Abstract
Purpose
Nitric oxide (NO), a short-lived gaseous free radical, is a potent mediator of biological responses involved in the pathogenesis of autoimmune rheumatic diseases, such as systemic lupus erythematosus (SLE) and rheumatoid arthritis (RA). Nitric oxide also serves as an important signal in physiological processes, including angiogenesis, thrombosis, and bone turnover, which are known to be related to the pathogenesis of osteonecrosis. We investigated whether NOS3 gene polymorphisms are associated with risk of osteonecrosis of the femoral head (ONFH).
Methods
Five polymorphisms in the NOS3 gene were genotyped using TaqMan assays in 306 controls, 150 SLE patients, and 50 SLE patients with ONFH (SLE_ONFH).
Results
We found that Asp258Asp and Glu298Asp (G894T) polymorphisms in the NOS3 gene were significantly associated with risk of ONFH. Additionally, we calculated haplotype frequencies of a linkage disequilibrium (LD) block in NOS3 (rs1799983 − rs1800780) and tested for haplotype associations. The haplotypes G-A and T-A showed significant protective (P = 1.6 × 10-3; OR 0.39, 95 % confidence intervals (CI) 0.22–0.7) and increased risk (P = 2.0 x 10-5–6.0 x 10-4; OR 3.17−3.73) effects for ONFH, respectively.
Conclusions
These results suggest that exonic NOS3 polymorphisms may increase the risk of ONFH in Korean SLE patients
Electronic supplementary material
The online version of this article (doi:10.1007/s00264-013-1966-6) contains supplementary material, which is available to authorized users.
Introduction
Osteonecrosis of the femoral head (ONFH) is a devastating bone disease in which cellular death within the femoral head occurs as a result of interruption of the blood supply to the anterior-superior-lateral region. ONFH is a complex and multifactorial disease that is influenced by a number of genetic factors with relatively small effects in addition to environmental factors. Diverse conditions, such as the use of corticosteroids, alcohol abuse, sickle cell anaemia, radiation, Gaucher disease, and others, are known to be implicated in the development of secondary osteonecrosis [1]. In particular, corticosteroid use and excessive alcohol intake are considered to be dominant risk factors, with high dose corticosteroid use (equating to ∼2 g of prednisone within two to three months) the most common risk factor, accounting for between ten and 30 % of osteonecrosis cases [2]. Steroid medications are often used in patients with systemic lupus erythematosus (SLE), rheumatoid arthritis (RA), and after renal transplant, all conditions known to be associated with susceptibility to osteonecrosis development [3]. However, only ∼5 % of patients with a history of high dose corticosteroid use, or alcohol abuse, develop osteonecrosis [4], indicating individual differences in sensitivity to these risk factors.
Although the pathogenesis and pathophysiology of osteonecrosis remain areas of controversy, the majority view is that the condition is the result of the combined effects of metabolic factors, local factors affecting blood supply, such as vascular damage, increased intraosseous pressure, and mechanical stresses [5]. The majority of association studies relating to osteonecrosis have concentrated on gene polymorphisms affecting the coagulation and fibrinolytic systems [6]. However, the association between genetic predisposition and thrombotic tendency may differ between ethnic groups. Moreover, neither the factor V Leiden (G1691A mutation in factor V), nor the prothrombin G20210A mutations have been found in the Korean population [7, 8].
Nitric oxide (NO) regulates a variety of biological processes involved in angiogenesis, thrombosis, and bone turnover, which are known to be related to the pathogenesis of ONFH [9]. Nitric oxide synthesised by endothelial nitric oxide synthase (eNOS) has vasodilatory effects on vascular tone, inhibits platelet aggregation, and modulates smooth muscle proliferation [10]. Overproduction of NO could contribute to tissue injury, given its capacity to increase vascular permeability, generate toxic free radicals, such as peroxynitrite, and induce cytotoxicity [11]. Deficiency of the NOS3 gene leads to reduced bone formation and impaired osteoblast function [12]. Many studies have been carried out to determine the associations between genetic polymorphisms in the NOS3 gene and vascular diseases, including coronary artery disease or myocardial infarction, hypertension, stroke, and renal diseases. A small number of studies have demonstrated association between NOS3 polymorphisms and ONFH; allele 4a of a variable number tandem repeat (VNTR) polymorphism in intron 4 and the T786C polymorphism in the NOS3 gene were associated with idiopathic ONFH [9, 13, 14].
In this study, we postulated that abnormal NO expression is associated with ONFH, and we investigated the influence of NOS3 gene polymorphisms on susceptibility to ONFH.
Materials and methods
Subjects
Blood samples and records were obtained from 306 healthy controls (28 male, 278 female), 150 SLE patients (13 male, 137 female), and 50 SLE patients with ONFH (SLE_ONFH; four male, 46 female). The healthy control, SLE, and SLE_ONFH participants were consecutively recruited from the Hanyang University Hospital for Rheumatic Diseases (Seoul, Korea). The study design was approved by the Institutional Review Board, and all individuals participating in the study gave their informed consent. Control subjects were defined by a lack of hip pain and the absence of any lesions with a sclerotic margin or subchondral collapse, consistent with ONFH, in anteroposterior and frog-leg lateral pelvic radiographs. The clinical characteristics of controls and patients are summarised in Table 1.
Table 1.
Clinical profiles of patient and control subjects in this study
| Characteristic | Control (n = 306) | SLE (n = 150) | SLE_ONFH (n = 50) | P values |
|---|---|---|---|---|
| Age (mean ± SD) | 32.76 ± 11.51 | 31.37 ± 9.97 | 31.28 ± 9.04 | NS |
| Gender | ||||
| Male | 28 | 13 | 4 | NS |
| Female | 278 | 137 | 46 | |
| BMI (kg/m2) | ||||
| ≥25 | 37 | 24 | 7 | NS |
| < 25 | 269 | 126 | 43 | |
SD standard deviation, NS not significant, BMI body mass index, SLE systemic lupus erythematosus, ONFH osteonecrosis of the femoral head
Genotyping
A total of five single nucleotide polymorphism (SNP) sites in the NOS3 gene were selected on the basis of their locations, allele frequencies, and potential relevance to disease. For genotyping of polymorphic sites, amplification primers and probes were designed for TaqMan assays (Applied Biosystems, Foster City, CA). Primer Express (Applied Biosystems) was used to design both the PCR primers and the minor groove binder (MGB) TaqMan probes. One allelic probe was labelled with the 6-carboxyfluorescein (FAM™) dye, and the other with fluorescent 2'-chloro-7'-phenyl-1,4-dichloro-6-carboxyfluorescein (VIC®) dye. All reactions were carried out following the manufacturer’s protocol. Detailed procedures regarding the PCR reaction and TaqMan assay have been described previously [15]. The fluorescence data files from each plate were collected and analysed using automated allele-calling software (SDS 2.2, Applied Biosystems).
Statistical analyses
For inclusion in subsequent analysis, SNPs were required to meet the minimum criteria of call rate (CR) > 95.0, minor allele frequency (MAF) > 0.05, and Hardy-Weinberg equilibrium (HWE) > 0.05. Chi-squared tests were used to determine whether individual variants were in equilibrium in the population at each locus.
Logistical regression analyses were used to calculate odds ratios (ORs), 95 % confidence intervals (CIs), and corresponding P-values for each SNP and haplotype, with age and sex as covariates, and using three alternative models (codominant, dominant, and recessive). Genotypes were given codes of 0, 1, and 2; 0, 1, and 1; and 0, 0, and 1 in the codominant, dominant, and recessive models, respectively. Linkage disequilibrium (LD) between loci was measured using the absolute value of Lewontin's D’ (|D’|) [16]. Haplotypes of each individual were inferred from the genotype data using the Haploview program 3.32 (http://www.broad.mit.edu/mpg/haploview/), which uses an accelerated expectation-maximisation (EM) algorithm. Haploview was also used to estimate haplotype structures and their frequencies within LD blocks. Haplotypes with frequencies < 5 % were excluded from further analysis. Continuous variables were compared by Student’s t-test or ANOVA. All analyses were two-tailed, and P-values < 0.05 were considered to be statistically significant. For multiple comparisons, a Bonferroni adjustment was used to adjust P values.
Results
To examine association of NOS3 gene polymorphisms with SLE and SLE_ONFH in Korean patients, we selected five polymorphic sites in the NOS3 gene, taking into consideration their location, allele frequencies, and known disease association as derived from public databases (Fig. 1). The genotypes of these five SNPs were analysed in 306 controls, 150 SLE patients, and 50 SLE_ONFH patients using TaqMan assays. The resulting SNP data, including location, amino acid substitution, genotype, MAF, and HWE of all studied polymorphisms are presented in Table 2. All of the genotyped SNPs fulfilled our criteria of a CR > 95.0, MAF > 0.05, and HWE > 0.05.
Fig. 1.
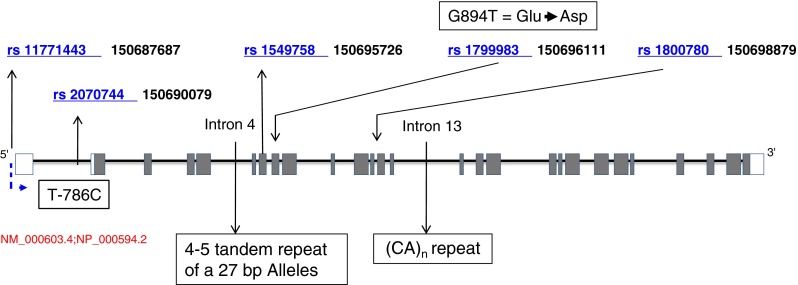
Polymorphisms identified in the NOS3 gene on chromosome 7q36. Coding exons are denoted by shadowed blocks and 5’ and 3’ UTRs by white blocks
Table 2.
SNP markers in the NOS3 gene genotyped in this case–control study
| rs No. (Alternative name) | Position | Amino acid substitution | Genotypea | MAF | HWEb | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| C/C | C/R | R/R | Control | Cased | Totald | Control | Casec | Totald | |||
| rs11771443 | 5’ UTR | CC | CT | TT | 0.461 | 0.490 | 0.472 | 1.000 | 0.253 | 0.530 | |
| rs2070744 (T786C) | promoter | TT | CT | CC | 0.113 | 0.065 | 0.094 | 0.149 | 0.581 | 0.292 | |
| rs1549758 | Exon6 | Asp258Asp (Synonym) | CC | CT | TT | 0.135 | 0.186 | 0.155 | 1.000 | 0.483 | 0.728 |
| rs1799983 (G894T) | Exon 7 | Glu298Asp (Non-synonym) | GG | GT | TT | 0.081 | 0.121 | 0.096 | 0.236 | 1.000 | 0.608 |
| rs1800780 | Intron 12 | CC | CT | TT | 0.400 | 0.351 | 0.381 | 0.123 | 0.424 | 0.089 | |
MAF minor allele frequency, SNP single nucleotide polymorphism, SLE systemic lupus erythematosus, ONFH osteonecrosis of the femoral head
a C/C: major homozygote, C/R: heterozygote, R/R: minor homozygote
b HWE: P values of deviation from Hardy-Weinberg equilibrium
c Case: SLE + SLE_ONFH
d Total: control + case
Table 3 presents a comparison of genotype frequencies between all the three groups. Compared to the control group, patients with SLE showed no significant differences after Bonferroni adjustment under any analysis model (Table 3, Supplementary Table 1). However, when genotype distributions between the control and SLE_ONFH groups were compared, the SNPs rs1549758 (Asp258Asp) in exon6 and rs1799983 (Glu298Asp) in exon7 of the NOS3 gene were significantly associated with risk of ONFH under all analysis models (P = 1.0 × 10−5–5.0 × 10−4; OR = 2.7-3.8) (Table 3, Supplementary Table 2). Next, we compared NOS3 SNP allele frequencies between SLE and SLE_ONFH, to determine whether the two associated SNPs are specifically associated with ONFH. The results showed that both SNPs, encoding Asp258Asp and Glu298Asp in the eNOS protein, were significantly associated with a risk of ONFH (P = 9.0 × 10−4–4.8 × 10−3; OR = 2.27–3.43) (Supplementary Table 3). Despite the application of stringent Bonferroni correction, which is considered to be a conservative method of adjustment for multiple testing, both associated SNPs retained significance.
Table 3.
Analyses of association between NOS3 gene polymorphisms and the risk of ONFH
| rs no. | Genotype | Frequencies (%) | Con vs SLE | Con vs SLE_ONFH | SLE vs SLE_ONFH | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Controls | SLE | SLE_ONFH | OR (95 % CI) | P | OR (95 % CI) | P | OR (95 % CI) | P | ||
| rs11771443 | CC | 89 (29.18) | 38 (25.68) | 9 (18.37) | 1.05 (0.79–1.40) | 0.735 | 1.40 (0.90 − 2.16) | 0.135 | 1.35 (0.83–2.20) | 0.222 |
| CT | 151 (49.51) | 80 (54.05) | 27 (55.1) | |||||||
| TT | 65 (21.31) | 30 (20.27) | 13 (26.53) | |||||||
| rs2070744 (T786C) | TT | 235 (77.81) | 130(86.67) | 45 (90) | 0.58 (0.34–0.98) | 0.043 p corr =1 |
0.39 (0.15 − 1.00) | 0.051 | 0.70 (0.26–1.91) | 0.485 |
| CT | 66 (21.85) | 19 (12.67) | 5 (10) | |||||||
| CC | 1 (0.33) | 1 ( 0.67) | 0 (0) | |||||||
| rs1549758 | CC | 226 (74.59) | 106 (72.6) | 24 (50) | 1.15 (0.77–1.71) | 0.493 | 2.76 (1.63 − 4.65) | 1.0 x 10-4
pcorr < 0.05 |
2.27 (1.31–3.96) | 3.6 x 10-3
pcorr < 0.05 |
| CT | 72 (23.76) | 36 (24.66) | 20 (41.67) | |||||||
| TT | 5 (1.65) | 4 (2.74) | 4 (8.33) | |||||||
| rs1799983 (G894T) | GG | 255 (83.88) | 124(83.22) | 30 (60) | 1.08 (0.64–1.84) | 0.775 | 3.80 (2.09 − 6.91) | 1.0 x 10-5
pcorr < 0.05 |
3.43 (1.77–6.67) | 3.0 x 10-4
pcorr < 0.05 |
| GT | 49 (16.12) | 25 (16.78) | 17 (34) | |||||||
| TT | 0 (0) | 0 (0) | 3 (6) | |||||||
| rs1800780 | GG | 103 (33.77) | 63 (43.15) | 16 (32.65) | 0.76 (0.56–1.03) | 0.079 | 0.94 (0.59 − 1.50) | 0.794 | 1.24 (0.76–2.04) | 0.386 |
| AG | 160 (52.46) | 67 (45.89) | 28 (57.14) | |||||||
| AA | 42 (13.77) | 16 (10.96) | 5 ( 10.2) | |||||||
OR odds ratio, CI confidence interval, SLE systemic lupus erythematosus, ONFH osteonecrosis of the femoral head
Genotype distributions are shown as number (%). Codominant p values and odds ratio (95 % CI) for logistical analyses, controlling for age and sex as covariates, are shown
*p corr values after Bonferroni correction
As LD is believed to be highly structured, with conserved blocks of sequence separated by hotspots of recombination, the function of a conserved haplotype may be the result of interaction between polymorphisms within a block. LD analysis revealed one haplotype block, including two SNPs: rs1799983 and rs1800780 (Fig. 2a). Three major haplotypes with frequencies > 0.05 were predicted, and the frequency of each was compared between controls and SLE and/or SLE_ONFH patients (Fig. 2b; Table 4). Haplotype 2 (ht2: G-A) and/or haplotype 3 (ht3: T-A) were significantly associated with a protective role (OR = 0.39) or an increased risk (OR = 3.73), respectively, with regard to development of ONFH under all analysis models, when compared with either control or SLE groups (Table 4, Supplementary Tables 5 and 6). However, comparison of SLE patients and controls at the haplotype level demonstrated no significant differences (Table 4). These results suggest that both exonic polymorphisms (rs1549758, rs1799983) may be functionally involved with increased susceptibility to ONFH in SLE patients.
Fig. 2.
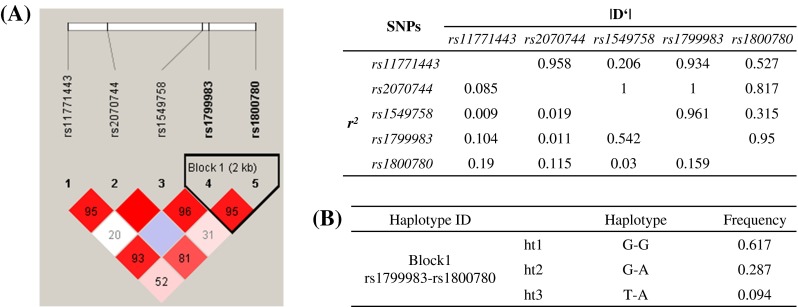
Linkage disequilibrium coefficients and haplotypes in the NOS3 gene (A) Linkage disequilibrium coefficients (|D'|) and an LD block among NOS3 polymorphisms (B) Haplotypes of NOS3 gene
Table 4.
Association of NOS3 gene haplotypes with ONFH
| Haplotype | Genotype | Frequencies (%) | Con vs SLE | Con vs SLE_ONFH | SLE vs SLE_ONFH | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Control | SLE | SLE_ONFH | OR (95 % CI) | P | OR (95 % CI) | P | OR (95 % CI) | P | ||
| Block1-ht1 G-G | −/− | 41 (13.49) | 16 (10.96) | 5 (10.2) | 1.32 (0.97–1.80) | 0.076 | 1.02 0.63–1.63) | 0.952 | 0.77 (0.47–1.26) | 0.291 |
| ht1/- | 161 (52.96) | 67 (45.89) | 29 (59.18) | |||||||
| ht1/ht1 | 102 (33.55) | 63 (43.15) | 15 (30.61) | |||||||
| Block1-ht2 G-A | −/− | 139 (45.72) | 82 (56.16) | 33 (67.35) | 0.72 (0.52–0.99) | 0.043 pcorr = 1.0 |
0.39 (0.22–0.7) | 1.6 x 10-3
pcorr < 0.05 |
0.57 (0.31–1.04) | 0.066 |
| ht2/- | 136 (44.74) | 54 (36.99) | 16 (32.65) | |||||||
| ht2/ht2 | 29 (9.54) | 10 (6.85) | 0 (0) | |||||||
| Block1-ht3 T-A | −/− | 256 (84.21) | 121 (82.88) | 30 (61.22) | 1.14 (0.67–1.95) | 0.625 | 3.73 (2.04–6.83) | 2.0 x 10-5
pcorr < 0.05 |
3.17 (1.64–6.13) | 6.0 x 10-4
pcorr < 0.05 |
| ht3/- | 48 (15.79) | 25 (17.12) | 16 (32.65) | |||||||
| ht3/ht3 | 0 (0) | 0 (0) | 3 (6.12) | |||||||
Haplotype Block1: rs1799983 (G > T)- rs1800780 (G > A)
Haplotype distributions are shown as number (%). Co-dominant P values and odds ratio (95 % CI) for logistic regression analyses, controlling for age and sex as covariates, are shown
OR odds ratio, CI confidence interval, SLE systemic lupus erythematosus, ONFH osteonecrosis of the femoral head
*pcorr indicates P values after Bonferroni correction
Discussion
ONFH is one of the most common diseases of the hip in Korea, with higher incidence than that in other countries; it is responsible for more than half of the cases of total hip arthroplasty, whereas it is relatively rare in the Unites States [8]. In recent years, the clinical significance of ONFH in hip diseases has received much attention, but the details of its pathogenesis and epidemiology are not well understood; although it is generally assumed that venous thrombosis with blood flow obstruction to the femoral head, mediated by thrombophilia and/or hypofibrinolysis, is important in the development of ONFH [17, 18]. At the genetic level, some genes have previously been reported as risk factors for ONFH, including a G1691A mutation in factor V (factor V Leiden), a G20210A mutation in prothrombin, and polymorphisms in 5, 10-methylenetetrahydrofolate reductase (MTHFR; C677T and A1298C) [8, 19, 20]. The presence of these genetic variations is associated with a hypercoagulable state, and increases the risk of thromboembolic events. However, the association between genetic predisposition and thrombotic tendency may differ between ethnic groups [8]. Moreover, neither factor V Leiden nor the prothrombin G20210A mutation has been found in the Korean population [7, 8].
Endothelial nitric oxide synthase is the predominant NOS isoform expressed in normal bone. In the skeletal system, eNOS modulates bone resorption and formation, and the anabolic effects of insulin-like growth factor and oestrogen are dependent on NO production via eNOS [21, 22]. Excessive NO production occurs during various rheumatic diseases, including SLE, RA, Sjögren's syndrome, vasculitis, and osteoarthritis [23]. Given the pleiotropic effects of NO, we have investigated whether there is a link between polymorphisms of the endothelial nitric oxide synthase (NOS3) gene and the development of ONFH.
In this study, we examined the frequency of five different polymorphisms and reconstructed haplotypes: rs11771443 (in 5’ UTR), rs2070744 (T786C, in the promoter), rs1549758 (encoding Asp258Asp, in exon 6), rs1799983 (encoding Glu298Asp, in exon 7), and rs1800780 (in intron 12) in patients and controls. We demonstrated that two exonic NOS3 gene polymorphisms, rs1549758 and rs1799983, and two haplotypes (rs1799983 − rs1800780: G-A and T-A) are statistically significantly associated with development of ONFH (Tables 3 and 4; Suppl. Tables 2, 3, 5 and 6). The haplotype rs1799983 − rs1800780, G-A showed a significant protective effect against ONFH development (P = 1.6 × 10−3; OR = 0.39, 95 % CI = 0.22–0.7). By contrast, the rs1799983-rs1800780 haplotype T-A was significantly associated with an increased risk of ONFH (P = 2.0 x 10−5–6.0 x 10−4, OR = 3.17–3.73) when SLE_ONFH was compared to healthy normal or SLE. Even using the stringent method of Bonferroni correction for multiple testing, the majority of the P-values of associated SNPs and haplotypes retained significance. These results suggest that the minor allele (T) of G894T (Glu298Asp) located in exon 7, which is associated with altered function of the protein encoded by this gene, may increase the risk of developing ONFH in the Korean population.
Previously, it was proposed that two polymorphisms of the NOS3 gene, G894T (Glu298Asp) in exon 7 and the VNTR in intron 4, may be associated with the altered function of this gene [24]. Such functional DNA variants in the NOS3 gene may lead to changes in eNOS expression and/or enzymatic activity. Recently, Koo et al. [9] investigated the association between two polymorphisms, the 27 bp repeat polymorphism in intron 4 and Glu298Asp in exon 7, and ONFH in Koreans. They reported that the 27 bp repeat polymorphism (4a/4b) in intron 4 of NOS3 was associated with idiopathic ONFH. The 4a allele is associated with lower synthesis of eNOS, suggesting a functional difference underlying the association of the 4a allele carrier state with ONFH. The G894T polymorphism is a non-synonymous common variant (Glu298Asp) in the coding region of the NOS3 gene, which is predicted to alter gene function. Our results showed that the Glu298Asp polymorphism was significantly associated with SLE_ONFH, but not with SLE alone. The synonymous polymorphism Asp258Asp in exon 6 was also associated with ONFH at a statistically significant level. In addition, the T786C NOS3 polymorphism has been reported to reduce the NOS3 gene promoter activity [25] and therefore may lead to a decrease in NO production. Decreased NO production leads to vasoconstriction, platelet aggregation, reduced angiogenesis, and reduced bone formation, all of which may be associated with osteonecrosis of the hip. Glueck et al. recently reported the T786C polymorphism in the NOS3 promoter was associated with idiopathic ONFH, but not with secondary osteonecrosis. [13]. Our results indicate that the T786C polymorphism was not associated with SLE_ONFH. The lack of consistency across these studies may be the result of geographic and ethnic variability of the populations included in them. Kim et al. [26] reported that three polymorphisms, T786C, 4a4b, and G894T, exhibit distinct genotype distribution profiles between Koreans and Caucasians. Approximately 50 % of Caucasians are heterozygotes for the T786C and G894T polymorphisms, whereas this is the case for less than 20 % of Koreans [26]. Therefore, it is possible that ethnic differences in NO-mediated effects may result from the proportional distribution of NOS3 variants among ethnic groups. Although our study showed positive relationship between eNOS polymorphisms and ONFH, there are some limitations. We had limited basic and clinical data of study samples. There was no information about family history of ONFH, onset of diseases, medication history, etc. Because we did not check MRI in the control, a few cases of early stage of osteonecrosis that can only be diagnosed with MRI may be included in control. The small sample size of ONFH with SLE patients and those factors may influence the results. Nevertheless, this study will strengthen our understanding of NO-mediated ONFH pathogenesis
In conclusion, we demonstrated that two polymorphisms, Asp258Asp in exon 6 and Glu298Asp in exon 7, appear to have a relationship with ONFH susceptibility in SLE patients. These findings may have important pharmacogenetic implications and their molecular basis must be addressed in further studies. To firmly establish the suggested association, well-designed, case–control or prospective studies with large sample size are required.
Electronic supplementary material
(DOC 65 kb)
(DOC 68 kb)
(DOC 69 kb)
(DOC 54 kb)
(DOC 55 kb)
(DOC 55 kb)
Acknowledgements
This work was supported by a Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology [2011–0021963].
Disclosures
The authors have declared no conflict of interest.
Contributor Information
Tae-Ho Kim, Phone: +82-53-4205436, FAX: +82-53-4205453, Email: kimth0929@ynu.ac.kr.
Shin-Yoon Kim, Phone: +82-53-4205454, FAX: +82-53-420-5453, Email: syukim@knu.ac.kr.
References
- 1.Assouline-Dayan Y, Chang C, Greenspan A, Shoenfeld Y, Gershwin ME. Pathogenesis and natural history of osteonecrosis. Semin Arthritis Rheum. 2002;32:94–124. [PubMed] [Google Scholar]
- 2.Wang GJ, Cui Q, Balian G. The Nicolas Andry award. The pathogenesis and prevention of steroid-induced osteonecrosis. Clin Orthop Relat Res. 2000;370:295–310. doi: 10.1097/00003086-200001000-00030. [DOI] [PubMed] [Google Scholar]
- 3.Abu-Shakra M, Buskila D, Shoenfeld Y. Osteonecrosis in patients with SLE. Clin Rev Allergy Immunol. 2003;25:13–24. doi: 10.1385/CRIAI:25:1:13. [DOI] [PubMed] [Google Scholar]
- 4.Jones LC, Hungerford DS. The pathogenesis of osteonecrosis. Instr Course Lect. 2007;56:179–196. [PubMed] [Google Scholar]
- 5.Mankin HJ. Nontraumatic necrosis of bone (osteonecrosis) N Engl J Med. 1992;326:1473–1479. doi: 10.1056/NEJM199205283262206. [DOI] [PubMed] [Google Scholar]
- 6.Ferrari P, Schroeder V, Anderson S, Kocovic L, Vogt B, Schiesser D, Marti HP, Ganz R, Frey FJ, Kohler HP. Association of plasminogen activator inhibitor-1 genotype with avascular osteonecrosis in steroid-treated renal allograft recipients. Transplantation. 2002;74:1147–1152. doi: 10.1097/00007890-200210270-00016. [DOI] [PubMed] [Google Scholar]
- 7.Kim SY, Suh JS, Park EK, Jung WB, Kim JW, Koo KH, Kim CY. Factor V Leiden gene mutation in femoral head osteonecrosis. J Korean Ortho Res Soc. 2003;6:259–264. [Google Scholar]
- 8.Chang JD, Hur M, Lee SS, Yoo JH, Lee KM. Genetic background of nontraumatic osteonecrosis of the femoral head in the Korean population. Clin Orthop Relat Res. 2008;466:1041–1046. doi: 10.1007/s11999-008-0147-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Koo KH, Lee JS, Lee YJ, Kim KJ, Yoo JJ, Kim HJ. Endothelial nitric oxide synthase gene polymorphisms in patients with nontraumatic femoral head osteonecrosis. J Orthop Res. 2006;24:1722–1728. doi: 10.1002/jor.20164. [DOI] [PubMed] [Google Scholar]
- 10.Cooke JP, Dzau VJ. Nitric oxide synthase: role in the genesis of vascular disease. Annu Rev Med. 1997;48:489–509. doi: 10.1146/annurev.med.48.1.489. [DOI] [PubMed] [Google Scholar]
- 11.Belmont HM, Levartovsky D, Goel A, Amin A, Giorno R, Rediske J, Skovron ML, Abramson SB. Increased nitric oxide production accompanied by the up-regulation of inducible nitric oxide synthase in vascular endothelium from patients with systemic lupus erythematosus. Arthritis Rheum. 1997;40:1810–1816. doi: 10.1002/art.1780401013. [DOI] [PubMed] [Google Scholar]
- 12.Aguirre J, Buttery L, O'Shaughnessy M, Afzal F, Fernandez de Marticorena I, Hukkanen M, Huang P, MacIntyre I, Polak J. Endothelial nitric oxide synthase gene-deficient mice demonstrate marked retardation in postnatal bone formation, reduced bone volume, and defects in osteoblast maturation and activity. Am J Pathol. 2001;158:247–257. doi: 10.1016/S0002-9440(10)63963-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Glueck CJ, Freiberg RA, Oghene J, Fontaine RN, Wang P. Association between the T-786C eNOS polymorphism and idiopathic osteonecrosis of the head of the femur. J Bone Joint Surg Am. 2007;89:2460–2468. doi: 10.2106/JBJS.F.01421. [DOI] [PubMed] [Google Scholar]
- 14.Gagala J, Buraczynska M, Mazurkiewicz T, Ksiazek A. Endothelial nitric oxide synthase gene intron 4 polymorphism in non-traumatic osteonecrosis of the femoral head. Int Orthop. 2013 doi: 10.1007/s00264-013-1892-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee HJ, Choi SJ, Hong JM, Lee WK, Baek JI, Kim SY, Park EK, Kim SY, Kim TH, Kim UK. Association of a polymorphism in the intron 7 of the SREBF1 gene with osteonecrosis of the femoral head in Koreans. Ann Hum Genet. 2009;73:34–41. doi: 10.1111/j.1469-1809.2008.00490.x. [DOI] [PubMed] [Google Scholar]
- 16.Hedrick PW. Gametic disequilibrium measures: proceed with caution. Genetics. 1987;117:331–341. doi: 10.1093/genetics/117.2.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jones LC, Mont MA, Le TB, Petri M, Hungerford DS, Wang P, Glueck CJ. Procoagulants and osteonecrosis. J Rheumatol. 2003;30:783–791. [PubMed] [Google Scholar]
- 18.Kerachian MA, Harvey EJ, Cournoyer D, Chow TY, Séguin C. Avascular necrosis of the femoral head: vascular hypotheses. Endothelium. 2006;13:237–244. doi: 10.1080/10623320600904211. [DOI] [PubMed] [Google Scholar]
- 19.Bjorkman A, Svensson PJ, Hillarp A, Burtscher IM, Runow A, Benoni G. Factor V Leiden and prothrombin gene mutation: risk factors for osteonecrosis of the femoral head in adults. Clin Orthop Relat Res. 2004;425:168–172. doi: 10.1097/00003086-200408000-00023. [DOI] [PubMed] [Google Scholar]
- 20.Glueck CJ, Freiberg RA, Fontaine RN, Tracy T, Wang P. Hypofibrinolysis, thrombophilia, osteonecrosis. Clin Orthop Relat Res. 2001;386:19–33. doi: 10.1097/00003086-200105000-00004. [DOI] [PubMed] [Google Scholar]
- 21.Samuels A, Perry MJ, Gibson RL, Colley S, Tobias JH. Role of endothelial nitric oxide synthase in estrogen-induced osteogenesis. Bone. 2001;29:24–29. doi: 10.1016/S8756-3282(01)00471-9. [DOI] [PubMed] [Google Scholar]
- 22.Lagumdzija A, Ou G, Petersson M, Bucht E, Gonon A, Pernow Y. Inhibited anabolic effect of insulin-like growth factor-I on stromal bone marrow cells in endothelial nitric oxide synthase-knockout mice. Acta Physiol Scand. 2004;182:29–35. doi: 10.1111/j.1365-201X.2004.01303.x. [DOI] [PubMed] [Google Scholar]
- 23.Clancy RM, Amin AR, Abramson SB. The role of nitric oxide in inflammation and immunity. Arthritis Rheum. 1998;41:1141–1151. doi: 10.1002/1529-0131(199807)41:7<1141::AID-ART2>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 24.Tsukada T, Yokoyama K, Arai T, Takemoto F, Hara S, Yamada A, Kawaguchi Y, Hosoya T, Igari J. Evidence of association of the ecNOS gene polymorphism with plasma NO metabolite levels in humans. Biochem Biophys Res Commun. 1998;245:190–193. doi: 10.1006/bbrc.1998.8267. [DOI] [PubMed] [Google Scholar]
- 25.Nakayama M, Yasue H, Yoshimura M, Shimasaki Y, Kugiyama K, Ogawa H, Motoyama T, Saito Y, Ogawa Y, Miyamoto Y, Nakao K. T-786– > C mutation in the 5'-flanking region of the endothelial nitric oxide synthase gene is associated with coronary spasm. Circulation. 1999;99:2864–2870. doi: 10.1161/01.CIR.99.22.2864. [DOI] [PubMed] [Google Scholar]
- 26.Kim IJ, Bae J, Lim SW, Cha DH, Cho HJ, Kim S, Yang DH, Hwang SG, Oh D, Kim NK. Influence of endothelial nitric oxide synthase gene polymorphisms (−786T > C, 4a4b, 894G > T) in Korean patients with coronary artery disease. Thromb Res. 2007;119:579–585. doi: 10.1016/j.thromres.2006.06.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOC 65 kb)
(DOC 68 kb)
(DOC 69 kb)
(DOC 54 kb)
(DOC 55 kb)
(DOC 55 kb)


