Abstract
Nm23-H1 has been identified as a metastasis suppressor gene but its protein interactions have yet to be understood with any mechanistic clarity. In this study, we evaluated the proteomic spectrum of interactions made by Nm23-H1 in 4T1 murine breast cancer cells derived from tissue culture, primary mammary tumors and pulmonary metastases. By this approach, we identified the actin-severing protein gelsolin as binding partner for Nm23-H1, verifying their interaction by co-immunoprecipitation in 4T1 cells as well as in human MCF7, MDA-MB-231T and MDA-MB-435 breast cancer cells. In gelsolin transfected cells, co-expression of Nm23-H1 abrogated the actin-severing activity of gelsolin. Conversely, actin severing by gelsolin was abrogated by RNAi-mediated silencing of endogenous Nm23-H1. Tumor cell motility was negatively affected in parallel with gelsolin activity, suggesting that Nm23-H1 binding inactivated the actin depolymerizing function of gelsolin to inhibit cell motility. Using indirect immunoflourescence to monitor complexes formed by gelsolin and Nm23-H1 in living cells, we observed their co-localization in a perinuclear cytoplasmic compartment that was associated with the presence of disrupted actin stress fibers. In vivo analyses revealed that gelsolin overexpression increased the metastasis of orthotopically implanted 4T1 or tail vein injected MDA-MB-231T cells (p=0.001, p=0.04, respectively), along with the proportion of mice with diffuse liver metastases, an effect ablated by co-expression of Nm23-H1. We observed no variation in proliferation among lung metastases. Our findings suggest a new actin-based mechanism that can suppress tumor metastasis.
Keywords: Nm23-H1, Gelsolin, interaction, actin, metastasis
Introduction
Metastatic disease, and the complications of its treatment, are the major causes of death in breast cancer patients (1). Metastasis suppressor genes have provided novel insights into the molecular control of metastasis. Upon overexpression, metastasis suppressors have no effect on primary tumor size, but significantly reduce metastasis formation (2).
Nm23 was the first metastasis suppressor gene to be identified (3). To date, ten different Nm23 genes (Nm23-H1 to Nm23-H10) have been identified in humans; the -H1 and -H2 forms are the most studied (4). In vitro, a hallmark of Nm23 is its suppression of motility to a variety of chemoattractants. Transfection of Nm23 genes significantly inhibited metastasis in multiple model systems (5); where studied, primary tumor size was unaffected. In leukemias, lymphomas and neuroblastoma, opposite trends were observed. Identifying the mechanism(s) of Nm23-H1 suppression of metastasis will be key to developing effective metastasis preventive strategies for this pathway, but remains incomplete. Potential contributors to Nm23-H1’s suppression of metastasis include its histidine kinase activity, nucleotide diphosphate kinase activity and 3′-5′ exonuclease activity(6), as well as protein-protein interactions (reviewed in (7)). The analysis of protein-protein interactions involving Nm23-H1 has been hampered by nonspecific associations, and has identified few interactions of obvious importance to the hallmark motility phenotype.
Here, we have taken an unbiased approach to identify potential binding partners for the human (-H1) and murine (-M1) forms of Nm23 in cultures, primary tumors and metastases formed by murine 4T1 mammary carcinoma cells. We report that Nm23-H1 and -M1 bind Gelsolin, the founding member of a family of actin-binding proteins involved in the remodeling of cellular actin filaments, regulating cell shape changes and movement (8). Structurally Gelsolin is composed of a Ca++-dependent regulatory C-terminal fragment, an actin-severing N-terminal fragment, and a 70 amino acid connector containing the consensus for Caspase3-mediated cleavage (9). After Ca++-dependent activation, Gelsolin binds and severs actin filaments and caps their fast-growing ends. Removal of Gelsolin from the actin filaments (uncapping) is mediated by phospholipids such as phosphatidyl inostil 4,5 bisphosphate (PIP2) (10) and lysophosphatidic acid (LPA) (11) permitting actin-polymerization. Gelsolin null mice were viable (12); in dermal fibroblasts from null mice, reduced membrane lamellipodia and motility were observed, with enhanced Rac expression and increased F-actin in stress fibers (13, 14). Gelsolin also participates in lipid signaling (15) and the apoptotic process (16, 17) and various protein-protein interactions (18–21).
The correlation between Gelsolin expression and tumorigenesis is still controversial (22–25). To date, the role of Gelsolin expression in metastasis is incompletely described. In vitro studies report that Gelsolin overexpression promoted tumor cell motility and invasion through modulation of several pathways, including EGFR, PI3K and Ras-PI3K-Rac (14, 26, 27). In vivo, Gelsolin suppressed the epithelial-mesenchymal transition in mammary epithelial cells (28) and acted as a metastasis suppressor in B16 melanoma cells (29).
Herein, we demonstrate that Nm23-H1 binds to Gelsolin in multiple tumor cell lines, and abrogates Gelsolin’s actin-depolymerization activity in vitro. In contrast with the limited published literature, Gelsolin overexpression in murine 4T1 and human MDA-MB-231T model systems had a significant effect on primary tumor formation and stimulated metastasis to the lungs and liver. Co-overexpression of Gelsolin and Nm23-H1 reduced Gelsolin’s promotion of motility in vitro and metastasis in vivo.
Materials and Methods
Cell culture conditions and treatments
Human breast cancer cell lines MDA-MB-435 and MCF7, and immortalized kidney cell line HEK293TN were obtained from American Type Culture Collection (ATCC, Manassas, VA). A subline of human MDA-MB-231 cells, designated MDA-MB-231T cells, was used after authentication by the Core Genotyping Facility (National Cancer Institute, Bethesda, MD) (30). Murine mammary adenocarcinoma cell line 4T1 expressing luciferase was obtained from Dr. Gary Sahagian, Tufts University. The cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM) (Invitrogen, Grand Island, NY) supplemented with 10% FBS, and antibiotics (100 U/ml penicillin, 100 μg/ml streptomycin), under a humidified 37°C incubator at 5% CO2. To reduce the presence of actin-filaments in cell lysates, cells were incubated 3 hours in presence of 0.33 μg/ml latrunculin B (Millipore, Billerica, MA).
Cell transfection
Nm23-H1 or Nm23-M1 cDNAs with an attached Flag tag were inserted into the BamH1 site in the MCS of pcDNA3.1. The 4T1 cells were transfected with empty vector or plasmid containing Nm23-H1 or -M1 using Lipofectamine (Invitrogen) according to the manufacturer’s recommended protocols. DMEM containing 600 μg/mL of G418 (Invitrogen) was used to select for clonal lines.
Gelsolin full-length or C- and N-terminus fragments (amino acids 1–360 and 361–742, respectively) cDNAs were amplified from pcDNA-Gelsolin, obtained from Dr. Shigeomi Shimizu, Medical Research Institute, Tokyo. The cDNAs were first cloned into pENTR/D-TOPO vector and then inserted into an N-terminal GFP fusion vector, pcDNA 6.2/N-EmGFP using LR recombination reaction according to manufacturer’s recommendations (Gateway vector system, Invitrogen). Plasmids were transfected into cells using Effectene (Bio-Rad, Hercules, CA). Cells were subsequently cultured in complete DMEM with blasticidin (5 μg/ml) for selection.
Gene silencing
MissionR TRC shRNA constructs were purchased from Sigma-Aldrich and included non-target control shRNA (SHC216), shRNA to Gelsolin (TRCN0000029727), shRNA to Nm23-H1 (TRCN0000010061). Cell infection procedure was followed according to the manufacturer’s protocol.
Immunoprecipitation and mass spectrometry
Lysates were prepared by washing cells twice in PBS, followed by incubation for 30 min on ice with a gentle lysis buffer (20 mM tris PH 8.0, 137 mM NaCl, 10% glycerol, 0.2% triton X-100, 1 mM PMSF, 20 μM leupeptin, 0.15 units/ml aprotinin, and 5 mM sodium vanadate). Frozen tumor samples were crushed prior to incubation in lysis buffer. Lysates were centrifuged at 13,000 g for 15 min. Protein concentration of the supernatants was measured using a bicinchoninic acid (BCA) protein assay (Thermo Fisher Scientific, Rockford, IL).
Protein G Sepharose beads (GE Healthcare, Pittsburgh, PA) were incubated with 10 μg of anti-Flag antibody (Sigma-Aldrich) for 1 hour at 4°C. After crosslinking the antibody to beads, they were washed with lysis buffer. The beads were incubated with 1mg of protein suspended in lysis buffer on a rotating mixer overnight at 4°C. The beads were then washed three times with lysis buffer and proteins were eluted by boiling with NuPage loading buffer and sample reducing agent (Invitrogen). The supernatant was collected and separated using SDS-PAGE.
Gel bands were excised from the gel and digested with trypsin. The resultant peptides were extracted and analyzed using reversed-phase liquid chromatography (Agilent 1200, Agilent Technologies, Wilmington, DE) coupled on-line with a LTQ XP mass spectrometer (Thermo Fisher Scientific). The seven most intense molecular ions in the mass spectrometry (MS) scan were sequentially selected for collision-induced dissociation using normalized collision energy of 35%. The MS/MS data were searched against UniProt Mus Musculus database from the European Bioinformatics Institute using BioWorks interfaced SEQUEST (Thermo Fisher Scientific).
Co-immunoprecipitation and immunoblotting
Lysates were prepared from cells in culture by washing them twice in PBS, followed by incubation for 30 min on ice with NP40 lysis buffer (20 mM Tris-HCl pH 8.0, 100 mM NaCl, 10% glycerol, 1% Nonidet P40, 2 mM EDTA). Lysates were centrifuged at 13,000 g for 15 min. Protein concentration of the supernatants was measured using BCA assay. Dynabeads Protein-A/-G (Invitrogen) were incubated 10 min with primary antibody and 30 min with 1 mg of protein lysates, suspended in lysis buffer on a rotating mixer at room temperature. The beads were washed three times with PBS and proteins were eluted by boiling with NuPage loading buffer and sample reducing agent (Invitrogen). The supernatant was collected and separated using SDS-PAGE and processed as a Western blot.
Primary antibodies used for either immunoprecipitation or Western blots are the following: mouse anti-Flag (Sigma-Aldrich), rabbit anti-Flag (Cell Signaling Technology, Danvers, MA), rabbit anti-GFP (Cell Signaling), mouse anti-GFP (Abcam, Cambridge, MA), rabbit anti-Gelsolin (Abcam), mouse anti-Gelsolin (Sigma-Aldrich), mouse anti-Nm23-H1 (BD Bioscience, San Jose, CA), rabbit anti-Nm23-H1 (Santa Cruz Biotechnology, Santa Cruz, CA), rabbit anti-PARP (Cell Signaling Technology), rabbit-anti-caspase3 (Cell Signaling Technology), rabbit anti-cleaved caspase3 (Cell Signaling Technology). Mouse anti-α-Tubulin (Sigma-Aldrich) was used as loading control in Western blots.
Transwell cell migration assay
Transwell migration assays were performed in Boyden chambers as described previously (31). The reported results represent the average of triplicate experiments.
Actin-severing and polymerization assays
The Gelsolin-severing activity was measured using the pyrene-actin polymerization Kit (Cytoskeleton, Denver, CO) following the manufacturer’s protocol. Briefly, 1 mg/ml purified rabbit muscle pyrene-actin (Cytoskeleton, Denver, CO) was diluted to 30 μg/ml and resuspended in polymerization buffer (50 mM KCl, 2 mM MgCl2, 0.5 mM ATP, 2 mM Tris, pH 8.0), incubated for 1 hour to form actin polymers. Cell lysates were prepared and 10 μl of equal amount of total protein was added to the final reaction volume (200 μl). The rate of florescence loss at 590 nm (530-nm excitation wavelength) was measured by fluorometry (VersaMax, Microplate Fluorescence Reader) using Softmax Pro 5.4 software. To measure the actin-polymerization activity, pyrene-actin was diluted to 2.3 μM in the general actin buffer (5 mM Tris-HCl PH 8.0, 0.2 mM CaCl2) containing 0.2 mM ATP and 0.5 mM DTT and stored on ice for 60 min to depolymerize actin oligomers. The tubes were centrifuged at 14 000× g force for 30 min at 4°C. The supernatants containing pyrene-actin were mixed with lysates, added into the 96-well plates, and held for 3 min. After the addition of 1/10 volume of 10X of polymerization buffer (500 mM KCl, 10 mM MgCl2), the kinetics of pyrene fluorescence upon initiation of actin-polymerization was monitored for 2 hours at 405 nm with an excitation wavelength of 355 nm using Microplate Fluorescence Reader.
Immunofluorescence microscopy
Approximately 1×104 cells were plated in each chamber of 4-well chamber slides (BD Bioscience) and grown for 24 hours. The cells were fixed with buffered formalin, pre-blocked in blocking buffer (PBS containing 2% FBS, 2% goat serum, and 0.2% TritonX-100) and incubated with the following primary antibodies diluted in blocking buffer overnight: mouse anti-Flag (Sigma-Aldrich), rabbit anti-Flag (Cell Signaling Technology), rabbit anti-Nm23-H1 (Santa Cruz Biotechnology), rabbit anti-Gelsolin (Abcam), mouse anti-Gelsolin (Sigma-Aldrich). Thereafter, cells were incubated with the following secondary antibodies diluted in blocking buffer: goat anti-mouse and goat anti-rabbit IgG antibodies conjugated to Alexa488 and Alexa546 (Molecular Probes, Eugene, OR), diluted 1:500. Nuclei and F-Actin were visualized by the addition of 200 μg/ml DAPI (4, 6-diamidino-2-phenylindole, dilactate, Molecular Probes) and rhodamine-phalloidin (Invitrogen), respectively, to the secondary antibody mixture. After washing four times with PBS, sections were mounted in Fluorescent Mounting Medium (DakoCytomation, Carpinteria, CA), and photographed using either Zeiss AxioSkope 2 microscope or confocal microscope Zeiss 710UV and ZEN2009 software.
Spontaneous and experimental metastasis assays
Experiments were performed under an approved National Cancer Institute Animal Use Agreement. Six-week-old Balb/c and athymic nude females were obtained from Charles River Laboratories (National Cancer Institute-Frederick Animal Production Area). The spontaneous metastasis assay was performed with 4T1 cells at a concentration of 5×105 cells/50 μl, which were injected under anesthetic into the #4 mammary fat pad of Balb/c mice. Primary tumors were measured by caliper every third day. On day 18 post-injection, primary tumors were resected under isoflurane anesthesia. On week 10 post-injection, mice were sacrificed and all relevant tumor and organ materials were fixed and embedded in OCT. Ten mice per experimental arm were used. Mice were weighed weekly and monitored for signs of morbidity and labored breathing.
The experimental pulmonary metastasis study was conducted injecting 5×105 MDA-MB-231T cells into the lateral tail vein of athymic nude mice. Nine weeks post-injection, at necropsy, the lungs were collected and fixed in Bouins’ solution. Surface metastatic lesions were counted on all lungs prior to embedding in paraffin, reported as a median for each group.
Histological analysis
OCT-embedded lungs and livers of mice from the spontaneous metastasis 4T1 model were sectioned on a cryostat.
Lungs collected from the experimental metastasis MDA-MB-231T model were paraffin-embedded and sectioned (10 μm).
Hematoxylin and eosin (H&E) staining was performed to count the metastatic lesions in two sections from each mouse, 200 μm apart. To measure proliferation rate in metastasis, lungs from both models were stained with rabbit anti-Ki67 antibody, following manufacturer’s protocol (Vector Laboratories, Burlingame, CA).
Statistical analysis
Either analysis of variance (ANOVA) or Kruskal-Wallis test was performed on the raw data. Statistical significance of difference between each sample and vector control arm was assessed using either Student’s t-test or Mann Whitney test. All statistical tests were two-sided and p-values are considered statistically significant for p<0.05. GraphPad Prism 4 software was used to analyze the data.
Results
Identification of Nm23-H1 binding proteins
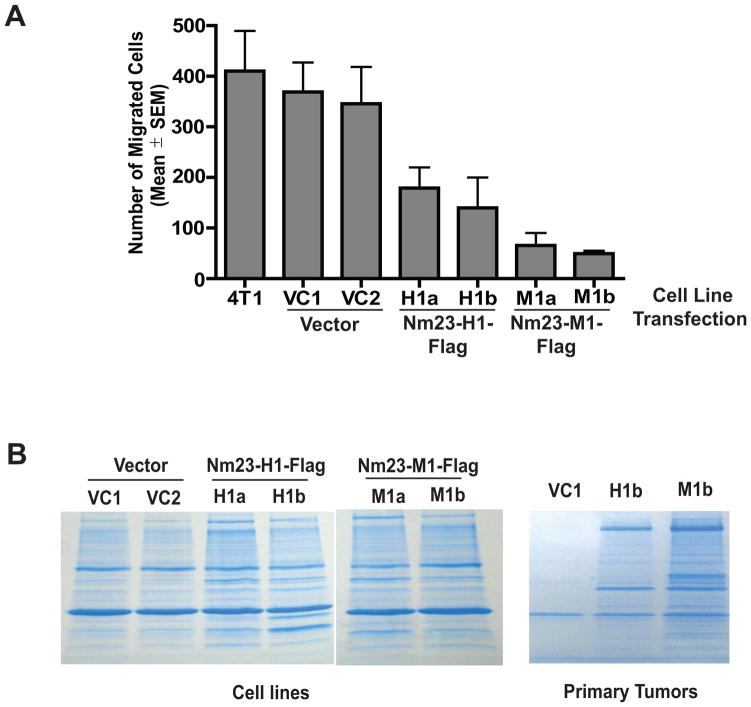
To identify Nm23 co-immunoprecipitating proteins, we utilized murine 4T1 mammary carcinoma cells, previously transfected with a vector, nm23-H1 (human isoform 1) or nm23-M1 (murine), the latter two fused to a C-terminal Flag epitope (32). In agreement with other Nm23 transfections (33), overexpression of either human or murine Nm23 reduced in vitro motility in Boyden chambers by 57% (p = 0.0002) and 85% (p < 0.0001), respectively, compared to vector (Fig. 1A). Implantation of the vector or Nm23 transfected 4T1 cells into the mammary fat pads (mfp) of syngeneic mice resulted in comparable primary tumor sizes ten days post-injection (32). When primary tumors were surgically removed and mice were kept until week 10 post-injection, Nm23 overexpression reduced the number of quantifiable metastases to the liver by 60–70% and lungs by 84–95% (32). Thus, in the 4T1 model system, Nm23-H1 or -M1 overexpression exhibited typical characteristics of a motility- and metastasis suppressor gene.
Figure 1. Nm23 inhibits the in vitro motility of 4T1 mammary carcinoma cells.
A. Boyden chamber motility assay using parental 4T1 cells, two independent clonal vector transfectants (VC1, VC2), and two independent clonal transfectants each of Nm23-H1 (human homolog, H1a and H1b) and Nm23-M1 (murine homolog, M1a, M1b). Cells migrated to 1% FBS for 4 hours. Nm23-H1 overexpression significantly inhibited motility as compared to vector control transfectants, P= 0.0002 and P < 0.0001, respectively. Data from three experiments are shown. B. Using anti-Flag antibody, proteins binding transfected Nm23-H1-Flag or Nm23-M1-Flag were pulled-down from lysates of transfected 4T1 cells in vitro (left panel) and from mfp primary tumors (right panel), and separated by electrophoresis. After elution, Nm23 binding proteins were identified using mass spectrometry.
To identify Nm23 binding proteins, Flag-tagged Nm23-H1 and -M1 proteins were immunoprecipitated from lysates of transfected 4T1 cells in vitro, from mfp primary tumors, and from grossly dissected lymph node-, spleen- and liver metastases. Electrophoresed co-immunoprecipitating proteins (Fig. 1B) were eluted and identified using mass spectrometry (MS). Table 1 lists selected potential co-immunoprecipitating proteins, noting the number of peptides sequenced from each cell line or organ preparation. This list included several proteins previously documented to bind Nm23, confirming the procedure used. Co-immunoprecipitation of Nm23 and the identified proteins was limited to tissue culture and primary tumors, and was largely lost in actual metastases. Notable candidates included several cytoskeletal proteins, which might underlie Nm23 function in tumor motility, invasion and metastasis.
Table 1.
Selected Nm23-H1 and –M1 binding proteins in 4T1 murine mammary carcinoma cells in vitro and in vivo1.
| Protein: | Accession #: | # Peptides Sequenced in Nm23-M1/Nm23-H1 Co-immunoprecipitations2: | Related Co-immunoprecipitating Proteins Also Identified: | Validation4: | ||
|---|---|---|---|---|---|---|
| Cell Culture | Primary Tumor | Metastasis3 | ||||
| Ezrin | P26040 | 2/5 | 17/12 | 0/0/0 | Co-IP | |
| Gelsolin | P13020 | 52/75 | 6/0/0 | Co-IP | ||
| Actin related protein (Arp) 3 | Q99JY9 | 3/32 | 11/21 | 1/0/0 | Arp 2/3 complex subunits 1b, 2, 4, 5 | |
| Eukaryotic initiation factor 4A-1 | P60843 | 3/0 | 9/25 | 0/0/0 | EIFs 2C2, 2-1, 3A, 3E. 3F, 3H, 5A1 | Nonspecific |
| Heat shock protein 90α | P07901 | 0/0 | 11/14 | 1/1/0 | (45) (indirectly) | |
| Casein kinase II, subunit α | Q60737 | 2/6 | 0/4 | 0/0/0 | (46) | |
| Tubulin β6 | Q922F4 | 0/0 | 6/2 | 0/0/0 | (47) | |
| Rab GDI α | P50396 | 0/0 | ½ | 0/0/0 | Rab GDIβ | |
| Rho GEF19 | Q8BWA8 | 1/0 | 1/3 | 0/0/0 | (48) | |
| Guanine nucleotide binding G(i), α2 | P08752 | 5/10 | 3/6 | 0/0/0 | Subunit β1 | (49) |
| Ser/Thr protein phosphatase PPI-α | P62137 | 2/16 | 2/7 | 0/0/2 | PPI 12A | |
| Fatty acid synthase | P19096 | 6/36 | 17/45 | 12/0/0 | Nonspecific | |
| AP-2 complex, a2 subunit | P17427 | 3/0 | 5/16 | 0/0/0 | Subunit β1 | |
| Histone H4 | P62806 | 1/0 | 9/23 | 0/1/0 | H1.2, H2A.1F, H2B.1B | |
| Lyn | P25911 | 0/4 | ¼ | 0/0/0 | Nonspecific | |
| Syk | P48025 | 0/0 | 0/2 | 0/0/0 | Nonspecific | |
| ATP-citrate lyase | Q91V92 | 6/12 | 1/5 | 0/0/7 | Other metabolic enzymes: isocitrate dehydrogenase, 6-phosphofructokinase, lactate dehydrogenase, hexokinase, transketolase | |
| Phosphoglycerate mutase | Q8BX10 | 1/5 | ½ | 0/0/0 | (50) | |
Co-immunoprecipitating proteins listed when multiple peptides were identified as binding to a Nm23 protein lysate.
Number of peptides sequenced from Nm23-M1/Nm23-H1 expressing transfectants, minus peptides sequenced from control vector transfectant.
Lymph node/Spleen/Liver metastasis peptides from Nm23-H1 and –M1 combined.
Published reports or unpublished co-immunoprecipitation (Co-IP) experiments. Nonspecific indicates a lack of a co-immunoprecipitation in unpublished experiments.
Nm23 proteins bind Gelsolin
One of the candidate proteins resulting from this analysis was the actin-binding protein Gelsolin. Notably, 52 and 75 peptides of Gelsolin were sequenced from anti-Flag -Nm23-H1 and -M1 immunoprecipitated primary tumor lysates, respectively. Binding was reduced or lost in the lymph node, spleen and liver metastases.
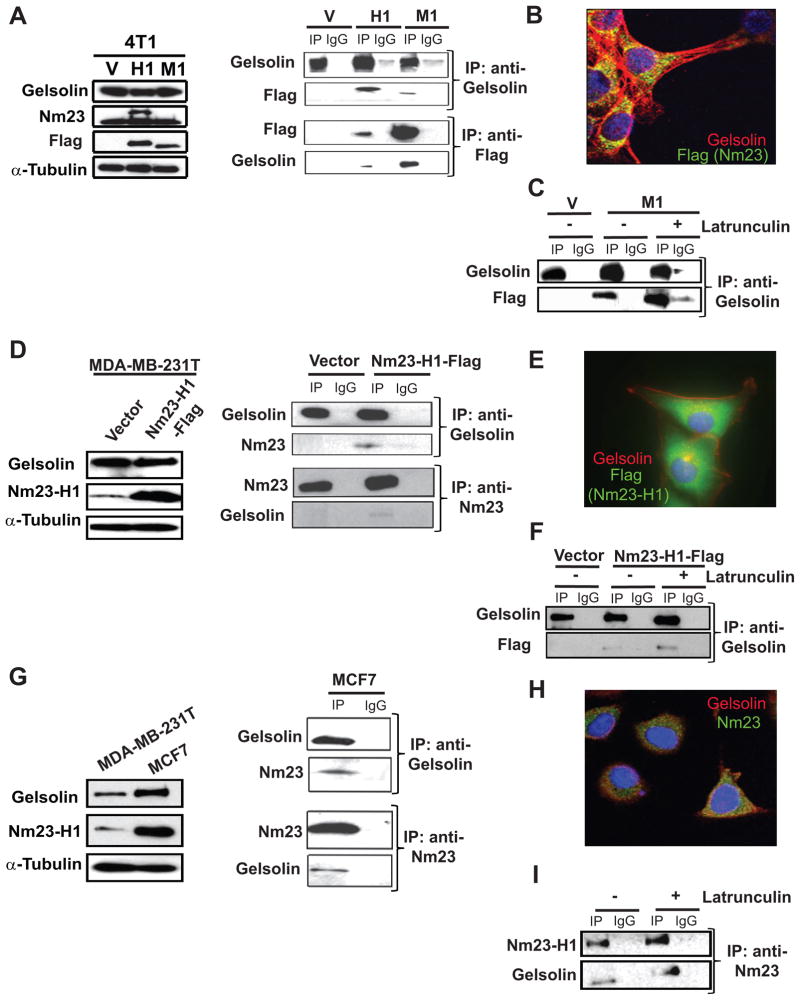
Co-immunoprecipitation assays were used to verify the interaction between Nm23 and Gelsolin in the murine mammary carcinoma cell line 4T1 overexpressing Flag-tagged Nm23-H1 and –M1 (Fig. 2A, left). Either anti-Flag or anti-Gelsolin antibodies were used to pull down the target protein from the cell lysates (Fig. 2A, right). Co-immunoprecipitation of Gelsolin and Nm23 was observed in both Nm23-H1 and M-1 samples. Co-immunofluorescence of Nm23 and Gelsolin was performed in 4T1 cells overexpressing Nm23-M1 along the edge of a scratch motility assay (Fig. 2B). Although Gelsolin was expressed in both cell edges and cytoplasm along the actin-cytoskeleton structure, co-localization of the two proteins was observed exclusively in the cytoplasmatic compartment of the cells. Since Gelsolin is an actin-binding protein, the interaction Nm23-Gelsolin may be an indirect result of Nm23 binding to the actin filaments. Treatment of the 4T1 cells with latrunculin B, an actin-depolymerization agent (34), destroyed the F-actin cytoskeleton but did not eliminate the Nm23-Gelsolin co-imunoprecipitation (Fig. 2C, Supplementary Fig. S1).
Figure 2. Gelsolin interacts with Nm23-H1 and -M1 in murine and human cell lines.
A. Left panel, western blot of lysates of 4T1 cells expressing either a vector (V), Flag-tagged Nm23-H1 (H1) or Flag-Nm23-M1 (M1). Right panel, co-immunoprecipitation of Gelsolin and Nm23 with either rabbit anti-Gelsolin or mouse anti-Flag antibody respectively. Either rabbit or mouse IgG was used as a control. B. Confocal microscopy image showed cytoplasmic co-immunoflorescence of Gelsolin (red) and Flag for Nm23-M1-Flag (green). C. Vector or Nm23-M1-Flag 4T1 cells were incubated 3 hours with either DMSO or 0.33μg/ml latrunculin B, and co-immunoprecipitation experiments performed to demonstrate Nm23/Gelsolin complex formation under actin-polymerization independent conditions. D. MDA-MB-231T human breast carcinoma cells were transfected with a vector or Flag-tagged Nm23-H1. Western blot (left) and co-immunoprecipitation (right) of Gelsolin and Flag-tagged Nm23-H1 from lysates was performed using mouse anti-Gelsolin and rabbit anti-Nm23 antibodies. E. Co-immunofluorescent staining was performed on MDA-MB-231T cells overexpressing Nm23-H1-Flag using mouse anti-Gelsolin red) and rabbit anti-Flag (green). F. MDA-MB-231T cells expressing either vector or Nm23-H1-Flag were incubated with either DMSO or 0.33μg/ml latrunculin B, and the lysates used for a co-immunoprecipitation experiment. G. Gelsolin and Nm23-H1 expression levels in MDA-MB-231T and MCF7 cell lines was analyzed by Western blot using mouse anti-Nm23-H1 and mouse anti-Gelsolin antibodies (left). The α-tubulin antibody was used as loading control. Protein lysate from MCF7 cells was used to pull down the either the endogenous Nm23-H1 with rabbit anti-Nm23-H1 antibody or Gelsolin with mouse anti-Gelsolin antibody. Western blots of the immunoprecipitates indicate complex formation between the endogenous Nm23-H1 and Gelsolin (right). H. Cytoplasmatic co-localization of Nm23-H1 and Gelsolin was identified in the immunoflorescent staining using mouse anti-Gelsolin (red) and rabbit anti-Nm23-H1 (green). I. Co-immunoprecipitation experiment was performed on MCF7 protein lysate after either DMSO or latrunculin B treatment of the cells in panel G.
In order to validate the Gelsolin/Nm23 interaction observed in the mammary cell line 4T1, a second cellular model, the human breast carcinoma cell line MDA-MB-231T, was used. Empty vector and Flag-tagged Nm23-H1 constructs were transfected in MDA-MB-231T cells (Fig. 2D, left). Anti-Nm23 and anti-Gelsolin antibodies were used to pull-down the endogenous proteins together with exogenous Flag-tagged Nm23-H1 (Fig. 2D, right). The Gelsolin/Nm23 complex was detected only in Nm23-H1 overexpressing MDA-MB-231T cells, which was consistent with our inability to detect even low levels of Nm23-H1 immunoprecipitation in lysates in the vector expressing controls. Immunoflorescence confirmed the co-localization of the two proteins in the perinuclear compartment (Fig. 2E), and the complex remained after destruction of the actin-cytoskeleton by latrunculin B (Fig. 2F, Supplementary Fig. S1). The data confirm an interaction of transfected Nm23-H1 and Gelsolin in two breast carcinoma cell lines. When Gelsolin was fragmented into N- and C-terminal subunits, Nm23-H1 co-immunoprecipitated with the C-terminal fragment (Supplementary Fig. S2). The co-immunoprecipitation results were validated in other two cell lines, MDA-MB-435 and HEK293TN, overexpressing the empty vector and Nm23-H1 or both Nm23-H1 and Gelsolin, respectively (Supplementary Fig. S3).
To evaluate the interaction of Nm23 and Gelsolin at endogenous levels, we used the MCF7 human breast carcinoma cell line. This cell line contains a higher Nm23 level compared to MDA-MB-231T cells (Fig. 2G, left). Both the immunoprecipitation and immunoflorescence assays demonstrated that Nm23 and Gelsolin interacted in an F-actin-independent manner and colocalized in the cytoplasmic compartment (Fig. 2H–I, Supplementary Fig. S1).
Nm23 overexpression inhibits the actin-severing activity of Gelsolin
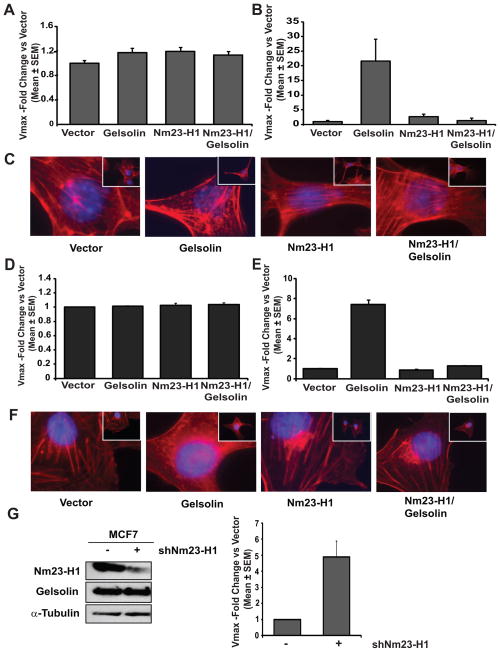
Gelsolin is a potent F-actin severing protein that, after filament cleavage, remains bound to the newly formed end (capping). In this way, Gelsolin facilitates depolymerization by blocking actin monomer addition at the growing end, while actin disassembly proceeds at the opposite end of the filament (35). Gelsolin also has two contrasting functions in apoptosis; it has a pro-apoptotic role after cleavage by caspases leading to uncontrolled F-actin severing; it can also prevent apoptosis by stabilizing mitochondria (9, 17). We hypothesized that Gelsolin’s interaction with Nm23 would inhibit its actin-depolymerization function, given Nm23’s profound effects on motility. Clonal cell lines overexpressing either a vector or Gelsolin, the latter with a GFP tag at its N-terminus, and either a vector or Nm23 (-H1 and -M1)-Flag were created in both the 4T1 and MDA-MB-231T cell lines. The GFP tag did not interfere with Gelsolin interaction with Nm23 (Supplementary Fig. S4). To evaluate whether Nm23 overexpression and interaction could affect actin-cytoskeleton remodeling by Gelsolin, actin-polymerization and depolymerization rates were measured using an in vitro, fluorescence-based assay for F-actin formation or severing activity. In 4T1 cells, transfection of either Gelsolin or Nm23-H1 had no effect on actin-polymerization (Fig. 3A), in keeping with the lack of reported Gelsolin effect on actin-polymerization (36). However, a decrease in fluorescent signal, corresponding to an increase in F-actin-severing, was observed in the lysate from Gelsolin overexpressing cells when the actin-depolymerization assay was performed (Fig. 3B), consistent with the reported literature (35). In contrast, no florescence decay was observed in the reaction with lysates containing 4T1 overexpressing Nm23 alone or together with Gelsolin. Phalloidin staining was used to analyze the actin cytoskeleton structure (Fig. 3C). Severing activity associated with Gelsolin expression in 4T1 cells was visualized by the decreased number of actin filaments (stress fibers) and appearance of short filaments or actin monomer accumulation (red dots). Nm23 overexpression together with Gelsolin reversed this phenotype as the actin cytoskeleton appeared similar to that in the vector cells. Data were validated using lysates from MDA-MB-231T cells (Fig. 3D, E–F). Similar results were obtained when actin-severing activity was measured in protein lysates from a scratch-wounded confluent layer of either 4T1 or MDA-MB-231T cells (Supplementary Fig. S5). While Nm23 showed only minor effect on reducing actin-severing as compared to the control, its co-overexpression with Gelsolin resulted in a significant inhibition of Gelsolin actin-severing activity (39% in 4T1, p=0.04; 47% in MDA-MB-231T, p=0.05). These results indicate that Nm23 overexpression and interaction with Gelsolin prevented its actin-severing activity and modified stress fiber formation. Moreover, when endogenous Nm23-H1 expression was knocked down in MCF7 cells, actin-severing increased 5-fold as compared to the control cells (Figure 3G), thus confirming its involvement in this process.
Figure 3. Nm23-H1 impairs Gelsolin- depolymerization activity.
A. A pyrene-actin polymerization assay was used to measure the actin-polymerization activity in protein lysates from 4T1 cells expressing either vector, Flag-tagged Nm23-H1 (Nm23-H1), GFP-tagged Gelsolin (Gelsolin) or both proteins (Nm23-H1/Gelsolin). Protein lysate was incubated for 2 hours in presence of pyrene-actin monomers and a polymerization buffer. The increase in fluorescence intensity at 590 nm was measured using VersaMax Microplate Fluorescence Reader and Softmax Pro 5.4 software to obtain the Vmax of each reaction. Data represent the ratio of Vmax of each sample versus Vmax of the vector transfectants. B. F-actin-depolymerization activity of the same samples was measured by incubating protein lysate in presence of pyrene-actin filaments for 2 hours. The decrease of fluorescence intensity at 590 nm was measured and Vmax ratio relative to the vector samples was graphed. C. Actin cytoskeleton structure in 4T1 cells was visualized by staining the cells with rhodamine-phalloidin (red) and DAPI (blue). 400X magnification images are shown. D. Actin-polymerization activity in protein lysates from MDA-MB-231T cells expressing either vector, Flag-tagged Nm23-H1 (Nm23-H1), GFP-tagged Gelsolin (Gelsolin) or both proteins (Nm23-H1/Gelsolin) was measured as previously described for Figure 3A. E. F-actin-depolymerization activity in protein lysates from MDA-MB-231T cells was determined as previously described for Figure 3B. F. Actin cytoskeleton structure in MDA-MB-231T cells was visualized by staining the cells with rhodamine-phalloidin (red) and DAPI (blue). 400X magnification images are shown. G. Western blot (left panel) and F-actin-depolymerization activity (right panel) of lysates from MCF7 expressing either non-target control shRNA (−) or shRNA specific to Nm23-H1 (+).
Effects on tumor motility
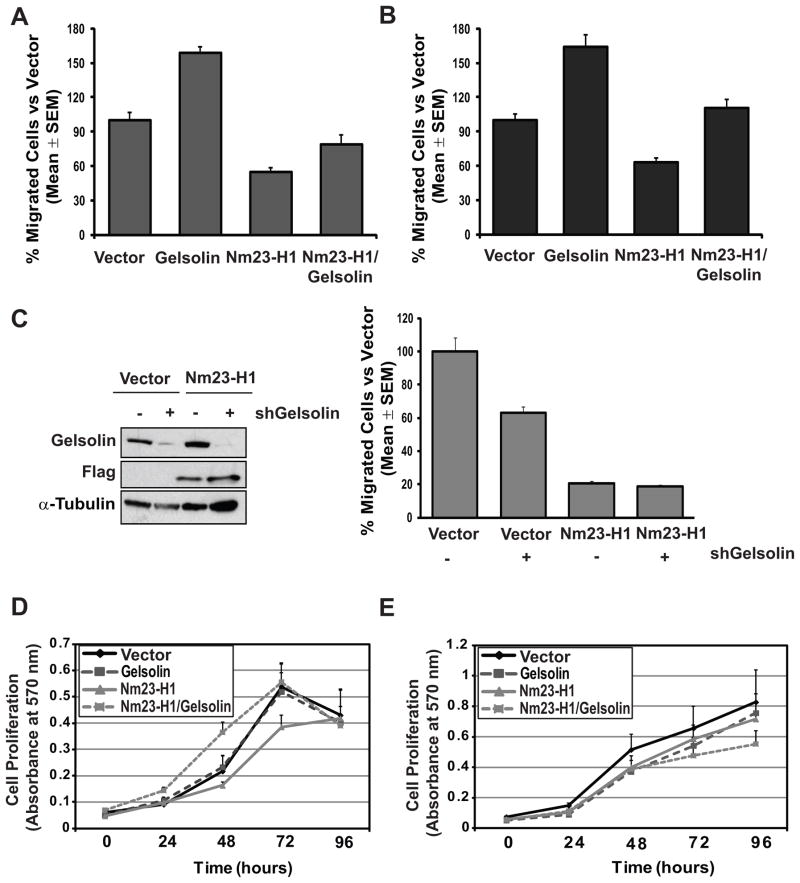
Because actin cytoskeleton reorganization is important for cell shape and motility, Gelsolin has crucial roles in the control of these cellular functions. The effects of Gelsolin on tumor cell motility are complex (29, 37). We observed distinct patterns of in vitro motility upon Gelsolin overexpression. Gelsolin overexpression promoted cell migration in presence of growth factors such as EGF (27) and insulin (26), while no effect was observed when other chemoattractants such as LPA (38) and low FBS were used (Supplementary Fig. S6 A–B). Motility to EGF was selected for further experiments, using 4T1 and MDA-MB-231T cells overexpressing Nm23, Gelsolin or both proteins (Fig. 4). Overexpression of Gelsolin in 4T1 cells enhanced cell migration by 58% (p=0.03), while Nm23 reduced it by 46% (p=0.02), compared to vector transfected cells. Cells overexpressing both proteins showed 50% reduction in motility compared the Gelsolin expressing cells (p=0.03) (Fig. 4A). MDA-MB-231T cells expressing Nm23, Gelsolin or both proteins were also assayed. Cell migration was increased by 64% after Gelsolin overexpression while was reduced by 37% after Nm23-H1 overexpression compared the vector (p=0.005 and p=0.0009, respectively). The co-overexpression of Nm23-H1/Gelsolin reversed the Gelsolin phenotype, reducing motility by 53% compared to Gelsolin expressing cells (p=0.02) (Fig. 4B). Thus, Nm23 inhibits the motility-promoting effects of Gelsolin, consistent with its abrogation of Gelsolin’s actin severing ability. In addition, knockdown of endogenous Nm23-H1 from MCF7 cells resulted in 4.8 fold increase in motility (p=0.04) (Supplementary Fig. S6 C). Knockdown of Gelsolin in the vector expressing MDA-MB-231T showed a 37% decreased motility (p= 0.03); it had no effect on motility in the Nm23-H1 expressing cells (Fig. 4C).
Figure 4. Nm23-H1 reversed Gelsolin-stimulated tumor cell motility.
A. Cell motility toward EGF (10ng/ml) of 4T1 cells expressing either vector, Flag-tagged Nm23-H1 (Nm23-H1), GFP-tagged Gelsolin (Gelsolin) or both proteins (Nm23-H1/Gelsolin) was measured using Boyden chamber system. The percent of migrated cells for each sample relative to vector control transfectant was graphed, based on four experiments. B. Cell motility toward EGF of MDA-MB-231T cells expressing either vector, Flag-tagged Nm23-H1 (Nm23-H1), GFP-tagged Gelsolin (Gelsolin) or both proteins (Nm23-H1/Gelsolin) was measured as previously described for Figure 4A. C. Western blot (left panel) and cell motility assay (right panel) performed on vector- and Flag-tagged Nm23-H1 (Nm23-H1) transfected MDA-MB-231T cells, expressing either non-target control shRNA (−) or shRNA specific to Gelsolin (+). D. Proliferation rate of the 4T1 cells was measured using colorimetric MTT assay (Sigma-Aldrich). E. Cell proliferation of MDA-MB-231T cells was measured using colorimetric MTT assay.
Other in vitro assays were performed. No significant effect of Nm23 or Gelsolin overexpression on proliferation of 4T1 or MDA-MB-231T cells was observed in MTT assays (Fig. 4D–E). No effect of the Nm23 or Gelsolin overexpression on apoptosis was observed (Supplementary Fig. S7).
Nm23 overexpression reduced Gelsolin’s potentiation of tumor metastasis
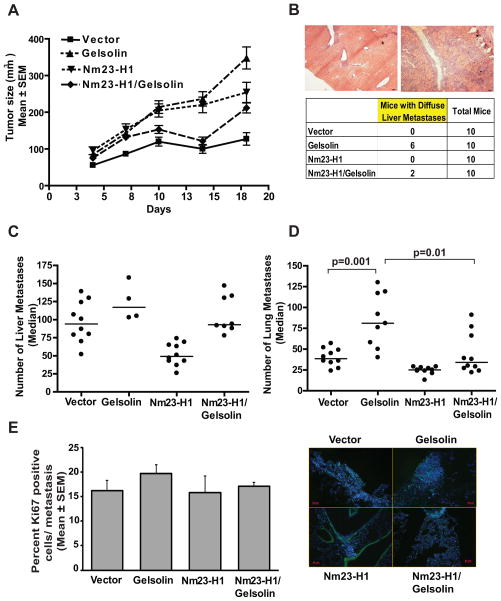
To evaluate the in vivo effects of Nm23-H1 and Gelsolin overexpression, 4T1 cells overexpressing either a vector or Gelsolin, and either a vector or Nm23-H1 were implanted into the mfp of syngeneic Balb/c mice. Differences in tumor growth between the cells were observed (Fig. 5A). In accord with its tumorigenic property in breast cancer (22, 23), Gelsolin overexpressing 4T1 cells demonstrated increased tumor growth, which was reversed when Nm23-H1 was co-overexpressed. Primary tumors were removed surgically on day 18 post-injection and the mice were sacrificed on day 69 post-injection to quantify metastasis (Fig. 5B–D). In the liver, two histologic forms of metastatic colonization were observed. In some mice discreet metastatic lesions were quantifiable, while other mice contained florid metastatic disease throughout the liver (Fig. 5B); these two patterns of metastasis were quantified separately as previously reported (32). In the liver, Gelsolin overexpression increased distinct metastatic lesions by 24% (p=0.187); diffuse liver metastases were observed in 60% (6 out of 10 mice) of mice as compared to none in the vector transfectants. Nm23-H1 overexpression reduced distinct metastasis formation by 48% (p=0.0002) and no mice had diffuse metastases. Co-overexpression of Gelsolin and Nm23-H1 reduced the percentage of mice with diffuse liver metastases to 20% (2 out of 10 mice).
Figure 5. Nm23-H1 reduces the pro-metastatic effect of Gelsolin in a 4T1 spontaneous metastasis assay.
A. 5×105 4T1 cells expressing either vector, Flag-tagged Nm23-H1 (Nm23-H1), GFP-tagged Gelsolin (Gelsolin) or both proteins (Nm23-H1/Gelsolin) were implanted into the mammary fat pads of Balb/c mice. Primary tumor size through day 18 post-injection was determined, at which time tumors were removed. Difference in tumor growth was observed between the arms. B. Representative images (50X magnification) of discreet (right) and diffuse (left) liver metastases obtained from mice 10 weeks post-injection with 4T1 cells overexpressing Gelsolin. Scale bars = 100μm. Number of mice showing liver with diffuse metastases was reported in the table. C. Discreet liver metastases in each arm were reported as median values. A dot represents the number of liver metastases indentified in each mouse. Mice presenting diffuse liver metastases (see figure 5B) were excluded from this analysis. Comparison of Gelsolin and vector arms (p=0.18), vector and Nm23-H1 arms (p=0.0002). The Nm23-H1/Gelsolin arm was not significantly different from the vector. D. Lung metastases in each arm, reported as median values. While Gelsolin overexpression increased metastasis by 107%, Nm23-H1overexpression reduced it by 36% (p=0.001 and p=0.0015, respectively) compared to the vector arm. Nm23-H1/Gelsolin arm showed a metastasis formation similar to the vector and significantly decreased metastasis compared the Gelsolin arm (59%, p=0.01). E. Ki67 staining of lungs from 4T1 injected mice. Representative pictures are shown in the right panel. Scale bar=50μm. The graphs show the percentage of Ki67 positive cells in metastasis expressed as the mean ± SEM.
Data in the lungs showed similar trends (Fig. 5D). Gelsolin overexpression increased metastasis formation by 107% (median number of metastases 39 to 81, p=0.001) as compared to the vector transfectants. Nm23-H1 overexpression reduced metastasis formation by 36% (median number of metastases 39 to 25, p=0.0015). Co-overexpression of Gelsolin and Nm23-H1 reduced lung metastasis formation to levels significantly different from the Gelsolin transfectant (median number of metastases 81 to 34, p=0.01). The data indicate a pro-metastatic function of Gelsolin in the 4T1 model system that is inhibited by Nm23-H1 overexpression.
To evaluate potential influence of proliferation on metastasis formation, Ki67 staining was performed on pulmonary metastatic lesions. No difference in proliferation was observed between the experimental groups (Fig. 5E).
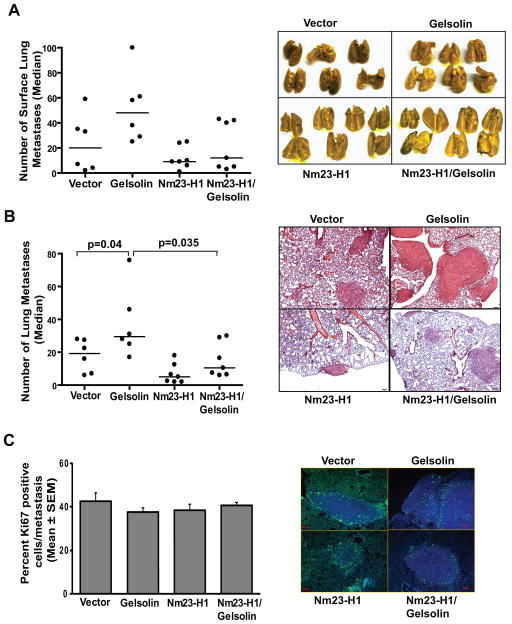
To exclude any influence on metastasis formation related to the differences observed in tumor growth, an experimental metastatic lung assay was performed. To confirm an inhibitory effect of Nm23 on Gelsolin’s pro-metastatic activity, MDA-MB-231T cells overexpressing either Nm23, Gelsolin, or both proteins were injected via the tail vein to produce lung metastases. Representative pictures of the lungs collected from mice after 9 weeks from the cell injection are shown on Fig. 6A, left. The median number of surface lung metastases/mouse was 20 in the vector arm and increased to 48 in the Gelsolin arm, confirming Gelsolin’s metastasis promoting activity in two model systems. Surface experimental metastases declined 55% to 9 in Nm23-H1 overexpressing arm. Co-overexpression of Nm23-H1 and Gelsolin produced an average of 12 surface lung metastases, not statistically different from the vector transfectant (p=0.83) (Fig. 6A, right).
Figure 6. Nm23-H1 reduced the pro-metastatic effect of Gelsolin in an MDA-MB-231T experimental metastasis assay.
A. 5×105 MDA-MB-231T cells expressing either vector (n=6), Flag-tagged Nm23-H1 (Nm23-H1) (n=7), GFP-tagged Gelsolin (Gelsolin) (n=6) or both proteins (Nm23-H1/Gelsolin) (n=7) were injected into the tail veins of athymic nude mice. At 9 weeks post-injection, the mice were sacrificed and the lungs were collected and fixed in Bouin’s solution. Surface lung metastases were counted. The Nm23-H1/Gelsolin arm showed a 75% reduction in metastases as compared to the Gelsolin arm (p=0.101). B. H&E stained sections were evaluated for lung metastases. Each dot represents a single mouse. Gelsolin expression increased metastasis by 53.2% while Nm23-H1 reduced metastasis by 74.1%, the number of lung metastases compared to the vector arm (p=0.041 and p=0.035 respectively). Nm23-H1/Gelsolin arm showed a 65% reduction in metastases as compared to the Gelsolin arm (p=0. 035). C. Ki67 staining was performed on lungs from MDA-MB-231T injected mice. Representative pictures are shown in the right panel. Scale bar =50μm. The graph shows the percentage of Ki67 positive cells in metastasis expressed as the mean ± SEM.
After paraffin-embedding and sectioning the lungs, H&E staining was performed to visualize and count histologic metastases. This quantification of metastatic burden confirmed the surface metastasis findings: Gelsolin expression induced a 53.2% increase in metastasis formation (p=0.041), while Nm23 reduced the number of lung metastases by 74.1% (p=0.035) as compared to the vector (Fig. 6B). Co-overexpression of Nm23-H1 and Gelsolin significantly reduced metastases by 75% as compared to Gelsolin arm (p=0.035).
No difference in proliferation was observed in metastatic lesions between the experimental groups (Fig. 6C).
Discussion
Although Nm23 has been thoroughly validated as a metastasis suppressor in solid tumors, its mechanism of action remains elusive (5). Protein-protein interactions involving Nm23 have been widely reported (7) but few have been independently confirmed and, likewise, few modulate Nm23’s hallmark phenotype of tumor cell motility. We conducted an unbiased search for Nm23-H1 and –M1 binding proteins using MS/MS sequencing of co-immunoprecipitating proteins from 4T1 lysates, primary tumors and metastases. Of the Nm23-binding proteins identified from primary tumors, several are intimately connected to cellular motility including Ezrin, Arp2/3 and Gelsolin. The interaction of Nm23 and each of these proteins was undetectable in lymph nodes, liver and spleen metastases from the same model system, which may reflect the down-regulation of Nm23-H1 expression in metastatic lesions (39), leading to the loss of an association and unhindered function of the binding protein. Two-way co-immunoprecipitation experiments using transfected Nm23-H1 in 4T1 cells and human MDA-MB-231T breast carcinoma cells, MDA-MB-435 carcinoma cells and HEK293TN immortalized kidney cells confirmed the association of Nm23 and Gelsolin. Co-immunoprecipitation of the two proteins at endogenous levels was observed in human MCF7 breast carcinoma cells, which express relatively high levels of Nm23-H1. It remains possible that additional proteins are part of the Nm23-H1-Gelsolin complex; the pro-metastatic protein Prune has been reported to be a binding partner of each protein, for instance (40, 41).
Gelsolin is traditionally understood as an actin binding protein. It severs and caps ends of actin filaments in a Ca++ regulated manner, providing for dynamic actin assembly and disassembly in normal cell elasticity and in the leading and lagging aspects of cell motility. Less well understood are Gelsolin’s protein-protein interactions with p53 (21), hormone receptors (18, 20) and other proteins (19), a lipid regulatory activity (15), and the apoptosis-inducing activity of its N-terminal fragment (16). Using lysates of 4T1 and MDA-MB-231T breast carcinoma cells transfected with either vector, Nm23-H1, Gelsolin or Nm23-H1/Gelsolin, enzymatic assays for actin-polymerization and depolymerization were conducted. Consistent with the literature, Gelsolin overexpression had no effect on actin-polymerization, but augmented its depolymerization. It is not known whether Gelsolin directly severed the actin filaments in the assay, or capped filaments severed by other enzymes. In agreement with an interaction between Nm23-H1 and Gelsolin, co-overexpression of both proteins abrogated Gelsolin’s depolymerization ability. Furthermore, after knocking down endogenous Nm23-H1 in MCF7 cells, an increase in actin-severing was observed, confirming that Nm23-H1 may affect Gelsolin function. The data indicate a functional consequence of the Nm23-H1-Gelsolin interaction. Our co-immunoflourescence data point to an interaction of Gelsolin and Nm23-H1 in the perinuclear cytoplasm, permitting the hypothesis that Nm23-H1 and Gelsolin interact to regulate stress fiber formation, which occurs in this area. In experiments conducted using MDA-MB-231T cells, Nm23-H1 co-immunoprecipitated a transfected C-terminal portion of Gelsolin. This region has a single actin binding site and regulates Gelsolin’s activity (8), while its actual actin-severing activity is localized to Gelsolin’s N-terminus.
Consistent with Gelsolin’s role in actin dynamics, its overexpression has been reported widely to increase tumor cell motility and invasion or, conversely, its knockdown to inhibit motility (14, 26–28). In both the 4T1 and MDA-MB-231T model systems, we confirmed a stimulation of tumor cell motility by Gelsolin overexpression. The stimulatory effect of Gelsolin was abrogated by co-overexpression of Nm23, consistent with their binding and inactivation of actin-depolymerization. Furthermore, while knock down of endogenous Nm23 in MCF7 cells resulted in increased motility, the knockdown of Gelsolin in Nm23 overexpressing MDA-MB-231T showed no effect on cell migration, suggesting that the functional interaction is unidirectional in effect, i.e., that Gelsolin may not affect Nm23 function.
We did not observe any evidence for a pro-apoptotic role of the transfected Gelsolin, but cannot rule out that other of its functions could also be contributory.
In the face of Gelsolin’s role in actin dynamics and tumor cell motility, in vivo data in the literature on the phenotypic consequences of its altered expression are both sparse and puzzling. Three reports showed a suppressive effect of Gelsolin overexpression, either wild type or a Pro321→His mutation, on primary tumor formation (42–44). A lipid metabolism role of Gelsolin was implicated in its tumor suppressive phenotype. We observed an increased in primary tumor size upon Gelsolin overexpression (22, 23). For metastasis, only one report has been published and it shows a surprising metastasis suppressive role for Gelsolin overexpression in B16 melanoma cells (29).This report stands in sharp contrast to two model systems reported herein, in which Gelsolin overexpression augmented 4T1 spontaneous metastasis to the lungs and liver, and experimental MDA-MB-231T metastasis to the lungs. The reason(s) for the discrepancy are unclear. Gelsolin overexpression in the B16 model was modest, and it is possible that its relative expression level can alter actin dynamics to produce divergent phenotypes. Other Gelsolin family members exert similar functions (27). In agreement with the co-immunoprecipitation of Gelsolin and Nm23, and Nm23’s abrogation of Gelsolin’s actin-depolymerization and motility induction properties, co-overexpression of Nm23 reduced the metastasis stimulatory phenotype of Gelsolin. Taken together, the data permit the hypothesis on a mechanism of action for Nm23 metastasis suppression: An Nm23-H1 interaction with Gelsolin limits actin-depolymerization and dynamic function, thereby attenuating metastasis in breast cancer model systems.
Supplementary Material
Acknowledgments
The authors thank Dr. Gary Sahagian, Tufts University, for luciferase labeled 4T1 cells and Dr. Shigeomi Shimizu, Medical Research Institute Tokyo, for pcDNA-Gelsolin plasmid.
Grant Support
This project has been funded with federal funds from the National Cancer Institute, National Institutes of Health, under contract number HHSN261200800001E.
Footnotes
The authors disclose no potential conflicts of interest.
References
- 1.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ. Cancer statistics, 2009. CA: a cancer journal for clinicians. 2009;59:225–49. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- 2.Rinker-Schaeffer CW, O’Keefe JP, Welch DR, Theodorescu D. Metastasis suppressor proteins: discovery, molecular mechanisms, and clinical application. Clinical cancer research: an official journal of the American Association for Cancer Research. 2006;12:3882–9. doi: 10.1158/1078-0432.CCR-06-1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Steeg PS, Bevilacqua G, Pozzatti R, Liotta LA, Sobel ME. Altered expression of NM23, a gene associated with low tumor metastatic potential, during adenovirus 2 Ela inhibition of experimental metastasis. Cancer research. 1988;48:6550–4. [PubMed] [Google Scholar]
- 4.Lacombe ML, Milon L, Munier A, Mehus JG, Lambeth DO. The human Nm23/nucleoside diphosphate kinases. Journal of bioenergetics and biomembranes. 2000;32:247–58. doi: 10.1023/a:1005584929050. [DOI] [PubMed] [Google Scholar]
- 5.Marino N, Nakayama J, Collins JW, Steeg PS. Insights into the biology and prevention of tumor metastasis provided by the Nm23 metastasis suppressor gene. Cancer metastasis reviews. 2012;31:593–603. doi: 10.1007/s10555-012-9374-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Steeg PS, Zollo M, Wieland T. A critical evaluation of biochemical activities reported for the nucleoside diphosphate kinase/Nm23/Awd family proteins: opportunities and missteps in understanding their biological functions. Naunyn-Schmiedeberg’s archives of pharmacology. 2011;384:331–9. doi: 10.1007/s00210-011-0651-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marino N, Marshall JC, Steeg PS. Protein-protein interactions: a mechanism regulating the anti-metastatic properties of Nm23-H1. Naunyn-Schmiedeberg’s archives of pharmacology. 2011;384:351–62. doi: 10.1007/s00210-011-0646-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sun HQ, Yamamoto M, Mejillano M, Yin HL. Gelsolin, a multifunctional actin regulatory protein. The Journal of biological chemistry. 1999;274:33179–82. doi: 10.1074/jbc.274.47.33179. [DOI] [PubMed] [Google Scholar]
- 9.Kothakota S, Azuma T, Reinhard C, Klippel A, Tang J, Chu K, et al. Caspase-3-generated fragment of gelsolin: effector of morphological change in apoptosis. Science. 1997;278:294–8. doi: 10.1126/science.278.5336.294. [DOI] [PubMed] [Google Scholar]
- 10.Janmey PA, Stossel TP. Modulation of gelsolin function by phosphatidylinositol 4,5-bisphosphate. Nature. 1987;325:362–4. doi: 10.1038/325362a0. [DOI] [PubMed] [Google Scholar]
- 11.Meerschaert K, De Corte V, De Ville Y, Vandekerckhove J, Gettemans J. Gelsolin and functionally similar actin-binding proteins are regulated by lysophosphatidic acid. The EMBO journal. 1998;17:5923–32. doi: 10.1093/emboj/17.20.5923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Witke W, Sharpe AH, Hartwig JH, Azuma T, Stossel TP, Kwiatkowski DJ. Hemostatic, inflammatory, and fibroblast responses are blunted in mice lacking gelsolin. Cell. 1995;81:41–51. doi: 10.1016/0092-8674(95)90369-0. [DOI] [PubMed] [Google Scholar]
- 13.Lu M, Witke W, Kwiatkowski DJ, Kosik KS. Delayed retraction of filopodia in gelsolin null mice. The Journal of cell biology. 1997;138:1279–87. doi: 10.1083/jcb.138.6.1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De Corte V, Bruyneel E, Boucherie C, Mareel M, Vandekerckhove J, Gettemans J. Gelsolin-induced epithelial cell invasion is dependent on Ras-Rac signaling. The EMBO journal. 2002;21:6781–90. doi: 10.1093/emboj/cdf680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Biswas RS, Baker D, Hruska KA, Chellaiah MA. Polyphosphoinositides-dependent regulation of the osteoclast actin cytoskeleton and bone resorption. BMC cell biology. 2004;5:19. doi: 10.1186/1471-2121-5-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Geng YJ, Azuma T, Tang JX, Hartwig JH, Muszynski M, Wu Q, et al. Caspase-3-induced gelsolin fragmentation contributes to actin cytoskeletal collapse, nucleolysis, and apoptosis of vascular smooth muscle cells exposed to proinflammatory cytokines. European journal of cell biology. 1998;77:294–302. doi: 10.1016/S0171-9335(98)80088-5. [DOI] [PubMed] [Google Scholar]
- 17.Kusano H, Shimizu S, Koya RC, Fujita H, Kamada S, Kuzumaki N, et al. Human gelsolin prevents apoptosis by inhibiting apoptotic mitochondrial changes via closing VDAC. Oncogene. 2000;19:4807–14. doi: 10.1038/sj.onc.1203868. [DOI] [PubMed] [Google Scholar]
- 18.Nishimura K, Ting HJ, Harada Y, Tokizane T, Nonomura N, Kang HY, et al. Modulation of androgen receptor transactivation by gelsolin: a newly identified androgen receptor coregulator. Cancer research. 2003;63:4888–94. [PubMed] [Google Scholar]
- 19.Keller JW, Haigis KM, Franklin JL, Whitehead RH, Jacks T, Coffey RJ. Oncogenic K-RAS subverts the antiapoptotic role of N-RAS and alters modulation of the N-RAS:gelsolin complex. Oncogene. 2007;26:3051–9. doi: 10.1038/sj.onc.1210103. [DOI] [PubMed] [Google Scholar]
- 20.Kim CS, Furuya F, Ying H, Kato Y, Hanover JA, Cheng SY. Gelsolin: a novel thyroid hormone receptor-beta interacting protein that modulates tumor progression in a mouse model of follicular thyroid cancer. Endocrinology. 2007;148:1306–12. doi: 10.1210/en.2006-0923. [DOI] [PubMed] [Google Scholar]
- 21.An JH, Kim JW, Jang SM, Kim CH, Kang EJ, Choi KH. Gelsolin negatively regulates the activity of tumor suppressor p53 through their physical interaction in hepatocarcinoma HepG2 cells. Biochemical and biophysical research communications. 2011;412:44–9. doi: 10.1016/j.bbrc.2011.07.034. [DOI] [PubMed] [Google Scholar]
- 22.Liu J, Liu YG, Huang R, Yao C, Li S, Yang W, et al. Concurrent down-regulation of Egr-1 and gelsolin in the majority of human breast cancer cells. Cancer genomics & proteomics. 2007;4:377–85. [PubMed] [Google Scholar]
- 23.Thor AD, Edgerton SM, Liu S, Moore DH, 2nd, Kwiatkowski DJ. Gelsolin as a negative prognostic factor and effector of motility in erbB-2-positive epidermal growth factor receptor-positive breast cancers. Clinical cancer research: an official journal of the American Association for Cancer Research. 2001;7:2415–24. [PubMed] [Google Scholar]
- 24.Rao J, Seligson D, Visapaa H, Horvath S, Eeva M, Michel K, et al. Tissue microarray analysis of cytoskeletal actin-associated biomarkers gelsolin and E-cadherin in urothelial carcinoma. Cancer. 2002;95:1247–57. doi: 10.1002/cncr.10823. [DOI] [PubMed] [Google Scholar]
- 25.Dosaka-Akita H, Hommura F, Fujita H, Kinoshita I, Nishi M, Morikawa T, et al. Frequent loss of gelsolin expression in non-small cell lung cancers of heavy smokers. Cancer research. 1998;58:322–7. [PubMed] [Google Scholar]
- 26.Chen P, Murphy-Ullrich JE, Wells A. A role for gelsolin in actuating epidermal growth factor receptor-mediated cell motility. The Journal of cell biology. 1996;134:689–98. doi: 10.1083/jcb.134.3.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lader AS, Lee JJ, Cicchetti G, Kwiatkowski DJ. Mechanisms of gelsolin-dependent and -independent EGF-stimulated cell motility in a human lung epithelial cell line. Experimental cell research. 2005;307:153–63. doi: 10.1016/j.yexcr.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 28.Tanaka H, Shirkoohi R, Nakagawa K, Qiao H, Fujita H, Okada F, et al. siRNA gelsolin knockdown induces epithelial-mesenchymal transition with a cadherin switch in human mammary epithelial cells. International journal of cancer Journal international du cancer. 2006;118:1680–91. doi: 10.1002/ijc.21559. [DOI] [PubMed] [Google Scholar]
- 29.Fujita H, Okada F, Hamada J, Hosokawa M, Moriuchi T, Koya RC, et al. Gelsolin functions as a metastasis suppressor in B16-BL6 mouse melanoma cells and requirement of the carboxyl-terminus for its effect. International journal of cancer Journal international du cancer. 2001;93:773–80. doi: 10.1002/ijc.1413. [DOI] [PubMed] [Google Scholar]
- 30.Palmieri D, Halverson DO, Ouatas T, Horak CE, Salerno M, Johnson J, et al. Medroxyprogesterone acetate elevation of Nm23-H1 metastasis suppressor expression in hormone receptor-negative breast cancer. Journal of the National Cancer Institute. 2005;97:632–42. doi: 10.1093/jnci/dji111. [DOI] [PubMed] [Google Scholar]
- 31.Horak CE, Lee JH, Elkahloun AG, Boissan M, Dumont S, Maga TK, et al. Nm23-H1 suppresses tumor cell motility by down-regulating the lysophosphatidic acid receptor EDG2. Cancer research. 2007;67:7238–46. doi: 10.1158/0008-5472.CAN-07-0962. [DOI] [PubMed] [Google Scholar]
- 32.Marshall JC, Collins JW, Nakayama J, Horak CE, Liewehr DJ, Steinberg SM, et al. Effect of inhibition of the lysophosphatidic acid receptor 1 on metastasis and metastatic dormancy in breast cancer. Journal of the National Cancer Institute. 2012;104:1306–19. doi: 10.1093/jnci/djs319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kantor JD, McCormick B, Steeg PS, Zetter BR. Inhibition of cell motility after nm23 transfection of human and murine tumor cells. Cancer research. 1993;53:1971–3. [PubMed] [Google Scholar]
- 34.Spector I, Shochet NR, Kashman Y, Groweiss A. Latrunculins: novel marine toxins that disrupt microfilament organization in cultured cells. Science. 1983;219:493–5. doi: 10.1126/science.6681676. [DOI] [PubMed] [Google Scholar]
- 35.Gremm D, Wegner A. Gelsolin as a calcium-regulated actin filament-capping protein. European journal of biochemistry/FEBS. 2000;267:4339–45. doi: 10.1046/j.1432-1327.2000.01463.x. [DOI] [PubMed] [Google Scholar]
- 36.Ditsch A, Wegner A. Nucleation of actin polymerization by gelsolin. European journal of biochemistry/FEBS. 1994;224:223–7. doi: 10.1111/j.1432-1033.1994.tb20015.x. [DOI] [PubMed] [Google Scholar]
- 37.Van den Abbeele A, De Corte V, Van Impe K, Bruyneel E, Boucherie C, Bracke M, et al. Downregulation of gelsolin family proteins counteracts cancer cell invasion in vitro. Cancer letters. 2007;255:57–70. doi: 10.1016/j.canlet.2007.03.023. [DOI] [PubMed] [Google Scholar]
- 38.van Leeuwen FN, Giepmans BN, van Meeteren LA, Moolenaar WH. Lysophosphatidic acid: mitogen and motility factor. Biochemical Society transactions. 2003;31:1209–12. doi: 10.1042/bst0311209. [DOI] [PubMed] [Google Scholar]
- 39.Guan-Zhen Y, Ying C, Can-Rong N, Guo-Dong W, Jian-Xin Q, Jie-Jun W. Reduced protein expression of metastasis-related genes (nm23, KISS1, KAI1 and p53) in lymph node and liver metastases of gastric cancer. International journal of experimental pathology. 2007;88:175–83. doi: 10.1111/j.1365-2613.2006.00510.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.D’Angelo A, Garzia L, Andre A, Carotenuto P, Aglio V, Guardiola O, et al. Prune cAMP phosphodiesterase binds nm23-H1 and promotes cancer metastasis. Cancer cell. 2004;5:137–49. doi: 10.1016/s1535-6108(04)00021-2. [DOI] [PubMed] [Google Scholar]
- 41.Garzia L, Roma C, Tata N, Pagnozzi D, Pucci P, Zollo M. H-prune-nm23-H1 protein complex and correlation to pathways in cancer metastasis. Journal of bioenergetics and biomembranes. 2006;38:205–13. doi: 10.1007/s10863-006-9036-z. [DOI] [PubMed] [Google Scholar]
- 42.Sagawa N, Fujita H, Banno Y, Nozawa Y, Katoh H, Kuzumaki N. Gelsolin suppresses tumorigenicity through inhibiting PKC activation in a human lung cancer cell line, PC10. British journal of cancer. 2003;88:606–12. doi: 10.1038/sj.bjc.6600739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sakai N, Ohtsu M, Fujita H, Koike T, Kuzumaki N. Enhancement of G2 checkpoint function by gelsolin transfection in human cancer cells. Experimental cell research. 1999;251:224–33. doi: 10.1006/excr.1999.4552. [DOI] [PubMed] [Google Scholar]
- 44.Mullauer L, Fujita H, Ishizaki A, Kuzumaki N. Tumor-suppressive function of mutated gelsolin in ras-transformed cells. Oncogene. 1993;8:2531–6. [PubMed] [Google Scholar]
- 45.Salerno M, Palmieri D, Bouadis A, Halverson D, Steeg P. Nm23-H1 metastasis suppressor expression level influences the binding properties, stability and function of the Kinase Suppressor of Ras (KSR1) Erk scaffold in breast carcinoma cells. Mol Cell Biol. 2005;25:1379–88. doi: 10.1128/MCB.25.4.1379-1388.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Engel M, Issinger OG, Lascu I, Seib T, Dooley S, Zang KD, et al. Phosphorylation of nm23/nucleoside diphosphate kinase by casein kinase 2 in vitro. Biochemical and biophysical research communications. 1994;199:1041–8. doi: 10.1006/bbrc.1994.1334. [DOI] [PubMed] [Google Scholar]
- 47.Lombardi D, Sacchi A, D’Agostino G, Tibursi G. The association of the Nm23-M1 protein and beta-tubulin correlates with cell differentiation. Experimental cell research. 1995;217:267–71. doi: 10.1006/excr.1995.1086. [DOI] [PubMed] [Google Scholar]
- 48.Iwashita S, Miyamoto M. Nucleoside diphosphate kinase nm23-H2 binds to Rho and negatively regulates Rho activity. Cell Structure and Function. 2005:30. [Google Scholar]
- 49.Wieland T. Interaction of nucleoside diphosphate kinase B with heterotrimeric G protein betagamma dimers: consequences on G protein activation and stability. Naunyn Schmiedebergs Arch Pharmacol. 2007;374:373–83. doi: 10.1007/s00210-006-0126-6. [DOI] [PubMed] [Google Scholar]
- 50.Engel M, Mazurek S, Eigenbrodt E, Welter C. Phosphoglycerate mutase-derived polypeptide inhibits glycolytic flux and induces cell growth arrest in tumor cell lines. J Biol Chem. 2004;279:35803–12. doi: 10.1074/jbc.M402768200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.