Abstract
Epigenetic control of gene expression is a critical component of transcriptional regulation. Remarkably, the deposition of epigenetic modifications is often guided by noncoding RNAs. Although noncoding RNAs have been most often implicated in posttranscriptional gene silencing, these molecules are now emerging as critical regulators of gene expression and genomic stability at the transcriptional level. Here, we review recent efforts to understand the mechanisms by which RNA controls the expression or content of DNA. We discuss the role of both small RNAs and long noncoding RNAs in directing chromatin changes through histone modifications and DNA methylation. Furthermore, we highlight the function of RNA in mediating DNA cleavage during genome rearrangements and pathogen defense. In understanding the mechanisms of RNA control over DNA, the power of RNA may one day be harnessed to impact gene expression in a therapeutic setting.
Introduction
Since each cell within an organism contains an identical copy of the genome, regulation of the output of the genome is responsible for determining cellular identity and allowing complex organisms to develop and function. On a cellular level, organisms face two main challenges: to maintain genome integrity in the face of mutagens and mobile genetic elements, and to express a specific repertoire of genes at the proper level and with the appropriate timing. Disruptions of either of these two processes can have catastrophic consequences, such as infertility or malignant transformation. Therefore, organisms have evolved elegant mechanisms to monitor the stability of the genome and fine-tune gene expression. In recent years, it has become increasingly evident that many of these regulatory systems rely on RNA to mediate their effects. This review will discuss the various classes of noncoding RNAs that exert control over DNA, focusing on those that maintain genomic stability or regulate DNA structure and organization through chromatin modifications or DNA cleavage.
The catalog of functional noncoding RNAs is continuously expanding, due in part to the development of next-generation sequencing technologies. Two important classes of functional RNAs responsible for mediating effects on DNA are small RNAs and long noncoding RNAs (lncRNAs). In general, small RNAs are generated from longer precursors, which can derive from both endogenous and exogenous sources, including acute viral infections and transposable elements (TEs). Following biogenesis, small RNAs are loaded into an Argonaute family member within a large effector protein complex. Two classes of Argonaute proteins exist in most animals: the ubiquitously expressed Argonaute (Ago) clade proteins, which are defined by their relationship to Arabidopsis AGO1, and members of the Piwi clade, which bear similarity to Drosophila Piwi and whose expression is largely restricted to the germline (Hutvágner and Simard, 2008). In many organisms, small RNAs are amplified to promote a more robust response; this amplification can occur through a variety of mechanisms.
The canonical role of small RNAs is to mediate posttranscriptional gene silencing (PTGS) of target RNA transcripts. During PTGS, base pairing between the small RNA, bound to its effector complex, and the target results in target cleavage or translational repression. However, seminal studies in plants and yeast, as well as more recent work in other systems, have established that small RNAs are also capable of directing transcriptional gene silencing (TGS), which can be achieved through DNA methylation or the deposition of repressive histone modifications. In these cases, the function of TGS is often to protect genomic integrity by maintaining a repressive heterochromatic state in repetitive regions of the genome, most notably those regions which harbor mobile genetic elements. Arguably the most extreme mechanism by which the content and expression of DNA can be controlled by small RNAs is DNA elimination. In some ciliates, small RNAs guide the excision of DNA elements, such as transposons, during genome rearrangements. Moreover, small RNAs in bacteria and archaea orchestrate the clustered regularly interspaced short palindromic repeat (CRISPR) pathway, which directs sequence-specific DNA cleavage of plasmids or invading phage. In the following sections, we will describe the mechanistic details of these small RNA-guided pathways and the recent advances in our understanding of their functions.
In contrast to small RNAs, the study of lncRNAs as a defined class of molecules is still in its relative infancy; indeed, the fact that the human genome is pervasively transcribed, yet that protein coding genes comprise only ~10% of its content, is a relatively recent revelation. Unlike small RNAs, there appear to be no unifying structural, biochemical, or functional characteristics that define a given transcript as a lncRNA; rather, the simplest definition of a lncRNA is merely an RNA transcript greater than 200 nucleotides in length with no coding potential (Ponting et al., 2009). Over the last 10 years, RNA-Seq data and chromatin maps from a staggering number of cell types and tissues have been used in large-scale efforts to catalog thousands of novel lncRNAs in organisms from plants to humans (Hu et al., 2012). lncRNAs are often poorly conserved at the sequence level, initially leading to uncertainty as to whether they represent active entities or transcriptional noise. However, as a growing number of lncRNAs are characterized, it has been established that orthologs can be identified in other species through synteny; although these “syntelogs” bear little to no sequence similarity, their position relative to neighboring protein coding genes has been maintained through evolution (Ulitsky et al., 2011). Nevertheless, the functions of most of the identified lncRNAs remain largely uncharacterized.
One theme that has emerged during large-scale characterization efforts is that lncRNAs are commonly involved in mediating chromatin-level gene regulation through interactions with histone modifiers, the DNA methylation machinery, or transcriptional regulators. Many other lncRNAs have been identified as key regulators of essential cellular processes but are not known to be involved in chromatin regulation and structure, and we will omit these from our discussion. However, we invite readers to refer to several excellent reviews on the subject (Ponting et al., 2009; Hu et al., 2012; Rinn and Chang, 2012). In the following sections, we highlight the functions of several diverse classes of small and large noncoding RNAs according to the mechanism by which the RNAs exert their control over DNA, specifically through histone modifications, DNA methylation, and DNA cleavage.
RNA-Directed Histone Modifications
Histones are responsible for the organization and regulation of DNA structure and are subject to a variety of modifications on their N-terminal tails, such as methylation, acetylation, and ubiquitination, that dynamically influence chromatin function. Histone modifications can function in activating or repressing gene expression; a common mark associated with active chromatin is the trimethylation of histone 3 (H3) at lysine 4 (K4), or H3K4me3, which is often found at promoters of actively transcribed genes (Black et al., 2012). Conversely, marks associated with silenced heterochromatin include di- and trimethylated H3K9, as well as trimethylation of H3K27 (Black et al., 2012). Histone methyltransferase (HMT) enzymes deposit methyl marks onto histone tails, while chromodomain-containing proteins, which directly bind to methylated lysines, often work in concert with HMTs and are involved in assembling protein complexes on DNA. Table 1 lists selected orthologs of HMTs and chromodomain-containing proteins operating within the organisms discussed in this review.
Table 1.
Chromatin Regulatory Machinery
| Organism | H3K9 Methyltransferases |
H3K27 Methyltransferases |
H3K4 Methyltransferases |
Chromodomain Proteins |
De Novo DNMTs | Maintenance DNMTs |
|---|---|---|---|---|---|---|
| S. pombe | Clr4 | Activity is present, no HMT identified | Set1 | Swi6 | No cytosine methylation | No cytosine methylation |
| Chp1 | ||||||
| Chp2 | ||||||
| Arabidopsis | KYP | CLF | ATX1 | LHP1 | DRM2 | MET1 (CG sites) |
| SUVH5 | SWN | ATX2 | CMT3 (CHG sites) | |||
| SUVH6 | MEA | ATXR7 | ||||
| Drosophila | Su(var)3–9 | E(z) | trx | HP1 | No cytosine methylation | No cytosine methylation |
| egg | ||||||
| Mammals | Suv39h1 | EZH2 | MLL | CBX7 | DNMT3A | DNMT1 |
| G9a | DNMT3B | |||||
| Tetrahymena | Ezl1p | Ezl1p | Activity is present, no HMT identified | Pdd1p | ? | ? |
| Pdd3p | ||||||
This table lists known orthologs of histone and DNA methyltransferases, as well as chromodomain-containing proteins responsible for binding methylated histones and nucleating protein complexes on chromatin.
Small RNA-Mediated Heterochromatin Formation in Yeast
A defining characteristic of eukaryotic centromeres is their high density of repetitive DNA elements, such as satellite repeats and transposons. Repressive histone modifications, particularly methylated H3K9 (H3K9me), are essential for controlling these mobile genetic elements within centromeric regions. The cores of fission yeast centromeres are flanked by tRNA genes and tandem copies of repetitive elements termed dg and dh repeats (Castel and Martienssen, 2013). Seminal work by Volpe and colleagues first established a role for small interfering (si) RNAs in directing transcriptional silencing of centromeric repeats in Schizosaccharomyces pombe. Mutants in RNA silencing factors are defective in centromeric silencing and accumulate centromere-derived sense and antisense transcripts (Volpe et al., 2002). Furthermore, loss of RNAi components leads to a loss of both H3K9me and the chromodomain-containing protein Swi6 from centromeric repeats, coupled with an increase in H3K4me3 and transcriptional activation of repeat sequences (Volpe et al., 2002).
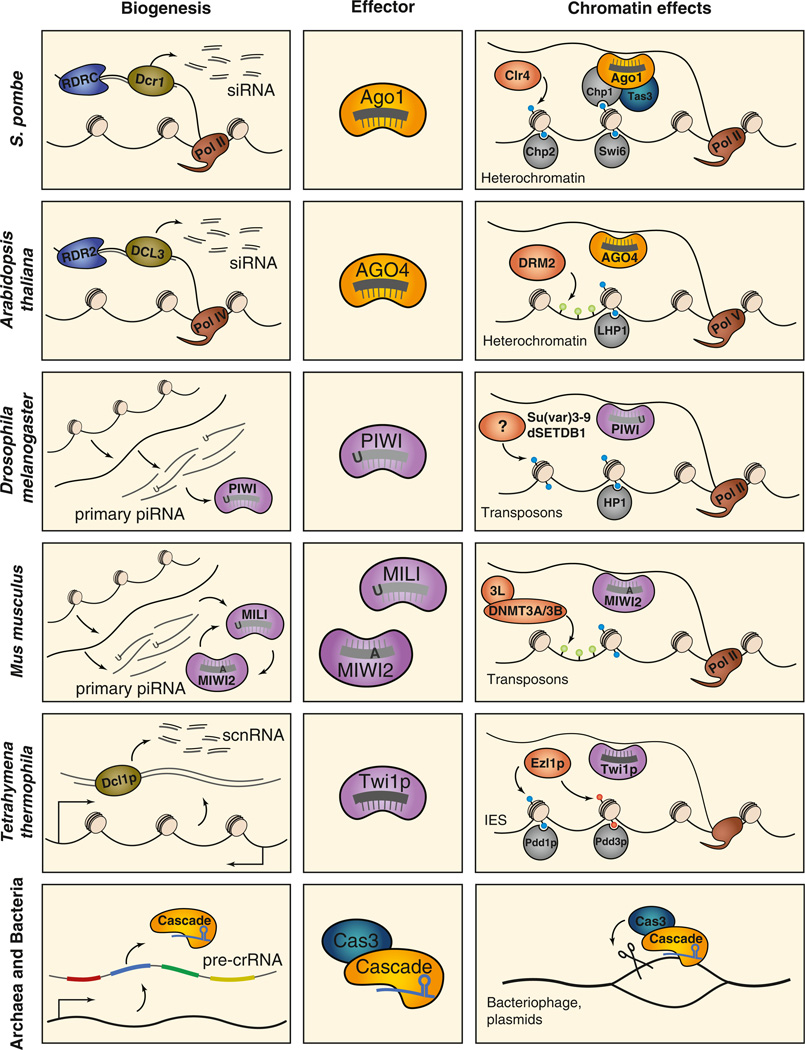
It is now well-established that centromeric transcripts are processed into siRNAs by Dcr1, which are then loaded into Ago1, the core component of the RNA-induced initiation of transcriptional silencing (RITS) complex (Verdel et al., 2004; Castel and Martienssen, 2013) (Figure 1). Other protein components of RITS include Chp1, a centromere-associated chromodomain protein that binds dimethylated H3K9, and Tas3, a GW domain-containing protein (Verdel et al., 2004).GW motifs are a common feature of Argonaute interactors; proteins containing this domain can also be found in Argonaute complexes in plants, flies, and mammals (Hutvágner and Simard, 2008). According to the current model for TGS in S. pombe, and in most other examples of small RNA-directed TGS in other species, target identification occurs through base pairing interactions between the bound siRNA and nascent transcripts at the target locus; in fission yeast, RITS associates with long, centromere-derived RNAs in a Dcr1-dependent manner (Motamedi et al., 2004). Moreover, RITS remains tethered to silent loci by the interaction of Chp1 with H3K9me, which is deposited by the HMT Clr4 (Noma et al., 2004). Clr4-dependent RITS tethering to heterochromatin is essential for maintaining transcriptional silencing and for generating new siRNAs (Noma et al., 2004).
Figure 1. Parallels in Small RNA-Mediated Chromatin Effects.
DNA silencing or cleavage is initiated by the biogenesis of small RNAs from host-encoded precursors, which are then bound by effector proteins and lead to the transcriptional silencing or elimination of complementary sequences. Orthologs are indicated as follows: Dicers, green; RNA-dependent RNA polymerase complexes, blue; polymerases, brown; Ago clade Argonautes, yellow; Piwi clade Argonautes, purple; histone and DNA methyltransferases, orange; and chromodomain-containing proteins, gray. Epigenetic modifications are represented as follows: H3K9me, blue; H3K27me, orange; and DNA methylation, green. The targets of small RNA-mediated TGS in each organism are indicated in the right panels.
RITS also physically interacts with another complex named RDRC, or the RNA-directed RNA polymerase complex, which contains Rdp1, an RNA-dependent RNA polymerase (Motamedi et al., 2004). The physical interaction between the RDRC and RITS complexes requires Dcr1 and Clr4 (Motamedi et al., 2004), suggesting that siRNA-driven RITS targeting to centromeric repeats, and stable H3K9 binding by Chp1, allows the recruitment of RDRC to chromatin. Moreover, the RDRC physically interacts with Dcr1, suggesting that siRNA biogenesis is coupled to dsRNA synthesis (Castel and Martienssen, 2013). Thus, the tethering of biogenesis, effector, and amplification complexes at the silenced loci likely organizes a self-perpetuating feedforward loop (Figure 1). The fundamental principles of siRNA-directed centromeric silencing that have been uncovered in S. pombe provide an excellent framework for understanding RNA-guided heterochromatin formation in other systems.
piRNA-Directed Transcriptional Silencing in Flies
Piwi-interacting RNAs (piRNAs) are a germline-specific class of small RNAs that bind to Piwi clade Argonaute proteins and constitute a transposon defense system in a variety of organisms, including humans. Defects in the piRNA pathway lead to uncontrolled transposition of mobile genetic elements, which can cause genomic instability and sterility (Malone and Hannon, 2009). piRNA-mediated transposon silencing has been studied most extensively in the Drosophila ovary. Although the mechanisms by which piRNAs repress transposon expression are still being elucidated, recent work in flies suggests that piRNAs induce and maintain heterochromatic silencing of these repetitive elements through the deposition of repressive histone modifications.
Drosophila piRNAs are between 23 and 29 nt in length and often bear a 5' uridine (1U), compared to endogenous siRNAs and miRNAs, which tend to be 20–23 nt and show little sequence bias (Malone and Hannon, 2009). Drosophila encodes three nonredundant Piwi clade proteins that bind piRNAs: Piwi, the founding member of the clade, Aubergine (Aub), and Argonaute 3 (Ago3) (Hutvágner and Simard, 2008). Importantly, primary piRNAs are not generated by a Dicer protein. Although the precise mechanisms are only now beginning to be understood, it is clear that piRNA biogenesis is initiated by the transcription and processing of piRNA clusters, which are unique loci comprised of inactive fragments of several classes of transposons (Brennecke et al., 2007) (Figure 1). Following primary biogenesis, piRNAs enter into the ping-pong amplification cycle, which generates secondary piRNAs and is predominantly mediated by Aub and Ago3 in Drosophila ovarian germ cells. During ping-pong, a Piwi- or Aub-bound antisense piRNA recognizes the sense strand of an active transposon, leading to transcript cleavage. This cleavage generates the 5' end of a new sense piRNA, which is bound by Ago3 and can recognize antisense transposon transcripts, generating new antisense piRNAs and thus continuing the amplification cycle (Brennecke et al., 2007; Gunawardane et al., 2007).
This elegant ping-pong amplification mechanism silences active transposons at the posttranscriptional level. How, then, does transposon repression occur in the somatic follicle cells of the ovary, where only Piwi and the primary piRNA pathway are known to operate? Several lines of evidence now point to a role for Piwi in mediating TGS in Drosophila. Piwi localizes to the nucleus, and cytoplasmic retention of Piwi results in massive transposon upregulation and sterility, which hints at a role in TGS rather than PTGS (Klenov et al., 2011). In addition, piRNA pathway mutants, including piwi, suppress position effect variegation (Malone and Hannon, 2009). Germline Piwi knockdown or cytoplasmic retention results in the depletion of H3K9me2 and HP1, the Drosophila homolog of Swi6, from a specific set of repetitive elements (Klenov et al., 2011; Wang and Elgin, 2011). In addition, nuclear run-on experiments in Piwi-depleted ovaries measured increased transcription of particular transposons as compared to wild-type tissue (Shpiz et al., 2011). Unexpectedly, nuclear run-on analysis of armitage mutants, in which Piwi does not bind piRNAs or enter the nucleus, did not detect differences in the transcription of repetitive elements between mutant and control ovaries, leading to doubts surrounding the role of Piwi in TGS (Malone and Hannon, 2009). Therefore, a direct comparison of chromatin states, nascent transcription, and steady-state transposon levels in the presence or absence of Piwi was necessary to unambiguously determine its role in TGS.
Recently, Sienski and colleagues reported such a study in an ovarian somatic follicle cell line, OSC, which does not express Aub or Ago3 and therefore allows the dissection of the Piwi-driven piRNA pathway in isolation. Following Piwi depletion, the authors found that transposon transcripts are upregulated, and Piwi-regulated transposons display enhanced Pol II occupancy and increased rates of nascent transcription (Sienski et al., 2012). By examining de novo insertions of individual TEs in the OSC genome, Sienski et al. found that active elements display Piwi-dependent H3K9me3. Moreover, two additional studies report similar effects on chromatin states and transposon suppression when Piwi is depleted in vivo (Le Thomas et al., 2013; Rozhkov et al., 2013).These data provide strong evidence that Piwi restricts transposon expression via TGS by directing the deposition of repressive histone modifications on TEs. Similar to heterochromatin formation in yeast, the recruitment of Piwi to TEs is likely mediated by base-pairing interactions between Piwi-bound piRNAs and nascent transposon transcripts, leading to the recruitment of H3K9 methyltransferases and associated chromatin binding factors to enforce a silent, heterochromatic state (Figure 1). Whether Piwi, like the yeast RITS complex, remains tethered to heterochromatic loci following H3K9 methylation has not yet been addressed.
Interactions of Long Noncoding RNAs with Chromatin Remodeling Machinery
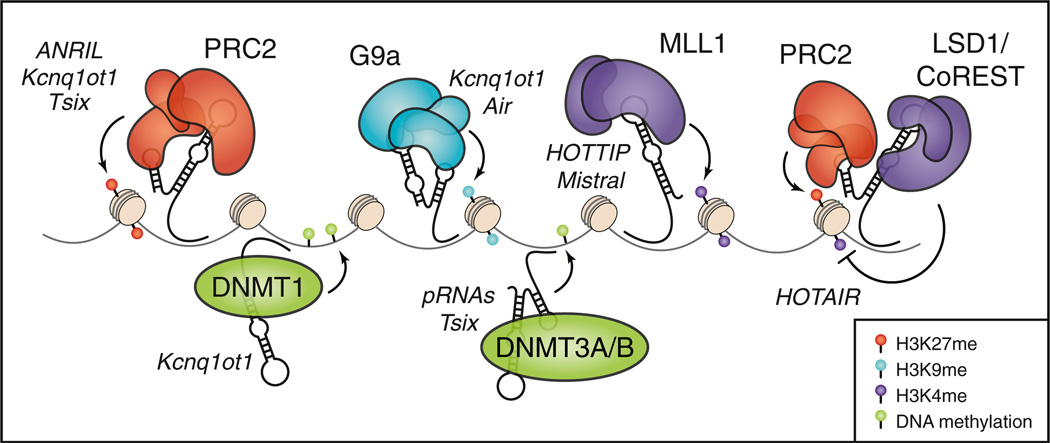
Although the diverse functions of lncRNAs are only beginning to be uncovered, their potential ability to interact with and modulate the activity of chromatin regulatory complexes may allow lncRNAs to affect gene expression on a genome-wide scale. Several of the well-characterized lncRNAs, such as Xist, are involved in X chromosome inactivation (XCI) and imprinting, which require changes in chromatin structure on a massive scale (Lee, 2011). Moreover, many recently identified lncRNAs copurify with chromatin remodeling complexes, suggesting that they may function in targeting these complexes to genomic loci, or may serve as molecular scaffolds for complex assembly (Figure 2). Whether genomic targeting of protein complexes by lncRNAs occurs in cis, through interactions with nascent lncRNA transcripts, or in trans, perhaps through RNA:DNA duplexes or triplexes, is an important facet of lncRNA biology that future work must address. In a recent study, Guttman and colleagues characterized lncRNAs bound to 12 chromatin-associated complexes in mouse embryonic stem cells (Guttman et al., 2011). Interestingly, several lncRNAs purified with more than one complex, supporting the notion that lncRNAs serve as molecular scaffolds that bridge multiple regulatory units. One of these complexes, PRC2, has emerged as a common binding partner for lncRNAs in multiple organisms (Figure 2). EZH2 is the catalytic subunit of PRC2 in mammals and methylates H3K27 to enforce repressive heterochromatin (Table 1). Studies in multiple human and mouse cell types have found that hundreds of lncRNAs interact with PRC2, although the specificity of each of these interactions has not been rigorously explored (Khalil et al., 2009; Zhao et al., 2010).
Figure 2. Effects of lncRNAs at the Chromatin Level.
lncRNAs interact with several activating and repressive chromatin-modifying complexes. Representative lncRNAs are grouped according to the protein complex with which they interact. Depending on the lncRNA, genomic targeting of these chromatin modifiers can occur in cis or in trans.
One of the most well-known PRC2-interacting lncRNAs is HOTAIR, a 2.1 kb transcript derived from the human HOXC locus that represses the expression of genes within the HOXD locus, as well as other targets throughout the genome (Rinn et al., 2007). HOTAIR binds EZH2 and is required for PRC2-mediated H3K27 trimethylation and silencing of the HOXD locus in humans (Rinn et al., 2007). However, HOTAIR may play a different role in mice, since a deletion within the HOXC locus that encompasses mHOTAIR has no effect on expression of the HOXD locus (Schorderet and Duboule, 2011). HOTAIR overexpression has been linked to increased invasiveness and poorer outcomes in several human cancers (Rinn and Chang, 2012). Interaction of HOTAIR with PRC2 is mediated through a region in its 5' terminus, while the 3' terminus binds LSD1, a H3K4 demethylase that functions within the CoREST/REST complexes (Tsai et al., 2010). Overexpression of HOTAIR results in global changes in PRC2 occupancy and H3K27me3 marks (Rinn and Chang, 2012). Conversely, knockdown of HOTAIR alters the chromatin occupancy of PRC2 and LSD1 genome-wide, leading to reduced H3K27me3 and increased H3K4me2 at target loci (Tsai et al., 2010). Thus, HOTAIR appears to serve as a scaffolding molecule, bridging PRC2 with LSD1 (Figure 2).
Another lncRNA associated with human cancers is ANRIL, a long, antisense transcript found in the INK4a/Arf locus. ANRIL is overexpressed in human leukemias and prostate cancers, and its expression leads to epigenetic silencing of the nearby tumor suppressor p15 (Yu et al., 2008; Yap et al., 2010). H3K27 trimethylation of the INK4a/Arf locus requires ANRIL, which was found to interact with SUZ12, a component of PRC2, and CBX7, a PRC1-associated chromodomain-containing protein (Rinn and Chang, 2012).
The Air and Kcnq1ot1 lncRNAs are both involved in imprinting in mammals and use similar mechanisms to induce the deposition of repressive marks at silenced alleles. Air is transcribed from the Igf2r locus and mediates H3K9 trimethylation of the nearby Slc22a3 promoter by recruitment of the repressive G9a chromatin-modifying complex in cis (Ponting et al., 2009). Similarly, Kcnq1ot1, which is transcribed antisense to the silenced paternal allele of Kcnq1, binds and recruits G9a and PRC2 to direct H3K9 and H3K27 trimethylation of the locus in cis (Pandey et al., 2008). Defects in silencing by either of these lncRNAs result in biallelic expression of their normally imprinted targets, underscoring their importance in maintaining monoallelic heterochromatic silencing.
In addition to the HOXC cluster-derived lncRNA HOTAIR, three HOXA locus-associated lncRNAs have been described. HOTTIP is expressed from the 5' end of the locus, Mistral is encoded between Hoxa6 and a7, and HOTAIRM1 is located at the distal 3' end of the cluster (Hu et al., 2012). Little is known about HOTAIRM1, apart from the fact that it is induced during myelopoiesis and is required for expression of HOXA genes during myeloid differentiation. HOTTIP and Mistral, on the other hand, mediate the transcriptional activation of HOXA genes through physical interactions with the MLL1 complex, a H3K4 methyltransferase complex which is known to bind to HOX gene promoters (Bertani et al., 2011; Wang et al., 2011). Depletion of either HOTTIP or Mistral results in a loss of MLL1 occupancy on HOXA target genes. Therefore, in the case of HOTTIP and Mistral, interaction of the lncRNA with chromatin modifiers results in the deposition of activating chromatin marks rather than repressive marks (Figure 2).
In general, the ability of lncRNAs to recruit the activities of protein complexes to genomic loci may allow the cell to impart specificity to broadly acting chromatin-modifying machineries. It is clear that much work remains to be done in order to fully understand the complex interactions between lncRNAs and chromatin-modifying complexes, particularly the acetylating or ubiquitinating complexes. However, by characterizing the functions of these molecules through dissecting their protein partners and identifying the genomic loci with which they interact, the regulatory power of lncRNAs may one day be harnessed for use in chromatin-targeted therapeutic applications.
RNA-Directed DNA Methylation
DNA methylation was the first RNA-guided epigenetic modification to be discovered, and is utilized by organisms ranging from plants to mammals. In most systems, methylation occurs on cytosine residues in the context of a CG dinucleotide, or CpG motif, by the action of DNA methyltransferase (DNMT) proteins. DNA methylation is a dynamic epigenetic modification that often functions as a repressive mark to silence transcription (Law and Jacobsen, 2010). Two types of DNMT proteins perform DNA methylation, and differ in their preferred substrates: de novo DNMTs methylate completely unmethylated CpG dinucleotides, while maintenance methyltransferases act on hemimethylated DNA during DNA replication to methylate the newly replicated strand (Law and Jacobsen, 2010). Table 1 lists the DNMTs present in species relevant to this review. Notably, many species have lost the ability to methylate their DNA; these include S. pombe and D. melanogaster.
Small RNA-Directed DNA Methylation in Plants
RNA-directed DNA methylation (RdDM) was first described in plants, following the observation that viroid cDNAs become specifically methylated upon integration into the tobacco genome (Law and Jacobsen, 2010). Since its initial discovery, the pathway has been characterized extensively in Arabidopsis and now serves as the defining model for RdDM in eukaryotes. Genome-wide mapping of methylation in Arabidopsis has revealed that most methylation occurs on transposons and repetitive elements, which are concentrated near centromeres (Castel and Martienssen, 2013). RNA Pol IV, a specialized polymerase essential for RdDM, produces long RNA transcripts from these heterochromatic regions (Henderson and Jacobsen, 2007). Pol IV transcripts are substrates for RDR2, an RNA-dependent RNA polymerase, which produces the complementary strand to generate dsRNA (Law and Jacobsen, 2010). The RNase III enzyme DCL3 processes the dsRNA from repetitive regions into 24 nt heterochromatic siRNAs, which are bound by the Argonaute family member AGO4 (Henderson and Jacobsen, 2007). AGO4 then targets the de novo methyltransferase DRM2 to the corresponding genomic locus for methylation and silencing (Henderson and Jacobsen, 2007) (Figure 1).
Further molecular details of this process were uncovered following the identification of a second specialized polymerase complex in Arabidopsis: RNA polymerase V. Wierzbicki and colleagues found that Pol V transcribes intergenic noncoding transcripts within heterochromatin, which serve as scaffolds for AGO4 recruitment (Wierzbicki et al., 2008). AGO4 not only binds to nascent Pol V transcripts through siRNA-mediated base pairing but also interacts directly with the largest Pol V subunit, NRPE1 (Law and Jacobsen, 2010). In a sense, the mechanism guiding AGO4 to repetitive loci parallels the recruitment of S. pombe RITS or Drosophila Piwi to their targets: each of these processes is driven by an interaction with nascent transcripts at the locus.
The mechanism by which the methyltransferase DRM2 is recruited to AGO4-targeted regions has not been fully elucidated, although several recent studies hint that RDM1, a novel regulator of RdDM, may play a role in the process (Castel and Martienssen, 2013). RDM1 specifically binds to single-stranded methylated DNA and is required for the accumulation of Pol V transcripts. Astonishingly, RDM1 interacts not only with Pol V subunits but also with AGO4 and DRM2 (Castel and Martienssen, 2013). Therefore, RDM1 may recruit Pol V to heterochromatic loci, while mediating subsequent interactions between components of the RNA silencing pathway and methylation machinery at the locus. However, the sequence of these events and the molecular mechanisms that drive them have not yet been established.
piRNA-Mediated Methylation of Mobile Genetic Elements in Mammals
Although many of the fundamental principles of piRNA pathway function have been characterized in Drosophila, including the recent discovery of piRNA-guided histone modifications, the role of piRNAs in TGS was first described in mammals, where piRNAs direct the methylation of transposons in order to enforce their transcriptional repression. Mammalian piRNA pathways operate in male germ cells, and the expression of piRNAs occurs at two distinct stages during development. Pre-pachytene piRNAs, and their interacting Piwi proteins, Mili and Miwi2, are expressed in primordial germ cells and map to transposons and repetitive elements (Malone and Hannon, 2009). Conversely, pachytene piRNAs are expressed during adult spermatogenesis, and although some pachytene piRNAs align to repetitive genomic regions, the majority map uniquely throughout the mouse genome, and their function remains unknown (Aravin et al., 2007a). The mouse Piwi proteins expressed at this stage are Miwi and, to a lesser extent, Mili, which do not appear to direct transposon repression via TGS or to engage in ping-pong amplification (Malone and Hannon, 2009).
During mammalian embryonic germ cell development, the genome undergoes a process of global demethylation, followed by the reestablishment of methylation at repetitive elements. Prior to the identification of piRNAs, it was known that transposon silencing in primordial germ cells was dependent upon methylation of repetitive elements by de novo methyltransferases DNMT3A and 3B (Okano et al., 1999). DNMT3L, which is related to DNMT3A and DNMT3B in sequence but lacks their catalytic motifs, stimulates the activity of DNMT3A and DNMT3B and specifically regulates transposon silencing in the germline; DNMT3L is required for the re-establishment of DNA methylation on repetitive elements, and its mutation results in transposon upregulation and sterility (Malone and Hannon, 2009). Due to the overlapping phenotypes between mili, miwi2, and dnmt3l mutants, investigators asked whether piRNAs were involved in the process of de novo methylation of transposons in developing germ cells. Indeed, transposon methylation is lost in mili and miwi2 mutants, inducing a massive upregulation of TEs, which eventually leads to sterility (Carmell et al., 2007; Aravin et al., 2007b; Kuramochi-Miyagawa et al., 2008). DNMT3L acts downstream of Mili and Miwi2, since dnmt3l mutants display only minor defects in piRNA populations (Malone and Hannon, 2009). Therefore, transposon suppression in primordial germ cells depends on TGS, which is achieved through methylation of repetitive elements via Mili- and Miwi2-bound piRNAs and the action of de novo DNMTs (Figure 1).
Although a functional link between the piRNA pathway and transposon methylation is clear, the molecular mechanisms by which the mammalian DNA methylation machinery is recruited to target loci remain unknown. Given the well-characterized pathways governing small RNA-mediated TGS in other organisms, it is likely that Mili and Miwi2 complexes are recruited to repetitive elements through piRNA-guided base-pairing interactions with nascent TE transcripts. Once tethered to the locus, the complexes may recruit the methylation machinery directly. Alternatively, it is also possible that histone methylation serves as a signal for DNA methylation. Indeed, H3K9 methylation is a prerequisite for DNA methylation in Neurospora crassa and guides maintenance DNA methylation in Arabidopsis (Malone and Hannon, 2009; Law and Jacobsen, 2010). Mammalian DNMT proteins have been reported to interact with EZH2, as well as H3K9 methyltransferases SUV39H1, G9a, and SETDB1 (Jin et al., 2011) (Table 1). Thus, methylation of histones and DNA may synergistically enforce and perpetuate a repressive, heterochromatic state at target loci. Investigation of potential interactions between the piRNA machinery and histone-modifying complexes will be an important next step in understanding piRNA-directed transcriptional repression in mammalian systems.
DNA Methylation Mediated by Long Noncoding RNAs
Given the complex interplay between DNA methylation and histone modifications, combined with rapidly mounting evidence supporting lncRNA-mediated effects on chromatin structure, it is not surprising that several examples of lncRNA-guided DNA methylation have been reported in the literature (Figure 2). However, the interaction of lncRNAs with the DNA methylation machinery has not been investigated on a global scale.
Tsix is a lncRNA that regulates the expression of Xist during XCI. In addition to binding and recruiting PRC2 to the Xist locus (Lee, 2011), Tsix physically interacts with the de novo methyltransferase DNMT3A and recruits the protein to the Xist promoter in order to induce DNA methylation and silencing (Sun et al., 2006). Similarly, recent studies have uncoupled the role of Kcnq1ot1 in tissue-specific imprinting from its role in regulating ubiquitously imprinted genes. A 890 nt domain within Kcnq1ot1 regulates differential methylation of somatically im-printed genes through a physical interaction with the maintenance methyltransferase DNMT1 (Mohammad et al., 2010). Loss of this domain does not abolish interaction of Kcnq1ot1 with G9a or EZH2, suggesting that the lncRNA utilizes a multifaceted approach to regulate its targets through interactions with both chromatin modifiers and DNA methylation machinery.
Noncoding RNAs are also involved in the silencing of clustered ribosomal RNA genes (rDNA) within mammalian genomes. rDNA is transcriptionally silenced through repressive histone modifications and DNA methylation of the promoter region, which is mediated by NoRC, a chromatin remodeling complex (Schmitz et al., 2010). RNA Pol I-derived transcripts from rDNA promoters, termed pRNAs, play an essential role in this process by binding to NoRC. Interestingly, pRNAs were recently found to mediate NoRC-independent de novo methylation and heterochromatic silencing of rDNA promoters. Through a series of elegant structural and functional studies, Schmitz and colleagues demonstrate that pRNAs bind to an element within the rDNA promoter termed T0. In fact, pRNAs form an RNA:DNA: DNA triplex structure with T0 in vitro (Schmitz et al., 2010). Moreover, DNMT3B, and to a lesser extent DNMT3A, preferentially binds RNA:DNA:DNA triplexes rather than DNA duplexes in vitro (Schmitz et al., 2010). Although the presence of this triplex has not been validated in vivo, the current model suggests that the triplex formed between pRNAs and T0 directly recruits DNMT3B to the rDNA promoter to induce DNA methylation and silencing.
This study has important implications for the study of lncRNA-induced chromatin changes; not only does it suggest that lncRNAs may be capable of forming triplex structures with DNA in vivo, but it also demonstrates that cellular proteins, such as DNMTs, are able to specifically recognize and bind to RNA:DNA:DNA triplex structures. The development of predictive tools such as the Triplexator (Buske et al., 2012), to predict triplex structures genome-wide, will hopefully aid in determining whether lncRNAs commonly utilize triplexes when locating target loci. The recent generation of genome-wide DNA methylation maps for a diverse number of organisms and cell types supports the notion that differentially methylated regions may be important in orchestrating gene expression across tissues. Considering how little we currently understand about the role of lncRNAs in directing DNA methylation genome-wide, it will be of interest to investigate the extent to which lncRNAs control differential methylation in specific cell types, and whether depletion or ectopic expression of lncRNAs is capable of modulating these patterns.
RNA-Directed DNA Cleavage
Yeast, plants, and animals have evolved elegant small RNA-directed strategies to suppress the potentially catastrophic activity of mobile genetic elements harbored within their own genomes, as discussed above. Perhaps not surprisingly, small RNA-driven pathways have also evolved in bacteria, archaea, and protists, and share many features with their analogous counterparts in other systems. However, as we will discuss in the following sections, RNA-directed targeting by these ancient silencing pathways leads to DNA cleavage rather than transcriptional repression. Accordingly, small RNA-driven silencing must be carefully regulated, since DNA cleavage and elimination, unlike chromatin silencing, is an irreversible process.
DNA Elimination in Ciliates
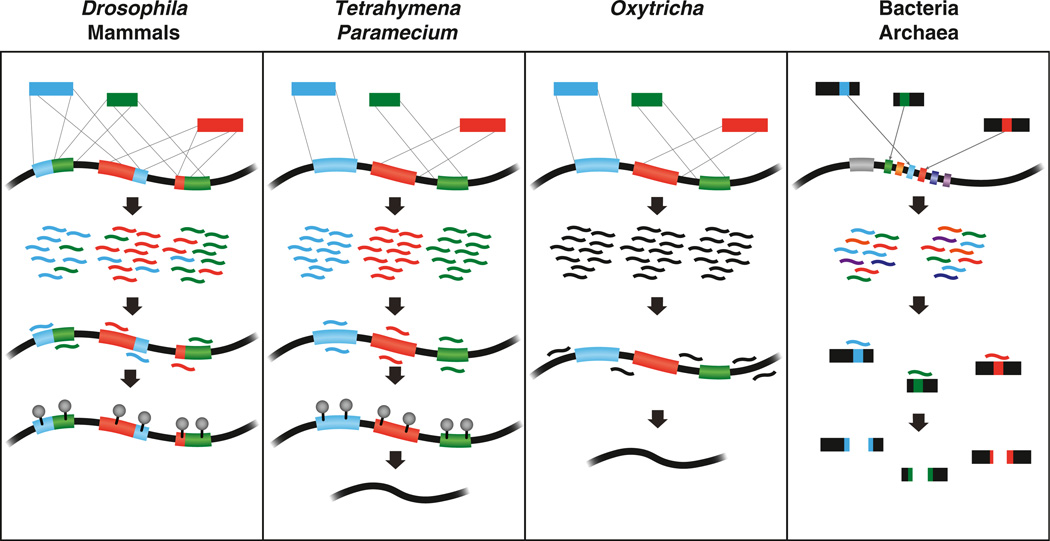
Ciliated protozoans have historically served as fascinating models for the study of noncoding RNAs and programmed DNA rearrangements. Ciliates compartmentalize their DNA into two distinct nuclei: the germline micronucleus (MIC) and the somatic macronucleus (MAC). The diploid MIC stores all of the genetic information necessary for reproduction and development and is transcriptionally silent during vegetative growth. The MAC, on the other hand, contains a polyploid, stripped-down copy of the MIC genome that functions as a transcriptional machine to produce the cellular factors critical for vegetative growth. Within the MAC, much of the intergenic or repetitive content that is present in the MIC has been eliminated, and split genes have been reassembled. During the sexual process of conjugation, a new MIC and MAC are generated from the zygotic nucleus. As the details of conjugation are complex and lie beyond the scope of this review, we refer the reader to two excellent reviews (Matzke and Birchler, 2005; Nowacki et al., 2011). As the new MAC differentiates, it undergoes an extensive series of gene rearrangements that result in elimination of a substantial portion of the parental genome. The eliminated sequences are termed internal eliminated segment (IES) sequences, many of which are believed to be remnants of inactivated transposons (Nowacki et al., 2011). Different ciliate species undergo varying degrees of DNA elimination; Tetrahymena thermophila discards ~6,000 unique sequences, or ~15% of its genome, while Paramecium tetraurelia expels an estimated 60,000 IES regions. Finally, Oxytricha trifallax discards an astounding 95%–98% of its genome during MAC differentiation while simultaneously unscrambling the retained segments in order to assemble full-length, properly ordered genes from discontinuous fragments (Nowacki et al., 2011).
How can such a process occur in a controlled, programmable manner? The first molecular insight into this question was the observation that IES retention or elimination was a homology-dependent phenomenon that was maternally transmitted in both Tetrahymena and Paramecium (Duharcourt et al., 1995; Chalker and Yao, 1996). That is, sequences present in the parental MAC were retained in the offspring, while elements that were absent in the parent were eliminated from the new MAC. In short, retention of a given sequence in the new MAC is dictated by its presence in the parental MAC.
We now know that the process of DNA elimination in Tetrahymena and Paramecium is orchestrated by small RNAs, termed scanRNAs (scnRNAs), which are expressed during conjugation and direct chromatin modifications to mark IES sequences for elimination. scnRNAs are generated from long, overlapping MIC transcripts and scan the parental MAC for homologous sequences. If a scnRNA identifies a complementary sequence in the MAC, it is degraded, while unpaired scnRNAs are transported into the developing MAC to mark homologous sequences for elimination (Mochizuki et al., 2002) (Figure 3). Interestingly, scnRNA selection occurs through the scanning of long RNA transcripts derived from the parental MAC rather than through scanning the DNA itself (Mochizuki, 2010).
Figure 3. Small RNA-Mediated Genome Defense.
Foreign transposable and repetitive sequences, represented by blue, green, and red segments, are present in the genomes of mammals, insects, and ciliates. Mammals and Drosophila utilize piRNAs to silence active transposons, either by DNA methylation or by repressive histone modifications (gray circles). In Tetrahymena and Paramecium, scnRNAs recognize IES regions, which include mobile genetic elements, and guide their genomic excision by marking them with repressive histone modifications. Oxytricha eliminates IES sequences using an orthogonal mechanism; piRNAs correspond to the retained sequences (black). Chromatin modifications that guide this process have not yet been identified. Bacteria and archaea incorporate sequences from foreign pathogens or plasmids (colored segments) into CRISPR loci. Expression of crRNAs from these loci results in cleavage of infecting bacteriophages and plasmid DNA.
Genetic and biochemical characterization of the protein factors that drive scnRNA-mediated DNA elimination in Tetrahymena and Paramecium have revealed remarkable parallels with cognate RNA silencing pathways in other systems (Figure 1). For simplicity, we will now focus on the mechanisms known to operate within Tetrahymena, although an analogous pathway also exists in Paramecium. Similar to siRNA biogenesis in yeast, plants, and higher organisms, the production of scnRNAs requires the activity of a Dicer protein, Dcl1p, which generates ~27–30 nt products (Malone et al., 2005; Mochizuki and Gorovsky, 2005). Following biogenesis, mature scnRNAs bind Twi1p, a Piwi clade Argonaute protein (Mochizuki et al., 2002), and therefore resemble mammalian and Drosophila piRNAs.
Similar to transposon silencing in S. pombe and Drosophila, repressive histone modifications are a critical component of DNA rearrangements in ciliates. Both H3K9me and H3K27me are required for proper genome rearrangements and mark IES sequences for elimination (Mochizuki, 2010). The HMT Ezl1p, which is related to Drosophila E(z) and EZH2 in mammals, is responsible for methylating both H3K9 and H3K27 in Tetrahymena, and is required for DNA elimination (Liu et al., 2004; 2007). Importantly, Dcl1p and Twi1p are also required for the methylation of H3K9 and H3K27 (Mochizuki, 2010). These chromatin marks are bound by the chromodomain-containing proteins Pdd1p and Pdd3p, which are required for proper genome rearrangement (Mochizuki, 2010). Tethering Pdd1 to an inactive IES is sufficient to induce its excision (Taverna et al., 2002), providing strong evidence that repressive histone modifications serve as a signal to promote DNA elimination. Taken together, these discoveries suggest a model for DNA elimination whereby selected scnRNAs, which are generated by Dcl1p and bound by Twi1p, enter the nucleus of the developing MAC, and locate homologous sequences within IES regions through interactions with nascent transcripts. Tethered Twi1p then recruits Ezl1p to deposit repressive methyl marks on the proximal histones, which are bound by Pdd1p and Pdd3p and trigger elimination by the DNA excision machinery (Figure 1).
The process of DNA elimination in Oxytricha resembles that of Paramecium and Tetrahymena in a number of ways, but one stark contrast between them has been the lack of evidence supporting a role for small RNAs in Oxytricha conjugation. However, recent work by Fang and colleagues has uncovered a surprising twist: Oxytricha expresses small RNAs during conjugation, but their function is orthogonal to scnRNAs; Oxytricha small RNAs mark complementary regions for retention, rather than elimination (Fang et al., 2012) (Figure 3). These conjugation-specific small RNAs, or piRNAs, are 27 nt in length and display a 1U bias, similar to piRNAs in other organisms (Fang et al., 2012; Zahler et al., 2012). The Piwi protein Otiwi1 binds Oxytricha piRNAs and is critical for the development of the new MAC. Accordingly, injection of a synthetic piRNA complementary to an eliminated IES programs its retention in the offspring (Fang et al., 2012). To unscramble split genes, Oxytricha utilizes a mechanism similar to scnRNA selection, in which transcription of the parental MAC produces long RNA transcripts that are used as templates to guide proper segment orientation for the reassembly of full-length genes in the developing MAC (Nowacki et al., 2011).
These insights into DNA elimination in Oxytricha resolve many of the uncertainties that have plagued the community for years, but they also raise new questions. For instance, are piRNAs generated from single- or double-stranded precursors, and which enzyme(s) is responsible for their biogenesis? Moreover, what are the signals deposited on macronuclear-destined sequences that mark them for retention? Intriguingly, conjugation triggers the methylation of Oxytricha DNA (Bracht et al., 2012). Cytosine methylation is enriched within the repetitive sequences that are eliminated during MAC development, suggesting that DNA methylation may be involved in marking specific elements for degradation. Further investigation of DNA methylation and other epigenetic marks present on retained and eliminated sequences during Oxytricha conjugation will be a critical next step.
CRISPR-Mediated Defense against Foreign DNA in Prokaryotes
Prokaryotic organisms face a constant barrage of viral pathogens; bacteriophages are the most abundant viruses on Earth and have the ability to rapidly evolve and adapt to their environments, creating the need for an antiviral defense strategy that is equally flexible and adaptable. Bacteria and archaea have responded by employing a small RNA-driven pathway that not only protects against invading phage but also retains a molecular memory of the pathogen through the incorporation of small portions of the foreign DNA into host loci termed CRISPRs. CRISPR loci can be found in~40% of bacterial species, and in most archaea (Marraffini and Sontheimer, 2010). A CRISPR locus is a tandem array of short direct-repeat sequences, which are separated by unique spacer regions. On average, there are 20 repeat-spacer units in a locus. Repeat lengths range between 21 and 47 nt, while spacer lengths can be 20–72 nt (Karginov and Hannon, 2010). CRISPR-dependent immunity is mediated by small RNAs termed CRISPR RNAs (crRNAs) and is achieved in three phases: adaptation, expression, and interference (Makarova et al., 2011). CRISPR loci are flanked by a diverse array of cas genes, which are responsible for mediating these three phases of immunity.
During the process of adaptation, short pieces of phage and plasmid sequences, termed protospacers, are identified and incorporated into the CRISPR loci (Barrangou et al., 2007). Although the molecular mechanism by which novel spacer sequences are integrated into CRISPR loci remains unclear, in most systems, their selection relies on the presence of a protospacer-adjacent motif (PAM). PAMs are short sequences encoded within the phage or plasmid genome that lie adjacent to the region destined for integration into a CRISPR locus (Makarova et al., 2011). These sequences likely serve as recognition motifs for the as-yet-unidentified protospacer integration machinery, and aid in target recognition and cleavage during the interference phase of immunity. The highly conserved cas proteins Cas1 and Cas2 are suspected tobeinvolvedin protospacer acquisition (Barrangou et al., 2007; Brouns et al., 2008), but no direct evidence linking them to this process has been reported.
The expression phase of CRISPR-mediated antiviral defense involves the transcription of CRISPR loci to yield long precursor RNAs, which are then processed by specialized Cas proteins into mature crRNAs (Marraffini and Sontheimer, 2010). Although the mechanism of crRNA biogenesis varies between different types of CRISPR systems, the expression phase ultimately results in the generation of small RNAs bearing phage- or plasmid-derived spacer sequences, which are then funneled into the interference phase of the pathway (Figure 1 and Figure 3).
CRISPR-mediated interference is responsible for targeting and cleaving the DNA of invading phage or plasmids through crRNA-mediated base-pairing interactions, reminiscent of miRNA-mediated seed pairing in other systems. In order to mediate pathogen interference, crRNAs recruit effector complexes to complementary sequences within the foreign DNA, which are then cleaved within the spacer sequence (Wiedenheft et al., 2012) (Figure 1 and Figure 3).
Three types of CRISPR loci have been characterized to date, and more than one type of locus can be found within a single organism (Makarova et al., 2011). Although the general phases of the pathway remain the same, the cas proteins responsible for carrying out these phases differ between CRISPR types. The hallmark of type I CRISPR-Cas systems is the presence of a cas3 gene, along with a large, multisubunit effector complex known as the CRISPR-associated complex for antiviral defense, or Cascade (Brouns et al., 2008; Makarova et al., 2011). Cascade processes long, pre-crRNA transcripts into mature crRNAs through endonucleolytic cleavage (Brouns et al., 2008) (Figure 1). Next, the complex recognizes and binds a PAM motif within the target dsDNA, and recruits Cas3, which catalyzes target cleavage (Marraffini and Sontheimer, 2010).
Type II CRISPR-Cas systems, such as those found in S. pyogenes, rely heavily on a large cas protein, Cas9, and a trans-activating CRISPR RNA (tracrRNA) (Wiedenheft et al., 2012). The tracrRNA bears 24 nt of perfect sequence complementarity to the repeat regions within the CRISPR locus, and base pairs with crRNA precursor RNAs. This duplex is bound by Cas9 and cleaved by a host-encoded RNase III protein to generate mature crRNAs (Deltcheva et al., 2011). Apart from its role in crRNA biogenesis, Cas9 was recently implicated in crRNA-guided DNA cleavage. Cas9, in complex with a crRNA and tracrRNA, identifies the dsDNA target and catalyzes two ssDNA cleavage events within the target. Most importantly, Jinek and colleagues demonstrate that Cas9 can be programmed for dsDNA cleavage using a single hybrid crRNA-tracrRNA molecule, which combines the essential features from both RNAs and can, in theory, be engineered to cleave any DNA of interest (Jinek et al., 2012). Indeed, several recent studies have demonstrated that programmed Cas9 can mediate specific DNA cleavage in human cells, providing tangible evidence that this technology mayone day be used clinically for genome editing or gene therapy (Cong et al., 2013; Mali et al., 2013).
The final type of CRISPR-Cas system is the type III pathway. Present in organisms such as P. furiosis, crRNA biogenesis is carried out by Cas6, which then transfers the small RNA to an effector complex to mediate target recognition and cleavage (Makarova et al., 2011). While some type III systems have been shown to target DNA, others appear to cleave complementary RNAs (Hale et al., 2009). By targeting RNA, these CRISPR-Cas systems bear an even closer resemblance to eukaryotic RNA-silencing pathways and may potentially be functioning in the degradation of viral transcripts.
It is clear that bacteria, archaea, and viral pathogens are engaged in a dynamic interplay that has shaped their evolutionary history. In order to gain an upper hand, bacteria and archaea have developed an intricate repertoire of small RNA-driven effector mechanisms based upon long-term immunological memory of previous viral challenge. Moreover, it seems that viruses have responded to the restrictive pressures placed upon them by CRISPR-Cas systems by evolving protein-based suppressors that inactivate CRISPR-mediated defenses during infection (Bondy-Denomy et al., 2013). Bacteriophage also engage in rapid sequence evolution by shuffling their genomes in 25 nt blocks (Andersson and Banfield, 2008). Given the size of the shuffled units, which approximate the size of many protospacers, it is tempting to speculate that the process is in some way linked to spacer acquisition in some CRISPR-Cas systems.
Molecular Connections between RNA and Chromatin: Future Perspectives
As connections between noncoding RNAs and chromatin-level regulatory mechanisms are uncovered, a number of striking similarities between diverse species have emerged. Although the RNAs themselves differ between organisms and cellular pathways, they converge on a common challenge: to regulate the content and expression of DNA. Both small and large non-coding RNAs are able to direct chromatin-modifying machinery to specific targets, often through base pairing with nascent transcripts at the locus. In many cases, interaction with genomic targets leads to the deposition of covalent modifications, or in the most extreme cases, DNA cleavage. Interactions between noncoding RNAs and chromatin-modifying machinery are key components of these regulatory systems; lncRNAs in particular can interact directly with both HMTs and DNMTs, which implicates this novel class of molecules in physically guiding regulatory protein complexes to genomic targets (Figure 2). Small RNAs, on the other hand, often associate with effector protein complexes, which then recruit chromatin modifiers to targets based on small RNA-mediated base pairing with nascent transcripts (Figure 1). In many small RNA-driven silencing pathways, the regulatory RNAs identify and mark potentially dangerous “nonself” elements for transcriptional silencing or elimination. However, in other systems, homology between the regulatory RNA and the target locus marks the region as “self” and protects it from elimination (Figure 3). Nevertheless, these complementary mechanisms both rely on signals directed by host-encoded small RNAs to maintain genomic stability.
Although many of theRNA-mediated pathways that we describe are responsible for preventing genetic lesions caused by activated transposons, another essential facet of genome stability is the ability to repair breaks inDNAonce they arise. Perhaps not surprisingly, noncoding RNAs were recently implicated in double-strand break repair in plants and animals (Francia et al., 2012; Wei et al., 2012). Although the mechanistic details of this process have not been fully characterized, particularly in mammals, these findings highlight the fact that we likely have only scratched the surface of the myriad ways in which RNA can influence DNA. While non-coding RNAs clearly mediate the methylation of histones and DNA, the role of RNA in directing other covalent marks, such as acetylation or ubiquitination, remains largely unexplored. As we expand our understanding of chromatin structure and modifications, we will undoubtedly continue to uncover exciting new mechanisms of epigenetic regulation by noncoding RNAs.
ACKNOWLEDGMENTS
Given the broad scope of this review and the limitations in space, we apologize to our colleagues whose work we were unable to describe or reference. L.R.S is a Damon Runyon Fellow supported by the Damon Runyon Cancer Research Foundation (DRG-2016-12). M.J.D. is supported by a graduate studies fellow-ship from “la Caixa” Foundation and by the Watson School of Biological Sciences. G.J.H. is supported by grants from the NIH and by a kind gift from Kathryn W. Davis. G.J.H. is an investigator of the HHMI.
Footnotes
Note Added in Proof
Since the submission of this manuscript, several additional papers have demonstrated the use of Cas9 as a programmable nuclease for genome editing in multiple organisms. These include the following:
Jinek, M., East, A., Cheng, A., Lin, S., Ma, E., and Doudna, J. (2013). RNA-programmed genome editing in human cells. Elife 2, e00471. http://dx.doi.org/10.7554/eLife.00471.
Cho, S.W., Kim, S., Kim, J.M., and Kim, J.-S. (2013). Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat. Biotechnol. Published online January 29, 2013. http://dx.doi.org/10.1038/nbt.2507.
Hwang, W.Y., Fu, Y., Reyon, D., Maeder, M.L., Tsai, S.Q., Sander, J.D., Peterson, R.T., Yeh, J.-R.J., and Joung, J.K. (2013). Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat. Biotechnol. Published online January 29, 2013. http://dx.doi.org/10.1038/nbt.2501.
Jiang, W., Bikard, D., Cox, D., Zhang, F., and Marraffini, L.A. (2013). RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat. Biotechnol. Published online January 29, 2013. http://dx.doi.org/10.1038/nbt.2508.
REFERENCES
- Andersson AF, Banfield JF. Virus population dynamics and acquired virus resistance in natural microbial communities. Science. 2008;320:1047–1050. doi: 10.1126/science.1157358. [DOI] [PubMed] [Google Scholar]
- Aravin AA, Hannon GJ, Brennecke J. The Piwi-piRNA pathway provides an adaptive defense in the transposon arms race. Science. 2007a;318:761–764. doi: 10.1126/science.1146484. [DOI] [PubMed] [Google Scholar]
- Aravin AA, Sachidanandam R, Girard A, Fejes-Toth K, Hannon GJ. Developmentally regulated piRNA clusters implicate MILI in transposon control. Science. 2007b;316:744–747. doi: 10.1126/science.1142612. [DOI] [PubMed] [Google Scholar]
- Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- Bertani S, Sauer S, Bolotin E, Sauer F. The noncoding RNA Mistral activates Hoxa6 and Hoxa7 expression and stem cell differentiation by recruiting MLL1 to chromatin. Mol Cell. 2011;43:1040–1046. doi: 10.1016/j.molcel.2011.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Black JC, Van Rechem C, Whetstine JR. Histone lysine methylation dynamics: establishment, regulation, and biological impact. Mol Cell. 2012;48:491–507. doi: 10.1016/j.molcel.2012.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bondy-Denomy J, Pawluk A, Maxwell KL, Davidson AR. Bacteriophage genes that inactivate the CRISPR/Cas bacterial immune system. Nature. 2013;493:429–432. doi: 10.1038/nature11723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracht JR, Perlman DH, Landweber LF. Cytosine methylation and hydroxymethylation mark DNA for elimination in Oxytricha trifallax. Genome Biol. 2012;13:R99. doi: 10.1186/gb-2012-13-10-r99. http://dx.doi.org/10.1186/gb-2012-13-10-r99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon GJ. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128:1089–1103. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- Brouns SJJ, Jore MM, Lundgren M, Westra ER, Slijkhuis RJH, Snijders APL, Dickman MJ, Makarova KS, Koonin EV, van der Oost J. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008;321:960–964. doi: 10.1126/science.1159689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buske FA, Bauer DC, Mattick JS, Bailey TL. Triplexator: detecting nucleic acid triple helices in genomic and transcriptomic data. Genome Res. 2012;22:1372–1381. doi: 10.1101/gr.130237.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmell MA, Girard A, vandeKant HJG, Bourc’his D, Bestor TH, deRooij DG, Hannon GJ. MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Dev Cell. 2007;12:503–514. doi: 10.1016/j.devcel.2007.03.001. [DOI] [PubMed] [Google Scholar]
- Castel SE, Martienssen RA. RNA interference in the nucleus: roles for small RNAs in transcription, epigenetics and beyond. Nat. Rev. Genet. 2013;14:100–112. doi: 10.1038/nrg3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalker DL, Yao MC. Non-Mendelian, heritable blocks to DNA rearrangement are induced by loading the somatic nucleus of Tetrahymena thermophila with germ line-limited DNA. Mol. Cell. Biol. 1996;16:3658–3667. doi: 10.1128/mcb.16.7.3658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deltcheva E, Chylinski K, Sharma CM, Gonzales K, Chao Y, Pirzada ZA, Eckert MR, Vogel J, Charpentier E. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 2011;471:602–607. doi: 10.1038/nature09886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duharcourt S, Butler A, Meyer E. Epigenetic self-regulation of developmental excision of an internal eliminated sequence on Paramecium tetraurelia. Genes Dev. 1995;9:2065–2077. doi: 10.1101/gad.9.16.2065. [DOI] [PubMed] [Google Scholar]
- Fang W, Wang X, Bracht JR, Nowacki M, Landweber LF. Piwi-interacting RNAs protect DNA against loss during Oxytricha genome re-arrangement. Cell. 2012;151:1243–1255. doi: 10.1016/j.cell.2012.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francia S, Michelini F, Saxena A, Tang D, de Hoon M, Anelli V, Mione M, Carninci P, d’Adda di Fagagna F. Site-specific DICER and DROSHA RNA products control the DNA-damage response. Nature. 2012;488:231–235. doi: 10.1038/nature11179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunawardane LS, Saito K, Nishida KM, Miyoshi K, Kawamura Y, Nagami T, Siomi H, Siomi MC. A slicer-mediated mechanism for repeat-associated siRNA 5' end formation in Drosophila. Science. 2007;315:1587–1590. doi: 10.1126/science.1140494. [DOI] [PubMed] [Google Scholar]
- Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, Munson G, Young G, Lucas AB, Ach R, Bruhn L, et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477:295–300. doi: 10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale CR, Zhao P, Olson S, Duff MO, Graveley BR, Wells L, Terns RM, Terns MP. RNA-guided RNA cleavage by a CRISPR RNA-Cas protein complex. Cell. 2009;139:945–956. doi: 10.1016/j.cell.2009.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson IR, Jacobsen SE. Epigenetic inheritance in plants. Nature. 2007;447:418–424. doi: 10.1038/nature05917. [DOI] [PubMed] [Google Scholar]
- Hu W, Alvarez-Dominguez JR, Lodish HF. Regulation of mammalian cell differentiation by long non-coding RNAs. EMBO Rep. 2012;13:971–983. doi: 10.1038/embor.2012.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutvágner G, Simard MJ. Argonaute proteins: key players in RNA silencing. Nat Rev. Mol. Cell Biol. 2008;9:22–32. doi: 10.1038/nrm2321. [DOI] [PubMed] [Google Scholar]
- Jin B, Li Y, Robertson KD. DNA methylation: superior or subordinate in the epigenetic hierarchy? Genes Cancer. 2011;2:607–617. doi: 10.1177/1947601910393957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karginov FV, Hannon GJ. The CRISPR system: small RNA-guided defense in bacteria and archaea. Mol. Cell. 2010;37:7–19. doi: 10.1016/j.molcel.2009.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, Thomas K, Presser A, Bernstein BE, van Oudenaarden A, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc. Natl. Acad. Sci. USA. 2009;106:11667–11672. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klenov MS, Sokolova OA, Yakushev EY, Stolyarenko AD, Mikhaleva EA, Lavrov SA, Gvozdev VA. Separation of stem cell maintenance and transposon silencing functions of Piwi protein. Proc. Natl. Acad. Sci. USA. 2011;108:18760–18765. doi: 10.1073/pnas.1106676108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuramochi-Miyagawa S, Watanabe T, Gotoh K, Totoki Y, Toyoda A, Ikawa M, Asada N, Kojima K, Yamaguchi Y, Ijiri TW, et al. DNA methylation of retrotransposon genes is regulated by Piwi family members MILI and MIWI2 in murine fetal testes. Genes Dev. 2008;22:908–917. doi: 10.1101/gad.1640708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law JA, Jacobsen SE. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat. Rev. Genet. 2010;11:204–220. doi: 10.1038/nrg2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JT. Gracefully ageing at 50, X-chromosome inactivation becomes a paradigm for RNA and chromatin control. Nat. Rev.Mol. Cell Biol. 2011;12:815–826. doi: 10.1038/nrm3231. [DOI] [PubMed] [Google Scholar]
- Le Thomas A, Rogers AK, Webster A, Marinov GK, Liao SE, Perkins EM, Hur JK, Aravin AA, Toth KF. Piwi induces piRNA-guided transcriptional silencing and establishment of a repressive chromatin state. Genes Dev. 2013;27:390–399. doi: 10.1101/gad.209841.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Mochizuki K, Gorovsky MA. Histone H3 lysine 9 methylation is required for DNA elimination in developing macronuclei in Tetrahymena. Proc. Natl. Acad. Sci. USA. 2004;101:1679–1684. doi: 10.1073/pnas.0305421101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Taverna SD, Muratore TL, Shabanowitz J, Hunt DF, Allis CD. RNAi-dependent H3K27 methylation is required for heterochromatin formation and DNA elimination in Tetrahymena. Genes Dev. 2007;21:1530–1545. doi: 10.1101/gad.1544207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Haft DH, Barrangou R, Brouns SJJ, Charpentier E, Horvath P, Moineau S, Mojica FJM, Wolf YI, Yakunin AF, et al. Evolution and classification of the CRISPR-Cas systems. Nat. Rev. Microbiol. 2011;9:467–477. doi: 10.1038/nrmicro2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, Yang L, Esvelt KM, Aach J, Guell M, Dicarlo JE, Norville JE, Church GM. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone CD, Hannon GJ. Small RNAs as guardians of the genome. Cell. 2009;136:656–668. doi: 10.1016/j.cell.2009.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone CD, Anderson AM, Motl JA, Rexer CH, Chalker DL. Germ line transcripts are processed by a Dicer-like protein that is essential for developmentally programmed genome rearrangements of Tetrahymena thermophila. Mol. Cell. Biol. 2005;25:9151–9164. doi: 10.1128/MCB.25.20.9151-9164.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marraffini LA, Sontheimer EJ. CRISPRinterference:RNA-directed adaptive immunity in bacteria and archaea. Nat. Rev. Genet. 2010;11:181–190. doi: 10.1038/nrg2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzke MA, Birchler JA. RNAi-mediated pathways in the nucleus. Nat. Rev. Genet. 2005;6:24–35. doi: 10.1038/nrg1500. [DOI] [PubMed] [Google Scholar]
- Mochizuki K. DNA rearrangements directed by non-coding RNAs in ciliates. Wiley Interdiscip. Rev. RNA. 2010;1:376–387. doi: 10.1002/wrna.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki K, Gorovsky MA. A Dicer-like protein in Tetrahymena has distinct functions in genome rearrangement, chromosome segregation, and meiotic prophase. Genes Dev. 2005;19:77–89. doi: 10.1101/gad.1265105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki K, Fine NA, Fujisawa T, Gorovsky MA. Analysis of a piwi-related gene implicates small RNAs in genome rearrangement in tetrahymena. Cell. 2002;110:689–699. doi: 10.1016/s0092-8674(02)00909-1. [DOI] [PubMed] [Google Scholar]
- Mohammad F, Mondal T, Guseva N, Pandey GK, Kanduri C. Kcnq1ot1 noncoding RNA mediates transcriptional gene silencing by interacting with Dnmt1. Development. 2010;137:2493–2499. doi: 10.1242/dev.048181. [DOI] [PubMed] [Google Scholar]
- Motamedi MR, Verdel A, Colmenares SU, Gerber SA, Gygi SP, Moazed D. Two RNAi complexes, RITS and RDRC, physically interact and localize to noncoding centromeric RNAs. Cell. 2004;119:789–802. doi: 10.1016/j.cell.2004.11.034. [DOI] [PubMed] [Google Scholar]
- Noma K-I, Sugiyama T, Cam H, Verdel A, Zofall M, Jia S, Moazed D, Grewal SIS. RITS acts in cis to promote RNA interference-mediated transcriptional and post-transcriptional silencing. Nat. Genet. 2004;36:1174–1180. doi: 10.1038/ng1452. [DOI] [PubMed] [Google Scholar]
- Nowacki M, Shetty K, Landweber LF. RNA-mediated epigenetic programming of genome rearrangements. Annu. Rev. Genomics Hum. Genet. 2011;12:367–389. doi: 10.1146/annurev-genom-082410-101420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- Pandey RR, Mondal T, Mohammad F, Enroth S, Redrup L, Komorowski J, Nagano T, Mancini-Dinardo D, Kanduri C. Kcnq1ot1 anti-sense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Mol. Cell. 2008;32:232–246. doi: 10.1016/j.molcel.2008.08.022. [DOI] [PubMed] [Google Scholar]
- Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu. Rev. Biochem. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, Chang HY. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozhkov NV, Hammell M, Hannon GJ. Multiple roles for Piwi in silencing Drosophila transposons. Genes Dev. 2013;27:400–412. doi: 10.1101/gad.209767.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz KM, Mayer C, Postepska A, Grummt I. Interaction of noncoding RNA with the rDNA promoter mediates recruitment of DNMT3b and silencing of rRNA genes. Genes Dev. 2010;24:2264–2269. doi: 10.1101/gad.590910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schorderet P, Duboule D. Structural and functional differences in the long non-coding RNA hotair in mouse and human. PLoS Genet. 2011;7:e1002071. doi: 10.1371/journal.pgen.1002071. http://dx.doi.org/10.1371/journal.pgen.1002071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shpiz S, Olovnikov I, Sergeeva A, Lavrov S, Abramov Y, Savitsky M, Kalmykova A. Mechanism of the piRNA-mediated silencing of Drosophila telomeric retrotransposons. Nucleic Acids Res. 2011;39:8703–8711. doi: 10.1093/nar/gkr552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sienski G, Dönertas D, Brennecke J. Transcriptional silencing of transposons by Piwi and maelstrom and its impact on chromatin state and gene expression. Cell. 2012;151:964–980. doi: 10.1016/j.cell.2012.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun BK, Deaton AM, Lee JT. A transient heterochromatic state in Xist preempts X inactivation choice without RNA stabilization. Mol. Cell. 2006;21:617–628. doi: 10.1016/j.molcel.2006.01.028. [DOI] [PubMed] [Google Scholar]
- Taverna SD, Coyne RS, Allis CD. Methylation of histone h3 at lysine 9 targets programmed DNA elimination in tetrahymena. Cell. 2002;110:701–711. doi: 10.1016/s0092-8674(02)00941-8. [DOI] [PubMed] [Google Scholar]
- Tsai M-C, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, Shi Y, Segal E, Chang HY. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulitsky I, Shkumatava A, Jan CH, Sive H, Bartel DP. Conserved function of lincRNAs in vertebrate embryonic development despite rapid sequence evolution. Cell. 2011;147:1537–1550. doi: 10.1016/j.cell.2011.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdel A, Jia S, Gerber S, Sugiyama T, Gygi S, Grewal SIS, Moazed D. RNAi-mediated targeting of heterochromatin by the RITS complex. Science. 2004;303:672–676. doi: 10.1126/science.1093686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volpe TA, Kidner C, Hall IM, Teng G, Grewal SIS, Martienssen RA. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science. 2002;297:1833–1837. doi: 10.1126/science.1074973. [DOI] [PubMed] [Google Scholar]
- Wang SH, Elgin SCR. Drosophila Piwi functions downstream of piRNA production mediating a chromatin-based transposon silencing mechanism in female germ line. Proc. Natl. Acad. Sci. USA. 2011;108:21164–21169. doi: 10.1073/pnas.1107892109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang KC, Yang YW, Liu B, Sanyal A, Corces-Zimmerman R, Chen Y, Lajoie BR, Protacio A, Flynn RA, Gupta RA, et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472:120–124. doi: 10.1038/nature09819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei W, Ba Z, Gao M, Wu Y, Ma Y, Amiard S, White CI, Rendtlew Danielsen JM, Yang Y-G, Qi Y. A role for small RNAs in DNA double-strand break repair. Cell. 2012;149:101–112. doi: 10.1016/j.cell.2012.03.002. [DOI] [PubMed] [Google Scholar]
- Wiedenheft B, Sternberg SH, Doudna JA. RNA-guided genetic silencing systems in bacteria and archaea. Nature. 2012;482:331–338. doi: 10.1038/nature10886. [DOI] [PubMed] [Google Scholar]
- Wierzbicki AT, Haag JR, Pikaard CS. Noncoding transcription by RNA polymerase Pol IVb/Pol V mediates transcriptional silencing of over-lapping and adjacent genes. Cell. 2008;135:635–648. doi: 10.1016/j.cell.2008.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap KL, Li S, Muñoz-Cabello AM, Raguz S, Zeng L, Mujtaba S, Gil J, Walsh MJ, Zhou M-M. Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Mol. Cell. 2010;38:662–674. doi: 10.1016/j.molcel.2010.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu W, Gius D, Onyango P, Muldoon-Jacobs K, Karp J, Feinberg AP, Cui H. Epigenetic silencing of tumour suppressor gene p15 by its antisense RNA. Nature. 2008;451:202–206. doi: 10.1038/nature06468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahler AM, Neeb ZT, Lin A, Katzman S. Mating of the stichotrichous ciliate Oxytricha trifallax induces production of a class of 27 nt small RNAs derived from the parental macronucleus. PLoS ONE. 2012;7:e42371. doi: 10.1371/journal.pone.0042371. http://dx.doi.org/10.1371/journal.pone.0042371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Ohsumi TK, Kung JT, Ogawa Y, Grau DJ, Sarma K, Song JJ, Kingston RE, Borowsky M, Lee JT. Genome-wide identification of polycomb-associated RNAs by RIP-seq. Mol. Cell. 2010;40:939–953. doi: 10.1016/j.molcel.2010.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]