Abstract
Age is an important factor when considering phenotypic changes in health and disease. Currently, the use of age information in medicine is somewhat simplistic, with ages commonly being grouped into a small number of crude ranges reflecting the major stages of development and aging, such as childhood or adolescence. Here, we investigate the possibility of redefining age groups using the recently developed Age-Phenome Knowledge-base (APK) that holds over 35,000 literature-derived entries describing relationships between age and phenotype. Clustering of APK data suggests 13 new, partially overlapping, age groups. The diseases that define these groups suggest that the proposed divisions are biologically meaningful. We further show that the number of different age ranges that should be considered depends on the type of disease being evaluated. This finding was further strengthened by similar results obtained from clinical blood measurement data. The grouping of diseases that share a similar pattern of disease-related reports directly mirrors, in some cases, medical knowledge of disease–age relationships. In other cases, our results may be used to generate new and reasonable hypotheses regarding links between diseases.
Keywords: Age, Age groups, Clustering, Disease
Introduction
Age plays an important role in medicine and medical research, being an important factor when considering phenotypic changes in health and disease. A patient’s age can affect the course and progression of a disease (Diamond et al. 1989; Hasenclever and Diehl 1998) or can be important in determining the correct course of treatment (Vecht 1993). Despite this, current use of age information in medicine is somewhat simplistic and coarse.
Ages are commonly grouped into a small number of crude age ranges, reflecting the major stages of development and aging (Carol and Sigelman 2005). Evidence, however, suggests that not all biomedical processes mesh with the same age-grouping template. For example, whereas sexual maturation usually ends by the age of 19 (DeLamater and Friedrich 2002), other developmental processes, such as brain development, continue well into the 20s (Giedd et al. 1999). Moreover, standard age ranges such as those defined by the Medical Subject Headings (MeSH) which is the National Library of Medicine’s controlled vocabulary thesaurus (Medical Subject Headings (Mesh), [http://www.ncbi.nlm.nih.gov/mesh]), are disjoined and nonspecific. When considering disease prevalence and treatment for many types of disease, the important age ranges differ from ranges that are acceptable. For example, venereal disease infections are expected to be most prevalent between the ages of 16 and 35 (Syrjanen et al. 1984; Weinstock et al. 2004). The age range mentioned above includes individuals belonging to three age groups according to the commonly used MeSH vocabulary (i.e., children, adolescents, and adults). Such limitations of the existing age classification model raise the need to revisit how age ranges are defined in the context of disease and health. If ranges could instead be defined in such a way that allows for overlap, it could better suit the description of age in the context of different diseases. Furthermore, although many agree that different age ranges should be considered in the context of different types of disease, this possibility has yet to be systematically evaluated.
While much data concerning disease and age exist, such information was not systematically organized and only of late became available for research. Recently, we developed the Age-Phenome Knowledge-base (APK) that holds a structured representation of knowledge derived from the scientific literature and clinical data regarding clinically-relevant traits and trends that occur at different ages, such as disease symptoms and propensity (Geifman and Rubin 2011). The database underpinning the APK contains over 35,000 entries that describe relationships between age and disease and were mined from over 1.5 million PubMed abstracts (Geifman and Rubin 2012). The availability of such ordered information can lend itself to the examination of age–disease relationships.
One approach for exploring the definition of age ranges involves clustering ages based on patterns of disease occurrence. Accordingly, clustering techniques that group genes, diseases, ages, or other traits that share similar patterns have been repeatedly used in biomedical research to generate new hypotheses. Many such techniques have also been used in the study of biological and medical data, especially in the analysis of microarray and gene expression data (Ben-Dor et al. 1999; Sherlock 2001; Yin et al. 2006). In fact, we have previously shown that using hierarchical clustering based on common patterns in laboratory test values, ages could be grouped into consistent clusters (i.e., where continuous ages are grouped together) and that these clusters largely overlapped with existing age-range definitions (Fliss et al. 2008).
Here, using clustering methods, we explored the possibility of redefining age ranges based on their similarity in disease profiles, as captured in the APK.
Results
Towards new age groups
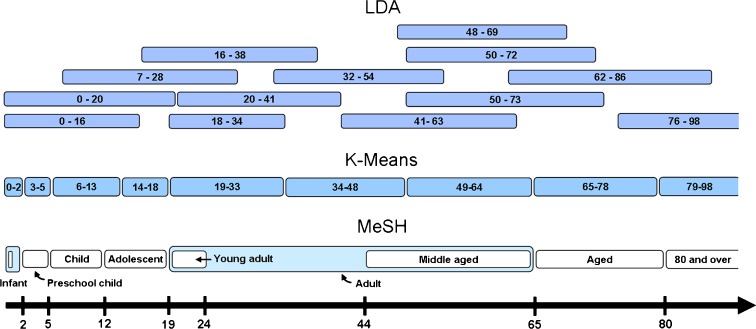
We initially conducted clustering analysis of the age–disease association data using a simple clustering method (k-means, see Methods). Using this approach, nine age groups were defined with the following ranges: 0–2, 3–5, 6–13, 14–18, 19–33, 34–48, 49–64, 65–78, and 79–98 years. These ranges closely match the accepted MeSH ranges (Fig. 1), with the exception of the young adults group, which according MeSH includes individuals 19–24 years of age; here, this range was extended to age 33. In addition, the newborn group (less than 1 month old) was absent from the k-means analysis results due to limitations of the data used (i.e., the use of 1 year resolution).
Fig. 1.
Age ranges as proposed by several methods, namely, the widely accepted MeSH, k-means, and LDA approaches. Age ranges defined by clustering of APK data by the k-means method strongly resemble those defined by MeSH. The LDA method offers several overlapping age ranges shown to be of biomedical significance
Our success in recapturing existing knowledge led us to seek new classifications, allowing for the possibility that multiple, overlapping age ranges better describe groupings pertinent to different diseases or disease classes. While the k-means algorithm could in principle allow for overlapping clusters, it is best suited for disjoined grouping. We thus chose to adopt the latent Dirichlet allocation (LDA) clustering method, a probabilistic “soft clustering” method that allows a given age to belong to multiple ranges. Using LDA with hyper-parameter optimization, 13 age clusters were identified (Table 1 and Fig. 1). These results were supported by our validation techniques (see “Methods”). Twelve of the 13 clusters were successfully recovered when discarding 20 or 40 % of the data. Even when 60 % of the data was discarded, nine of the clusters remain. When repeating the analysis, setting the maximum number of clusters to 13, we obtained highly similar results within the limits of what is expected of a stochastic algorithm (see “Supplementary material”).
Table 1.
Summary of the LDA clustering results
| Cluster # | Range | Representative age |
|---|---|---|
| 1 | 0–16 | 1 |
| 2 | 0–20 | 12 |
| 3 | 7–28 | 16 |
| 4 | 16–38 | 21 |
| 5 | 18–34 | 22 |
| 6 | 20–41 | 35 |
| 7 | 32–54 | 47 |
| 8 | 41–63 | 53 |
| 9 | 48–69 | 61 |
| 10 | 50–73 | 62 |
| 11 | 50–72 | 67 |
| 12 | 62–86 | 74 |
| 13 | 76–98 | 91 |
Ages were clustered with the LDA algorithm based on disease co-occurrence (see “Methods”). For each cluster, the ages belonging to that cluster and the most significant age are presented. We mark the most probable ages in a given cluster as the representative age of the resulting age range
As expected, LDA clustering yielded very different clusters from k-means clustering as well as existing age-range definitions. Importantly, the LDA method-derived clusters overlap. Cluster 2, for example, extends into cluster 1 (spanning the 1–16 and the 1–20 year age ranges, respectively), yet has a later representative age (12 years as compared to 1).
An examination of the resulting age groups suggests that many might be of biological significance. LDA cluster 6, which spans ages 20–41 years, contained a large proportion of instances of a variety of female and male fertility-related diseases (e.g., spermatocytoma, male infertility, anovulation, female infertility, etc.). This age range corresponds to the child-bearing and rearing years and likely reflects those ages when patients try to conceive (Dunson et al. 2004), rather than the actual age of fertility which is likely to begin earlier. Surprisingly, substance abuse-related conditions, such as chronic alcohol intoxication, heroin dependence, and cocaine addiction, were also associated with the same cluster, possibly reflecting related social, psychological, and/or biological processes that co-occur in the same age group.
LDA cluster 12, which spans the ages of 62–86 years, was associated with conditions such as hip fractures, amnesia, and arterial stenosis, all of which are recognized age-related diseases that tend to occur in the later stages of life. Another cluster, covering the ages of 76–98 years (LDA cluster 13), was highly associated with other age-related diseases, such as Alzheimer’s disease, dementia, Parkinson’s disease, tooth attrition, age-related macular degeneration, cataracts, contusions, and DNA fragmentation. This scenario may reflect a subdivision of the older age-associated diseases into separate age groups (i.e., “old” versus “older”). In addition, a few of the resulting clusters span similar age ranges and have a similar representative age (clusters 4 and 5 as well as clusters 9 and 10). Careful examination of the clusters reveals that these are indeed distinct; differences in the diseases which define them are evident (see “Supplementary material”). Moreover, when we looked for a smaller number of clusters as part of the validation process, these clusters remained distinct.
Finally, LDA cluster 1 that includes ages 0–16 years (with age 1 year being the most significant age in the cluster) was highly associated with diseases of childhood and infancy, such as otitis media, childhood leukemia, rota virus infection, sudden infant death syndrome and infantile spasms, as well as a variety of genetic conditions and birth defects. This cluster suggests a definition of childhood that overlaps with existing definitions but only partially coincides with them.
The association of disease with age groups (clusters) also corresponds to known ages of incidence. Carcinoma of the nasopharynx, shown to peak at ages 45–64 years (Ho 1978), was associated with LDA cluster 7, which spans ages 32–54 years (with the most significant age being 47), in 56 % of the reported instances of the diseases listed in the APK and with LDA cluster 10, ranging from the age of 50 to 73 years (with the most significant age being 62 years), accounting for an additional 35 % of the reported instances. The incidence of vaginitis peaked in two age groups (Foxman et al. 2000), the first spanning the ages of 18–24 years and a second peak spanning the 35–44 year range. According to our analysis, vaginitis was found to be highly associated with two LDA clusters, namely cluster 4 (with the most significant age being 21 years), accounting for 34 % of the APK-listed instances of the disease, and cluster 7, accounting for 60 % of the listed cases. HTLV-I, a virus known to cause several common cancers, had a low prevalence (~10 %) in ages under 39 years but became more common with age, reaching a prevalence of nearly 50 % by age 70 (Mueller 1991). In our analysis, HTLV-1 associated with five LDA clusters, with a low proportion of instances associated with clusters 2, 3, and 6 (5–7 %) and with a high proportion of cases being grouped in clusters 8 and 12 (36–39 %). Since clusters 2, 3, and 6 include young adults while clusters 8 and 12 include adults and older individuals, these associations follow the rise of detection with advanced age.
Different types of disease divide the human lifespan differently
The existence of overlapping age ranges, as the LDA results suggest, further supports the hypothesis that different disease types divide life into different numbers of groups. To test this hypothesis, diseases were grouped to form classes of diseases based on the Disease Ontology. Ages within each disease class were clustered on the basis of the hierarchical clustering algorithm, using the pvclust R package (Suzuki and Shimodaira 2006) to define statistically significant divisions into clusters (p value < 0.05).
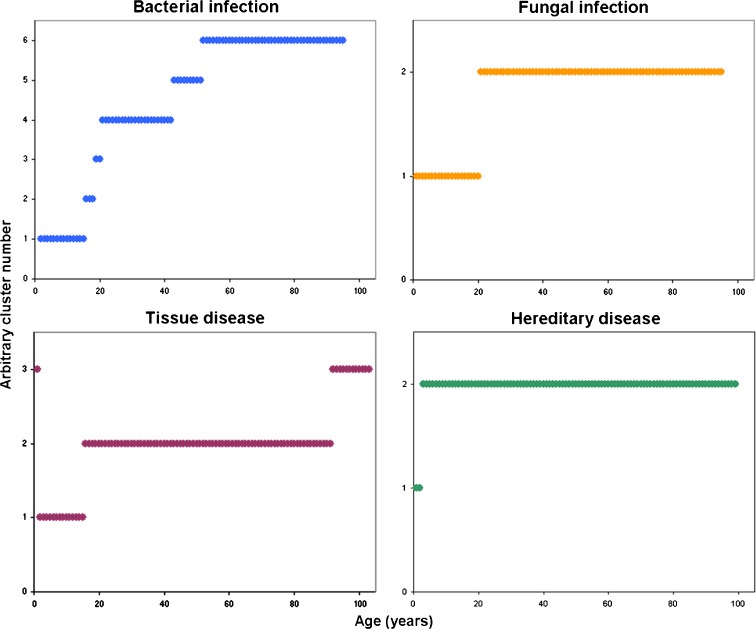
Our results suggest that different disease classes divide the human lifespan into different numbers of age ranges (see Fig. 2 for selected disease classes and “Online supplementary material” for all disease classes). “Fungal Infection”, for example, defined two age ranges, namely 1–20 and 21–95 years. “Bacterial infections”, in comparison, divided life into six segments, while “tissue diseases” divided life into three sections. We note, however, that for many of the disease classes evaluated, a break was observed in the late teens or around age 20. This observation coincides with common medical knowledge; many changes occur in the late teens. The switch from adolescence to adulthood possibly transcends the age divisions associated with specific disease classes. However, the need to consider age differently for the different disease classes was further demonstrated when age distribution within each class was considered. When the median value per class was visualized (Fig. 3a), disease classes clearly differed in terms of their characteristic age ranges.
Fig. 2.
Different disease classes divide life into different numbers of age clusters. The age clusters for four disease classes are illustrated (p value < 0.05)
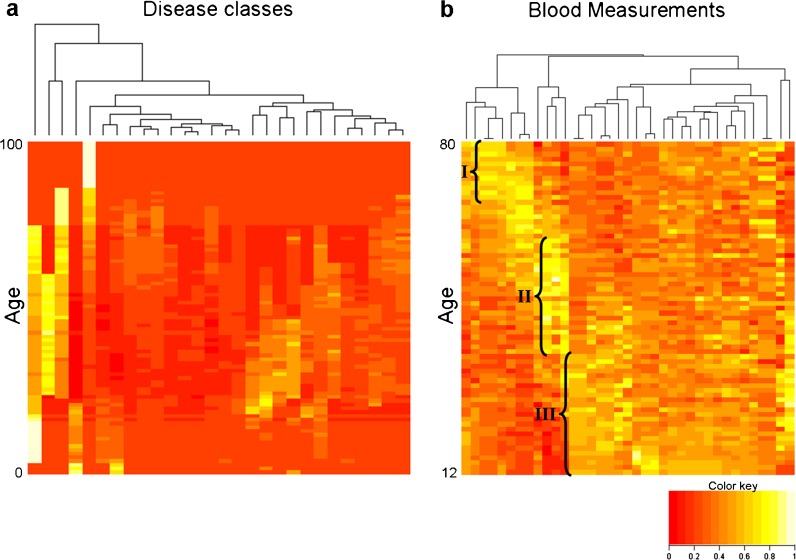
Fig. 3.
Age-related changes in clinical parameters. a The distribution of APK values across 28 disease classes. For each disease class (columns), the median values of instances across all the diseases in that class and that age are shown. b Heat-map of the number of patients with abnormal values per age and blood measurement (ages range from 12 to 80 years). This heat-map was generated using data from the NHANES survey (2007–2010)
We further evaluated clustering of ages based on abnormal blood measurements obtained from the National Health and Nutrition Examination Survey (NHANES) (see “Methods”). Briefly, the survey provides, among other things, laboratory measurements of a random sample of nonhospitalized US residents. We calculated the fraction of individuals in the 2007–2010 surveys that had abnormal values, using the definitions of normal values provided in the NHANES survey. A heat-map of these data illustrates how different abnormal blood measurements divide the human lifespan differently (Fig. 3b). Abnormal values are clearly enriched in some blood measurements in older individuals aged 67 and above (I), in middle aged subjects (II), and in adolescents and young adults (III).
Grouping diseases according to their age-related pattern
We next set out to further demonstrate that multiple processes occur in aging and development by reversing the process, namely using hierarchal clustering of disease by age. We hypothesized that diseases that are governed by similar age-related processes should share similar age patterns.
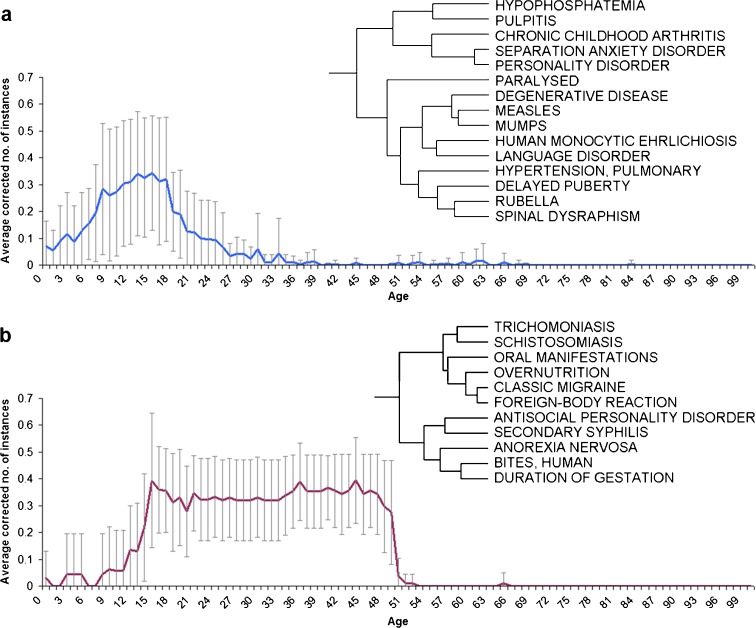
Several of the clusters thus generated both demonstrate the fact that these clusters are likely to represent actual biomedical knowledge and propose new disease associations. Two representative clusters are described in detail (Fig. 4). The first cluster (Fig. 4a) involves several childhood-related diseases (such as rubella, mumps, measles, and chronic childhood arthritis), as well as some neuropsychological-related disorders (such as separation anxiety disorder, personality disorder, and language disorder). The second cluster (Fig. 4b) shows the clustering of a wide array of disorders ranging from parasitic diseases, such as schistosomiasis and trichomoniasis, to anorexia nervosa and classic migraines. Interestingly, neither of the clusters can be well described by the existing age-range definitions. The first cluster mostly involves ages 6–21 years, while the second involves ages 13–51 years.
Fig. 4.
Hierarchical clustering results. a A phylogram and graphical representation for a cluster of diseases that the literature reports peaks in childhood and early teens. b A phylogram and graphical representation for a cluster of diseases that the literature reports peaks in the late teens and up to mid-life. Graphical representation of the average of corrected number of instances per age and disease
Discussion
In this study, several analytic techniques were used to analyze data stored in the Age-Phenome Knowledge-base. Our goal was to test two hypotheses: (1) that the current definition of age ranges can be refined by mining existing knowledge of age–disease associations with advanced analysis methods and (2) that age groups are context-specific, such that different biological process divide life into different ranges.
To examine the first hypothesis, we initially showed that by using a simple, standard technique, data from the APK reflects the currently accepted medical age-range definition. This attests that despite limitations of the APK (Geifman and Rubin 2012), APK data is of sufficient quality to be useful. By allowing overlapping age ranges, we show that new, biologically relevant age ranges can be defined. Indeed, using the LDA clustering method, we generated age clusters based on age–disease relationships, as described in the APK.
The clustering of ages based on disease co-occurrence using a soft clustering method, namely LDA, defined several age intervals, many of which have clear biological significance. For example, a cluster which contains ages 76–98 years was highly associated with diseases such as Alzheimer’s disease, dementia, and Parkinson’s disease, all of which are known to have a high prevalence in later stages of life. On the other hand, a cluster containing ages 0 to 16 years was highly associated with known childhood conditions. Interestingly, LDA suggests that several age-related diseases can be divided over older-ages clusters. For instance, cluster 12 (ages 66–86 years) is highly associated with hip fractures, amnesia, and arterial stenosis, while LDA cluster 13 (containing ages 76 to 98 years) is highly associated with neurodegenerative diseases, tooth attrition, age-related macular degeneration, cataract, contusions, and DNA fragmentation. These results suggest that there might be more than one process occurring at advanced ages, and that different diseases are associated with different processes.
We next tested the hypothesis that multiple processes underlie aging and development and that the number of different age ranges that should be considered depends on the type of disease which is being evaluated. By considering the optimal age ranges of different disease classes, we showed that different types of disease divide life into different intervals. Moreover, some of the intervals defined by clustering specific disease classes are not currently used, to the best of our knowledge, to classify patient ages (for example, ages 43–51 years when considering bacterial infection). Similar results were obtained by clustering ages based on abnormal blood measurements, showing that as with different disease classes, various abnormal blood measurements differ in the number of age ranges they define. Moreover, we show that the clustering of diseases based on age patterns groups together related diseases. Finally, the LDA analysis allowed us to define multiple overlapping age ranges, possibly reflecting parallel yet different underlying processes. Taken together, our results suggest that the current universal division of life (i.e., division in into childhood, adulthood, etc.) might need to be revisited, and that disease- or process-specific classification should be considered.
The hierarchal clustering of diseases, in addition to demonstrating context-specific age patterns, allowed us to investigate possible links between diseases. The disease clusters were found to be useful in generating new hypotheses regarding links between diseases that share similar age-dependant, literature-derived patterns. Take for example, the clustering of hypophosphatemia and pulpitis seen in Fig. 4a. Although no causative link necessarily exists between the two, there is evidence linking the two via X-linked hypophosphatemia (XLH), an X-linked dominant form of rickets. Patients with XLH are highly susceptible to dental caries and attrition, leading to bacterial invasion into the dental pulp which can result in pulpitis (Su et al. 2007). It should be noted that the clustering of hypophosphatemia and pulpitis is not due to both being mentioned in the same PubMed abstracts from which the APK data were mined. Instead, their similar pattern of occurrence with age is independent of this.
A second example derived from our approach is the clustering of overnutrition and two parasitic diseases, namely schistosomiasis and trichomoniasis. Substantial evidence links parasitic infections and malnutrition. The role of the immune system as a major factor in limiting and eliminating parasitic invasion is well known. Malnutrition, known to impair immunity, leads to a lowered ability to fight off parasitic infections (Chandra 1984). In truth, both undernutrition and overnutrition can lead to reduced immunity. For example, obesity was associated with alterations in cellular immunity (Samartín and Chandra 2001). Hence, it is plausible that overnutrition that can hamper the immune system could increase susceptibility to parasitic infections.
These two examples were chosen in order to demonstrate how our results can be used to generate hypotheses regarding the possible association of different diseases. However, considering the huge number of hypotheses which can be derived from these clusters, not all are equally probable. For several of the co-clustered diseases, no neat, straight-forward hypothesis could be formulated. For example, the disease anorexia nervosa was clustered with human bites and duration of gestation (Fig. 4b). We could find some studies that link anorexia and duration of gestation: girls who were born preterm are more likely to develop anorexia later in life (Cnattingius et al. 1999), and women who suffer from anorexia are likely to deliver their babies prematurely (Ekeus et al. 2006). However, it is highly improbable that anorexia and duration of gestation are linked to human bites. Thus, while some interesting hypothesis can be generated from our results, others may be invaluable.
Based on our clustering results, additional hypotheses regarding linkage between diseases could be made. For example, since rubella and delayed puberty share a similar pattern in age and were, therefore, clustered together, it could be hypothesized that a medical/biological connection between the two exists. Such possible links between diseases that emerge from the clustering results should be further investigated to draw meaningful conclusions.
The main weaknesses of this study lies in its sensitivity to research biases. Since our knowledge-base mostly contains data extracted from published papers, it may be influenced by the way clinical research perceives age–disease relationships. We note, however, that as these perceptions have a strong influence on clinical care, capturing them is a useful goal unto itself. At the same time, the main strength of this work comes with the use of a novel, data-driven approach to generate hypotheses about age, possibly leading to new research directions. To the best of our knowledge, this is the first attempt at defining overlapping age ranges based on knowledge mining techniques. Our strategy utilizes a novel knowledge resource, namely the APK, together with advanced data analysis techniques. Further research using improved knowledge-bases and other clinical and biological data may be used to redefine age intervals and to secure the definition of multiple, overlapping age ranges that are context-specific yet clinically and biologically relevant.
Conclusions
To conclude, in this work, we demonstrate that meaningful age groups can be redefined based on data derived from the biomedical literature. These new age ranges are potentially better suited to describe important ages in the context of patient health. We further show that the age groups are context-specific and differ between disease types. Furthermore, we show that by grouping diseases together based on their occurrence in age, new hypotheses regarding links between diseases can be generated.
Methods
A quantitative age–disease matrix
For each disease mapped in the APK database, a count of the number of instances per age was obtained. Evidence linked to inferred age ranges (e.g., inferring 0–50 from the sentence “Under 50 years old”) were excluded. A matrix of disease over age was generated such that each cell contained the number of instances reported for that age (i.e., the age 42 years has a value of 22 in the atherosclerosis column as it is associated with atherosclerosis through 22 database instances). The matrix was normalized by dividing the cells for each disease by the disease total instances count to control for diseases over- or underrepresented in the literature.
Clustering ages by disease co-occurrence
Three clustering algorithms were used to examine age clustering: The Latent Dirichlet Allocation algorithm, the k-means algorithm, and a hierarchical clustering algorithm.
The LDA is described in detail in Blei et al. (2003). Briefly, LDA assumes that each observation is the probabilistic product of a number of underlying processes. In the case of ages, the underlying process is the correlation between a group of ages and a disease. Based on observations (association of specific ages and disease), this method creates age clusters representing the underlying processes (age groups) and learns to correlate between each disease and the underlying age groups. Thus, the method assigns a probability for each age to belong to each age cluster, and another probability for each disease to be associated with each cluster.
In this study, we used the combined list of all the ages in all the studies found for each disease in the APK. Each disease is described by a list of all the ages that are associated with that disease. “Renal carcinoma”, for example, is linked to the age “14 year” twice (i.e., the age range in two studies included 14 year olds), “18 years” once, “47 years” thrice, etc.
We trained a topic model using Mallet with hyper-parameter optimization and the number of clusters set to 25. Ages associated with each cluster were chosen to include all the ages with a probability of 0.01 or better. Low (less than 1 %) abundance clusters were discarded as suggested in by Wallach and McCallum (2009). We marked the most probable ages in a given cluster as the representative ages of the resulting age range.
To validate the results, we sampled a fraction of the data and repeated the analysis. A cluster was considered to be robust if it had an equivalent cluster after sampling, using a tool developed for this purpose (Cohen et. al., unpublished results; the relevant code is available at http://sourceforge.net/projects/topicmodelalig/). This analysis was preformed with 80, 60, and 40 % of the data. Moreover, we repeated the analysis setting the number of clusters to the number of clusters obtained after removing the low abundance clusters.
For k-means clustering (Hartigan and Wong 1979), implementation in Matlab was used for learning the clusters. The number of clusters was chosen empirically as the number yielding the highest mean silhouette (as calculated by Matlab).
For hierarchical clustering of ages (Johnson 1967), the R implementation (R versions 2.13.1) was used. The disease ontology was employed to define disease classes by selecting classes three steps from the root and all of their children. Diseases were filtered such that only diseases linked to at least ten ages and only disease classes which contain more than three such diseases were used. The resulting 28 disease classes were then used to cluster each disease class separately by extracting the sub-matrix of the quantitative age–disease matrix associated with diseases from that class and applying hierarchical clustering to this subset. To identify statistically significant clusters, the pvclust package for R (Suzuki and Shimodaira 2006) was used with the following parameters: The method was set to “average”, the alpha variable was set to 0.95 (p value < 0.05) and the number of bootstrapping was set to 1,000. Out of the 28 disease classes, 24 had age clusters which passed the statistical threshold.
A heat-map of the median values of instances per age and disease class was generated using a script implemented in R.
Clustering diseases by age co-occurrence
Hierarchical clustering (Johnson 1967) of diseases was performed with Expander 5 (Sharan et al. 2003), using the Pearson correlation option for distance calculation.
Clinical data
Data from the NHANES survey was obtained (years 2007–2010). Using blood test data (chemistry and complete blood count), we generated a data matrix as follows. For each subject, measurements were interpreted as normal or abnormal according to the normal ranges defined in the NHANES study. Subjects aged 12 to 80 years were selected (N = 13493), given that a large proportion of the measurement were not performed for children under 12 years of age. A matrix of blood measurements over age was generated such that each cell contained the fraction of patients of that age who presented an abnormal value for that measurement.
Availability
The detailed results, disease classes used for clustering, and all the scripts used to generate the results and images presented in this work are available at http://rubinlab.med.ad.bgu.ac.il/APK/APK_clustering_supplementary.html.
Acknowledgements
This project was funded by The National Institute for Biotechnology in the Negev.
Footnotes
All supplementary material is available at: http://rubinlab.med.ad.bgu.ac.il/APK/APK_clustering_supplementary.html (URL appears in the manuscript under Methods - availability).
References
- Ben-Dor A, Shamir R, Yakhini Z. Clustering gene expression patterns. J Comput Biol. 1999;6(3–4):281–297. doi: 10.1089/106652799318274. [DOI] [PubMed] [Google Scholar]
- Blei DM, Ng AY, Jordan MI. Latent Dirichlet allocation. J Mach Learn Res. 2003;3:993–1022. [Google Scholar]
- Carol K, Sigelman EAR (2005) Life-span human development. 6. Wadsworth Publishing, 3–4
- Chandra RK. Parasitic infection, nutrition, and immune response. Fed Proc. 1984;43(2):251–255. [PubMed] [Google Scholar]
- Cnattingius S, Hultman CM, Dahl M, Sparen P. Very preterm birth, birth trauma, and the risk of anorexia nervosa among girls. Arch Gen Psychiatry. 1999;56(7):634–638. doi: 10.1001/archpsyc.56.7.634. [DOI] [PubMed] [Google Scholar]
- DeLamater J, Friedrich WN. Human sexual development. J Sex Res. 2002;39(1):10–14. doi: 10.1080/00224490209552113. [DOI] [PubMed] [Google Scholar]
- Diamond SG, Markham CH, Hoehn MM, McDowell FH, Muenter MD. Effect of age at onset on progression and mortality in Parkinson’s disease. Neurology. 1989;39(9):1187–1190. doi: 10.1212/WNL.39.9.1187. [DOI] [PubMed] [Google Scholar]
- Dunson DB, Baird DD, Colombo B. Increased infertility with age in men and women. Obstet Gynecol. 2004;103(1):51–56. doi: 10.1097/01.AOG.0000100153.24061.45. [DOI] [PubMed] [Google Scholar]
- Ekeus C, Lindberg L, Lindblad F, Hjern A. Birth outcomes and pregnancy complications in women with a history of anorexia nervosa. Bjog. 2006;113(8):925–929. doi: 10.1111/j.1471-0528.2006.01012.x. [DOI] [PubMed] [Google Scholar]
- Fliss A, Ragolsky M, Rubin E. Reverse translational bioinformatics: a bioinformatics assay of age, gender and clinical biomarkers. CA: AMIA summit on translation bioinformatics San Francisco; 2008. [PMC free article] [PubMed] [Google Scholar]
- Foxman B, Barlow R, D’Arcy H, Gillespie B, Sobel JD. Candida vaginitis: self-reported incidence and associated costs. Sex Transm Dis. 2000;27(4):230–235. doi: 10.1097/00007435-200004000-00009. [DOI] [PubMed] [Google Scholar]
- Geifman N, Rubin E. Towards an age-phenome knowledge-base. BMC Bioinforma. 2011;12:229. doi: 10.1186/1471-2105-12-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geifman N, Rubin E (2012) The age-phenome database. SpringerPlus 1(4) [DOI] [PMC free article] [PubMed]
- Giedd JN, Blumenthal J, Jeffries NO, Castellanos FX, Liu H, Zijdenbos A, Paus T, Evans AC, Rapoport JL. Brain development during childhood and adolescence: a longitudinal MRI study. Nat Neurosci. 1999;2(10):861–863. doi: 10.1038/13158. [DOI] [PubMed] [Google Scholar]
- Hartigan JA, Wong MA. A K-means clustering algorithm. Appl Stat. 1979;28:100–108. doi: 10.2307/2346830. [DOI] [Google Scholar]
- Hasenclever D, Diehl V. A prognostic score for advanced Hodgkin’s disease. International prognostic factors project on advanced hodgkin’s disease. N Engl J Med. 1998;339(21):1506–1514. doi: 10.1056/NEJM199811193392104. [DOI] [PubMed] [Google Scholar]
- Ho JH. An epidemiologic and clinical study of nasopharyngeal carcinoma. Int J Radiat Oncol Biol Phys. 1978;4(3–4):182–198. [PubMed] [Google Scholar]
- Johnson SC. Hierarchical clustering schemes. Psychometrika. 1967;32(3):241–254. doi: 10.1007/BF02289588. [DOI] [PubMed] [Google Scholar]
- MALLET: A Machine Learning for Language Toolkit [http://mallet.cs.umass.edu]
- Mueller N. The epidemiology of HTLV-I infection. Cancer Causes Control. 1991;2(1):37–52. doi: 10.1007/BF00052359. [DOI] [PubMed] [Google Scholar]
- Samartín S, Chandra RK. Obesity, overnutrition and the immune system. Nutr Res. 2001;21(1–2):243–262. doi: 10.1016/S0271-5317(00)00255-4. [DOI] [Google Scholar]
- Sharan R, Maron-Katz A, Shamir R. CLICK and EXPANDER: a system for clustering and visualizing gene expression data. Bioinformatics. 2003;19(14):1787–1799. doi: 10.1093/bioinformatics/btg232. [DOI] [PubMed] [Google Scholar]
- Sherlock G. Analysis of large-scale gene expression data. Brief Bioinform. 2001;2(4):350–362. doi: 10.1093/bib/2.4.350. [DOI] [PubMed] [Google Scholar]
- Su JM, Li Y, Ye XW, Wu ZF. Oral findings of hypophosphatemic vitamin D-resistant rickets: report of two cases. Chin Med J (Engl) 2007;120(16):1468–1470. [PubMed] [Google Scholar]
- Suzuki R, Shimodaira H. Pvclust: an R package for assessing the uncertainty in hierarchical clustering. Bioinformatics. 2006;22(12):1540–1542. doi: 10.1093/bioinformatics/btl117. [DOI] [PubMed] [Google Scholar]
- Syrjanen K, Vayrynen M, Castren O, Yliskoski M, Mantyjarvi R, Pyrhonen S, Saarikoski S. Sexual behaviour of women with human papillomavirus (HPV) lesions of the uterine cervix. Br J Vener Dis. 1984;60(4):243–248. doi: 10.1136/sti.60.4.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vecht CJ. Effect of age on treatment decisions in low-grade glioma. J Neurol Neurosurg Psychiatry. 1993;56(12):1259–1264. doi: 10.1136/jnnp.56.12.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallach HM, McCallum DMA (2009) Rethinking LDA: Why Priors Matter. NIPS
- Weinstock H, Berman S, Cates W., Jr Sexually transmitted diseases among American youth: incidence and prevalence estimates, 2000. Perspect Sex Reprod Health. 2004;36(1):6–10. doi: 10.1363/3600604. [DOI] [PubMed] [Google Scholar]
- Yin L, Huang CH, Ni J. Clustering of gene expression data: performance and similarity analysis. BMC Bioinforma. 2006;7(Suppl 4):S19. doi: 10.1186/1471-2105-7-S4-S19. [DOI] [PMC free article] [PubMed] [Google Scholar]