J Clin Virol. 2011;51(2):100–104. doi: 10.1016/j.jcv.2011.03.003. [DOI] [PubMed] [Google Scholar]
- 49.WHO/ICO Information Centre on HPV and cervical cancer. Human Papillomavirus and Related Cancers: South Africa. Summary Report Update. September 15, 2010. Available from: http://apps.who.int/hpvcentre/statistics/dynamic/ico/country_pdf/ZAF.pdf?CFID=6593267&CFTOKEN=21680899. Accessed 3/1/2012.
- 50.Walboomers JM, Jacobs MV, Manos MM, et al. Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J Pathol. 1999;189:12–19. doi: 10.1002/(SICI)1096-9896(199909)189:1<12::AID-PATH431>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 51.Chen Z, Schiffman M, Herrero R, et al. Evolution and taxonomic classification of human papillomavirus 16 (HPV16)-related variant genomes: HPV31, HPV33, HPV35, HPV52, HPV58, and HPV67. PLoS ONE. 2011;6(5):e20183. doi: 10.1371/journal.pone.0020183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bernard HU, Burk RD, Chen Z, et al. Classification of papillomaviruses (PVs) based on 189 PV types and proposal of taxonomic amendments. Virology. 2010;401(1):70–79. doi: 10.1016/j.virol.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.CANSA—Cancer Association of South Africa. Available from: http://www.cansa.org.za/cause_data/images/1056/NCRTables_2001.pdf. Accessed 6/20/2011.
- 54.Altini M, Kola AH. Age-specific and age-standardised incidence rates for intraoral squamous cell carcinoma in blacks on the Witwatersrand, South Africa. Community Dent Oral Epidemiol. 1985;13:334–339. doi: 10.1111/j.1600-0528.1985.tb00467.x. [DOI] [PubMed] [Google Scholar]
- 55.Walbeek C. Recent trends in smoking prevalence in South Africa: some evidence from AMPS data. Available from: http://archive.idrc.ca/ritc//sarelease3.pdf. Accessed 6/18/2011. [PubMed]
- 56.Zhao N, Ang M-K, Yin X-Y, et al. Different cellular p16INK4a localisation may signal different survival outcomes in head and neck cancer. Br J Cancer. 2012;107:482–490. doi: 10.1038/bjc.2012.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schache AG, Liloglou T, Risk JM, et al. Validation of a novel diagnostic standard in HPV-positive oropharyngeal squamous cell carcinoma. Br J Cancer. 2013;108:1332–1339. doi: 10.1038/bjc.2013.63. [DOI] [PMC free article] [PubMed] [Google Scholar]