Abstract
Saccharopolyspora erythraea, a mycelium-forming actinomycete, produces a clinically important antibiotic erythromycin. Extensive investigations have provided insights into erythromycin biosynthesis in S. erythraea, but knowledge of its morphogenesis remains limited. By gene inactivation and complementation strategies, the TetR-family transcriptional regulator SACE_0012 was identified to be a negative regulator of mycelium formation of S. erythraea A226. Detected by quantitative real-time PCR, the relative transcription of SACE_7115, the amfC homolog for an aerial mycelium formation protein, was dramatically increased in SACE_0012 mutant, whereas erythromycin biosynthetic gene eryA, a pleiotropic regulatory gene bldD, and the genes SACE_2141, SACE_6464, SACE_6040, that are the homologs to the sporulation regulators WhiA, WhiB, WhiG, were not differentially expressed. SACE_0012 disruption could not restore its defect of aerial development in bldD mutant, and also did not further accelerate the mycelium formation in the mutant of SACE_7040 gene, that was previously identified to be a morphogenesis repressor. Furthermore, the transcriptional level of SACE_0012 had not markedly changed in bldD and SACE_7040 mutant over A226. Taken together, these results suggest that SACE_0012 is a negative regulator of S. erythraea morphogenesis by mainly increasing the transcription of amfC gene, independently of the BldD regulatory system.
Electronic supplementary material
The online version of this article (doi:10.1007/s00284-013-0410-x) contains supplementary material, which is available to authorized users.
Introduction
During its life cycle, the soil-inhabiting Actinomycetes undergoes a complex morphological differentiation to adapt to adverse environments [1]. Growth of Actinomycetes begins with spore germination and hyphal outgrowth, leading to the formation of a vegetative, or substrate mycelium. Sensing of nutrient deprivation stimulates reproductive growth resulting in the development of aerial hyphae and spore chains [2]. Saccharopolyspora erythraea could form the aerial hyphae, and produce erythromycin, which is a macrolide antibiotic with broad-spectrum antimicrobial activity. Extensive genetic and biochemical studies have provided detailed insights into the genes involved in erythromycin biosynthesis in S. erythraea [3, 4], yet its morphological differentiation remains poorly understood.
In recent years, the availability of the complete genome sequence of S. erythraea allowed a deeper exploration of the molecular processes controlling its morphogenesis [5]. Guided by in vitro and in vivo investigations, BldD (SACE_2077) was discovered to be a key developmental regulator in actinomycetes [1], controlling erythromycin biosynthesis and morphological differentiation in S. erythraea [6]. Furthermore, we identified a TetR-family transcriptional regulator (SACE_7040) involving in S. erythraea mycelium formation, and established genetic evidence for the crosstalk between SACE_7040 and BldD [7]. In this study, we have used gene inactivation, complementation, and transcriptional analysis to delineate the role of a new TetR-family regulator (SACE_0012) in the development differentiation of S. erythraea. Deletion of SACE_0012 principally influences the transcription of a putative aerial mycelium formation gene SACE_7115, that is homologous to amfC of Streptomyces.
Materials and Methods
Strains and Growth Conditions
Saccharopolyspora erythraea A226 and its derivatives were incubated in TSB medium at 30 °C for DNA extraction, protoplast preparation, and in liquid fermentation medium R5 for analysis of erythromycin production. R3M agar medium was used for protoplast regeneration, phenotypic observations, and RNA extraction [7]. Escherichia coli DH5α was the host for plasmid construction [8]. Bacillus subtilis PUB110 was used for an inhibition test of erythromycin production in S. erythraea.
Plasmid, DNA Isolation, and Manipulation
Plasmid pUCTSR [9] was a pUC18 derivative containing a 1.36 kb fragment of a thiostrepton resistance cassette (tsr) cloned into the BamHI/SmaI sites. The E. coli- S. erythraea integrative shuttle expression vector pZMW [4, 10] was used for the gene complementation. DNA isolation and manipulation in E. coli and S. erythraea were carried out according to the standard methods [8, 11].
Disruption of SACE_0012 in S. erythraea A226, bldD, and SACE_7040 Mutant
Two 1.5 kb fragments flanking the SACE_0012 gene were amplified from genomic DNA of S. erythraea A226 by PCR using the primer pairs P1/P2 (5′-TGC GAA TTC CTC CTC GGC CGG TGA GCA GC-3′; 5′-GAT GGT ACC ATA CGA GCG GCC CCA ACC CGA AAG CCC-3′) and P3/P4 (5′-ATT TCT AGA ACA CGC CCG CCA CCG GCT TCG C-3′; 5′-ACC AAG CTT AAG GGC TCG ATC GAC TCC TGG CGG-3′). Then, the two DNA PCR products were inserted into the EcoRI/KpnI and XbaI/HindIII sites of pUCTSR, respectively, yielding pUCTSRΔ0012. By linearized fragment homologous recombination [7], the SACE_0012 gene was replaced with the thiostrepton resistance gene in the S. erythraea A226 chromosome, and the selected mutants were verified by PCR using the primers P1/P4 (Fig. 1a, b). Similarly, SACE_0012 disruption was formed in the bldD and SACE_7040 mutant strains.
Fig. 1.
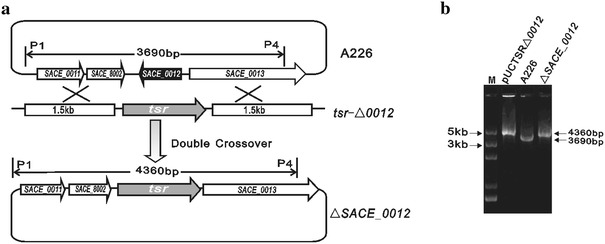
Inactivation of the TetR-family regulatory gene SACE_0012. a Schematic deletion of SACE_0012 in S. erythraea A226. b Confirmation of SACE_0012 deletion mutant by PCR analysis with the primer pair P1/P4. The size of 3.69 kb for the PCR-amplified bands was observed in wild-type A226, while a band of the size 4.36 kb was observed in mutant ΔSACE_0012, suggesting that the SACE_0012 gene was completely deleted
Genetic Complementation of the SACE_0012 Mutant
For complementation, the SACE_0012 gene was amplified by the primers P5 (5′-TAA CAT ATG TTG AAA ACG GCG TCA ATC CTC ATC CCG-3′) and P6 (5′-CGC GAT ATC TCA GCG ATC GGC GGT AGT CG-3′) from genomic DNA of S. erythraea A226, and was ligated into the NdeI/EcoRV sites of pZMW [9] to generate pZMW-0012. Then, pZMW-0012 was introduced into SACE_0012 mutant by PEG-mediated protoplast transformation, generating the complemented strain ΔSACE_0012/pZMW-0012.
Quantitative Real-Time PCR (qRT-PCR)
The transcriptional levels of eryA, bldD, SACE_0012, and homologous genes of whiA, whiB, whiG, and amfC associated with morphogenesis in Streptomyces (Table S1) [12], were determined by qRT-PCR. Specific primers were designed as listed in Table S2. Total RNA was isolated from S. erythraea A226 and the mutants of SACE_0012, bldD, and SACE_7040 after 2 or 4 days growth on R3M agar medium. Then, extracted RNA was treated with DNase I (Fermentas), and reverse transcription was accomplished using a cDNA synthesis kit (Fermentas). qRT-PCR reactions were performed on the Applied Biosystems StepOnePlus system with Maxima™ SYBR Green/ROX qPCR Master Mix (Fermentas). The hrdB gene encoding the major sigma factor in S. erythraea was used as an internal control, and relative quantification was evaluated using a comparative cycle threshold method as described by Livak and Schmittgen [13].
Fermentation and Analysis
Wild-type strain A226, ΔSACE_0012, and ΔbldD were grown in 30 ml R5 liquid medium in 250 ml baffled flasks for 6 days at 30 °C. 5 μl fermentation supernatant from these cultures was added to LB agar plates, which was sprayed with an overnight culture of B. subtilis PUB110. The plates were incubated at 37 °C for 12 h, and the erythromycin production was estimated by scoring the growth-inhibition zones. Furthermore, erythromycin A produced by these cultures were quantitatively analyzed by high performance liquid chromatograph (HPLC) as described previously [14]. Erythromycin was isolated from the fermentation culture, and quantified by a standard curve [15].
Results and Discussion
Characterization of the SACE_0012 Gene Deletion Mutant
Given a key role of the TetR-family regulator in morphological differentiation in actinomycetes [16], by gene inactivation and phenotype observation, we have identified several TetR-family regulators related to morphogenesis in S. erythraea, including the SACE_7040 gene previously reported [7] and the SACE_0012 gene currently studied. Bioinformatic analysis shows that the SACE_0012 gene has a full-length of 690 bp (GenBank Accession No. NC-009142.114,813–115,502 nt) and is a member of the TetR-family regulators that consists of 229 amino acids with a molecular mass of 25 kDa. To investigate its function, SACE_0012 was inactivated by replacing the 690 bp gene with a thiostrepton resistance cassette in S. erythraea A226 by the linearized fragment homologous recombination. A thiostrepton resistant mutant ΔSACE_0012 was formed and confirmed by PCR analysis (Fig. 1a, b).
When grown on R3M medium, the mutant ΔSACE_0012 formed aerial hypha earlier than original strain A226 in a three-day assay (Fig. 3b). When complemented with a cloned SACE_0012 under the control of the PermE* constitutive promoter (pZMW-0012), the ΔSACE_0012/pZMW-0012 strain had restored the timing of aerial mycelium on R3M agar medium (data not shown). After a longer cultivation to the sixth day, no significant phenotypic difference was observed between the wild-type strain A226, mutant ΔSACE_0012, and ΔSACE_0012/pZME-0012 (data not shown), revealing that SACE_0012 was responsible for the early aerial hypha formation of S. erythraea. Moreover, ΔSACE_0012 and A226 strains had comparable inhibition activity for B. subtilis, and produced similar amount of erythromycin A by HPLC analysis of fermentation products (Fig. S1A–B), confirming that SACE_0012 was specifically involved in the morphological differentiation of S. erythraea.
Fig. 3.
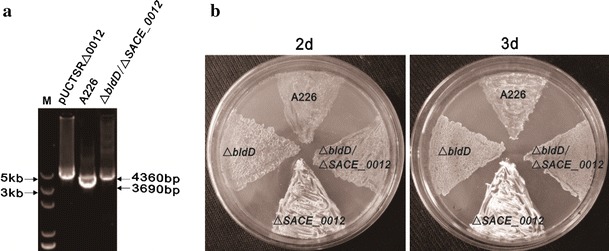
Confirmation and phenotype of the ΔbldD/ΔSACE_0012 double mutant. a Confirmation of ΔbldD/ΔSACE_0012 mutant by PCR analysis with the primer pair P1/P4. b Phenotype of the ΔbldD/ΔSACE_0012 mutant
Effect of SACE_0012 Disruption on Transcription of the Genes for Morphogenesis and Erythromycin Biosynthesis
To test the effect of SACE_0012 disruption on the expression of morphogenesis and erythromycin biosynthesis genes, we compared A226 and mutant ΔSACE_0012 for the transcriptional change to sporulation genes (whi and bldD), an aerial mycelium formation gene amfC [17], and the erythromycin structure gene eryA (Table S1). The homologous genes to whiA, whiB, whiG involved in the regulation of sporulation in Streptomyces [18] (SACE_2141, SACE_6464, SACE_6040, respectively) were examined by qRT-PCR. SACE_2141, SACE_6464, SACE_6040 transcriptions were slightly increased but not statistically different in mutant ∆SACE_0012 over strain A226. bldD and eryA were also not differentially expressed. However, the transcriptional levels of the amfC homolog SACE_7115 (Table S1), an aerial mycelium formation gene conserved presented in Streptomyces [17], were approximately 3.0-fold higher in the mutant ∆SACE_0012 (Fig. 2).
Fig. 2.
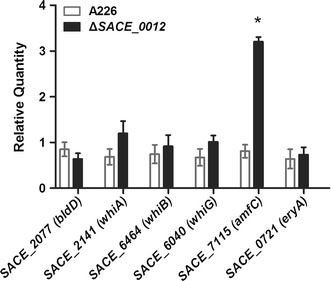
Transcriptional analysis of the genes for morphogenesis and erythromycin biosynthesis in the SACE_0012 mutant. Mean values of three independent experiments are shown, with the standard deviation indicated by error bars. Statistical significance (*P < 0.05) compared to culture of wild-type strain A226
SACE_0012 Disruption Failed to Restore the Defect in the Mycelium Formation of a bldD Mutant
bldD is required for development differentiation in S. erythraea [1]. To examine the relationship of SACE_0012 and bldD, SACE_0012 was disrupted in the bldD mutant to form a double knockout mutant strain, ΔbldD/ΔSACE_0012 (Fig. 3a). No significant phenotypic difference was observed between the mutant ΔbldD/ΔSACE_0012 and ΔbldD (Fig. 3b), indicating that SACE_0012 disruption could not restore the defect of aerial hyphae in bldD mutant. qRT-PCR analysis showed that SACE_0012 transcriptions were slightly decreased but not obviously different in mutant ∆bldD (Fig. 5). These indicate that SACE_0012, although influencing morphological differentiation, seems to have no connection with the BldD regulatory system.
Fig. 5.
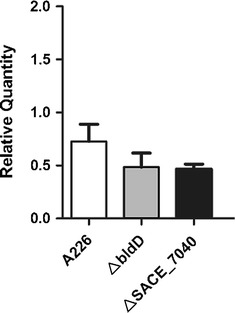
Relative transcriptional levels of SACE_0012 in the deletion mutants of bldD and SACE_7040 over A226. Mean values of three independent experiments are shown, with the standard deviation indicated by error bars
SACE_0012 Disruption Did Not Further Accelerate the Mycelium Formation of SACE_7040 Mutant
We identified a TetR-family regulator SACE_7040 negatively involving in the morphological differentiation of S. erythraea, in which an interplay with the bldD gene was previously established [7]. Therefore, we inactivated the SACE_0012 gene in the SACE_7040 mutant to study combined effect upon the morphogenesis (Fig. 4a). It appears that the mycelium formation of SACE_7040 mutant was earlier than SACE_0012 mutant, but no obvious change of aerial mycelia was detected in the double knockout mutant ΔSACE_7040/ΔSACE_0012 relative to the SACE_7040 mutant (Fig. 4b). Likewise, SACE_0012 transcriptions were slightly decreased but not statistically different in mutant ∆SACE_7040 (Fig. 5).
Fig. 4.
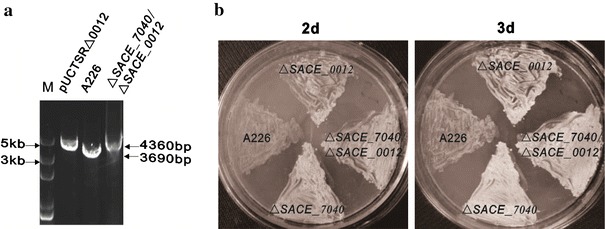
Confirmation and phenotype of the ΔSACE_7040/ΔSACE_0012 double mutant. a Confirmation of ΔSACE_7040/ΔSACE_0012 mutant by PCR analysis with the primer pair P1/P4. b Phenotype of the ΔSACE_7040/ΔSACE_0012 mutant
In conclusion, these results indicated that compared with original strain A226, aerial hypha formation initiates earlier in SACE_0012 mutant. The likely cause of an early aerial hypha formation is the higher transcriptional level of amfC. Previous genetic evidences revealed that amfC positively controlled aerial mycelium formation in Streptomyces coelicolor and Streptomyces griseus, and was distributed widely in this genus [17], implying that amfC might be under the control of SACE_0012 to affect the early aerial hypha formation of S. erythraea. In addition, we found that SACE_0012 disruption could not restore the defect of aerial development in a bldD mutant, and also could not further accelerate the mycelium formation in a mutant of SACE_7040 gene. Further qRT-PCR analysis showed that SACE_0012 transcriptions were slightly decreased but not obviously different in mutant ∆bldD and ∆SACE_7040 relative to A226. Thus, SACE_0012 was likely independent of the BldD regulatory system for controlling S. erythraea morphogenesis, distinct from the TetR-family regulator SACE_7040 previously reported [7].
With structural and sequence conserved analysis [19], homologous of SACE_0012 are predominantly distributed in rare actinomycetes, such as Amir_6428 from Actinosynnema mirum (identities 50 %), BN6_77090 from Saccharothrix espanaensis (identities 50 %), AMED_0889 from Amycolatopsis mediterranei (identities 48 %), etc. (Fig. S3). However, functional analysis of the TetR-family regulator has been never reported, signifying a new regulatory mechanism for mycelial formation in actinomycetes, such as how it works with its ligand and target [10]. Therefore, these findings provide novel insights into S. erythraea developmental biology. In the future, more detailed regulatory mechanism of the SACE_0012 gene will likely be valuable to deepening the understanding of the modulation of S. erythraea morphogenesis.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
We are grateful to Professor Yiguang Wang (Chinese Academy of Medical Sciences, Beijing, China) for providing S. erythraea A226 and Professor David T. Weaver (Anhui University, Hefei, China) for reviewing the manuscript. This study was supported by Grants from the National Basic Research Program (973) (Grant No. 2013CB734001), the National Natural Science Foundation of China (Grant Nos. 30870069) and The Natural Science Foundation of Anhui Province (Grant No. 1208085MC46).
Footnotes
Xiaojuan Yin and Xinqiang Xu contributed equally to this study.
Contributor Information
Hang Wu, Email: wuh2007@gmail.com.
Buchang Zhang, Email: zhbc@ahu.edu.cn.
References
- 1.Elliot M, Damji F, Passantino R, et al. The bldD gene of Streptomyces coelicolor A3(2): a regulatory gene involved in morphogenesis and antibiotic production. J Bacteriol. 1998;180(6):1549–1555. doi: 10.1128/jb.180.6.1549-1555.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kelemen GH, Buttner MJ. Initiation of aerial mycelium formation in Streptomyces. Curr Opin Microbiol. 1998;1:656–662. doi: 10.1016/S1369-5274(98)80111-2. [DOI] [PubMed] [Google Scholar]
- 3.Cane DE. Programming of erythromycin biosynthesis by a modular polypeptide synthase. J Biol Chem. 2010;285(36):27517–27523. doi: 10.1074/jbc.R110.144618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weber JM, Leung JO, Maine GT, et al. Organization of a cluster of erythromycin genes in Saccharopolyspora erythraea. J Bacteriol. 1990;172:2372–2383. doi: 10.1128/jb.172.5.2372-2383.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Oliynyk M, Samborskyy M, Lester JB, et al. Complete genome sequence of the erythromycin-producing bacterium Saccharopolyspora erythraea NRRL23338. Nat Biotechnol. 2007;25(4):447–453. doi: 10.1038/nbt1297. [DOI] [PubMed] [Google Scholar]
- 6.Chng C, Lum AM, Vroom JA, et al. A key developmental regulator controls the synthesis of the antibiotic erythromycin in Saccharopolyspora erythraea. PNAS. 2008;105(32):11346–33351. doi: 10.1073/pnas.0803622105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Han S, Song P, Ren T, et al. Identification of SACE_7040, a member of TetR family related to the morphological differentiation of Saccharopolyspora erythraea. Curr Microbiol. 2011;63(2):121–125. doi: 10.1007/s00284-011-9943-z. [DOI] [PubMed] [Google Scholar]
- 8.Sambrook J, Russell DW. Molecular cloning: a laboratory manual. New York: Cold Spring Harbor Laboratory; 2001. [Google Scholar]
- 9.Zhang BC, Li LL, Yu XQ, et al. Construction of Saccharopolyspora erythraea expression vector pZMW. Bull Acad Mil Med Sci. 2003;27(3):176–179. [Google Scholar]
- 10.Corre C. In search of the missing ligands for TetR family regulators. Chem Biol. 2013;20(2):140–142. doi: 10.1016/j.chembiol.2013.02.005. [DOI] [PubMed] [Google Scholar]
- 11.Bierman M, Logan R, O’Brien K, et al. Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene. 1992;116(1):43–49. doi: 10.1016/0378-1119(92)90627-2. [DOI] [PubMed] [Google Scholar]
- 12.McCormick JR, Flärdh K. Signals and regulators that govern Streptomyces development. FEMS Microbiol Rev. 2012;36(1):206–231. doi: 10.1111/j.1574-6976.2011.00317.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT. Method. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 14.Tsuji K, Goetz JF. HPLC as a rapid means of monitoring erythromycin and tetracycline fermentation processes. J Antibiot (Tokyo) 1978;31(4):302–308. doi: 10.7164/antibiotics.31.302. [DOI] [PubMed] [Google Scholar]
- 15.Wu J, Zhang Q, Deng W, et al. Toward improvement of erythromycin A production in an industrial Saccharopolyspora erythraea strain via facilitation of genetic manipulation with an artificial attB site for specific recombination. Appl Environ Microbiol. 2011;77(21):7508–7516. doi: 10.1128/AEM.06034-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ramos JL, Martinez-Bueno M, Molina-Henares AJ, et al. The TetR family of transcriptional repressors. Microbiol Mol Biol Rev. 2005;69(2):326–356. doi: 10.1128/MMBR.69.2.326-356.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yonekawa T, Ohnishi Y, Horinouchi S, et al. Involvement of amfC in physiological and morphological development in Streptomyces coelicolor A3(2) Microbiology. 1999;145:2273–2280. doi: 10.1099/00221287-145-9-2273. [DOI] [PubMed] [Google Scholar]
- 18.Flardh K, Findlay KC, Chater KF, et al. Association of early sporulation genes with suggested developmental decision points in Streptomyces coelicolor A3(2) Microbiology. 1999;145:2229–2243. doi: 10.1099/00221287-145-9-2229. [DOI] [PubMed] [Google Scholar]
- 19.Yu Z, Reichheld SE, Savchenko A, et al. A comprehensive analysis of structural and sequence conservation in the TetR family transcriptional regulators. J Mol Biol. 2010;23(4):847–864. doi: 10.1016/j.jmb.2010.05.062. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


