Abstract
Gene–environment interactions need to be studied to better understand the obesity. We aimed at determining whether genetic susceptibility to obesity associates with diet intake levels and whether diet intakes modify the genetic susceptibility. In 29,480 subjects of the population-based Malmö Diet and Cancer Study (MDCS), we first assessed association between 16 genome-wide association studies identified obesity-related single-nucleotide polymorphisms (SNPs) with body mass index (BMI) and associated traits. We then conducted association analyses between a genetic risk score (GRS) comprising of 13 replicated SNPs and the individual SNPs, and relative dietary intakes of fat, carbohydrates, protein, fiber and total energy intake, as well as interaction analyses on BMI and associated traits among 26,107 nondiabetic MDCS participants. GRS associated strongly with increased BMI (P = 3.6 × 10−34), fat mass (P = 6.3 × 10−28) and fat-free mass (P = 1.3 × 10−24). Higher GRS associated with lower total energy intake (P = 0.001) and higher intake of fiber (P = 2.3 × 10−4). No significant interactions were observed between GRS and the studied dietary intakes on BMI or related traits. Of the individual SNPs, after correcting for multiple comparisons, NEGR1 rs2815752 associated with diet intakes and BDNF rs4923461 showed interaction with protein intake on BMI. In conclusion, our study does not provide evidence for a major role for macronutrient-, fiber- or total energy intake levels in modifying genetic susceptibility to obesity measured as GRS. However, our data suggest that the number of risk alleles as well as some of the individual obesity loci may have a role in regulation of food and energy intake and that some individual loci may interact with diet.
Electronic supplementary material
The online version of this article (doi:10.1007/s12263-013-0352-8) contains supplementary material, which is available to authorized users.
Keywords: Obesity susceptibility loci, Fat mass, Fat-free mass, Gene–diet interactions, Macronutrients, Genetic risk score
Introduction
Obesity is a globally increasing major health problem, and considerable evidence from twin, adoption and family studies demonstrates that around 40–70 % of population variation in body mass index (BMI) is accounted by genetic factors (Herrera and Lindgren 2010; Maes et al. 1997; Stunkard et al. 1990). However, the rapid rises in the prevalence of overweight and obesity worldwide argue against a solely genetic etiology. The risk is more likely determined by environmental changes and complex interactions between genetic and environmental factors.
Recently, genome-wide association studies (GWAS) have discovered a large number of genetic loci robustly associated with various obesity-related traits (Frayling et al. 2007; Loos et al. 2008; Meyre et al. 2009; Thorleifsson et al. 2009; Willer et al. 2009; Heid et al. 2010; Speliotes et al. 2010). However, the mechanisms by which the identified variants affect the risk of obesity are mostly unclear. Several studies of the fat mass and obesity-associated gene (FTO) have indicated that environmental factors such as diet composition and physical activity level may modify the genetic susceptibility to develop obesity (Andreasen et al. 2008; Li et al. 2012; Sonestedt et al. 2011; Sonestedt et al. 2009). Apart from those in FTO, several of the identified SNPs are located in or near genes clearly related to appetite regulation (Willer et al. 2009), and recently, five of the novel obesity loci (SH2B1, KCTD15, MTCH2, NEGR1 and BDNF) were indicated to be associated with dietary macronutrient intake levels (Bauer et al. 2009). Another study reported that the obesity-associated risk variants may affect the pattern and content of food consumption (McCaffery et al. 2012). On the other hand, the MONICA/KORA study comprising of 12,462 individuals found no evidence of nutritional intake or energy expenditure as mediators of genetic variant-BMI association (Holzapfel et al. 2010).
Apart from the studies that looked for the association between individual obesity-associated SNPs and diet intakes, studies investigating interaction between genetic predisposition score and dietary intakes or other environmental factors are lacking. A couple of studies have actually investigated interactions between genetic predisposition score for obesity and physical activity (Li et al. 2010) and/or television watching (Qi et al. 2012b), but with the best of our knowledge, no study so far has investigated interaction between genetic risk score (GRS) of obesity SNPs and dietary intakes.
In this study, we wanted to clarify whether dietary factors play a role in modifying the genetic susceptibility to obesity. For this, we first tested association between 16 GWAS identified obesity susceptibility variants and body composition traits in the population-based Malmö Diet and Cancer Study cohort (MDCS). We then created a GRS of the replicated variants and investigated associations with BMI and related traits and with dietary intake levels of fats, carbohydrates, protein, fiber as well as intake of total energy. Further, we studied interaction between the diet intakes and GRS on BMI and related traits. Finally, in post hoc analyses, we studied interactions between the individual SNPs and macronutrient intakes on BMI and body composition.
Subjects and methods
Study population
The Malmö Diet and Cancer Study (MDCS) is a large population-based cohort of Southern Sweden comprising of men born between 1923 and 1945 and women born between 1923 and 1950 who were invited to a baseline examination during 1991–1996 (Berglund et al. 1993; Manjer et al. 2002). Mental incapacity and limited Swedish language skills were the only exclusion criteria. All participants visited the study center on two occasions. During the first visit, anthropometric measures were taken, blood samples were collected, and the participants were provided with the questionnaires and detailed instructions about the dietary data collection procedure. During the second visit, after nearly 10 days, trained dietary interviewers conducted individual interviews to complete the diet history and to check the correctness of completed questionnaires. The MDCS study protocols were approved by the ethics committee of Lund University. All participants provided written informed consent. Full details on recruitment of participants and study procedures have been described previously (Manjer et al. 2001). In total, 30,447 individuals participated in baseline examinations (1991–1996). After excluding individuals without DNA or other crucial basic phenotypic information (n = 58) or lacking genotype information for more than 40 % of the SNPs (n = 909), 29,480 individuals remained and constituted the study sample for the genetic analyses. For the dietary analyses, we further excluded individuals with incomplete dietary data (n = 2,258) and/or with diabetes at baseline (n = 1,115) and were left with 26,107 individuals. Diabetes at baseline was identified through the use of antidiabetic therapy or self-reported diabetes diagnosis.
Anthropometric measures
Weight was measured in kilograms using a balance-beam scale with subjects wearing light clothes and no shoes, height was measured in centimeters using a fixed stadiometer, and waist circumference was measured in centimeters by taking measurements midway between the lowest rib margin and iliac crest. Total body FM and fat-free mass (FFM) were determined using the body composition analyzer (BIA 103; JRL Systems, Mt. Clemens, MI, USA), which employs bioelectric impedance principle. Obesity was defined according to the criteria set by World Health Organization, i.e., individuals with BMI ≥ 30 kg/m2 were considered obese and those with BMI ≥ 25 kg/m2 but <30 kg/m2 were considered overweight (World Health Organization 2000).
Dietary assessment
A modified dietary history method specifically designed for the MDCS was used and is described in detail earlier (Wirfalt et al. 2002). Shortly, the dietary history method combined a 7-day menu book (covering cooked lunches, dinner meals and cold beverages), a 168-item dietary questionnaire (covering foods regularly consumed during the past year not covered by the menu book) and a 1-h interview (concerning food preparation methods and portion sizes of foods collected in the menu book). The coding routines of the dietary data were slightly altered in September 1994 in order to shorten the interview time. Although the change did not have any major influence on the ranking of individuals, a variable for diet assessment method version has been constructed and is used as a covariate when appropriate. The average daily intake of foods was calculated based on the information available in the menu book, the questionnaire and interview. Food intakes were converted to nutrient intake data using the MDC Food and Nutrient Database, which was specifically developed for the MDC study and originated from PC KOST2-93 of the Swedish National Food Administration. The relative validity of the dietary method had been evaluated with 18 days of weighted food records as the reference method. Energy-adjusted Pearson correlation coefficients in women and men, respectively, were 0.53/0.54 for protein, 0.69/0.64 for fat, 0.70/0.66 for carbohydrates and 0.69/0.74 for fiber (Riboli et al. 1997).
Genotyping
DNA was extracted from whole blood samples using Qiagen Maxipreps (Qiagen, Valencia, CA, USA). We selected 16 single-nucleotide polymorphisms (SNPs) identified through recent GWAS showing significant associations with BMI and/or obesity (Meyre et al. 2009; Thorleifsson et al. 2009; Willer et al. 2009). Of these, six SNPs (MC4R rs17782313, SH2B1 rs7498665, GNPDA2 rs10938397, BDNF rs4923461, NCR3/AIF1/BAT2 rs2844479 and FTO rs9939609) were genotyped by Taqman allelic discrimination assay-by-design method using an ABI 7900 PCR system (Applied Biosystems, Foster City, CA, USA). Six SNPs (MTCH2 rs10838738, BCDIN3D/FAIM2 rs7138803, SEC16B/RASAL2 rs10913469, TMEM18 rs6548238, MAF rs1424233 and NPC1 rs1805081) were genotyped using a Sequenom iPLEX platform (Sequenom, San Diego, CA, USA). Each of the remaining four SNPs was genotyped by two different methods. Two SNPs (NEGR1 rs2815752 and PTER rs10508503) were genotyped by Sequenom and Taqman. The KCTD15/CHST8 rs29941 and SFRS10 rs7647305 were genotyped by a KASPar allelic discrimination method (KBioscience, Hoddesdon, UK) (rs7647305 and rs29941) or by Taqman and Sequenom methods, respectively. Average successful genotype call rate was 98.4 %. All SNPs were in Hardy–Weinberg equilibrium with P > 0.0031 (Bonferroni correction for 16 independent tests at α = 0.05), as determined by a chi-square test.
Genetic risk score calculation
The GRS comprising of 13 out of the 16 genotyped SNPs was calculated by using PLINK (version 1.05). Three of the SNPs (AIF1 rs2844479, PTER rs10508503 and MAF rs1424233) were not replicated in our study and were therefore not included in the GRS. Individual genotypes were recoded as 0, 1 and 2 according to the number of alleles associating with higher BMI and/or risk of obesity for each SNP defined on the basis of robust observations in GWAS (Meyre et al. 2009; Thorleifsson et al. 2009; Willer et al. 2009).
Statistical analysis
All statistical analyses were performed with SPSS version 20 (IBM Corp., Armonk, NY, USA). To normalize the distributions, all variables were logarithmically transformed. Assuming additive model and adjusting for age and sex, we used logistic regression to analyze association between each SNP or GRS with overweight and obesity and linear regression to analyze association between the SNPs or GRS and quantitative variables (height, weight, waist and hip circumference, FM, FFM, body fat percentage and dietary variables including total energy intake (kcal/day), percentage of energy [E%] from fat, carbohydrates and protein or according to fiber density [g/1,000 kcal]). The analyses of associations with dietary variables were adjusted for age, sex, season, diet assessment method version and total energy intake.
The associations between GRS and BMI, FM and FFM were evaluated in strata of population-specific quintiles of macronutrients (E%]) or fiber (g/1,000 kcal). Interactions were assessed by including diet quintiles, GRS (treated as continuous variables), or the individual SNPs, and a multiplicative term of them to the analysis. Categorized dietary variables were used in order to handle skewness of the variables and to minimize the influence of extreme and possibly less reliably reported intakes. As dietary habits and diet reporting may differ and body fat composition differs between males and females, in addition to the main analyses of the whole cohort, we additionally report all GRS × diet and SNP × diet interaction analyses separately in males and females using gender-specific diet quintiles. Finally, in sensitivity analyses, we excluded 5,053 (18.6 %) individuals identified as putative nonadequate reporters of energy intake (under and overreporters, hereafter called as misreporters). Such misreporters were identified by comparing total energy intake and expenditure, and this method is described in detail elsewhere (Mattisson et al. 2005).
Power calculations were performed using Quanto (Quanto version 1.2.4: http://hydra.usc.edu/gxe). We estimated that with the sample size of this study and assuming an average minor allele frequency of 0.32, we were able to detect an effect size of 0.17 BMI units for each additional risk allele in GRS. Moreover, with an estimated effect size of 0.028 for dietary variables and 0.11 for GRS, we were able to detect a gene × diet interaction of at least 0.022 on BMI with 80 % power at α level of 0.05.
A P value of <0.05 was considered significant in the analyses between the SNPs or the GRS and the BMI and body composition variables as well as in the interaction analyses between the GRS and diet variables on BMI and associated traits. In contrast, the P values from association analyses of the individual SNPs with the dietary intakes were corrected for multiple testing adjusting for the number of the analyzed SNPs (n = 16), but not with the number of diet variables due to them being highly correlated. Similarly, we corrected the secondary interaction analyses between the individual SNPs and dietary intakes on BMI and body composition variables with the number of the analyzed SNPs (n = 16), but not with the number of diet variables nor with the number of BMI and related traits due to high correlation between the analyzed diet variables as well as between the BMI-related traits. A P value of ≤0.003125 was considered significant in the analyses corrected for multiple testing.
Results
The characteristics of the study participants are reported in Table 1. Of the 29,480 individuals, 40.2 % were overweight, 14.1 % were obese, and 4.2 % had diabetes at the time of baseline examinations during 1991–1996.
Table 1.
Characteristics of the Malmö Diet and Cancer Study (MDCS) participants
| Variables | All | Men | Women |
|---|---|---|---|
| n | 29,480 | 11,754 | 17,726 |
| Age (years) | 58.0 ± 7.6 | 59.1 ± 7.0 | 57.3 ± 7.9 |
| Height (cm) | 168.6 ± 8.9 | 176.3 ± 6.7 | 163.5 ± 6.1 |
| Weight (kg) | 73.6 ± 13.8 | 81.9 ± 12.4 | 68.2 ± 11.8 |
| BMI (kg/m2) | 25.8 ± 4.1 | 26.3 ± 3.6 | 25.5 ± 4.3 |
| Waist (cm) | 84.4 ± 13.1 | 93.9 ± 10.3 | 78.1 ± 10.7 |
| Hip (cm) | 98.6 ± 8.9 | 99.4 ± 7.3 | 98.0 ± 9.7 |
| Fat mass (kg) | 19.8 ± 6.9 | 17.4 ± 6.2 | 21.5 ± 6.9 |
| Fat-free mass (kg) | 53.2 ± 11.1 | 63.9 ± 8.3 | 46.2 ± 5.7 |
| Body fat (%) | 26.8 ± 7.0 | 20.8 ± 5.1 | 30.8 ± 5.0 |
| Overweight (%) | 40.2 | 49.6 | 33.9 |
| Obesity (%) | 14.1 | 13.7 | 14.3 |
| Diabetes (%) | 4.2 | 5.7 | 3.3 |
| Total energy intake (kcal/day) | 2,275 ± 655 | 2,644 ± 683 | 2,035 ± 507 |
| Carbohydrate intake (E%) | 45.2 ± 6.1 | 44.7 ± 6.2 | 45.5 ± 6.0 |
| Protein intake (E%) | 15.8 ± 2.6 | 15.5 ± 2.5 | 16.0 ± 2.6 |
| Fat intake (E%) | 39.0 ± 6.1 | 39.8 ± 6.3 | 38.5 ± 6.0 |
| Fiber density (g/1,000 kcal) | 9.1 ± 2.8 | 8.3 ± 2.5 | 9.6 ± 2.8 |
| Misreporters of energya (%) | 18.6 | 15.6 | 20.5 |
Obese: BMI ≥ 30 kg/m2; overweight: 25 kg/m2 ≤ BMI < 30 kg/m2. Data are presented as mean ± SD, unless otherwise indicated
BMI body mass index
aMisreporters of energy are individuals identified to report a nonadequately low or high energy intake or physical activity level
Association of the obesity susceptibility SNPs with obesity-related traits
Of the tested 16 SNPs, 14 were directionally consistent with the results reported in the original GWAS concerning association with BMI, waist or hip circumference, and 11 SNPs reached statistical significance with at least one of these traits (Table 2). Variants in/near FTO and TMEM18 showed the largest effect sizes for all of the three continuous traits. Variants in/near FTO, MC4R, FAIM2, SEC16B and AIF1 associated with greater height and 11 SNPs associated with increased weight (Table 2).
Table 2.
Association of 16 BMI or obesity-associated variants with anthropometric measures in the Malmö Diet and Cancer Study
| Chr | Gene or nearby gene | SNP (1/2) | BMI (kg/m2) | Height (cm) | Weight (kg) | Waist (cm) | Hip (cm) | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| β ± SE | P | β ± SE | P | β ± SE | P | β ± SE | P | β ± SE | P | |||
| 16 | FTO | rs9939609 (A/T) | 0.32 ± 0.04 | 7.0 × 10−20 | 0.11 ± 0.06 | 0.039 | 0.99 ± 0.11 | 2.6 × 10−21 | 0.70 ± 0.09 | 3.1 × 10−14 | 0.62 ± 0.08 | 3.5 × 10−16 |
| 18 | MC4R | rs17782313 (C/T) | 0.13 ± 0.04 | 0.002 | 0.18 ± 0.06 | 0.004 | 0.53 ± 0.12 | 2.3 × 10−5 | 0.30 ± 0.11 | 0.005 | 0.29 ± 0.09 | 0.001 |
| 16 | SH2B1 | rs7498665 (G/A) | 0.086 ± 0.04 | 0.009 | 0.019 ± 0.005 | 0.72 | 0.27 ± 0.11 | 0.009 | 0.30 ± 0.09 | 0.001 | 0.27 ± 0.08 | 2.6 × 10−4 |
| 4 | GNPDA2 | rs10938397 (G/A) | 0.12 ± 0.03 | 0.001 | −0.026 ± 0.05 | 0.64 | 0.29 ± 0.10 | 0.003 | 0.28 ± 0.09 | 0.001 | 0.22 ± 0.07 | 0.002 |
| 11 | MTCH2 | rs10838738 (G/A) | 0.038 ± 0.03 | 0.26 | −0.14 ± 0.05 | 0.014 | −0.002 ± 0.10 | 0.95 | 0.15 ± 0.09 | 0.11 | 0.075 ± 0.08 | 0.32 |
| 1 | NEGR1 | rs2815752 (T/C) | 0.15 ± 0.03 | 1.3 × 10−5 | −0.27 ± 0.05 | 2.3 × 10−7 | 0.17 ± 0.10 | 0.091 | 0.30 ± 0.09 | 0.001 | 0.25 ± 0.07 | 4.6 × 10−4 |
| 3 | SFRS10 | rs7647305 (C/T) | 0.11 ± 0.04 | 0.005 | 0.040 ± 0.07 | 0.55 | 0.37 ± 0.13 | 0.004 | 0.38 ± 0.11 | 0.001 | 0.21 ± 0.09 | 0.022 |
| 12 | BCDIN3D/FAIM2 | rs7138803 (A/G) | 0.085 ± 0.03 | 0.017 | 0.16 ± 0.05 | 0.002 | 0.38 ± 0.10 | 2.8 × 10−4 | 0.26 ± 0.09 | 0.005 | 0.25 ± 0.07 | 0.001 |
| 1 | SEC16B/RASAL2 | rs10913469 (C/T) | 0.18 ± 0.03 | 2.9 × 10−5 | 0.19 ± 0.06 | 0.003 | 0.66 ± 0.12 | 1.2 × 10−7 | 0.38 ± 0.11 | 0.001 | 0.36 ± 0.09 | 9.8 × 10−5 |
| 11 | BDNF | rs4923461 (A/G) | 0.14 ± 0.04 | 0.001 | 0.012 ± 0.07 | 0.84 | 0.40 ± 0.13 | 0.001 | 0.29 ± 0.11 | 0.008 | 0.25 ± 0.09 | 0.007 |
| 19 | KCTD15/CHST8 | rs29941 (C/T) | 0.065 ± 0.04 | 0.063 | −0.053 ± 0.05 | 0.35 | 0.14 ± 0.11 | 0.19 | 0.081 ± 0.09 | 0.40 | 0.084 ± 0.08 | 0.27 |
| 2 | TMEM18 | rs6548238 (C/T) | 0.19 ± 0.04 | 1.8 × 10−5 | 0.021 ± 0.07 | 0.69 | 0.59 ± 0.13 | 2.9 × 10−5 | 0.53 ± 0.12 | 9.0 × 10−6 | 0.34 ± 0.10 | 0.001 |
| 6 | NCR3/AIF1/BAT2 | rs2844479 (T/G) | −0.039 ± 0.04 | 0.25 | 0.40 ± 0.05 | 1.1 × 10−13 | 0.26 ± 0.10 | 0.019 | −0.037 ± 0.09 | 0.55 | 0.10 ± 0.08 | 0.20 |
| 10 | PTER locus | rs10508503 (C/T) | −0.004 ± 0.06 | 0.95 | −0.095 ± 0.09 | 0.36 | −0.099 ± 0.10 | 0.72 | 0.038 ± 0.15 | 0.74 | −0.036 ± 0.13 | 0.82 |
| 16 | MAF locus | rs1424233 (A/G) | 0.023 ± 0.03 | 0.34 | −0.18 ± 0.05 | 4.8 × 10−4 | −0.099 ± 0.10 | 0.48 | 0.028 ± 0.09 | 0.62 | 0.055 ± 0.07 | 0.34 |
| 18 | NPC1 | rs1805081 (A/G) | 0.097 ± 0.03 | 0.002 | 0.003 ± 0.05 | 0.97 | 0.28 ± 0.10 | 0.004 | 0.23 ± 0.09 | 0.008 | 0.21 ± 0.07 | 0.002 |
All anthropometrics (logarithmically transformed variables) are adjusted for age and sex
Chr chromosome number, SNP single-nucleotide polymorphism, (1/2) 1 refers to the risk allele, 2 refers to other allele; BMI, body mass index
SNPs in the FTO, GNPDA2, SEC16B, BDNF, TMEM18 and NPC1 loci associated with increased risk of both overweight and obesity (Table 3). SNPs in SH2B1, SFRS10 and KCTD15 associated with increased risk of overweight, but not with obesity, while the risk alleles in MC4R and NEGR1 associated with increased risk of obesity, but not with overweight (Table 3).
Table 3.
Association of 16 BMI or obesity-associated variants with overweight and obesity among participants of Malmö Diet and Cancer Study
| Gene or nearby gene | SNP (1/2) | Lean versus overweight | Lean versus obese | ||
|---|---|---|---|---|---|
| OR (95 % CI) | P | OR (95 % CI) | P | ||
| FTO | rs9939609 (A/T) | 1.12 (1.08–1.16) | 4.8 × 10−9 | 1.21 (1.14–1.27) | 3.9 × 10−12 |
| MC4R | rs17782313 (C/T) | 1.03 (0.99–1.08) | 0.14 | 1.09 (1.03–1.16) | 0.004 |
| SH2B1 | rs7498665 (G/A) | 1.06 (1.02–1.10) | 0.005 | 1.05 (0.99–1.10) | 0.091 |
| GNPDA2 | rs10938397 (G/A) | 1.07 (1.03–1.11) | 0.001 | 1.10 (1.05–1.16) | 1.4 × 10−4 |
| MTCH2 | rs10838738 (G/A) | 1.00 (0.97–1.04) | 0.86 | 1.03 (0.98–1.09) | 0.25 |
| NEGR1 | rs2815752 (T/C) | 1.04 (1.00–1.08) | 0.051 | 1.09 (1.03–1.14) | 0.002 |
| SFRS10 | rs7647305 (C/T) | 1.06 (1.01–1.11) | 0.021 | 1.04 (0.98–1.11) | 0.20 |
| BCDIN3D/FAIM2 | rs7138803 (A/G) | 1.02 (0.98–1.05) | 0.37 | 1.05 (0.99–1.10) | 0.086 |
| SEC16B/RASAL2 | rs10913469 (C/T) | 1.05 (1.00–1.10) | 0.032 | 1.12 (1.05–1.19) | 3.8 × 10−4 |
| BDNF | rs4923461 (A/G) | 1.06 (1.01–1.11) | 0.012 | 1.08 (1.01–1.15) | 0.026 |
| KCTD15/CHST8 | rs29941 (C/T) | 1.05 (1.02–1.10) | 0.006 | 1.04 (0.99–1.10) | 0.13 |
| TMEM18 | rs6548238 (C/T) | 1.06 (1.01–1.11) | 0.027 | 1.16 (1.08–1.24) | 2.2 × 10−5 |
| NCR3/AIF1/BAT2 | rs2844479 (T/G) | 1.00 (0.96–1.03) | 0.79 | 0.98 (0.93–1.03) | 0.46 |
| PTER locus | rs10508503 (C/T) | 1.00 (0.94–1.06) | 0.92 | 1.01 (0.92–1.10) | 0.84 |
| MAF locus | rs1424233 (A/G) | 1.00 (0.96–1.03) | 0.89 | 0.98 (0.93–1.03) | 0.43 |
| NPC1 | rs1805081 (A/G) | 1.06 (1.02–1.10) | 0.001 | 1.09 (1.04–1.15) | 0.001 |
Adjusted for age and sex
BMI body mass index, SNP single-nucleotide polymorphism, (1/2) 1 refers to risk allele, and 2 refers to other allele; lean: BMI < 25 kg/m2, overweight: 25 kg/m2 ≥ BMI < 30 kg/m2, obese: BMI ≥ 30 kg/m2, OR odds ratio, 95 % CI 95 % confidence interval
SNPs in FTO, GNPDA2, SFRS10, TMEM18 and NPC1 loci associated with increased FM, FFM and body fat percentage (BF%). SH2B1 and NEGR1 SNPs associated with increased FM and BF%, while the MTCH2 variant associated only with increased BF%. Variants in MC4R, FAIM2, SEC16B and BDNF loci associated with both higher FM and FFM. The risk allele of the SNP in AIF1 locus associated with increased FFM and decreased BF% (Table 4).
Table 4.
Association of 16 BMI or obesity-associated variants with measures of body composition among participants of Malmö Diet and Cancer Study
| Gene or nearby gene | SNP (1/2) | Body fat (%) | Fat mass (kg) | Fat-free mass (kg) | |||
|---|---|---|---|---|---|---|---|
| β ± SE | P | β ± SE | P | β ± SE | P | ||
| FTO | rs9939609 (A/T) | 0.24 ± 0.04 | 4.3 × 10−8 | 0.47 ± 0.06 | 5.6 × 10−16 | 0.52 ± 0.06 | 1.8 × 10−18 |
| MC4R | rs17782313 (C/T) | 0.071 ± 0.05 | 0.076 | 0.19 ± 0.07 | 0.002 | 0.33 ± 0.07 | 3.5 × 10−6 |
| SH2B1 | rs7498665 (G/A) | 0.14 ± 0.04 | 0.005 | 0.20 ± 0.06 | 0.001 | 0.098 ± 0.06 | 0.077 |
| GNPDA2 | rs10938397 (G/A) | 0.11 ± 0.04 | 0.031 | 0.16 ± 0.06 | 0.005 | 0.13 ± 0.06 | 0.024 |
| MTCH2 | rs10838738 (G/A) | 0.11 ± 0.04 | 0.006 | 0.094 ± 0.06 | 0.090 | −0.087 ± 0.06 | 0.17 |
| NEGR1 | rs2815752 (T/C) | 0.17 ± 0.04 | 3.7 × 10−5 | 0.18 ± 0.06 | 0.001 | 0.007 ± 0.06 | 0.88 |
| SFRS10 | rs7647305 (C/T) | 0.12 ± 0.05 | 0.028 | 0.18 ± 0.07 | 0.006 | 0.19 ± 0.07 | 0.013 |
| BCDIN3D/FAIM2 | rs7138803 (A/G) | 0.068 ± 0.04 | 0.18 | 0.17 ± 0.06 | 0.006 | 0.21 ± 0.06 | 2.2 × 10−4 |
| SEC16B/RASAL2 | rs10913469 (C/T) | 0.076 ± 0.05 | 0.21 | 0.25 ± 0.07 | 0.001 | 0.43 ± 0.07 | 8.8 × 10−10 |
| BDNF | rs4923461 (A/G) | 0.065 ± 0.05 | 0.30 | 0.15 ± 0.07 | 0.021 | 0.25 ± 0.07 | 0.001 |
| KCTD15/CHST8 | rs29941 (C/T) | 0.047 ± 0.04 | 0.37 | 0.076 ± 0.06 | 0.21 | 0.069 ± 0.06 | 0.24 |
| TMEM18 | rs6548238 (C/T) | 0.11 ± 0.06 | 0.037 | 0.24 ± 0.07 | 0.001 | 0.33 ± 0.08 | 1.9 × 10−5 |
| NCR3/AIF1/BAT2 | rs2844479 (T/G) | −0.12 ± 0.04 | 0.003 | −0.025 ± 0.06 | 0.56 | 0.29 ± 0.06 | 5.0 × 10−6 |
| PTER locus | rs10508503 (C/T) | 0.049 ± 0.07 | 0.35 | 0.016 ± 0.10 | 0.58 | −0.10 ± 0.10 | 0.47 |
| MAF locus | rs1424233 (A/G) | 0.063 ± 0.04 | 0.21 | 0.006 ± 0.06 | 0.67 | −0.11 ± 0.06 | 0.072 |
| NPC1 | rs1805081 (A/G) | 0.091 ± 0.04 | 0.034 | 0.14 ± 0.06 | 0.005 | 0.14 ± 0.06 | 0.017 |
Adjusted for age and sex
BMI body mass index, SNP single-nucleotide polymorphism, (1/2) 1 refers to risk allele, and 2 refers to other allele
Association of genetic risk score (GRS) with obesity-related traits
To examine the cumulative associated effect of the obesity susceptibility SNPs, a GRS comprising of 13 SNPs was created. Three SNPs (AIF1 rs2844479, PTER rs10508503 and MAF rs1424233) were not replicated in our study and were therefore not included in the GRS. Of these, AIF1 was originally reported as a weight locus and the PTER and MAF as loci for morbid obesity. The AIF1 rs2844479 allele that associated with higher weight in the original GWAS was observed to associate with lower BMI in the present study, while the PTER and MAF SNPs did not associate with BMI or obesity in the present study.
Each additional BMI-increasing allele of GRS associated with a 0.12 kg/m2 (standard error [SE] 0.01, P = 3.6 × 10−34) increase in BMI, which corresponds to a 347 g increase in body weight per allele for a person 170 cm tall. GRS associated with weight, waist circumference and hip circumference but did not show any association with height (Table 5). In addition, comparing the individuals having a low number of risk alleles (GRS of ≤11) with those having a high number (GRS ≥16) indicated a difference of 2.2 kg of total weight, 1.2 kg of FM and 1.0 kg of FFM between the groups (Electronic Supplementary Material, Fig. 2).
Table 5.
Association of genetic risk score (GRS) comprising of 13 obesity SNPs with anthropometric measures, body composition traits and obesity in the Malmö Diet and Cancer Study
| Trait | β ± SE | P |
|---|---|---|
| BMI (kg/m2) | 0.12 ± 0.01 | 3.6 × 10−34 |
| Weight (kg) | 0.35 ± 0.03 | 2.6 × 10−31 |
| Height (cm) | 0.007 ± 0.02 | 0.59 |
| Waist (cm) | 0.29 ± 0.03 | 2.5 × 10−29 |
| Hip (cm) | 0.24 ± 0.02 | 4.2 × 10−29 |
| Body fat (%) | 0.10 ± 0.01 | 2.4 × 10−16 |
| Fat mass (kg) | 0.18 ± 0.02 | 6.3 × 10−28 |
| Fat-free mass (kg) | 0.17 ± 0.02 | 1.3 × 10−24 |
| OR (95 % CI) | P | |
|---|---|---|
| Overweight | 1.05 (1.04–1.06) | 1.4 × 10−19 |
| Obesity | 1.08 (1.06–1.10) | 1.2 × 10−22 |
Adjusted for age and sex
BMI body mass index, SNP single-nucleotide polymorphism, overweight: 25 kg/m2 ≥ BMI < 30 kg/m2, obese: BMI ≥ 30 kg/m2, OR odds ratio, 95 % CI 95 % confidence interval
Each additional risk allele of GRS associated with 5 % increased odds of being overweight and 8 % increased odds of being obese (Table 5). We observed an increase in odds ratios for obesity at baseline with the increasing GRS against the group with a GRS of ≤11 which included 21 % of the study participants and was used as a reference group. In total, 18 % of all individuals had a GRS of ≥16 and they had a 1.38-fold higher risk of being overweight (95 % CI 1.27–1.49, P = 1.6 × 10−14) and 1.64-fold higher risk of being obese (95 % CI 1.47–1.84, P = 5.1 × 10−18) compared with the reference group (Electronic Supplementary Material, Fig. 3).
Association of the obesity susceptibility SNPs and GRS with total energy intake and intake levels of macronutrients and fiber
When the 16 SNPs were tested for association with dietary intakes, the obesity risk alleles of four of them associated nominally significantly with lower total energy intake (FTO, GNPDA2, NEGR1 and NPC1) and one (AIF1) with higher energy intake. Only the association with FTO remained significant after correction for multiple comparisons (P = 0.001). Four loci (FTO, MTCH2, NEGR1 and MAF) associated nominally significantly with intake levels of at least one of the macronutrients, but only the associations with NEGR1 locus remained significant after multiple comparisons. The obesity risk allele of the NEGR1 SNP associated significantly with lower fat intake (P = 3.2 × 10−5) but with higher carbohydrate (P = 3.3 × 10−5) and fiber intakes (P = 1.1 × 10−4) (Electronic Supplementary Material, Table S3). In sensitivity analyses after excluding misreporters, the association of the obesity risk allele of FTO rs9939609 with total energy intake became nonsignificant (P = 0.083), but the associations between the obesity risk allele of NEGR1 rs2815752 with fat (P = 5.5 × 10−5), carbohydrates (P = 1.8 × 10−4) and fiber (P = 4.0 × 10−6) intakes remained unchanged.
Mean total energy intake and intake levels of protein and fiber, but not of carbohydrates or fat, differed significantly across GRS groups. Individuals with higher GRS had on average lower total energy intake (P = 0.001) and higher intake of protein (P = 0.011) and fiber (P = 2.3 × 10−4) (Electronic Supplementary Material, Fig. 4). However, after further adjustment for BMI, the association of protein intake with GRS was no more significant (P = 0.25), while the associations with lower total energy intake and higher fiber intake by higher GRS were unchanged. In sensitivity analyses after excluding misreporters, the association of GRS with protein intake became nonsignificant (P = 0.11), but the associations with total energy (P = 0.019) and fiber intakes (P = 2.1 × 10−4) remained significant.
Interaction between GRS and dietary intakes on obesity-related traits
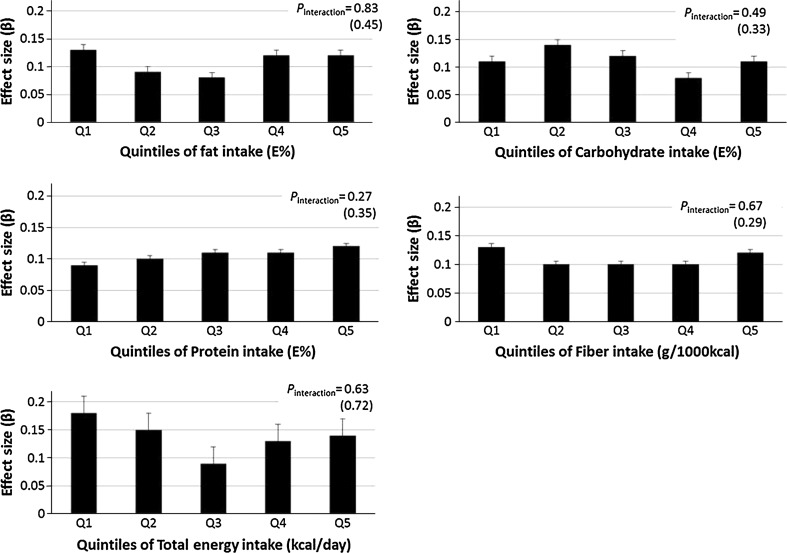
GRS was significantly associated with BMI in each diet and energy intake quintile with P values varying between P = 0.001 and P = 5.3 × 10−10 (Electronic Supplementary Material, Table S1a). However, the associated effect sizes did not differ significantly across the quintiles of fat (P interaction = 0.82), carbohydrate (P interaction = 0.49), protein (P interaction = 0.27), fiber (P interaction = 0.67) or total energy (P interaction = 0.63) intakes (Fig. 1 and Electronic Supplementary Material, Table S1a). We also observed no significant interactions between GRS and dietary intakes on odds of being overweight or obese (Electronic Supplementary Material, Table S2a). Further adjustments with fiber intake or with physical activity levels did not affect any of the results, and results remained similar in sensitivity analysis by excluding potential misreporters.
Fig. 1.
Associated effect sizes of the genetic risk score (GRS) on BMI according to intake quintiles of fat (E%), carbohydrate (E%), protein (E%), fiber (g/1,000 kcal) and total energy intake (kcal/day). Error bars represent standard error (SE). P values for interaction (P interaction) were adjusted for age, sex, season, diet assessment method version and total energy intake. The P interaction values in parenthesis refer to sensitivity analyses after excluding misreporters
In line with the BMI results, GRS associated with both FM and FFM in each diet quintile with P values varying between P = 0.017 and P = 1.7 × 10−10 (Electronic Supplementary Material, Table S1a), and no significant interactions were observed between GRS and quintiles of dietary or total energy intakes on neither FM nor FFM. The results remained similar in sensitivity analyses.
When repeating all analyses separately in both sexes, we did not observe any significant interactions between the diet variables and GRS on BMI, FM, FFM, overweight or obesity. All the results were similar using gender-specific diet quintiles as well as after excluding misreporters except that nominally significant interactions were observed between GRS and protein intake on obesity and FM among women in sensitivity analyses after excluding misreporters (P interaction = 0.039 and 0.030, respectively) (Electronic Supplementary Material, Tables S1b, S1c, S2b and S2c).
Interaction between the obesity susceptibility SNPs and dietary intakes on obesity-related traits
Of the studied SNPs, 11 indicated nominally significant interactions with at least one of the dietary intakes on BMI, FM or FFM (Electronic Supplementary Material, Table S4). However, after correction for multiple testing, only the interaction between the SNP rs4923461 in BDNF locus and protein intake on BMI remained significant (P interaction = 0.001). Higher protein intake associated significantly with higher BMI in all BDNF genotype groups, but this association was significantly stronger among individuals with the obesity susceptibility allele A of BDNF rs4923461 (P = 1.0 × 10−33 and 1.1 × 10−60 among GA- and AA-genotype carriers, respectively) as compared to carriers of the GG-genotype (P = 2.7 × 10−4) (Electronic Supplementary Material, Fig. 5). In sensitivity analyses, after excluding misreporters, these associations remained similar (P = 1.1 × 10−25 and 1.3 × 10−43 for GA and AA-genotype carriers, respectively, as compared to P = 3.9 × 10−4 for GG-genotype carriers).
Discussion
In our population-based cohort of 29,480 individuals, we replicated association between 13 of the 16 studied obesity susceptibility variants with BMI, overweight, obesity and body fat distribution. The SNPs at AIF1, PTER and MAF loci were not included in the GRS as they were not replicated in our study and have not been replicated in other studies either (den Hoed et al. 2012; Sandholt et al. 2011). When information of the 13 replicated SNPs was combined to GRS, each additional BMI-increasing allele associated with an average increase of 347 g of body weight. High GRS was found to associate with higher fiber intake but with lower total energy intake, but the dietary macronutrient, fiber or total energy intakes modified the association between GRS and BMI, FM or FFM, i.e., the associated obesogenic effects per allele of GRS did not significantly differ between the different diet intake groups. Analyses of individual SNPs revealed several nominally significant associations with dietary intakes and interactions with dietary intakes on BMI and associated traits of which association between the NEGR1 locus and intake levels of fat, carbohydrates and fiber and interaction between the BDNF locus and protein intake levels remained significant after correction for multiple testing.
Genetic factors play an important role in obesity and its comorbidities. It is, however, not genes but environmental factors, i.e., increased energy intake and reduced energy expenditure that are considered primarily responsible for the worldwide rapid increase in obesity rate during the last decades (Hill et al. 2003; Slyper 2004). Consequently, better understanding of genetic and environmental determinants of body weight regulation and interactions between them can aid in determining the role of dietary habits in the prevalence and pathogenesis of obesity and be invaluable in facilitating identification of new tools to fight against the obesity epidemic.
It is evident that the studied BMI- and obesity-associated variants only explain a small proportion of heritability of obesity and population variance in BMI. Ignorance of complex pathways together with the scarcity of information on the role of rare variants and copy number variants may cover part of the unexplained heritability. Another potential source is gene–environment interactions. We hypothesized that the relative content of dietary fats, carbohydrates, protein or fiber could influence the genetic risk of being obese. However, our results suggest that the genetic associations between GRS and BMI and associated traits were not significantly modified by dietary macronutrient contents. A couple of studies have earlier reported interaction between an obesity SNP GRS and physical activity and/or television watching in relation to BMI. In these studies, GRS was composed of 12 and 32 BMI-associated SNPs, respectively (Li et al. 2010; Qi et al. 2012b). However, our study is the first to investigate the interaction between an obesity GRS and dietary intakes.
Majority of earlier studies have either been analyzing individual SNPs and their association with lifestyle factors, including diet intake levels or dietary patterns, or analyzed whether lifestyle factors mediate part of the association between the genetic risk variants and obesity (Holzapfel et al. 2010). We found nominal associations between total energy intake and five of the analyzed SNPs, but only the association with lower total energy intake and the obesity risk allele of FTO remained significant after correcting for multiple tests. However, in sensitivity analyses after excluding misreporters, the association of FTO with total energy intake became nonsignificant. In line with our findings, another study that investigated 12 obesity susceptibility SNPs among 1,700 European female participants did not find any significant associations between any of the studied SNPs and total energy intake (Bauer et al. 2009). We observed nominal associations between six of the obesity susceptibility SNPs and dietary intake levels of macronutrients, fiber or total energy intakes, and after correcting for multiple tests, the NEGR1 locus remained significantly associated with lower fat, but with higher carbohydrate-and fiber intake levels. Similar to our results, Bauer et al. (2009) reported significant association of the NEGR1 rs2568958 (proxy for rs2815752; r 2 = 1) with lower fat intake. Further, Lee et al. (2012) recently reported evidence for NEGR1 being involved in body weight control and food intake in mice, which is in line with our results (Lee et al. 2012).
We did not find any significant interactions between GRS and total energy intake or macronutrient intakes on BMI or any of the associated traits, but we observed lower total energy intake and higher intake levels of protein and fiber among the individuals with higher GRS. As these associations could be secondary to the associations between GRS and BMI, or consequences of underreporting, we performed sensitivity analyses by excluding misreporters of energy as well as by additional adjustments for BMI and physical activity, but the associations of lower total energy intake and higher fiber intake with higher GRS remained significant in all these analyses. As the GRS, and risk alleles of four of the SNPs indicated association with lower total energy intake, it is possible that some of the obesity susceptibility variants could be involved in regulation of basal metabolic rate but more studies are needed to challenge this question. The question whether the association between higher GRS and higher fiber intake reflects involvement of fiber intake in appetite or weight regulation or the fact that overweight and obese individuals may eat differently remains unanswered in our study. However, additional adjustment with fiber intake did not influence the interaction between GRS and fats, carbohydrates or protein intakes on BMI, body composition or obesity.
The only significant interactions we detected were between GRS and protein intake on obesity and FM among women after excluding potential misreporters of energy intake. These interactions remained significant even after additionally adjusting for fiber intake and it would be interesting to see whether this finding would be replicated in other studies. Although we did not observe any significant interactions between GRS and dietary intakes on BMI or associated phenotypes, we observed a significant interaction between the BDNF locus and protein intake on BMI, the risk allele carriers having a stronger association between higher protein intake and higher BMI. We cannot exclude the possibility that some interactions may have left undetected due to insufficient power of our study to detect weaker interactions. Further, we need to emphasize that all the detected associations and the observed interaction between the BDNF locus and protein intake on BMI need to be confirmed in other studies to exclude by chance findings. Earlier studies have demonstrated that BDNF is involved in regulation of food intake, obesity and energy homeostasis (Rosas-Vargas et al. 2011). A recent study found that the association between BDNF rs6265 variant and obesity-related traits was modified by polyunsaturated fatty acid intakes among men in the Boston Puerto Rican population, which provides some further support for a role for BDNF in energy balance regulation and obesity (Ma et al. 2012). In addition to the interaction with BDNF, we observed several nominal interactions with some of the other susceptibility SNPs and diet, and we have earlier in MDCS reported interaction between dietary fat- and carbohydrate levels and the FTO genotype on FM (Sonestedt et al. 2011). Recently, a high-protein diet was found to be beneficial for weight loss and improvement of body composition and fat distribution among risk allele carriers of FTO variant rs1558902 evaluated in a 2-year diet intervention trial (Zhang et al. 2012). Another study conducted a randomized nutritional intervention with a Mediterranean-style-diet. After 3 years, the risk A-allele carriers of FTO rs9939609 variant had lower body weight gain than those without the risk allele. However, they did not find any significant interaction between nutritional intervention and the FTO variant (Razquin et al. 2010).
It is tempting to speculate that different obesity loci can partially act through different physiological mechanisms, and when the risk SNPs are simply combined to a GRS, the potential individual SNP interaction might get diluted or neutralized by interactions with specific dietary factors on specific directions. In addition, we only investigated interaction with intake levels of total energy, macronutrients and fiber and cannot exclude interaction with intake levels of other dietary factors like foods rich in carbohydrates or fats or with dietary patterns. Indeed, a recent study has reported that the genetic association of obesity SNP GRS with adiposity was more pronounced among individuals with greater consumption of sugar-sweetened beverages (Qi et al. 2012a). Obviously, further interaction and functional studies are required to understand the individual causal effects of these loci and variants.
The strength of our study is that MDCS is a large cohort with unique high-quality diet data as compared to most other epidemiological diet studies. A limitation of our study is the lack of BMI data at different time points so that we could follow the change during time. Secondly, so far at least 50 SNPs have been conclusively associated with obesity or related traits, but our study only included 16 SNPs which is another limitation of the current study. Furthermore, although our study is large and can be estimated to have sufficient power to detect relatively weak interactions, the evaluation of interactions between genotypes × dietary intakes on obesity is apparently difficult because of the complicated epidemiology of obesity and the very small effect sizes of the so far identified common genetic variants.
It is a generally accepted assumption that genetic and environmental factors that contribute to obesity interact in some way although very little is known about how such interactions take place. Our study suggests that higher susceptibility to obesity estimated as a GRS of 13 obesity-associated SNPs associates with lower total energy intake but with higher fiber intake and that some of the obesity loci like the NEGR1 locus may regulate food intake. Further, we did not find evidence for a role of macronutrient, fiber or total energy intake levels in modifying the association between GRS and obesity traits although some individual obesity loci like the BDNF may interact with diet intakes. It is important that our results are replicated in other studies with dietary data of good quality and that more studies focus on the role and mechanisms of interactions between genes and diet in obesity.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
This study is supported by the Swedish Research Council, the Swedish Heart and Lung Foundation, the Region Skåne, the Novo Nordic Foundation, the Albert Påhlsson Research Foundation, the equipment grant from the Knut and Alice Wallenberg Foundation and the Linnéus for the Lund University Diabetes Center (LUDC). MOM is a senior scientist at the Swedish Research Council. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. We thank Mrs. Malin Svensson and Mrs. Widet Tas for excellent technical assistance.
Conflict of interest
The authors declare that there is no duality of interest associated with this manuscript.
References
- Andreasen CH, Stender-Petersen KL, Mogensen MS, Torekov SS, Wegner L, Andersen G, Nielsen AL, Albrechtsen A, Borch-Johnsen K, Rasmussen SS, Clausen JO, Sandbaek A, Lauritzen T, Hansen L, Jorgensen T, Pedersen O, Hansen T. Low physical activity accentuates the effect of the FTO rs9939609 polymorphism on body fat accumulation. Diabetes. 2008;57(1):95–101. doi: 10.2337/db07-0910. [DOI] [PubMed] [Google Scholar]
- Bauer F, Elbers CC, Adan RA, Loos RJ, Onland-Moret NC, Grobbee DE, van Vliet-Ostaptchouk JV, Wijmenga C, van der Schouw YT. Obesity genes identified in genome-wide association studies are associated with adiposity measures and potentially with nutrient-specific food preference. Am J Clin Nutr. 2009;90(4):951–959. doi: 10.3945/ajcn.2009.27781. [DOI] [PubMed] [Google Scholar]
- Berglund G, Elmstahl S, Janzon L, Larsson SA. The Malmo Diet and Cancer Study. Design and feasibility. J Intern Med. 1993;233(1):45–51. doi: 10.1111/j.1365-2796.1993.tb00647.x. [DOI] [PubMed] [Google Scholar]
- den Hoed M, Luan J, Langenberg C, Cooper C, Sayer AA, Jameson K, Kumari M, Kivimaki M, Hingorani AD, Grontved A, Khaw KT, Ekelund U, Wareham NJ, Loos RJ. Evaluation of common genetic variants identified by GWAS for early onset and morbid obesity in population-based samples. Int J Obes (Lond) 2012 doi: 10.1038/ijo.2012.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frayling TM, Timpson NJ, Weedon MN, Zeggini E, Freathy RM, Lindgren CM, Perry JR, Elliott KS, Lango H, Rayner NW, Shields B, Harries LW, Barrett JC, Ellard S, Groves CJ, Knight B, Patch AM, Ness AR, Ebrahim S, Lawlor DA, Ring SM, Ben-Shlomo Y, Jarvelin MR, Sovio U, Bennett AJ, Melzer D, Ferrucci L, Loos RJ, Barroso I, Wareham NJ, Karpe F, Owen KR, Cardon LR, Walker M, Hitman GA, Palmer CN, Doney AS, Morris AD, Smith GD, Hattersley AT, McCarthy MI. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007;316(5826):889–894. doi: 10.1126/science.1141634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heid IM, Jackson AU, Randall JC, Winkler TW, Qi L, Steinthorsdottir V, Thorleifsson G, Zillikens MC, Speliotes EK, Magi R, Workalemahu T, White CC, Bouatia-Naji N, Harris TB, Berndt SI, Ingelsson E, Willer CJ, Weedon MN, Luan J, Vedantam S, Esko T, Kilpelainen TO, Kutalik Z, Li S, Monda KL, Dixon AL, Holmes CC, Kaplan LM, Liang L, Min JL, Moffatt MF, Molony C, Nicholson G, Schadt EE, Zondervan KT, Feitosa MF, Ferreira T, Lango Allen H, Weyant RJ, Wheeler E, Wood AR, Magic EK, Goddard ME, Lettre G, Mangino M, Nyholt DR, Purcell S, Smith AV, Visscher PM, Yang J, McCarroll SA, Nemesh J, Voight BF, Absher D, Amin N, Aspelund T, Coin L, Glazer NL, Hayward C, Heard-Costa NL, Hottenga JJ, Johansson A, Johnson T, Kaakinen M, Kapur K, Ketkar S, Knowles JW, Kraft P, Kraja AT, Lamina C, Leitzmann MF, McKnight B, Morris AP, Ong KK, Perry JR, Peters MJ, Polasek O, Prokopenko I, Rayner NW, Ripatti S, Rivadeneira F, Robertson NR, Sanna S, Sovio U, Surakka I, Teumer A, van Wingerden S, Vitart V, Zhao JH, Cavalcanti-Proenca C, Chines PS, Fisher E, Kulzer JR, Lecoeur C, Narisu N, Sandholt C, Scott LJ, Silander K, Stark K, Tammesoo ML, Teslovich TM, Timpson NJ, Watanabe RM, Welch R, Chasman DI, Cooper MN, Jansson JO, Kettunen J, Lawrence RW, Pellikka N, Perola M, Vandenput L, Alavere H, Almgren P, Atwood LD, Bennett AJ, Biffar R, Bonnycastle LL, Bornstein SR, Buchanan TA, Campbell H, Day IN, Dei M, Dorr M, Elliott P, Erdos MR, Eriksson JG, Freimer NB, Fu M, Gaget S, Geus EJ, Gjesing AP, Grallert H, Grassler J, Groves CJ, Guiducci C, Hartikainen AL, Hassanali N, Havulinna AS, Herzig KH, Hicks AA, Hui J, Igl W, Jousilahti P, Jula A, Kajantie E, Kinnunen L, Kolcic I, Koskinen S, Kovacs P, Kroemer HK, Krzelj V, Kuusisto J, Kvaloy K, Laitinen J, Lantieri O, Lathrop GM, Lokki ML, Luben RN, Ludwig B, McArdle WL, McCarthy A, Morken MA, Nelis M, Neville MJ, Pare G, Parker AN, Peden JF, Pichler I, Pietilainen KH, Platou CG, Pouta A, Ridderstrale M, Samani NJ, Saramies J, Sinisalo J, Smit JH, Strawbridge RJ, Stringham HM, Swift AJ, Teder-Laving M, Thomson B, Usala G, van Meurs JB, van Ommen GJ, Vatin V, Volpato CB, Wallaschofski H, Walters GB, Widen E, Wild SH, Willemsen G, Witte DR, Zgaga L, Zitting P, Beilby JP, James AL, Kahonen M, Lehtimaki T, Nieminen MS, Ohlsson C, Palmer LJ, Raitakari O, Ridker PM, Stumvoll M, Tonjes A, Viikari J, Balkau B, Ben-Shlomo Y, Bergman RN, Boeing H, Smith GD, Ebrahim S, Froguel P, Hansen T, Hengstenberg C, Hveem K, Isomaa B, Jorgensen T, Karpe F, Khaw KT, Laakso M, Lawlor DA, Marre M, Meitinger T, Metspalu A, Midthjell K, Pedersen O, Salomaa V, Schwarz PE, Tuomi T, Tuomilehto J, Valle TT, Wareham NJ, Arnold AM, Beckmann JS, Bergmann S, Boerwinkle E, Boomsma DI, Caulfield MJ, Collins FS, Eiriksdottir G, Gudnason V, Gyllensten U, Hamsten A, Hattersley AT, Hofman A, Hu FB, Illig T, Iribarren C, Jarvelin MR, Kao WH, Kaprio J, Launer LJ, Munroe PB, Oostra B, Penninx BW, Pramstaller PP, Psaty BM, Quertermous T, Rissanen A, Rudan I, Shuldiner AR, Soranzo N, Spector TD, Syvanen AC, Uda M, Uitterlinden A, Volzke H, Vollenweider P, Wilson JF, Witteman JC, Wright AF, Abecasis GR, Boehnke M, Borecki IB, Deloukas P, Frayling TM, Groop LC, Haritunians T, Hunter DJ, Kaplan RC, North KE, O’Connell JR, Peltonen L, Schlessinger D, Strachan DP, Hirschhorn JN, Assimes TL, Wichmann HE, Thorsteinsdottir U, van Duijn CM, Stefansson K, Cupples LA, Loos RJ, Barroso I, McCarthy MI, Fox CS, Mohlke KL, Lindgren CM. Meta-analysis identifies 13 new loci associated with waist-hip ratio and reveals sexual dimorphism in the genetic basis of fat distribution. Nat Genet. 2010;42(11):949–960. doi: 10.1038/ng.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrera BM, Lindgren CM. The genetics of obesity. Curr Diabet Rep. 2010;10(6):498–505. doi: 10.1007/s11892-010-0153-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill JO, Wyatt HR, Reed GW, Peters JC. Obesity and the environment: where do we go from here? Science. 2003;299(5608):853–855. doi: 10.1126/science.1079857. [DOI] [PubMed] [Google Scholar]
- Holzapfel C, Grallert H, Huth C, Wahl S, Fischer B, Doring A, Ruckert IM, Hinney A, Hebebrand J, Wichmann HE, Hauner H, Illig T, Heid IM. Genes and lifestyle factors in obesity: results from 12,462 subjects from MONICA/KORA. Int J Obes (Lond) 2010;34(10):1538–1545. doi: 10.1038/ijo.2010.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AW, Hengstler H, Schwald K, Berriel-Diaz M, Loreth D, Kirsch M, Kretz O, Haas CA, de Angelis MH, Herzig S, Brummendorf T, Klingenspor M, Rathjen FG, Rozman J, Nicholson G, Cox RD, Schafer MK. Functional inactivation of the genome-wide association study obesity gene neuronal growth regulator 1 in mice causes a body mass phenotype. PLoS One. 2012;7(7):e41537. doi: 10.1371/journal.pone.0041537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Zhao JH, Luan J, Ekelund U, Luben RN, Khaw KT, Wareham NJ, Loos RJ (2010) Physical activity attenuates the genetic predisposition to obesity in 20,000 men and women from EPIC-Norfolk prospective population study. PLoS Med 7(8). doi:10.1371/journal.pmed.1000332 [DOI] [PMC free article] [PubMed]
- Li H, Kilpelainen TO, Liu C, Zhu J, Liu Y, Hu C, Yang Z, Zhang W, Bao W, Cha S, Wu Y, Yang T, Sekine A, Choi BY, Yajnik CS, Zhou D, Takeuchi F, Yamamoto K, Chan JC, Mani KR, Been LF, Imamura M, Nakashima E, Lee N, Fujisawa T, Karasawa S, Wen W, Joglekar CV, Lu W, Chang Y, Xiang Y, Gao Y, Liu S, Song Y, Kwak SH, Shin HD, Park KS, Fall CH, Kim JY, Sham PC, Lam KS, Zheng W, Shu X, Deng H, Ikegami H, Krishnaveni GV, Sanghera DK, Chuang L, Liu L, Hu R, Kim Y, Daimon M, Hotta K, Jia W, Kooner JS, Chambers JC, Chandak GR, Ma RC, Maeda S, Dorajoo R, Yokota M, Takayanagi R, Kato N, Lin X, Loos RJ. Association of genetic variation in FTO with risk of obesity and type 2 diabetes with data from 96,551 East and South Asians. Diabetologia. 2012;55(4):981–995. doi: 10.1007/s00125-011-2370-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loos RJ, Lindgren CM, Li S, Wheeler E, Zhao JH, Prokopenko I, Inouye M, Freathy RM, Attwood AP, Beckmann JS, Berndt SI, Prostate LC, Ovarian Cancer Screening T, Jacobs KB, Chanock SJ, Hayes RB, Bergmann S, Bennett AJ, Bingham SA, Bochud M, Brown M, Cauchi S, Connell JM, Cooper C, Smith GD, Day I, Dina C, De S, Dermitzakis ET, Doney AS, Elliott KS, Elliott P, Evans DM, Sadaf Farooqi I, Froguel P, Ghori J, Groves CJ, Gwilliam R, Hadley D, Hall AS, Hattersley AT, Hebebrand J, Heid IM, Kora, Lamina C, Gieger C, Illig T, Meitinger T, Wichmann HE, Herrera B, Hinney A, Hunt SE, Jarvelin MR, Johnson T, Jolley JD, Karpe F, Keniry A, Khaw KT, Luben RN, Mangino M, Marchini J, McArdle WL, McGinnis R, Meyre D, Munroe PB, Morris AD, Ness AR, Neville MJ, Nica AC, Ong KK, O’Rahilly S, Owen KR, Palmer CN, Papadakis K, Potter S, Pouta A, Qi L, Nurses’ Health S, Randall JC, Rayner NW, Ring SM, Sandhu MS, Scherag A, Sims MA, Song K, Soranzo N, Speliotes EK, Diabetes Genetics I, Syddall HE, Teichmann SA, Timpson NJ, Tobias JH, Uda M, Sardi NIAS, Vogel CI, Wallace C, Waterworth DM, Weedon MN, Wellcome Trust Case Control C, Willer CJ, Fusion, Wraight, Yuan X, Zeggini E, Hirschhorn JN, Strachan DP, Ouwehand WH, Caulfield MJ, Samani NJ, Frayling TM, Vollenweider P, Waeber G, Mooser V, Deloukas P, McCarthy MI, Wareham NJ, Barroso I, Jacobs KB, Chanock SJ, Hayes RB, Lamina C, Gieger C, Illig T, Meitinger T, Wichmann HE, Kraft P, Hankinson SE, Hunter DJ, Hu FB, Lyon HN, Voight BF, Ridderstrale M, Groop L, Scheet P, Sanna S, Abecasis GR, Albai G, Nagaraja R, Schlessinger D, Jackson AU, Tuomilehto J, Collins FS, Boehnke M, Mohlke KL (2008) Common variants near MC4R are associated with fat mass, weight and risk of obesity. Nat Genet 40(6):768–775. doi:10.1038/ng.140 [DOI] [PMC free article] [PubMed]
- Ma XY, Qiu WQ, Smith CE, Parnell LD, Jiang ZY, Ordovas JM, Tucker KL, Lai CQ. Association between BDNF rs6265 and obesity in the Boston Puerto Rican Health Study. J Obes. 2012;2012:102942. doi: 10.1155/2012/102942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maes HH, Neale MC, Eaves LJ. Genetic and environmental factors in relative body weight and human adiposity. Behav Genet. 1997;27(4):325–351. doi: 10.1023/A:1025635913927. [DOI] [PubMed] [Google Scholar]
- Manjer J, Carlsson S, Elmstahl S, Gullberg B, Janzon L, Lindstrom M, Mattisson I, Berglund G. The Malmo Diet and Cancer Study: representativity, cancer incidence and mortality in participants and non-participants. Eur J Cancer Prev. 2001;10(6):489–499. doi: 10.1097/00008469-200112000-00003. [DOI] [PubMed] [Google Scholar]
- Manjer J, Elmstahl S, Janzon L, Berglund G. Invitation to a population-based cohort study: differences between subjects recruited using various strategies. Scand J Public Health. 2002;30(2):103–112. doi: 10.1177/14034948020300020401. [DOI] [PubMed] [Google Scholar]
- Mattisson I, Wirfält E, Aronsson CA, Wallström P, Sonestedt E, Gullberg B, Berglund G. Misreporting of energy: prevalence, characteristics of misreporters and influence on observed risk estimates in the Malmö Diet and Cancer cohort. Br J Nutr. 2005;94(05):832–842. doi: 10.1079/BJN20051573. [DOI] [PubMed] [Google Scholar]
- McCaffery JM, Papandonatos GD, Peter I, Huggins GS, Raynor HA, Delahanty LM, Cheskin LJ, Balasubramanyam A, Wagenknecht LE, Wing RR, Genetic Subgroup of Look A. Look ARG. Obesity susceptibility loci and dietary intake in the Look Ahead Trial. Am J Clin Nutr. 2012;95(6):1477–1486. doi: 10.3945/ajcn.111.026955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyre D, Delplanque J, Chevre JC, Lecoeur C, Lobbens S, Gallina S, Durand E, Vatin V, Degraeve F, Proenca C, Gaget S, Korner A, Kovacs P, Kiess W, Tichet J, Marre M, Hartikainen AL, Horber F, Potoczna N, Hercberg S, Levy-Marchal C, Pattou F, Heude B, Tauber M, McCarthy MI, Blakemore AI, Montpetit A, Polychronakos C, Weill J, Coin LJ, Asher J, Elliott P, Jarvelin MR, Visvikis-Siest S, Balkau B, Sladek R, Balding D, Walley A, Dina C, Froguel P. Genome-wide association study for early-onset and morbid adult obesity identifies three new risk loci in European populations. Nat Genet. 2009;41(2):157–159. doi: 10.1038/ng.301. [DOI] [PubMed] [Google Scholar]
- Qi Q, Chu AY, Kang JH, Jensen MK, Curhan GC, Pasquale LR, Ridker PM, Hunter DJ, Willett WC, Rimm EB, Chasman DI, Hu FB, Qi L. Sugar-sweetened beverages and genetic risk of obesity. N Engl J Med. 2012;367(15):1387–1396. doi: 10.1056/NEJMoa1203039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Q, Li Y, Chomistek AK, Kang JH, Curhan GC, Pasquale LR, Willett WC, Rimm EB, Hu FB, Qi L. Television watching, leisure time physical activity, and the genetic predisposition in relation to body mass index in women and men. Circulation. 2012;126(15):1821–1827. doi: 10.1161/CIRCULATIONAHA.112.098061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razquin C, Martinez JA, Martinez-Gonzalez MA, Bes-Rastrollo M, Fernandez-Crehuet J, Marti A. A 3-year intervention with a Mediterranean diet modified the association between the rs9939609 gene variant in FTO and body weight changes. Int J Obes (Lond) 2010;34(2):266–272. doi: 10.1038/ijo.2009.233. [DOI] [PubMed] [Google Scholar]
- Riboli E, Elmstahl S, Saracci R, Gullberg B, Lindgarde F. The Malmo Food Study: validity of two dietary assessment methods for measuring nutrient intake. Int J Epidemiol. 1997;26(Suppl 1):S161–S173. doi: 10.1093/ije/26.suppl_1.S161. [DOI] [PubMed] [Google Scholar]
- Rosas-Vargas H, Martinez-Ezquerro JD, Bienvenu T. Brain-derived neurotrophic factor, food intake regulation, and obesity. Arch Med Res. 2011;42(6):482–494. doi: 10.1016/j.arcmed.2011.09.005. [DOI] [PubMed] [Google Scholar]
- Sandholt CH, Vestmar MA, Bille DS, Borglykke A, Almind K, Hansen L, Sandbaek A, Lauritzen T, Witte D, Jorgensen T, Pedersen O, Hansen T. Studies of metabolic phenotypic correlates of 15 obesity associated gene variants. PLoS One. 2011;6(9):e23531. doi: 10.1371/journal.pone.0023531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slyper AH. The pediatric obesity epidemic: causes and controversies. J Clin Endocrinol Metab. 2004;89(6):2540–2547. doi: 10.1210/jc.2003-031449. [DOI] [PubMed] [Google Scholar]
- Sonestedt E, Roos C, Gullberg B, Ericson U, Wirfalt E, Orho-Melander M. Fat and carbohydrate intake modify the association between genetic variation in the FTO genotype and obesity. Am J Clin Nutr. 2009;90(5):1418–1425. doi: 10.3945/ajcn.2009.27958. [DOI] [PubMed] [Google Scholar]
- Sonestedt E, Gullberg B, Ericson U, Wirfalt E, Hedblad B, Orho-Melander M. Association between fat intake, physical activity and mortality depending on genetic variation in FTO. Int J Obes (Lond) 2011;35(8):1041–1049. doi: 10.1038/ijo.2010.263. [DOI] [PubMed] [Google Scholar]
- Speliotes EK, Willer CJ, Berndt SI, Monda KL, Thorleifsson G, Jackson AU, Lango Allen H, Lindgren CM, Luan J, Magi R, Randall JC, Vedantam S, Winkler TW, Qi L, Workalemahu T, Heid IM, Steinthorsdottir V, Stringham HM, Weedon MN, Wheeler E, Wood AR, Ferreira T, Weyant RJ, Segre AV, Estrada K, Liang L, Nemesh J, Park JH, Gustafsson S, Kilpelainen TO, Yang J, Bouatia-Naji N, Esko T, Feitosa MF, Kutalik Z, Mangino M, Raychaudhuri S, Scherag A, Smith AV, Welch R, Zhao JH, Aben KK, Absher DM, Amin N, Dixon AL, Fisher E, Glazer NL, Goddard ME, Heard-Costa NL, Hoesel V, Hottenga JJ, Johansson A, Johnson T, Ketkar S, Lamina C, Li S, Moffatt MF, Myers RH, Narisu N, Perry JR, Peters MJ, Preuss M, Ripatti S, Rivadeneira F, Sandholt C, Scott LJ, Timpson NJ, Tyrer JP, van Wingerden S, Watanabe RM, White CC, Wiklund F, Barlassina C, Chasman DI, Cooper MN, Jansson JO, Lawrence RW, Pellikka N, Prokopenko I, Shi J, Thiering E, Alavere H, Alibrandi MT, Almgren P, Arnold AM, Aspelund T, Atwood LD, Balkau B, Balmforth AJ, Bennett AJ, Ben-Shlomo Y, Bergman RN, Bergmann S, Biebermann H, Blakemore AI, Boes T, Bonnycastle LL, Bornstein SR, Brown MJ, Buchanan TA, Busonero F, Campbell H, Cappuccio FP, Cavalcanti-Proenca C, Chen YD, Chen CM, Chines PS, Clarke R, Coin L, Connell J, Day IN, den Heijer M, Duan J, Ebrahim S, Elliott P, Elosua R, Eiriksdottir G, Erdos MR, Eriksson JG, Facheris MF, Felix SB, Fischer-Posovszky P, Folsom AR, Friedrich N, Freimer NB, Fu M, Gaget S, Gejman PV, Geus EJ, Gieger C, Gjesing AP, Goel A, Goyette P, Grallert H, Grassler J, Greenawalt DM, Groves CJ, Gudnason V, Guiducci C, Hartikainen AL, Hassanali N, Hall AS, Havulinna AS, Hayward C, Heath AC, Hengstenberg C, Hicks AA, Hinney A, Hofman A, Homuth G, Hui J, Igl W, Iribarren C, Isomaa B, Jacobs KB, Jarick I, Jewell E, John U, Jorgensen T, Jousilahti P, Jula A, Kaakinen M, Kajantie E, Kaplan LM, Kathiresan S, Kettunen J, Kinnunen L, Knowles JW, Kolcic I, Konig IR, Koskinen S, Kovacs P, Kuusisto J, Kraft P, Kvaloy K, Laitinen J, Lantieri O, Lanzani C, Launer LJ, Lecoeur C, Lehtimaki T, Lettre G, Liu J, Lokki ML, Lorentzon M, Luben RN, Ludwig B, Magic MP, Marek D, Marre M, Martin NG, McArdle WL, McCarthy A, McKnight B, Meitinger T, Melander O, Meyre D, Midthjell K, Montgomery GW, Morken MA, Morris AP, Mulic R, Ngwa JS, Nelis M, Neville MJ, Nyholt DR, O’Donnell CJ, O’Rahilly S, Ong KK, Oostra B, Pare G, Parker AN, Perola M, Pichler I, Pietilainen KH, Platou CG, Polasek O, Pouta A, Rafelt S, Raitakari O, Rayner NW, Ridderstrale M, Rief W, Ruokonen A, Robertson NR, Rzehak P, Salomaa V, Sanders AR, Sandhu MS, Sanna S, Saramies J, Savolainen MJ, Scherag S, Schipf S, Schreiber S, Schunkert H, Silander K, Sinisalo J, Siscovick DS, Smit JH, Soranzo N, Sovio U, Stephens J, Surakka I, Swift AJ, Tammesoo ML, Tardif JC, Teder-Laving M, Teslovich TM, Thompson JR, Thomson B, Tonjes A, Tuomi T, van Meurs JB, van Ommen GJ, Vatin V, Viikari J, Visvikis-Siest S, Vitart V, Vogel CI, Voight BF, Waite LL, Wallaschofski H, Walters GB, Widen E, Wiegand S, Wild SH, Willemsen G, Witte DR, Witteman JC, Xu J, Zhang Q, Zgaga L, Ziegler A, Zitting P, Beilby JP, Farooqi IS, Hebebrand J, Huikuri HV, James AL, Kahonen M, Levinson DF, Macciardi F, Nieminen MS, Ohlsson C, Palmer LJ, Ridker PM, Stumvoll M, Beckmann JS, Boeing H, Boerwinkle E, Boomsma DI, Caulfield MJ, Chanock SJ, Collins FS, Cupples LA, Smith GD, Erdmann J, Froguel P, Gronberg H, Gyllensten U, Hall P, Hansen T, Harris TB, Hattersley AT, Hayes RB, Heinrich J, Hu FB, Hveem K, Illig T, Jarvelin MR, Kaprio J, Karpe F, Khaw KT, Kiemeney LA, Krude H, Laakso M, Lawlor DA, Metspalu A, Munroe PB, Ouwehand WH, Pedersen O, Penninx BW, Peters A, Pramstaller PP, Quertermous T, Reinehr T, Rissanen A, Rudan I, Samani NJ, Schwarz PE, Shuldiner AR, Spector TD, Tuomilehto J, Uda M, Uitterlinden A, Valle TT, Wabitsch M, Waeber G, Wareham NJ, Watkins H, Procardis C, Wilson JF, Wright AF, Zillikens MC, Chatterjee N, McCarroll SA, Purcell S, Schadt EE, Visscher PM, Assimes TL, Borecki IB, Deloukas P, Fox CS, Groop LC, Haritunians T, Hunter DJ, Kaplan RC, Mohlke KL, O’Connell JR, Peltonen L, Schlessinger D, Strachan DP, van Duijn CM, Wichmann HE, Frayling TM, Thorsteinsdottir U, Abecasis GR, Barroso I, Boehnke M, Stefansson K, North KE, McCarthy MI, Hirschhorn JN, Ingelsson E, Loos RJ. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat Genet. 2010;42(11):937–948. doi: 10.1038/ng.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stunkard AJ, Harris JR, Pedersen NL, McClearn GE. The body-mass index of twins who have been reared apart. N Engl J Med. 1990;322(21):1483–1487. doi: 10.1056/NEJM199005243222102. [DOI] [PubMed] [Google Scholar]
- Thorleifsson G, Walters GB, Gudbjartsson DF, Steinthorsdottir V, Sulem P, Helgadottir A, Styrkarsdottir U, Gretarsdottir S, Thorlacius S, Jonsdottir I, Jonsdottir T, Olafsdottir EJ, Olafsdottir GH, Jonsson T, Jonsson F, Borch-Johnsen K, Hansen T, Andersen G, Jorgensen T, Lauritzen T, Aben KK, Verbeek AL, Roeleveld N, Kampman E, Yanek LR, Becker LC, Tryggvadottir L, Rafnar T, Becker DM, Gulcher J, Kiemeney LA, Pedersen O, Kong A, Thorsteinsdottir U, Stefansson K. Genome-wide association yields new sequence variants at seven loci that associate with measures of obesity. Nat Genet. 2009;41(1):18–24. doi: 10.1038/ng.274. [DOI] [PubMed] [Google Scholar]
- Willer CJ, Speliotes EK, Loos RJ, Li S, Lindgren CM, Heid IM, Berndt SI, Elliott AL, Jackson AU, Lamina C, Lettre G, Lim N, Lyon HN, McCarroll SA, Papadakis K, Qi L, Randall JC, Roccasecca RM, Sanna S, Scheet P, Weedon MN, Wheeler E, Zhao JH, Jacobs LC, Prokopenko I, Soranzo N, Tanaka T, Timpson NJ, Almgren P, Bennett A, Bergman RN, Bingham SA, Bonnycastle LL, Brown M, Burtt NP, Chines P, Coin L, Collins FS, Connell JM, Cooper C, Smith GD, Dennison EM, Deodhar P, Elliott P, Erdos MR, Estrada K, Evans DM, Gianniny L, Gieger C, Gillson CJ, Guiducci C, Hackett R, Hadley D, Hall AS, Havulinna AS, Hebebrand J, Hofman A, Isomaa B, Jacobs KB, Johnson T, Jousilahti P, Jovanovic Z, Khaw KT, Kraft P, Kuokkanen M, Kuusisto J, Laitinen J, Lakatta EG, Luan J, Luben RN, Mangino M, McArdle WL, Meitinger T, Mulas A, Munroe PB, Narisu N, Ness AR, Northstone K, O’Rahilly S, Purmann C, Rees MG, Ridderstrale M, Ring SM, Rivadeneira F, Ruokonen A, Sandhu MS, Saramies J, Scott LJ, Scuteri A, Silander K, Sims MA, Song K, Stephens J, Stevens S, Stringham HM, Tung YC, Valle TT, Van Duijn CM, Vimaleswaran KS, Vollenweider P, Waeber G, Wallace C, Watanabe RM, Waterworth DM, Watkins N, Wellcome Trust Case Control C, Witteman JC, Zeggini E, Zhai G, Zillikens MC, Altshuler D, Caulfield MJ, Chanock SJ, Farooqi IS, Ferrucci L, Guralnik JM, Hattersley AT, Hu FB, Jarvelin MR, Laakso M, Mooser V, Ong KK, Ouwehand WH, Salomaa V, Samani NJ, Spector TD, Tuomi T, Tuomilehto J, Uda M, Uitterlinden AG, Wareham NJ, Deloukas P, Frayling TM, Groop LC, Hayes RB, Hunter DJ, Mohlke KL, Peltonen L, Schlessinger D, Strachan DP, Wichmann HE, McCarthy MI, Boehnke M, Barroso I, Abecasis GR, Hirschhorn JN, Genetic Investigation of ATC (2009) Six new loci associated with body mass index highlight a neuronal influence on body weight regulation. Nat Genet 41(1):25–34. doi:10.1038/ng.287 [DOI] [PMC free article] [PubMed]
- Wirfalt E, Mattisson I, Johansson U, Gullberg B, Wallstrom P, Berglund G. A methodological report from the Malmo Diet and Cancer study: development and evaluation of altered routines in dietary data processing. Nutr J. 2002;1:3. doi: 10.1186/1475-2891-1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization (2000) Obesity: preventing and managing the global epidemic. Report of a WHO consultation. World Health Organ Tech Rep Ser 894:1–253 [PubMed]
- Zhang X, Qi Q, Zhang C, Smith SR, Hu FB, Sacks FM, Bray GA, Qi L. FTO genotype and 2-year change in body composition and fat distribution in response to weight-loss diets: the pounds lost trial. Diabetes. 2012;61(11):3005–3011. doi: 10.2337/db11-1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.