Abstract
Malnutrition inflammation syndrome (MIS) is common among ESRD patients. In the present study, we have investigated the association of genetic markers associated with appetite and energy regulation with malnutrition inflammation syndrome among end-stage renal disease (ESRD) patients. Two hundred and fifty-seven patients on maintenance hemodialysis and 200 normal healthy controls were included in the study. Nutritional assessment was done by subjective global assessment scores (SGA). Genotyping of leptin-2548 G/A (rs7799039), ghrelin Leu72Met (rs696217-408 C/A), Arg51Gln (rs34911341-346 G/A) and uncoupling protein 2 (UCP2) 45 bp insertion deletion was done using PCR–RFLP. Levels of leptin and acyl ghrelin were assessed using ELISA. Leptin-2548 AA genotype was associated with twofold higher risk of disease susceptibility while UCP2 insertion–deletion heterozygotes showed protective effect. Ghrelin Gln51Gln and Met72Met genotype were associated with 3.4- and 2.5-fold higher disease susceptibility. The Met72 and Gln51 allele showed 3.3- and 2.1-fold higher susceptibility to malnutrition in severe SGA group. Further, the levels of acyl ghrelin were significantly less in severe category of malnutrition and in poor appetite group. On combined analysis, the group 2 (presence of 3–4 risk alleles) showed 1.5- and twofold higher susceptibility to disease and malnutrition, respectively. On docking analysis, it was observed that higher receptor binding energy was associated with the mutant form of ghrelin (Gln51). Moderate and severe SGA were associated with 2.2- and 4.1-fold higher death hazard. Our study suggests that ghrelin may be major marker contributing to susceptibility to MIS among ESRD patients.
Electronic supplementary material
The online version of this article (doi:10.1007/s12263-013-0353-7) contains supplementary material, which is available to authorized users.
Keywords: End-stage renal disease, Malnutrition inflammation syndrome, Leptin, Ghrelin UCP2
Introduction
Protein energy malnutrition is frequently observed in end-stage renal disease (ESRD) and is a key predictor of survival in their survival. In ESRD patients on hemodialysis protein energy malnutrition is frequently observed and a key predictor of survival. There are diverse aspects which might cause malnutrition which includes anorexia, hindrance in gastric emptying, inflammation, weak protein assimilation, depression and comorbidity (Mitch 2002). Universal features of malnutrition are muscle wasting, loss of protein stores, hypoalbuminemia, increased energy expenditure and anorexia. A significant amount of mortality risk has been reported in peritoneal dialysis (PD) patients with poor appetite (Gama-Axelsson et al. 2013). Leptin is an important hormone produced by adipocytes, and belongs to IL6 family of cytokines, related to diminished food intake. It has been implied that TNF-α is related to cachexia (Tracey et al. 1988). A dose-dependent increase in leptin levels was detected on endotoxin or cytokines like IL-1 or TNF-α administration in experimental animals (Grunfeld et al. 1996). This suggests that weight loss and anorexia in chronic wasting diseases might be linked with leptin. The ESRD patients are associated with hyperleptinemia, suggesting hyperleptinemia stimulated feedback inhibition (Nordfors et al. 1998) as a cause of anorexia and decreased food intake (Young et al. 1997).
On the contrary, ghrelin is an appetite stimulating hormone and transduces signals to hypothalamic regulatory nuclei that control energy homeostasis. Ghrelin is shown to enhance food intake in dialysis patients with mild to moderate malnutrition in randomized, placebo-controlled trial (Wynne et al. 2005). It is found in acylated and des-acylated versions in the body. The acylated version attaches to growth hormone secretagogue receptor (GHS-R1a) and exerts biological function while des-acylated form is non-functional and may contribute to appetite loss in ESRD patients (Mak et al. 2012). Increase in plasma levels of total ghrelin correlates with fat mass in ESRD patients (Cheung and Mak 2010). Malnutrition is also linked with enhanced energy expenditure. UCP2 is an energy regulator which uncouples respiration from ATP synthesis by providing an alternative route for protons to enter in the mitochondrial matrix resulting into heat generation. The functions of UCP2 are thermogenesis and reduction of reactive oxygen species. UCP2 is also related to energy metabolism and obesity in human and rodent models (Schrauwen and Hesselink 2002). Increase in energy expenditure in ESRD patients is identified as a factor associated with malnutrition inflammation syndrome; hence, UCP2 is a potential marker of interest in this regard (Carrero et al. 2013).
These factors are genetically regulated which in turn may influence the severity of MIS. Recently, there are many studies in ESRD which have demonstrated association of the ghrelin and leptin levels with malnutrition (Caliskan et al. 2012) and mortality (Carrero et al. 2011). UCP2 expression level has not been explored in malnourished ESRD patients, but expression of UCP2 has been investigated in cancer cachectic patients and mice model (Julienne et al. 2012). In our previous study, we found that inflammatory markers like TNF-α, IL-6 and IL-10 have an important role in inducing malnutrition and inflammation at both genotypic and phenotypic level (published elsewhere). Interestingly, there is no study where the leptin, ghrelin and UCP2 gene polymorphisms have been collectively studied in malnutrition inflammation syndrome in ESRD in spite of the fact that they fall in the appetite-energy regulatory pathway.
In the present study, we investigated the genetic aspect of nutritional markers leptin (-2548 G/A-dbSNPID rs7799039), ghrelin (Leu72Met-dbSNP ID rs696217-406 C/A, Arg51Gln-dbSNP ID rs34911341-346 G/A) and UCP2 45 bp I/D in exon 8 and their association with malnutrition. The functional aspect of leptin and ghrelin is investigated by measuring their levels in the serum.
Materials and methods
Study participants
Two hundred and fifty-seven ESRD patients on maintenance hemodialysis were prospectively registered from December 2008–2011 in this study. These patients were under regular follow-up in the dialysis unit at Sanjay Gandhi Post Graduate Institute of Medical Sciences, Lucknow. The study was approved by the Ethical committee of SGPGIMS, Lucknow, India. The patients with any active viral infection were excluded from the study. Two hundred age, gender and ethnically matched healthy controls were included. Written informed consent was obtained from all the participants included in the study. Since our study comprised of more than 70 % male patients, care was taken to include more male controls to eliminate gender bias. The male–female ratio in patients and controls was 77.4:21.8 (199/56) and 74.5:24.5 (149/51), respectively. They were selected randomly from the hospital staff after evaluating their biochemical parameters. Both the patients and controls belonged to the state of Uttar Pradesh.
Nutritional assessment
Nutritional assessment was done using SGA scores (CANUSA 1996; Blumenkrantz et al. 1980). SGA scoring was done on the basis of weight change in last 6 months, dietary intake (solid, semi-solid, liquid, starvation), gastrointestinal symptoms (nausea, anorexia, vomit, diarrhea), functional impairment, physical examination (loss of subcutaneous fat, muscle wasting and edema). Resting metabolic rate of patients was assessed using bioimpedance analysis in resting state. The patients were asked to rate their appetite for past 1 week as good, average or poor (Bossola et al. 2009) for appetite assessment.
Biochemical profiling
Measurement of serum biochemical and lipid profiles was done using blood samples for in the morning after fasting of 8 h. These included renal function test, S. Albumin, S. Protein, urinary protein level, liver function test, lipid profile, iron profile, CRP and PTH.
Enzyme-linked immunosorbent assay
Levels were quantified using commercially available kit for leptin (Ray Biotech, Inc., Norcross GA) and acyl ghrelin (SPI BIO, Bertin Pharma Biotech, Montigny le Bretonneau, France). To prevent proteolysis, 200 KIU of aprotinin was added per ml of blood sample. Serum was isolated for leptin and plasma for measurement of acyl ghrelin. These aliquots were kept at −80 °C until use. The assay was performed in duplicates independently for each sample according to the manufacturer’s instructions. The result was expressed as nanograms per milliliter (ng/ml) for leptin and picograms per milliliter (pg/ml) for ghrelin, based on the standard provided with the kits. Based on SGA scoring, 25 patients with normal nutritional status, 20 mild, 25 moderate and 20 severe malnourished patients were included. These levels were compared with 30 healthy controls.
Genotyping
Three milliliter of peripheral blood was collected in EDTA vials, and genomic DNA was extracted using commercial kit (Qiagen). The polymorphisms assessed in the present study were leptin-2548 G/A (dbSNP ID rs7799039), ghrelin Leu72Met (dbSNP ID rs696217-408 C/A), ghrelin Arg51Gln (dbSNP ID rs34911341-346 G/A) in exon 2 and UCP2 45 bp I/D in exon 8. The detailed genotyping protocol is shown in Table S1. Thirty percent of samples were arbitrarily selected to be genotyped second time to ensure reproducibility. Genotyping of all subjects was performed blinded to clinical status.
In silico analysis
Potential functional impact of non-synonymous SNPs was identified using PolyPhen-2, SIFT, I-Mutant, Prop v.1.0 b propeptide cleavage site prediction and PopMusic. The docking analysis was performed for Arg51Gln mutation as this particular amino acid lies in the mature ghrelin protein. Molecular modeling of wild and mutant ghrelin was carried out using easy modeler (Sali et al. 1995). These were then octanoylated using AutoDock. Molecular modeling of ghrelin receptor was done using Galaxy TBM. Template used by Galaxy TBM server for modeling was 2ks9_A, 1u19_A and 2z73_A. Docking of octanoylated ghrelin over the ghrelin receptor was done using Hex software. The UCP2 I/D polymorphism is a potential site for miRNA binding which was assessed using Target Scan 6.
Statistical analysis
Sample size was calculated using Quanto (Ver. 1.1.) so as to achieve 80 % power of study. The biochemical data was compared by independent samples t test and ANOVA. The values are expressed as mean ± SD. Alleles and genotypic frequencies for were calculated by using gene counting method. Comparison of the categorical data, i.e., different leptin, ghrelin and UCP2 genotypes, among controls and patients was done by logistic regression analysis to calculate odds ratios (OR) and their 95 % confidence intervals (CI). P value of <0.05 was taken as a significant difference. Bonferroni correction was applied wherever applicable. All the statistical analysis was done by using SPSS 15.0 version and Graphpad Prism (version.5). Allele frequencies of both patients and controls were tested for Hardy–Weinberg equilibrium. Haplotypes were constructed using SNPStats (http://bioinfo.iconcologia.net/snpstats). Kaplan–Meier survival analysis and log rank test were used to assess clinical outcome, that is, overall survival (OS) in relation to genotypes studied.
Results
Study participants and demographics
The mean age was 39.3 ± 12.6 in patients and 38.8 ± 13.17 in controls. The BMI in our patients was in normal range but lower than normal healthy controls. On comparing patients and controls, almost all the biochemical parameters were significantly different between both the groups (Table S2). The period of dialysis in our patients was 2 to 36 months. Forty-eight percent of patients had elevated CRP levels. On the basis of SGA, the patients were classified into 4 sub-groups–normal (n = 78), mild (n = 38), moderate (n = 100) and severe (n = 41). The patients falling in the severe SGA group had significantly less BMI, while in this group of patients, the period on dialysis, systolic blood pressure and ESR were significantly increased (Table S3). More than 60 % patients reported poor appetite in severe malnutrition.
Patients were sub-grouped into inflamed and non-inflamed sets on the basis of CRP levels as they are predictor of acute phase response (Pearson et al. 2003) Inflamed group constituted patients who had CRP greater than 1 mg/dl (n = 156), and non-inflamed group were the patients who had CRP was less than 1 mg/dl (n = 101). At biochemical level, there was no significant difference between both the groups. Since the range of period of dialysis of patients was broad, the patients were sub-grouped into two groups as period of dialysis <1 year (n = 170, 66.1 %) and period of dialysis ≥1 year (n = 87, 33.9 %). The patients whose period of dialysis ≥1 year had decreased weight (P = 0.01) and increased serum levels of albumin (P = 0.003, Table S4).
Genotype distribution in patients and controls
Leptin-2548 AA genotype was associated with higher susceptibility to the disease (OR = 2.0, 95 % CI 1.2–3.3, P = 0.007). Further, ID genotype of UCP2 showed protective effect (OR = 0.5, 95 % CI 0.3–0.8, P = 0.003). Ghrelin 364-AA (Gln51Gln) genotype was associated with higher susceptibility (OR = 3.41, 95 % CI 1.2–7.5, P = 0.02). The allele frequency of G and A was 78.4 % and 21.6 % patients and 84.3 and 15.7 % in controls which showed significant difference between both the groups (OR = 1.6, 95 % CI 1.1–2.1, P = 0.02). Ghrelin 408-AA (Met72Met) showed higher susceptibility to the disease (OR = 2.5, 95 % CI 1.2–5.4, P = 0.02) as shown in Table 1. None of the ghrelin haplotypes were significant among cases or controls (Table S5).
Table 1.
Distribution of genotypes between patients and controls
| Marker | Genotype | Patient | Control | OR (95 % CI) | P value |
|---|---|---|---|---|---|
| UCP2 I/D | |||||
| DD | 149 (57.6 %) | 94 (47.0 %) | Reference | ||
| ID | 70 (27.6 %) | 82 (41.0 %) | 0.5 (0.3–0.8) | 0.003* | |
| II | 38 (14.8 %) | 24 (12.0 %) | 1.3 (0.7–2.2) | 0.4 | |
| D | 368 (71.6 %) | 270 (67.5 %) | Reference | ||
| I | 146 (28.4 %) | 130 (32.5 %) | 1.2 (0.9–1.6) | 0.2 | |
| Dominant model | |||||
| DD versus II + ID | 1.2 (0.8–1.7) | 0.2 | |||
| Recessive model | |||||
| II versus ID + DD | 0.7 (0.45–1.3) | 0.4 | |||
| Leptin-2548 G/A | |||||
| GG | 90 (35.0 %) | 82 (41.0 %) | Reference | ||
| GA | 99 (38.5 %) | 92 (46.0 %) | 1.2 (0.8–1.9) | 0.39 | |
| AA | 68 (26.5 %) | 26 (13.0 %) | 2.0 (1.2–3.3) | 0.007* | |
| G | 279 (54.3 %) | 256 (64.0 %) | Reference | ||
| A | 235 (45.7 %) | 144 (36.0 %) | 1.5 (1.1–1.9) | 0.003* | |
| Dominant model | |||||
| GG versus GA + AA | 0.27 (0.1–0.4) | 0.001* | |||
| Recessive model | |||||
| AA vs. GG + GA | 1.8 (1.1–2.9) | 0.012* | |||
| Ghrelin Arg51Gln | |||||
| GG | 164 (63.8 %) | 142 (71.0 %) | Reference | ||
| GA | 75 (29.2 %) | 53 (26.5 %) | 1.2 (0.8–1.8) | 0.33 | |
| AA | 18 (7.0 %) | 5 (2.5 %) | 3.4 (1.2–7.5) | 0.02* | |
| G | 403 (78.4 %) | 337 (84.3 %) | Reference | ||
| A | 111 (21.6 %) | 63 (15.7 %) | 1.6 (1.1–2.1) | 0.02* | |
| Dominant model | |||||
| GG versus GA + AA | 0.7 (0.4–1.0) | 0.1 | |||
| Recessive model | |||||
| AA versus GG + GA | 2.9 (1.0–8.0) | 0.003* | |||
| Ghrelin Leu72Met | |||||
| CC | 178 (69.3 %) | 141 (70.5 %) | Reference | ||
| CA | 51 (19.8 %) | 50 (25.0 %) | 0.8 (0.5–1.3) | 0.2 | |
| AA | 28 (10.9 %) | 9 (4.5 %) | 2.5 (1.2–5.4) | 0.02* | |
| C | 407 (79.2 %) | 332 (83.0 %) | Reference | ||
| A | 107 (20.8 %) | 68 (17.0 %) | 1.3 (0.9–1.8) | 0.15 | |
| Dominant model | |||||
| CC versus CA + AA | 1.07 (0.7–1.6) | 0.75 | |||
| Recessive model | |||||
| AA versus CC + CA | 2.58 (1.2–5.6) | 0.01* | |||
Logistic regression analysis was used for comparing the genotypes
Genotype distribution in SGA categories
The 408-CA genotype of ghrelin Leu72Met was associated with higher susceptibility to malnutrition in mild (OR = 2.6, 95 % CI 1.1–6.9, P = 0.04) and severe SGA groups (OR = 3.8, 95 % CI 1.5–7.6, P = 0.005). The 408A allele was significantly associated with severe SGA group (OR = 3.3, 95 % CI 1.7–6.5, P = 0.0004). Furthermore, the ghrelin 346-AA (Arg51Arg) genotype was associated with higher susceptibility to malnutrition in severe SGA group (OR = 3.3, 95 % CI 1.3–7.4, P = 0.04). This difference was significant at the allelic level (OR = 2.1, 95 % CI 1.1–3.6, P = 0.02) as shown in Table 2.
Table 2.
Comparison of genotype among patients on the basis of SGA scores
| Genotype | Normal | Mild | Moderate | Severe | Normal versus mild OR (95 % CI), P |
Normal versus moderate OR (95 % CI), P |
Normal versus severe OR (95 % CI), P |
|---|---|---|---|---|---|---|---|
| UCP2 I/D | |||||||
| DD | 47 (60.2 %) | 22 (58.0 %) | 56 (56.0 %) | 24 (58.5 %) | Reference | Reference | Reference |
| ID | 22 (28.2 %) | 8 (21.0 %) | 31 (31.0 %) | 9 (22.0 %) | 0.7 (0.2–2.0), 0.6 | 1.1 (0.6–2.3), 0.6 | 0.8 (0.3–2.0), 0.6 |
| II | 9 (11.5 %) | 8 (21.0 %) | 13 (13.0 %) | 8 (19.5 %) | 1.9 (0.6–5.5), 0.2 | 1.2 (0.4–3.08), 0.7 | 1.7 (0.5–5.0), 0.3 |
| D | 116 (74.3 %) | 52 (68.4 %) | 143 (71.5 %) | 57 (69.5 %) | Reference | Reference | Reference |
| I | 40 (25.7 %) | 24 (31.6 %) | 57 (28.5 %) | 25 (30.5 %) | 1.2 (0.7–2.3), 0.4 | 1.1 (0.69–1.79), 0.7 | 1.2 (0.68–2.2), 0.5 |
| Leptin-2548 G/A | |||||||
| GG | 26 (33.3 %) | 11 (28.9 %) | 38 (38.0 %) | 15 (36.5 %) | Reference | Reference | Reference |
| GA | 31 (39.7 %) | 16 (42.1 %) | 37 (37.0 %) | 15 (36.5 %) | 0.9 (0.4–1.9), 0.8 | 1.1 (0.6–2.0), 0.7 | 0.8 (0.4–1.9), 0.8 |
| AA | 21 (26.9 %) | 11 (28.9 %) | 25 (25.0 %) | 11 (26.8 %) | 0.9 (0.3–2.1), 0.8 | 1.1 (0.5–2.1), 0.8 | 1.0 (0.4–2.3), 1.0 |
| G | 83 (53.4 %) | 38 (50.0 %) | 113 (56.5 %) | 45 (54.8 %) | Reference | Reference | Reference |
| A | 73 (46.7 %) | 38 (50.0 %) | 87 (43.5 %) | 37 (45.2 %) | 1.1 (0.6–1.9), 0.6 | 0.8 (0.5–1.3), 0.6 | 0.9 (0.5–1.5), 0.8 |
| Ghrelin Arg51Gln | |||||||
| GG | 50 (64.1 %) | 22 (57.9 %) | 73 (73.0 %) | 19 (46.3 %) | Reference | Reference | Reference |
| GA | 24 (30.8 %) | 13 (34.2 %) | 23 (23.0 %) | 15 (36.6 %) | 1.2 (0.5–2.6), 0.8 | 0.7 (0.3–1.3), 0.3 | 1.3 (0.5–2.8), 0.5 |
| AA | 4 (5.1 %) | 3 (7.8 %) | 4 (4.0 %) | 7 (17.1 %) | 1.6 (0.3–7.4), 0.6 | 0.8 (0.2–3.2), 0.7 | 3.3 (1.2–13.8), 0.04 |
| G | 124 (79.5 %) | 57 (75.0 %) | 169 (84.5 %) | 53 (64.3 %) | Reference | Reference | Reference |
| A | 32 (29.5 %) | 19 (25.0 %) | 31 (15.5 %) | 29 (35.7 %) | 1.3 (0.7–2.4), 0.5 | 0.6 (0.3–1), 0.1 | 2.1 (1.1–3.8), 0.02* |
| Ghrelin Leu72Met | |||||||
| CC | 63 (80.8 %) | 22 (65.8 %) | 72 (72.0 %) | 21 (51.2 %) | Reference | Reference | Reference |
| CA | 11 (14.1 %) | 10 (26.3 %) | 16 (16.0 %) | 14 (34.1 %) | 2.6 (1.1–6.9), 0.04* | 1.2 (0.5–2.9), 0.5 | 3.8 (1.5–7.6), 0.005* |
| AA | 4 (5.1 %) | 6 (7.9 %) | 12 (12.0 %) | 6 (14.7 %) | 4.2 (1.1–16), 0.03 | 2.6 (0.8–8.5), 0.1 | 4.5 (1.1–10.5), 0.03 |
| C | 137 (87.8 %) | 54 (79.0 %) | 160 (80.0 %) | 56 (68.3 %) | Reference | Reference | Reference |
| A | 19 (12.2 %) | 22 (21.0 %) | 40 (20.0 %) | 26 (31.7 %) | 1.9 (0.9–3.9), 0.08 | 1.8 (0.9–3.25), 0.06 | 3.3 (1.7–6.5), 0.004* |
* P ≤ 0.05 significant, values in parenthesis are % frequencies, normal SGA category was taken as reference for comparison with other SGA categories by logistic regression
Genotype distribution in CRP categories
The ghrelin 346-GA (Arg51Gln) genotype was associated with higher susceptibility to inflammation (OR = 1.8, 95 % CI 1.1–3.1, P = 0.03, Table S6).
Genotype distribution in dialysis categories
None of the genotypes showed any significant risk association with period of dialysis (Table S7).
Serum level of leptin and acyl ghrelin
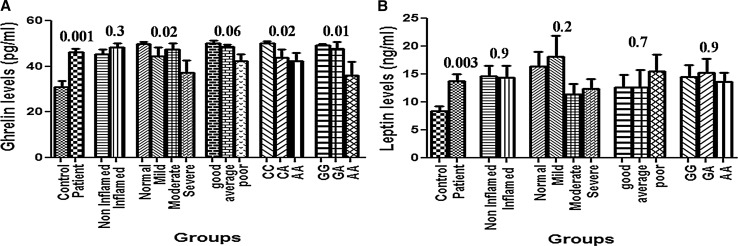
The levels of leptin (P = 0.003) and acyl ghrelin (P = 0.001) were significantly higher in the patients as compared to control. On comparing the levels between inflamed and non-inflamed groups, no significant difference was observed. On comparing the SGA groups, the acyl ghrelin levels were significantly lower in the severe category of SGA (P = 0.02). The levels of leptin were higher in normal and mild groups; however, the difference was not significant (Fig. 1a, b). The patients with good appetite had higher levels of acyl ghrelin (50.1 ± 6.3 pg/ml) followed by average (48.5 ± 4.3 pg/ml) and poor appetite group (43.4 ± 13.0 pg/ml, P = 0.06) but could not achieve significance. The levels of leptin and acyl ghrelin were almost similar and non-significant on comparing the dialysis categories.
Fig. 1.
a Serum leptin level in different groups and genotypes, b Plasma ghrelin level in different groups and genotypes (Leu72Met and Arg51Gln, respectively)
The resting metabolic rate was higher in II (1,513.8 ± 181.2) as compared to DD and ID (1,404.6 ± 158.0 and 1,375.4 ± 196.4, respectively, P value = 0.09). No significant differences in RMR (1,419.9 ± 265.2 vs. 1,399.2 ± 177.7, P = 0.65) were observed in dialysis categories.
Phenotype assignment—leptin-2549 A, ghrelin 51Gln (A), ghrelin 72Met (A), was coded low producer alleles, UCP2 I allele was associated with high Resting metabolic rate.
Combined analysis
A combined analysis was carried out by coding genotypes as low producer alleles of leptin (-2548-A) and ghrelin (51Gln-346-A, 72Met-408-A) and high resting metabolic rate UCP2 (I). The presence of 0–2 allele was grouped as group 1, and 3–4 alleles were coded in group 2. The combined effect model revealed a higher susceptibility in high-risk group 2 on comparing patients and controls (OR = 1.5, 95 % CI 1.1–2.1, P = 0.04). On comparing the normal SGA groups with other SGA groups (Mild + Moderate + Severe), higher susceptibility to malnutrition was associated with malnourished group (OR = 2.0, 95 % CI 1.2–3.5, P = 0.01, Table 3).
Table 3.
Combined analysis of various groups
| Group | Patient | Control | OR (95 % CI) | P value |
|---|---|---|---|---|
| Group 1a | 107 (41.6) | 102 (51.0) | Reference | |
| Group 2b | 150 (58.3) | 98 (49.0) | 1.5 (1.1–2.1) | 0.04* |
| Group | Normal | Malnourished | OR (95 % CI) | P value |
|---|---|---|---|---|
| Group 1a | 42 (53.8) | 65 (36.3) | Reference | |
| Group 2b | 36 (46.1) | 114 (63.7) | 2.0 (1.2–3.5) | 0.01* |
| Group | Inflamed | Non-inflamed | OR (95 % CI) | P value |
|---|---|---|---|---|
| Group 1a | 46 (45.5) | 61 (39.1) | Reference | |
| Group 2b | 55 (54.5) | 95 (60.9) | 1.3 (0.7–2.1) | 0.3 |
| Group | Period < 1 year | Period ≥ 1 year | OR (95 % CI) | P value |
|---|---|---|---|---|
| Group 1a | 69 (40.5) | 38 (43.6) | Reference | |
| Group 2b | 101 (59.5) | 49 (56.4) | 1.1 (0.6–1.9) | 0.68 |
Period is categories of period of dialysis in patients. Logistic regression analysis was used to compare groups
* P ≤ 0.05 significant, values in parenthesis are percent frequencies
aGroup 1 = 0–2 alleles of low producer of ghrelin and leptin and high energy expenditure of UCP2 (I)
bGroup 2 = 3–4 alleles of low producer of ghrelin and leptin and high energy expenditure of UCP2 (I)
Survival analysis
The mean survival of the patients was 19.6 ± 10.5 months. All the patients were followed for 3 years. On comparing the SGA groups, the moderate (HR = 2.2, 95 % CI 1.1–4.8, P = 0.03) and severe groups (HR = 4.1, 95 % CI 1.8–9.4, P = 0.001) were significantly associated with higher death hazard (Table 4).
Table 4.
Kaplan–Meier survival estimation of mean survival and hazard ratios (HRs) among different genotypes
| Genotype | N | Number of events | Mean survival (in months) | Log rank P value | HR (95 % CI) | P value |
|---|---|---|---|---|---|---|
| Ghrelin Arg51Gln | ||||||
| GG | 164 (63.8) | 37 (67.3) | 37.2 ± 1.4 | 0.691 | Reference | |
| GA | 75 (29.2) | 15 (27.3) | 36.1 ± 1.7 | 0.8 (0.4–1.5) | 0.557 | |
| AA | 18 (7.0) | 3 (5.4) | 36.5 ± 2.8 | 0.6 (02–2.1) | 0.489 | |
| Ghrelin Leu72Met | ||||||
| CC | 178 (69.3) | 38 (69.1) | 37.9 ± 1.3 | 0.061 | Reference | |
| CA | 51 (19.8) | 7 (12.7) | 36.6 ± 1.6 | 0.6 (0.2–1.3) | 0.208 | |
| AA | 28 (10.9) | 10 (18.2) | 29.7 ± 3.3 | 1.8 (0.8–3.6) | 0.09 | |
| UCP2 I/D | ||||||
| DD | 149 (57.6) | 28 (50.9) | 35.2 ± 1.2 | 0.345 | Reference | |
| ID | 70 (27.6) | 19 (34.5) | 35.5 ± 2.2 | 1.5 (0.8–2.7) | 0.151 | |
| II | 38 (14.8) | 8 (14.5) | 28.7 ± 1.95 | 1.2 (0.5–2.6) | 0.651 | |
| Leptin-2548 G/A | ||||||
| GG | 90 (35.0) | 22 (40.0) | 32.5 ± 1.6 | 0.582 | Reference | |
| GA | 99 (38.5) | 19 (34.5) | 38.9 ± 1.6 | 0.7 (0.4–1.4) | 0.338 | |
| AA | 68 (26.5) | 14 (25.5) | 35.2 ± 1.7 | 0.7 (0.3–1.5) | 0.456 | |
| Combined analysis | ||||||
| Group 1 | 107 (41.6) | 26 (47.3) | 32.5 ± 1.3 | 0.79 | Reference | |
| Group 2 | 150 (58.3) | 29 (52.7) | 38.3 ± 1.4 | 0.93 (0.5–1.5) | 0.792 | |
| CRP category | ||||||
| Non-inflamed | 156 (60.7) | 30 (54.5) | 35.9 ± 1.3 | 0.561 | Reference | 0.565 |
| Inflamed | 101 (39.3) | 25 (45.5) | 37.6 ± 1.5 | 1.1 (0.6–1.9) | ||
| SGA categories | ||||||
| Normal | 78 (30.4) | 9 (16.4) | 38.0 ± 1.2 | 0.004* | Reference | |
| Mild | 38 (14.8) | 7 (12.7) | 35.2 ± 2.3 | 1.8 (0.6–4.8) | 0.243 | |
| Moderate | 100 (38.9) | 24 (43.6) | 37.1 ± 1.7 | 2.2 (1.1–4.8) | 0.03* | |
| Severe | 41 (15.9) | 15 (27.3) | 23.5 ± 2.3 | 4.1 (1.8–9.4) | 0.001* | |
| Period of dialysis | ||||||
| <1 year | 170 (66.1) | 38 (69.1) | 36.3 ± 1.5 | 0.618 | Reference | |
| ≥1 year | 87 (33.9) | 16 (29.9) | 34.5 ± 1.8 | 0.8 (0.6–1.2) | 0.438 | |
* P ≤ 0.05 significant, values in parenthesis are percent frequencies, Kaplan–Meier survival analysis, and Cox regression was used for hazard estimation
In silico analysis
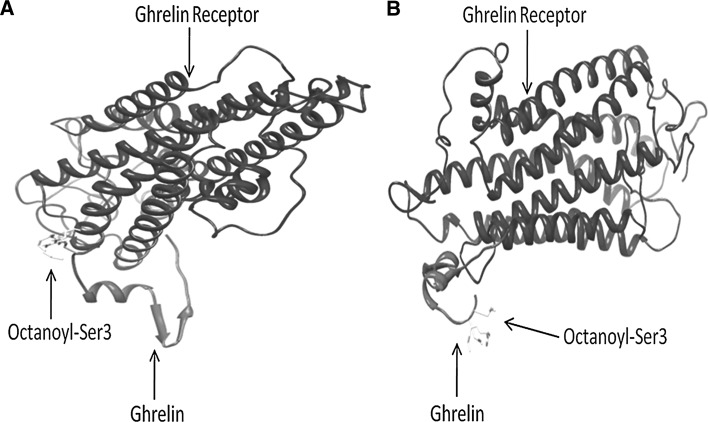
We performed molecular modeling and in silico docking analysis to explore the significance of Arg51Gln on structure and function of ghrelin protein. The wild-type ghrelin has Arg at 51st position which was mutated to Gln to construct the mutant version. Both of them were then octanoylated at 3rd serine residue using AutoDock. The mutant version had a little higher energy (−0.08) as compared to the wild version (−0.1). Furthermore, the octanoylated version was then docked on the ghrelin receptor (Fig. 2a, b). The Hex Score for the mutant ghrelin (−345.95) was higher as compared to wild-type version (−371.62).
Fig. 2.
a Hex generated GH-receptor-ghrelin complex for wild type, b Hex generated GH-receptor-ghrelin complex for mutant type
Polyphen predicted both Leu72Met and Arg51Gln as probably damaging which signifies deleterious effect. SIFT analysis reveals that Arg51Gln mutation is not tolerated, i.e., deleterious while Leu72Met was predicted to be tolerated, i.e., it has no significant effect of mutation. I-Mutant showed that both the SNPs affect the stability of the protein, however, Popmusic showed that only Arg51Gln is associated with decreased stability. Prop v.1.0 inferred that the cleavage site at position 51 is no longer recognized by the enzyme due to mutation while Leu72Met mutation has no effect on peptide cleavage as it is not the part of cleavage site and lies outside the cleavage site. The Target scan predicted binding of following miRNA–hsa-miR-3619-5p, hsa-miR-761, hsa-miR-214 and hsa-miR-761–may bind to the insertion allele at the 3′UTR of exon 8 of UCP2.
Discussion
In this study, we have investigated the association and interaction of SNP of leptin, ghrelin and UCP2 in 257 patients and 200 controls. Our results revealed low producer genotypes of leptin-2548 AA and ghrelin Met72Met and Gln51Gln were associated with higher disease susceptibility. The UCP2 ID genotype showed protective effect against the disease. Ghrelin 72Met and Gln51 allele was associated with 2.1- and 3.3-fold higher susceptibility to severe malnutrition, respectively. In silico analysis revealed that insertion allele of UCP2 may be potent miRNA binding site, and thus, it affects the transcription stability of UCP2 mRNA. Ghrelin Gln51Gln mutation is associated with higher binding energy to its receptor which signifies that mutation had induced conformational change in the ghrelin protein and thus affects its binding affinity with its receptor. The combined analysis revealed that the group 2 was associated with 1.5-fold higher risk of disease and twofold higher risk of malnutrition in patients.
Leptin inhibits food intake and stimulates energy expenditure and thus reported to be associated with appetite control in rodents and healthy subjects (Auwerx and Staels 1998; Jequier and Tappy 1999). It acts on peripheral tissue and stimulates the inflammatory markers like IL6, TNF-α and IL-12. The levels of leptin are elevated in uremia as compared to normal healthy controls (Atamer et al. 2008) which is in agreement with our results. The increase in the serum level of leptin might be due to decreased renal clearance. Leptin decreases hypothalamic NPY levels and enhances sympathetic activity with hyperinsulinemia, resulting in appetite suppression. Further, the levels of leptin are also associated with markers of poor nutritional status like low serum albumin and hypercatabolism as well as with decrease in GFR (Cheung et al. 2010). In our study, the levels of leptin were lower in severe and moderate SGA groups which are in agreement with previous findings (Scholze et al. 2007). The decreased serum levels of leptin in severe SGA group might be associated with acquired leptin receptor resistance. There are reports which reveal that low serum levels of leptin are also independent predictors of mortality (Scholze et al. 2007) while another study shows that serum leptin is associated with CVD but not with mortality. We observed that leptin-2548-AA genotype was associated with twofold higher risk of disease susceptibility which is in concordance with previous studies where the SNPs located in the leptin gene is associated with disease susceptibility (Okpechi et al. 2010). However, none of the leptin genotype showed any association with malnutrition and the levels of leptin. The leptin-2548 site does not map to any recognized regulatory site, but is proximal to a Sp1 binding motif which is regulated by Sp1 in an insulin-dependent fashion in MDA-MB- 231 cells (Bartella et al. 2008). Since ESRD is associated with hyperinsulinemia and insulin resistance (Atamer et al. 2008), this deregulation might be the cause of lack of association of the leptin-2548 G/A genotype in this regard.
Uncoupling protein 2 (UCP2) is an inner mitochondrial protein implicated in uncoupling and thermogenesis thus leading to generation of heat and resting energy expenditure. Various cytokines like TNF-α and leptin are associated with increase in mRNA expression of UCP2 thus contributing to thermogenesis. The ID gene polymorphism located in exon 8 of 3′UTR of UCP2 gene have been previously reported to be associated with energy expenditure (Walder et al. 1998), increased BMI (Cassell et al. 1999), and accumulation of fat tissue (Pedersen et al. 2005). In our study UCP2 ID showed protective effect. There are studies on ESRD patients where the D/D genotype is shown to be associated with acquired fat tissue in peritoneal dialysis patients (Nordfors et al. 2000). Another study on PD and MHD showed that DD and ID are associated with increased total and truncal fat mass in these patients (Wang et al. 2007) which is a protective trait as higher amount of fat signifies more energy stores in these patients. In a recent study, the UCP2 I/D genotypes were compared on the basis of resting energy expenditure in non-dialyzed patients and found no association which is in concordance with our study (Avesani et al. 2008). It has been previously reported that UCP2 ID at 3′UTR is associated with mRNA stability, so to confirm this finding, we carried out in silico analysis where it was observed that miRNA binding may cause down-regulation of this mitochondrial channel protein by affecting mRNA stability. There are other regulatory mechanisms that contribute to the overall expression like reactive oxygen species, fatty acids, hyperglycemia and hyperlipidemia along with transcriptional and translational regulation. It has also been reported that UCP2 mRNA is not necessarily associated with the amount of UCP2 protein in varying conditions (Pecqueur et al. 2001). Present study demonstrates that the ID genotype was protective as it is associated with intermediate resting energy expenditure in our patients but not with malnutrition.
Ghrelin is an appetite stimulating hormone. Increased total ghrelin has been reported to be associated with cachexia in chronic heart failure patients (Nagaya et al. 2001). Increased total ghrelin levels are also reported in dialysis patients (Perez-Fontan et al. 2004). There are two major forms of ghrelin–acylated and des-acylated. Acylated version is responsible for the appetite stimulating properties of ghrelin (Wren et al. 2001), down-regulates cytokines (Wu et al. 2007) and improves lean body mass (Deboer et al. 2008) whereas the des-acylated ghrelin induces negative energy balance and could be involved in anorexia (Yoshimoto et al. 2002). In our study, the acyl ghrelin levels were increased in patients as compared to control which is in concordance with other studies (Oner-Iyidogan et al. 2011). Further, the levels of acyl ghrelin were lowest in the severe SGA category of malnutrition and in patients with poor appetite. It has been reported that low acyl ghrelin is associated with poor nutritional status, loss of appetite (Oner-Iyidogan et al. 2011) and CVD (Chou et al. 2010).
The variants of ghrelin gene have not been extensively investigated in ESRD; however, it is largely studied in concern with diabetes and obesity. In our study, carriers of Met72Met and Gln51Gln showed 3.4- and 2.4-fold higher susceptibility to ESRD. Furthermore, the 72Met and 51Gln allele carriers were also associated with 2.1- and 3.3-fold higher susceptibility to severe malnutrition. The 51 Gln allele is reported to be associated with increased risk for type 2 diabetes and hypertension (Poykko et al. 2003). Leu72Met polymorphism was also found to be associated with serum creatinine and Lipoprotein(a) levels in diabetic patients (Ukkola and Kesaniemi 2003). It has been reported that the Arg51Gln was associated with low total plasma ghrelin levels (Steinle et al. 2005). In our study, the Met72Met and Gln51Gln carrier status was associated with lower acyl ghrelin secretion which also emphasizes the role of ghrelin genotypes in lower ghrelin production and malnutrition in these patients. Further Arg51Gln showed higher risk of inflammation in patients. The in silico analysis of Arg51Gln showed that the docking of mutated protein to its receptor was associated with higher binding energy in contrast to the wild-type protein which was associated with lower energy and higher stability binding to its receptor which suggests that mutation induces conformational change in the mature ghrelin molecule and thus affect the binding affinity with its receptor which in turn affect the function of the receptor as well as ghrelin. Combined analysis revealed twofold higher susceptibility to malnutrition. Moderate and severe SGA group revealed 2.0- and 4.0-fold higher death hazard in the patients.
In conclusion, ghrelin 72Met and 51Gln are major alleles and are not only associated with disease susceptibility but also with malnutrition. Arg51Gln showed higher risk of inflammation in patients. Insilco analysis revealed that mutated allele is associated with higher binding energy to the ghrelin receptor. The levels of acyl ghrelin were lowest in the severe group of malnutrition. Leptin levels were increased in the normal and mild category of SGA showing protective effect of higher leptin levels in malnutrition. Further studies on appetite and energy homeostasis regulating markers are required for establishing their role in malnutrition. The demographic and biochemical factors in patients whose period of dialysis was ≥1 year showed marginally improved serum albumin and decreased weight which might be due to achievement of dry weight in these patients. This shows the relevance of genetic background as revealed by the combined effect model of genotypes where group 2 (alleles associated with low leptin, high ghrelin and low RMR) showed significant association with disease susceptibility and poor nutritional status. The limitation of our study is we were not able to quantify des-acyl ghrelin levels as it may have a potential role in inducing anorexia in our patients. Our results are convincing as we have taken care of sample size, ethnicity, followed stringent conditions for the selection and classification of patients on the basis of SGA and we have considered only those P values and CI which reveal significance.
Electronic supplementary material
Acknowledgments
Richa Sharma is receiving SRF from the Department of Biotechnology, Government of India, New Delhi, India. The study was supported by intramural funding from Sanjay Gandhi Post Graduate Institute of Medical Sciences, Lucknow, India.
Conflict of interest
All the authors declare no conflict of interest.
Contributor Information
Richa Sharma, Email: richa2612@gmail.com.
Suraksha Agrawal, Phone: +91-522-249-4338, Email: sur_ksha_agrawal@yahoo.co.in.
Anita Saxena, Phone: +91-522-249-4354, Email: anitimmy@sgpgi.ac.in.
Manmohan Pandey, Email: way2manmohan@gmail.com.
R. K. Sharma, Phone: +91-522-249-4121, FAX: +91-522-6680572, Email: rksharma@sgpgi.ac.in
References
- (1996) Adequacy of dialysis and nutrition in continuous peritoneal dialysis: association with clinical outcomes. Canada-USA (CANUSA) Peritoneal Dialysis Study Group. J Am Soc Nephrol 198–207 [DOI] [PubMed]
- Atamer A, Alisir Ecder S, Akkus Z, Kocyigit Y, Atamer Y, Ilhan N, Ecder T. Relationship between leptin, insulin resistance, insulin-like growth factor-1 and insulin-like growth factor binding protein-3 in patients with chronic kidney disease. J Int Med Res. 2008;36:522–528. doi: 10.1177/147323000803600317. [DOI] [PubMed] [Google Scholar]
- Auwerx J, Staels B. Leptin. Lancet. 1998;351:737–742. doi: 10.1016/S0140-6736(97)06348-4. [DOI] [PubMed] [Google Scholar]
- Avesani CM, Kamimura MA, Utaka S, Pecoits-Filho R, Nordfors L, Stenvinkel P, Lindholm B, Draibe SA, Cuppari L. Is UCP2 gene polymorphism associated with decreased resting energy expenditure in nondialyzed chronic kidney disease patients? J Ren Nutr. 2008;18:489–494. doi: 10.1053/j.jrn.2008.08.009. [DOI] [PubMed] [Google Scholar]
- Bartella V, Cascio S, Fiorio E, Auriemma A, Russo A, Surmacz E. Insulin-dependent leptin expression in breast cancer cells. Cancer Res. 2008;68:4919–4927. doi: 10.1158/0008-5472.CAN-08-0642. [DOI] [PubMed] [Google Scholar]
- Blumenkrantz MJ, Kopple JD, Gutman RA, Chan YK, Barbour GL, Roberts C, Shen FH, Gandhi VC, Tucker CT, Curtis FK, Coburn JW. Methods for assessing nutritional status of patients with renal failure. Am J Clin Nutr. 1980;33:1567–1585. doi: 10.1093/ajcn/33.7.1567. [DOI] [PubMed] [Google Scholar]
- Bossola M, Giungi S, Luciani G, Tazza L. Appetite in chronic hemodialysis patients: a longitudinal study. J Ren Nutr. 2009;19:372–379. doi: 10.1053/j.jrn.2009.01.015. [DOI] [PubMed] [Google Scholar]
- Caliskan Y, Yelken B, Gorgulu N, Ozkok A, Yazici H, Telci A, Turkmen A, Yildiz A, Sever MS. Comparison of markers of appetite and inflammation between hemodialysis patients with and without failed renal transplants. J Ren Nutr. 2012;22:258–267. doi: 10.1053/j.jrn.2011.07.005. [DOI] [PubMed] [Google Scholar]
- Carrero JJ, Nakashima A, Qureshi AR, Lindholm B, Heimburger O, Barany P, Stenvinkel P. Protein-energy wasting modifies the association of ghrelin with inflammation, leptin, and mortality in hemodialysis patients. Kidney Int. 2011;79:749–756. doi: 10.1038/ki.2010.487. [DOI] [PubMed] [Google Scholar]
- Carrero JJ, Stenvinkel P, Cuppari L, Ikizler TA, Kalantar-Zadeh K, Kaysen G, Mitch WE, Price SR, Wanner C, Wang AY, Ter Wee P, Franch HA. Etiology of the protein-energy wasting syndrome in chronic kidney disease: a consensus statement from the international society of renal nutrition and metabolism (ISRNM) J Ren Nutr. 2013;23:77–90. doi: 10.1053/j.jrn.2013.01.001. [DOI] [PubMed] [Google Scholar]
- Cassell PG, Neverova M, Janmohamed S, Uwakwe N, Qureshi A, McCarthy MI, Saker PJ, Albon L, Kopelman P, Noonan K, Easlick J, Ramachandran A, Snehalatha C, Pecqueur C, Ricquier D, Warden C, Hitman GA. An uncoupling protein 2 gene variant is associated with a raised body mass index but not type II diabetes. Diabetologia. 1999;42:688–692. doi: 10.1007/s001250051216. [DOI] [PubMed] [Google Scholar]
- Cheung WW, Mak RH (2010) Ghrelin in chronic kidney disease. Int J Pept 2010. doi:10.1155/2010/567343 [DOI] [PMC free article] [PubMed]
- Cheung WW, Paik KH, Mak RH. Inflammation and cachexia in chronic kidney disease. Pediatr Nephrol. 2010;25:711–724. doi: 10.1007/s00467-009-1427-z. [DOI] [PubMed] [Google Scholar]
- Chou CC, Bai CH, Tsai SC, Wu MS. Low serum acylated ghrelin levels are associated with the development of cardiovascular disease in hemodialysis patients. Intern Med. 2010;49:2057–2064. doi: 10.2169/internalmedicine.49.3047. [DOI] [PubMed] [Google Scholar]
- Deboer MD, Zhu X, Levasseur PR, Inui A, Hu Z, Han G, Mitch WE, Taylor JE, Halem HA, Dong JZ, Datta R, Culler MD, Marks DL. Ghrelin treatment of chronic kidney disease: improvements in lean body mass and cytokine profile. Endocrinology. 2008;149:827–835. doi: 10.1210/en.2007-1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gama-Axelsson T, Lindholm B, Barany P, Heimburger O, Stenvinkel P, Qureshi AR. Self-rated appetite as a predictor of mortality in patients with stage 5 chronic kidney disease. J Ren Nutr. 2013;23:106–113. doi: 10.1053/j.jrn.2012.04.009. [DOI] [PubMed] [Google Scholar]
- Grunfeld C, Zhao C, Fuller J, Pollack A, Moser A, Friedman J, Feingold KR. Endotoxin and cytokines induce expression of leptin, the ob gene product, in hamsters. J Clin Invest. 1996;97:2152–2157. doi: 10.1172/JCI118653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jequier E, Tappy L. Regulation of body weight in humans. Physiol Rev. 1999;79:451–480. doi: 10.1152/physrev.1999.79.2.451. [DOI] [PubMed] [Google Scholar]
- Julienne CM, Dumas JF, Goupille C, Pinault M, Berri C, Collin A, Tesseraud S, Couet C, Servais S (2012) Cancer cachexia is associated with a decrease in skeletal muscle mitochondrial oxidative capacities without alteration of ATP production efficiency. J Cachexia Sarcopenia Muscle. doi:10.1007/s13539-012-0071-9 [DOI] [PMC free article] [PubMed]
- Mak RH, Cheung WW, Zhan JY, Shen Q, Foster BJ. Cachexia and protein-energy wasting in children with chronic kidney disease. Pediatr Nephrol. 2012;27:173–181. doi: 10.1007/s00467-011-1765-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitch WE. Malnutrition: a frequent misdiagnosis for hemodialysis patients. J Clin Invest. 2002;110:437–439. doi: 10.1172/JCI16494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaya N, Uematsu M, Kojima M, Date Y, Nakazato M, Okumura H, Hosoda H, Shimizu W, Yamagishi M, Oya H, Koh H, Yutani C, Kangawa K. Elevated circulating level of ghrelin in cachexia associated with chronic heart failure: relationships between ghrelin and anabolic/catabolic factors. Circulation. 2001;104:2034–2038. doi: 10.1161/hc4201.097836. [DOI] [PubMed] [Google Scholar]
- Nordfors L, Lonnqvist F, Heimburger O, Danielsson A, Schalling M, Stenvinkel P. Low leptin gene expression and hyperleptinemia in chronic renal failure. Kidney Int. 1998;54:1267–1275. doi: 10.1046/j.1523-1755.1998.00088.x. [DOI] [PubMed] [Google Scholar]
- Nordfors L, Heimburger O, Lonnqvist F, Lindholm B, Helmrich J, Schalling M, Stenvinkel P. Fat tissue accumulation during peritoneal dialysis is associated with a polymorphism in uncoupling protein 2. Kidney Int. 2000;57:1713–1719. doi: 10.1046/j.1523-1755.2000.00016.x. [DOI] [PubMed] [Google Scholar]
- Okpechi IG, Rayner BL, van der Merwe L, Mayosi BM, Adeyemo A, Tiffin N, Ramesar R. Genetic variation at selected SNPs in the leptin gene and association of alleles with markers of kidney disease in a Xhosa population of South Africa. PLoS ONE. 2010;5:e9086. doi: 10.1371/journal.pone.0009086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oner-Iyidogan Y, Gurdol F, Kocak H, Oner P, Cetinalp-Demircan P, Caliskan Y, Kocak T, Turkmen A. Appetite-regulating hormones in chronic kidney disease patients. J Ren Nutr. 2011;21:316–321. doi: 10.1053/j.jrn.2010.07.005. [DOI] [PubMed] [Google Scholar]
- Pearson TA, Mensah GA, Alexander RW, Anderson JL, Cannon RO, 3rd, Criqui M, Fadl YY, Fortmann SP, Hong Y, Myers GL, Rifai N, Smith SC, Jr, Taubert K, Tracy RP, Vinicor F. Markers of inflammation and cardiovascular disease: application to clinical and public health practice: a statement for healthcare professionals from the Centers for Disease Control and Prevention and the American Heart Association. Circulation. 2003;107:499–511. doi: 10.1161/01.CIR.0000052939.59093.45. [DOI] [PubMed] [Google Scholar]
- Pecqueur C, Alves-Guerra MC, Gelly C, Levi-Meyrueis C, Couplan E, Collins S, Ricquier D, Bouillaud F, Miroux B. Uncoupling protein 2, in vivo distribution, induction upon oxidative stress, and evidence for translational regulation. J Biol Chem. 2001;276:8705–8712. doi: 10.1074/jbc.M006938200. [DOI] [PubMed] [Google Scholar]
- Pedersen SB, Nyholm B, Kristensen K, Nielsen MF, Schmitz O, Richelsen B. Increased adiposity and reduced adipose tissue mRNA expression of uncoupling protein-2 in first-degree relatives of type 2 diabetic patients: evidence for insulin stimulation of UCP-2 and UCP-3 gene expression in adipose tissue. Diabetes Obes Metab. 2005;7:98–105. doi: 10.1111/j.1463-1326.2005.00365.x. [DOI] [PubMed] [Google Scholar]
- Perez-Fontan M, Cordido F, Rodriguez-Carmona A, Peteiro J, Garcia-Naveiro R, Garcia-Buela J. Plasma ghrelin levels in patients undergoing haemodialysis and peritoneal dialysis. Nephrol Dial Transplant. 2004;19:2095–2100. doi: 10.1093/ndt/gfh313. [DOI] [PubMed] [Google Scholar]
- Poykko S, Ukkola O, Kauma H, Savolainen MJ, Kesaniemi YA. Ghrelin Arg51Gln mutation is a risk factor for type 2 diabetes and hypertension in a random sample of middle-aged subjects. Diabetologia. 2003;46:455–458. doi: 10.1007/s00125-003-1058-z. [DOI] [PubMed] [Google Scholar]
- Sali A, Potterton L, Yuan F, van Vlijmen H, Karplus M. Evaluation of comparative protein modeling by MODELLER. Proteins. 1995;23:318–326. doi: 10.1002/prot.340230306. [DOI] [PubMed] [Google Scholar]
- Scholze A, Rattensperger D, Zidek W, Tepel M. Low serum leptin predicts mortality in patients with chronic kidney disease stage 5. Obesity (Silver Spring) 2007;15:1617–1622. doi: 10.1038/oby.2007.191. [DOI] [PubMed] [Google Scholar]
- Schrauwen P, Hesselink M. UCP2 and UCP3 in muscle controlling body metabolism. J Exp Biol. 2002;205:2275–2285. doi: 10.1242/jeb.205.15.2275. [DOI] [PubMed] [Google Scholar]
- Steinle NI, Pollin TI, O’Connell JR, Mitchell BD, Shuldiner AR. Variants in the ghrelin gene are associated with metabolic syndrome in the old order Amish. J Clin Endocrinol Metab. 2005;90:6672–6677. doi: 10.1210/jc.2005-0549. [DOI] [PubMed] [Google Scholar]
- Tracey KJ, Lowry SF, Cerami A. Cachetin/TNF-alpha in septic shock and septic adult respiratory distress syndrome. Am Rev Respir Dis. 1988;138:1377–1379. doi: 10.1164/ajrccm/138.6.1377. [DOI] [PubMed] [Google Scholar]
- Ukkola O, Kesaniemi YA. Preproghrelin Leu72Met polymorphism in patients with type 2 diabetes mellitus. J Intern Med. 2003;254:391–394. doi: 10.1046/j.1365-2796.2003.01208.x. [DOI] [PubMed] [Google Scholar]
- Walder K, Norman RA, Hanson RL, Schrauwen P, Neverova M, Jenkinson CP, Easlick J, Warden CH, Pecqueur C, Raimbault S, Ricquier D, Silver MH, Shuldiner AR, Solanes G, Lowell BB, Chung WK, Leibel RL, Pratley R, Ravussin E. Association between uncoupling protein polymorphisms (UCP2-UCP3) and energy metabolism/obesity in Pima Indians. Hum Mol Genet. 1998;7:1431–1435. doi: 10.1093/hmg/7.9.1431. [DOI] [PubMed] [Google Scholar]
- Wang X, Axelsson J, Nordfors L, Qureshi AR, Avesani C, Barany P, Schalling M, Heimburger O, Lindholm B, Stenvinkel P. Changes in fat mass after initiation of maintenance dialysis is influenced by the uncoupling protein 2 exon 8 insertion/deletion polymorphism. Nephrol Dial Transplant. 2007;22:196–202. doi: 10.1093/ndt/gfl504. [DOI] [PubMed] [Google Scholar]
- Wren AM, Seal LJ, Cohen MA, Brynes AE, Frost GS, Murphy KG, Dhillo WS, Ghatei MA, Bloom SR. Ghrelin enhances appetite and increases food intake in humans. J Clin Endocrinol Metab. 2001;86:5992. doi: 10.1210/jc.86.12.5992. [DOI] [PubMed] [Google Scholar]
- Wu R, Dong W, Cui X, Zhou M, Simms HH, Ravikumar TS, Wang P. Ghrelin down-regulates proinflammatory cytokines in sepsis through activation of the vagus nerve. Ann Surg. 2007;245:480–486. doi: 10.1097/01.sla.0000251614.42290.ed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wynne K, Giannitsopoulou K, Small CJ, Patterson M, Frost G, Ghatei MA, Brown EA, Bloom SR, Choi P. Subcutaneous ghrelin enhances acute food intake in malnourished patients who receive maintenance peritoneal dialysis: a randomized, placebo-controlled trial. J Am Soc Nephrol. 2005;16:2111–2118. doi: 10.1681/ASN.2005010039. [DOI] [PubMed] [Google Scholar]
- Yoshimoto A, Mori K, Sugawara A, Mukoyama M, Yahata K, Suganami T, Takaya K, Hosoda H, Kojima M, Kangawa K, Nakao K. Plasma ghrelin and desacyl ghrelin concentrations in renal failure. J Am Soc Nephrol. 2002;13:2748–2752. doi: 10.1097/01.ASN.0000032420.12455.74. [DOI] [PubMed] [Google Scholar]
- Young GA, Woodrow G, Kendall S, Oldroyd B, Turney JH, Brownjohn AM, Smith MA. Increased plasma leptin/fat ratio in patients with chronic renal failure: a cause of malnutrition? Nephrol Dial Transplant. 1997;12:2318–2323. doi: 10.1093/ndt/12.11.2318. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.