Abstract
Despite the clinical importance of bacterial-fungal interactions, their molecular details are poorly understood. A hallmark of such medically-important interspecies associations is the interaction between the two nosocomial pathogens Staphylococcus aureus and Candida albicans, which can lead to mixed biofilm-associated infections with enhanced antibiotic resistance. Here, we use single-cell force spectroscopy to quantify the forces engaged in bacterial-fungal co-adhesion, focusing on the poorly investigated S. epidermidis-C. albicans interaction. Force curves recorded between single bacterial and fungal germ tubes showed large adhesion forces (~5 nN) with extended rupture lengths (up to 500 nm). By contrast, bacteria poorly adhered to yeast cells, emphasizing the important role of the yeast-to-hyphae transition in mediating adhesion to bacterial cells. Analysis of mutant strains altered in cell wall composition allowed us to distinguish the main fungal components involved in adhesion, i.e. Als proteins and O-mannosylations. We suggest that the measured co-adhesion forces are involved in the formation of mixed biofilms, thus possibly as well in promoting polymicrobial infections. In the future, we anticipate that this SCFS platform will be used in nanomedicine to decipher the molecular mechanisms of a wide variety of pathogen-pathogen interactions and may help designing novel anti-adhesion agents.
Keywords: AFM, cell-cell adhesion, pathogens, single-cell force spectroscopy, Candida albicans, Staphyloccocus epidermidis, polymicrobial infections
INTRODUCTION
The interactions between bacterial and fungal pathogens are of high clinical importance as they may lead to higher morbidity and mortality.1–4 Polymicrobial infections generally involve the formation of mixed biofilms, i.e. attachment of various microbial species to a substrate and to each other.5–8 Therefore, knowledge of the molecular mechanisms behind bacterial-fungal co-adhesion is critical to our understanding of mixed infections and may aid in the development of novel anti-biofilm molecules.
A widely investigated example of such association is the interaction between Staphylococcus aureus and Candida albicans.9–11 It has been shown that co-inoculation of C. albicans and Staphylococcus aureus leads to mortality increases.2, 3, 12 In vivo, the synergistic effect of the two microorganisms has been observed on mouse.12 When grown in mixed biofilms, S. aureus has been shown to attach primarily to C. albicans hyphae.9, 10 Recent biochemical and microscopy studies have shown that the C. albicans adhesion proteins Als mediate fungal-bacterial interactions,13 in particular Als3 which is primarily expressed on germ tubes and is directly involved in the adhesion to Streptococcus gordonii14 and Staphylococcus aureus.15 Whether the other nosocomial Staphylococcus species S. epidermidis also interacts with C. albicans and can lead to mixed infections has been much less investigated. Both species showed extensive interactions when grown in mixed fungal-bacterial biofilms.16 In addition, it appeared that the two species could modulate the action of antibiotics and antifungals in mixed biofilms.16 So far, however, the adhesion forces engaged in the S. epidermidis - C. albicans interaction have never been investigated.
Single-cell microbiology is a fast-growing field that uses emerging technologies for single-cell analysis, thereby revealing population and cell heterogeneity, as well as rare events that were otherwise not accessible.17, 18 In cell adhesion and biofilm research, single-cell force spectroscopy (SCFS) offers unprecedented possibilities to quantify cell-cell and cell-solid interactions at the single-cell and single-molecule levels.19, 20 In this report, we used SCFS to quantify the forces engaged in the S. epidermidis-C. albicans interaction. As the yeast-to-hyphae transition is important for C. albicans adhesion and biofilm formation,4, 21 we measured the forces between single bacterial cells and fungal hyphae. The results emphasize the important role of cellular morphogenesis, Als proteins and O-mannosylations in controlling S. epidermidis-C. albicans co-adhesion.
Results and discussion
Experimental set-up
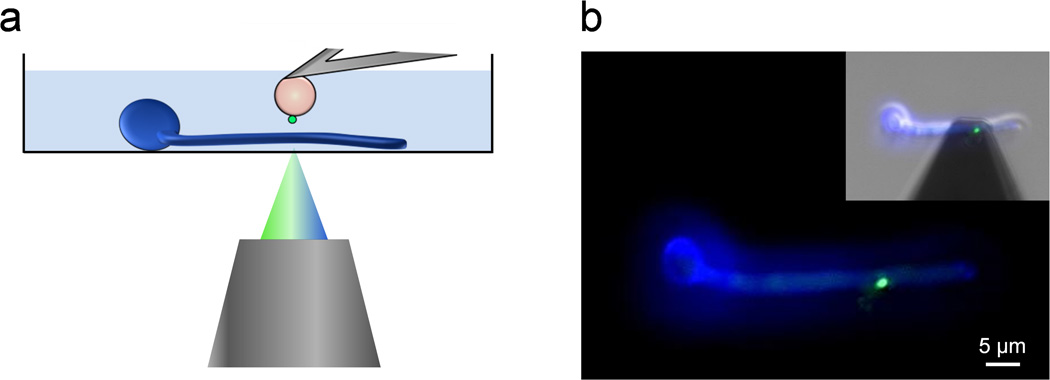
To probe bacterial-fungal interaction forces by SCFS, we used a recently developed protocol which combines the use of colloidal probe cantilevers and of a bioinspired polydopamine wet adhesive.22 Single S. epidermidis cells were picked up with a polydopamine-coated colloidal probe and approached towards a fungal cell immobilized on a hydrophobic substrate (Fig. 1a). Using an integrated AFM-inverted optical microscope, the bacterial probe was positioned on top of random spots across the fungal cell (Fig. 1b). Fluorescence imaging confirmed that single bacteria attached on the probe were alive (Baclight LIVE/DEAD stain; green color). Note that in Fig. 1b C. albicans was stained in blue (Calcofluor White) for better visualization, but as this dye alters cell surface properties it was not used in force experiments.
Fig. 1.
Single-cell force spectroscopy of bacterial-fungal interactions. (a) Schematic of the experimental set-up. (b) Using an integrated AFM-inverted optical microscope, the S. epidermidis probe (green) is approached towards a C. albicans hyphae (blue). The image was obtained using epifluorescence microscopy while the inset shows a merged phase/epifluorescence image.
Bacterial-fungal adhesion: germ tube vs. yeast region
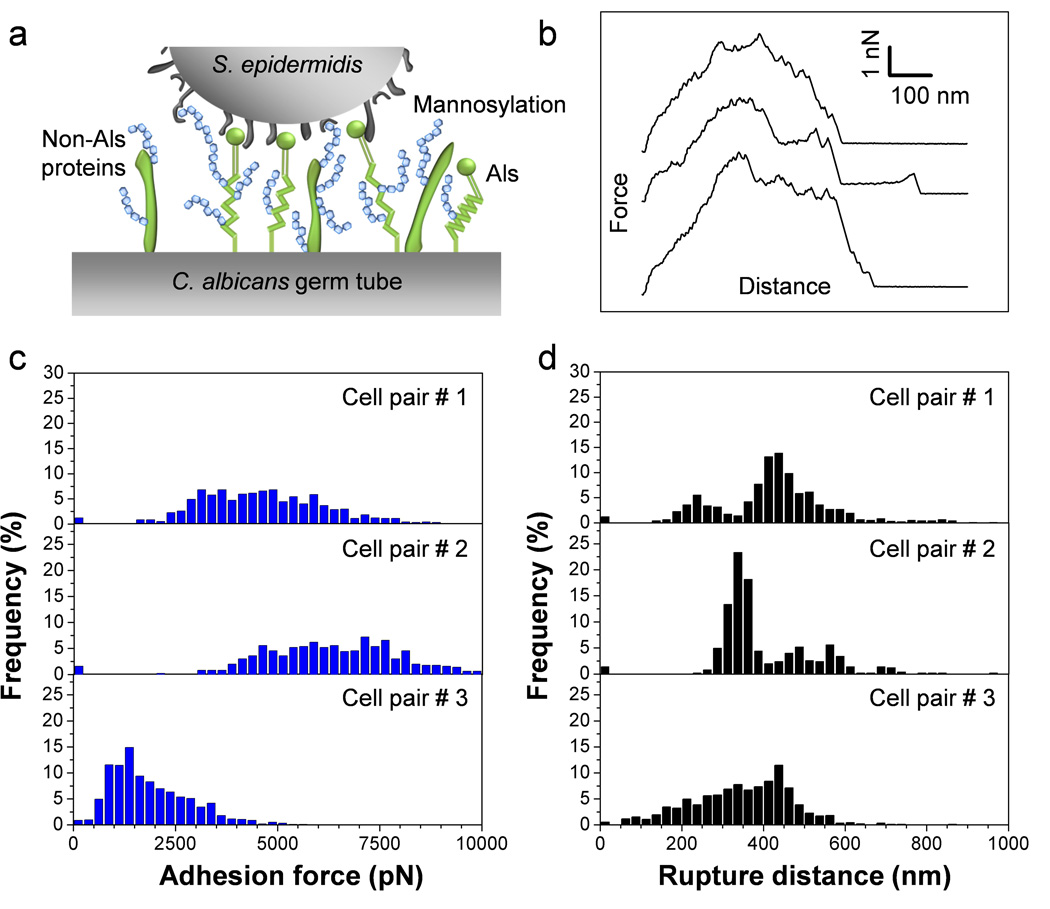
In C. albicans, the yeast-to-hyphae transition is associated with changes in cell wall composition that play important roles in promoting biofilm formation.4 Consistent with this, single-molecule analyses recently showed that cellular morphogenesis leads to a major increase in the distribution and biophysical properties (stickiness, extension) of Als adhesins on the fungal cell surface.23 With this in mind, we measured the adhesion between single S. epidermidis cells and C. albicans hyphae (Fig. 2a). Fig. 2b shows a set of representative force-distance curves recorded between individual bacteria and germ tubes. All curves showed large adhesion forces (4.6 nN ± 1.5 nN; n = 975 force curves corresponding to cell pair #1 in Figs. 2c and d) with multiple, sequential peaks and extended rupture lengths (419 ± 137 nm). The general features of the curves did not substantially change when recording consecutive force curves (up to several hundreds) on the same spot, indicating that force measurements did not alter the interacting cell surfaces. Also, similar force signatures were observed when probing different regions of the germ tubes (e.g. apex vs. center of the tube), suggesting that the adhesion properties of the tube were homogeneous. Figs. 2c and d show that probing bacterial-fungal interactions using cells from independent cultures generally revealed adhesion properties that were in the same range (pairs #1 and #2); however, in some cases weaker adhesion was observed (1.9 nN ± 1.0 nN, pair #3), an effect that we believe could reflect heterogeneity in the bacterial and/or fungal cell populations.
Fig. 2.
SCFS quantifies the adhesion forces between S. epidermidis and C. albicans germ tubes. (a) Key cell wall components that are involved in C. albicans surface interactions are cell-surface glycoproteins (in green) and mannose-rich glycoconjugates (in blue). (b) Typical force-distance curves recorded in Tris NaCl buffer between S. epidermidis and C. albicans hyphae. (c, d) Adhesion force (c) and rupture length (d) histograms obtained by recording force curves between 3 cell pairs from different cell cultures, and representative of a total of 7 cell pairs (n > 500 force-distances curves for each pair).
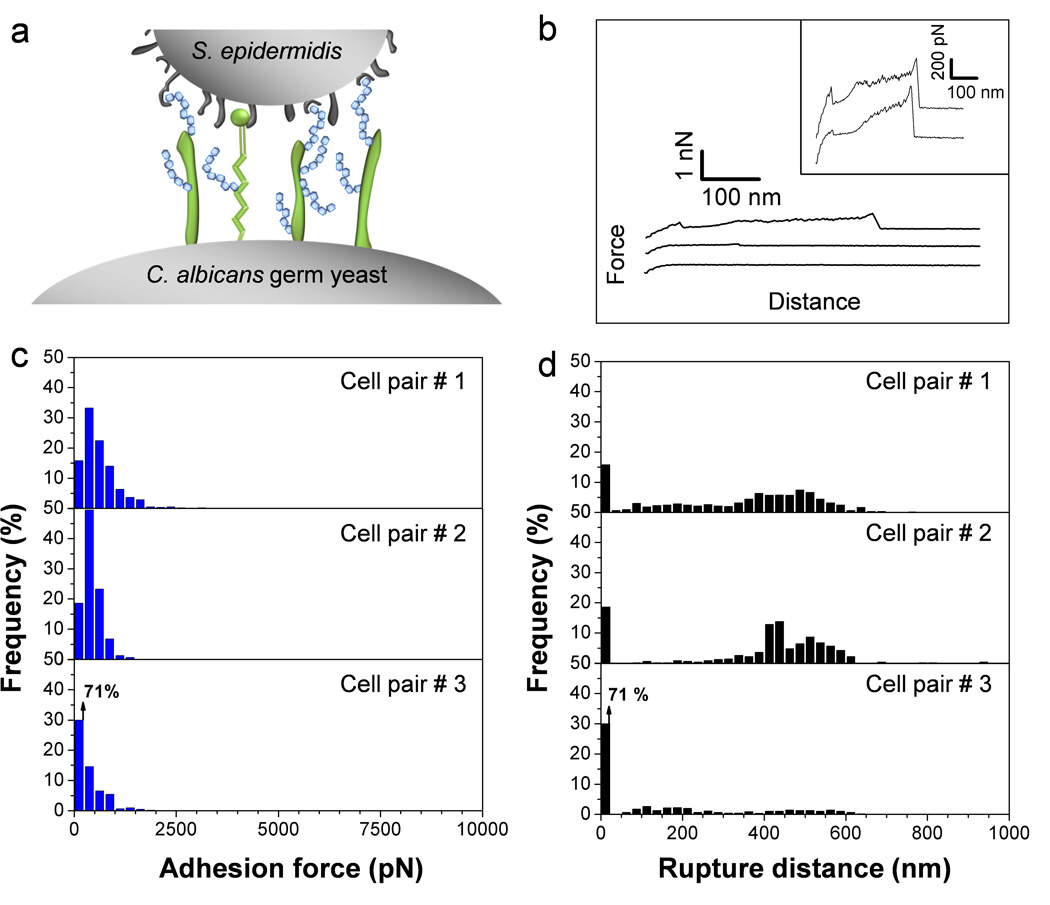
To determine whether the measured forces are specific to the C. albicans germ tube, we then probed the yeast region of the germinating cell (Fig. 3a). As can be seen in Fig. 3, a drop in adhesion frequency was observed (from 99 % on germ tube to 84 % on yeast; cell pair #1), together with a decrease in adhesion forces (from 4.6 nN ± 1.5 nN to 0.6 nN ± 0.5 nN). Sometimes, these effects were even more pronounced (cell pair #3). On close examination, a number of force curves showed sawtooth patterns with multiple large force peaks rupturing at around 500 pN and in the 300–600 nm range. In the light of earlier single-molecule work,23–25 we suggest these features reflect the sequential unfolding of the tandem repeat (TR) domains of Als proteins on the yeast surface. As the average forces measured on germ tubes (4.6 nN) are much larger than those associated with single protein unfolding, it is likely that the strongly adhesive signatures result from multiple Als unfolding interactions, thus explaining why they consist of multiple poorly defined peaks.
Fig. 3.
C. albicans germinating yeasts show much weaker adhesion than germ tubes. (a) Germinating yeasts express fewer Als proteins than germ tubes. (b) Typical force-distance curves recorded in Tris NaCl buffer between S. epidermidis and C. albicans germinating yeasts. (c, d) Adhesion force (c) and rupture length (d) histograms obtained by recording force curves between 3 cell pairs from different cell cultures, and representative of a total of 6 cell pairs (n > 500 force-distances curves for each pair).
Biological specificity of adhesion forces
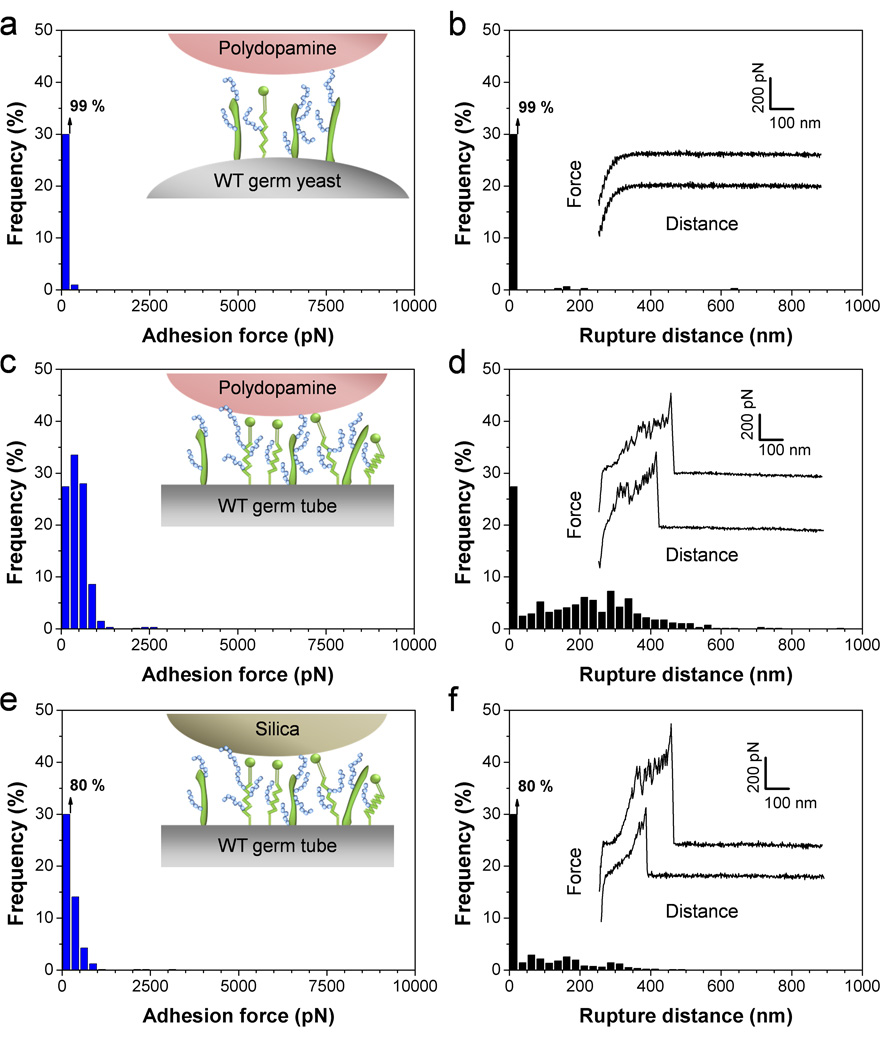
To determine the specificity of the measured adhesion forces and rule out the possibility of artifacts associated with the cell probe preparation, two control experiments were performed, i.e. use of polydopamine-coated probes or silica probes instead of bacterial probes. As can be seen in Fig. 4, use of these non-cellular probes led to a major reduction of adhesion frequency (down to 1 % between polydopamine and the germinating yeast, Fig. 4a) and mean adhesion force, indicating that the strong adhesion forces measured earlier indeed reflect bacterial-fungal interactions. These data also confirm that the polydopamine adhesive does not interfere with the measurements, e.g. through contamination of the bacterial probe.
Fig. 4.
Control experiments using polydopamine and silica probes. (a–f) Adhesion force (a, c, e) and rupture length (b, d, f) histograms, together with representative force curves, obtained by recording force curves between polydopamine-coated probes and C. albicans germinating yeasts (a, b) or germ tubes (c, d), and between silica probes and C. albicans germ tubes (e, f). For each probe, similar data were obtained in 3 independent experiments.
Als proteins and O-mannosylations on the C. albicans surface are required for bacterial adhesion
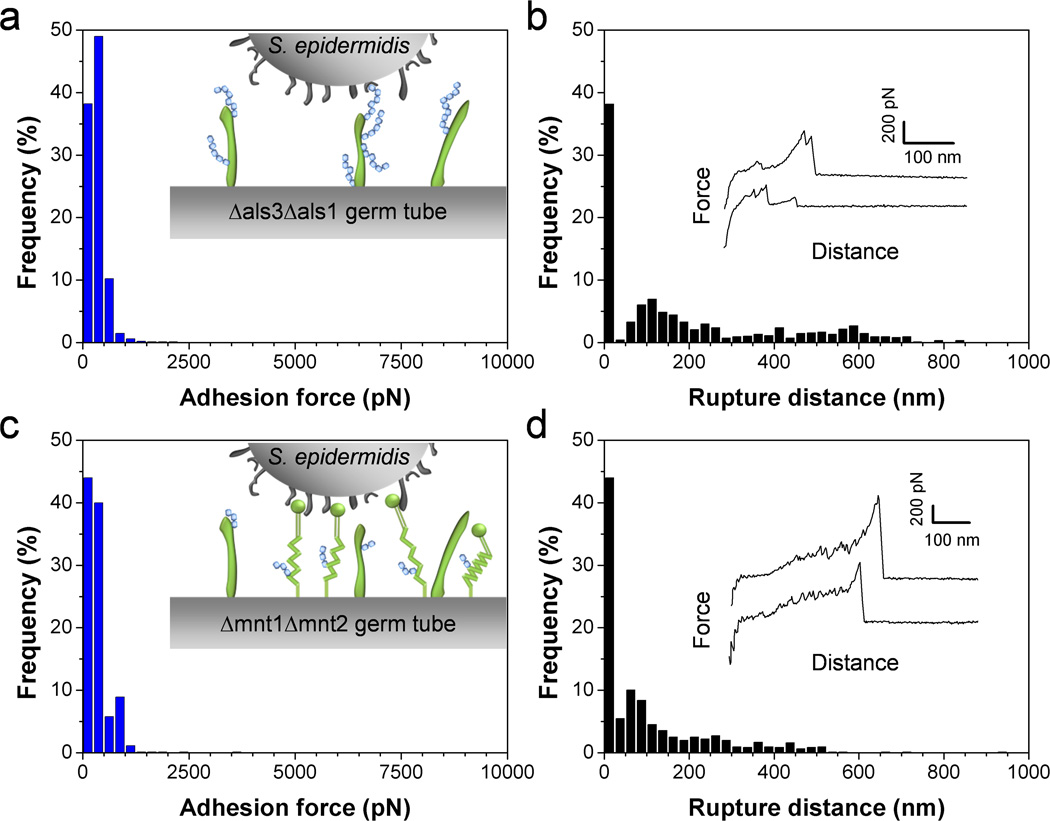
Cell-surface glycoproteins and mannose-rich glycoconjugates play key roles in C. albicans surface interactions.26 Specifically, Als adhesins mediate cell adhesion and biofilm formation, and mannose-rich polymers are recognized by a variety of lectin receptors on immune cells. We therefore reasoned that both compounds may be involved in bacterial-fungal adhesion. To test this hypothesis, we measured the forces between single S. epidermidis cells and C. albicans mutant strains altered in cell wall components. Figs 5a and b show the curves obtained on germ tubes from the double mutant als3Δ/als3Δ als1Δ/als1Δ, in which the genes coding for the expression of Als1 and Als3 proteins have been deleted. Adhesion forces and rupture distances that were much smaller than those on the WT were observed, thus demonstrating that Als3 and/or Als1 proteins, primarily expressed on germ tubes, are required for bacterial-fungal association. Similar observations have recently been reported for the interaction between S. aureus and C. albicans.15, 27 We suggest that N-terminal immunoglobulin-like regions of Als proteins specifically binds peptide ligands on the bacterial surface, i.e. peptide sequences containing the “τϕ+” motif.28
Fig. 5.
Als proteins and O-mannosylations on the C. albicans surface are required for bacterial adhesion. (a–d) Adhesion force (a, c) and rupture length (b, d) histograms, together with representative force curves, obtained by recording force curves in Tris NaCl buffer between a single S. epidermidis bacterium and a C. albicans germ tube from the mutant als3Δ/als3Δ als1Δ/als1Δ (Δals3Δals1) (a, b) or a C. albicans germ tube from the mutant mnt1Δ/mnt1Δ mnt2Δ/mnt2Δ (Δmnt1Δmnt2) (c, d). For each mutant, similar data were obtained in 3 independent experiments using 3 different cell pairs.
Can the measured adhesion forces be converted into a surface density of interacting molecules? As the specific binding force of single Als proteins was previously measured to be ~330 pN,23 we estimate that the 4.6 nN forces would correspond to ~14 Als bonds. The obtained values may be converted into protein surface densities, considering the cell-cell contact area. As a rough approximation, the contact zone of a deformable sphere (the bacterium) pressed on a more rigid flat surface (the fungus) may be estimated by the following equation29, 30 A = π R δ, in which A is the contact area, R the radius of the cell, and δ the cell deformation. Considering a cell radius of 0.5 µm and a deformation of 30 nm (estimated from indentation curves), we found a contact area of ~0.05 µm2, thus yielding a protein surface density of around 280 proteins/µm2. Note that this value is an upper estimate as we expect that the curvature of the fungal germ tube will lower the cell-cell contact area. Nevertheless, this density is roughly consistent with the value expected for fungal adhesins, and with numbers estimated from single-molecule imaging experiments.23
Another important finding is that the C. albicans double mutant strain mnt1Δ/mnt1Δ mnt2Δ/mnt2Δ defective in O-linked mannosylations31 showed similar reduction in adhesion probability and adhesion strength (Figs. 5c and d), suggesting strongly that fungal mannosylations are recognized by lectins on the bacterial surface. In addition, smaller rupture distances were observed, consistent with the notion that mannosylations in the mutant are shorter. That O-linked mannosylations are important for bacterial-fungal adhesion agrees well with earlier reports showing their involvement in adhesion to host cells31 and to Pseudomonas aeruginosa bacteria.32 This result is also consistent with the notion that adhesion of bacterial pathogens, such as Pseudomonas aeruginosa, involves mannose-binding lectins on the bacterial surface.33
Conclusions
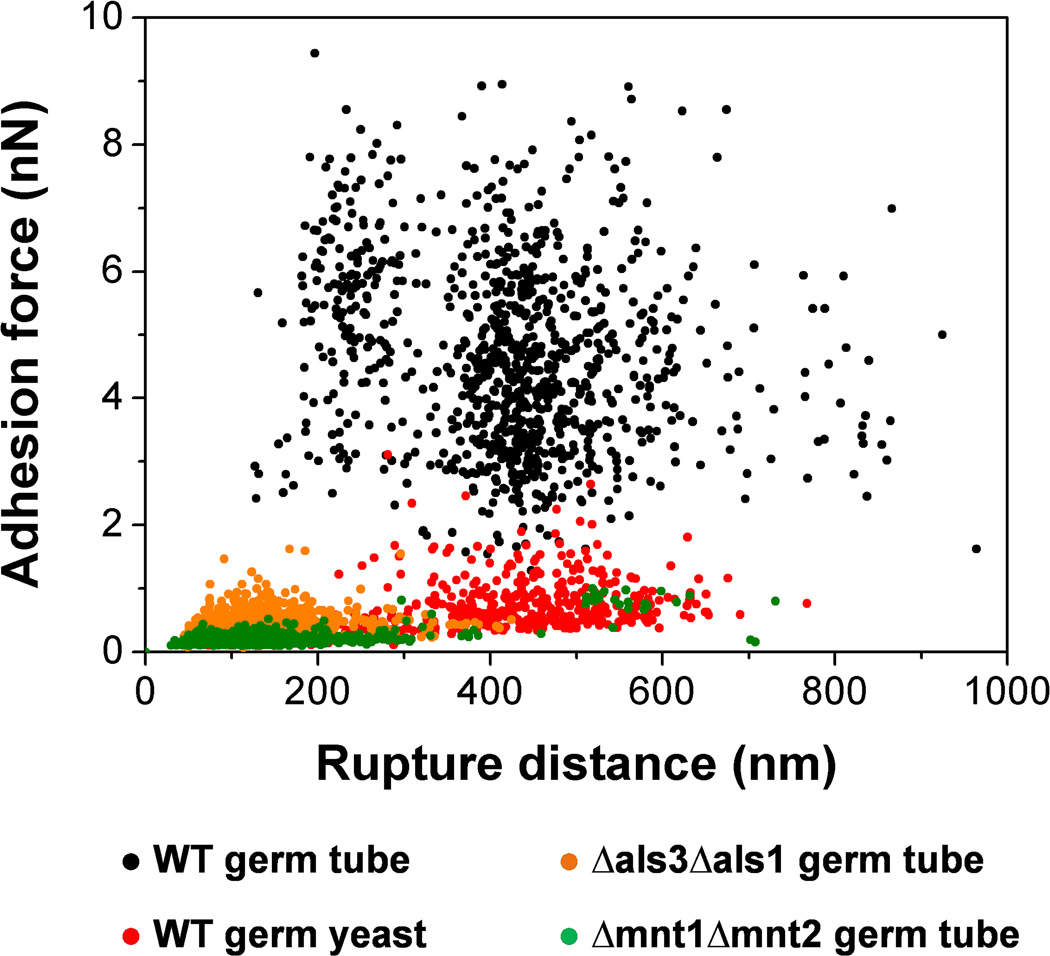
In recent years, there have been several attempts to apply AFM force spectroscopy to probe the adhesion forces engaged in bacterial-fungal interactions, including the important S. aureus – C. albicans interaction.15, 27 These results are difficult to interpret at the molecular level because of a poorly-controlled methodology: cells are attached on the cantilever using protocols that may lead to cell surface denaturation or cell death, multiple cells are attached and probed together, cell positioning and cell-substrate contact area are poorly controlled. We have shown that SCFS with polydopamine-coated colloidal probes is a valuable approach for quantifying the adhesion forces of medically-important bacterial-fungal interactions. Unlike most other protocols used in microbiology, this method is non-destructive (living cells are probed), guarantees true single-cell measurements and affords precise positioning of the interacting cells, thereby ensuring true and reliable single-bacterial cell analysis. Fig. 6 summarizes our main findings, that is: i) S. epidermidis strongly binds to C. albicans germ tubes but poorly adheres to yeast cells, emphasizing the important role of the yeast-to-hyphae transition in mediating adhesion to bacterial cells; ii) co-adhesion primarily involves two types of highly adhesive and extended macromolecules, i.e. Als proteins and O-mannosylations, that we believe bind to Als ligands and lectins on the bacterial surface. When subjected to mechanical force, the interacting cell surfaces will detach but the cells will remain bridged through these extended polymers. Our finding of strong S. epidermidis-C. albicans adhesion forces is reminiscent of the well-known S. aureus-C. albicans interaction3, 9, 15, 27, 34 thus suggesting that the S. epidermidis-C. albicans co-adhesion quantified here will favor the formation of mixed biofilms, and in turn promote polymicrobial infections.
Fig. 6.
Role of cellular morphogenesis, Als proteins and O-mannosylations in S. epidermidis-C. albicans adhesion. Plots of the adhesion forces versus rupture distances measured between S. epidermidis and WT germ tubes (black symbols), WT germinated yeasts (red symbols), als3Δ/als3Δ als1Δ/als1Δ (Δals3Δals1) germ tubes (orange symbols) and mnt1Δ/mnt1Δ mnt2Δ/mnt2Δ (Δmnt1Δmnt2) germ tubes (green symbols). Strong co-adhesion is only observed on germ tubes and involves two types of highly adhesive and extended macromolecules, i.e. Als proteins and O-mannosylations.
Methods
Microorganisms and cultures
C. albicans SC531435 was cultivated in YPD medium (1% yeast extract, 2% Bacto-peptone, 2% D-glucose, supplemented with 2% agar) at 30°C. A few colonies were inoculated in YPD liquid medium and incubated overnight (30°C, 200 rpm). For hyphae formation, germination was induced by inoculating 250 µL of cell suspension in 8 mL of RPMI 1640 medium buffered with MOPS (Sigma) at pH 7, and incubated at 37°C, 200 rpm, for 90 min unless otherwise stated.36 We used two C. albicans mutant strains, i.e. als3Δ/als3Δ als1Δ/als1Δ (designated as Δals3Δals1) with deletions of both alleles of ALS genes (kindly provided by Aaron Mitchell, Carnegie Mellon University, Pittsburgh, PA)37 and mnt1Δ/mnt1Δ mnt2Δ/mnt2Δ (designated as Δmnt1Δmnt2) yielding defective O-linked mannosylations (kindly provided by Neil Gow, University of Aberdeen, UK).31 S. epidermidis ATCC 35984 cells were grown in Trypto-Caseine-Soy (Bio-rad) at 37°C, 150 rpm. Overnight cultures were diluted in fresh media to an OD600 nm of 0.1. The cells were harvested in the exponential growth phase (5 hours at 37°C, 150 rpm), and washed 3 times in 50 mM Tris-NaCl buffer. For cell probe preparation, 50 µL of a 100-fold diluted solution were transferred in a glass petri dish and the bacteria were let to settle for 15 min.
Immobilization of C. albicans
Germinating yeast cells of C. albicans were immobilized through hydrophobic attachment on solid substrata. To this end, glass coverslips coated with a thin layer of gold were immersed overnight in a 1 mM solution of 1-dodecanethiol (Sigma), rinsed with ethanol and dried under N2. After induction of germ tube formation in RPMI, the cells were harvested and rinsed three times in Tris-NaCl buffer, pH 7.5. Drops (200 µL) of the concentrated suspension were deposited on the hydrophobic substrates and let stand for 3 h. The substrate was then rinsed to remove unattached yeast and fixed on a glass-bottom petri dish using double-sided tape. A droplet of buffer was then deposited on the substrate to avoid drying of the immobilized yeast.
Bacterial cell probes
Using a Nanoscope VIII Multimode AFM (Bruker corporation, Santa Barbara, CA), triangular shaped tipless cantilevers (NP-O10, Microlevers, Veeco Metrology Group) were slowly immersed in a very thin layer of UV-curable glue (NOA 63, Norland Edmund Optics) spread on a glass slide, and slowly brought into contact with a silica microsphere (6.1 µm diameter, bangs laboratories). After 3 min of contact, the colloidal probe was cured for 10 min under a UV-lamp. The cantilever was then immersed for 1 hr in a 10 mM Tris Buffer solution (pH 8.5) containing 4 mg/mL dopamine hydrochloride (99%, Sigma). The probe was then washed and dried under N2.
Proper attachment and positioning of bacteria on the colloidal probe were achieved using a Bioscope Catalyst (Bruker Corporation, Santa Barbara, CA) equipped with a Zeiss Axio Observer Z1 and a Hamamatsu camera C10600. To check the viability of the bacteria, a Live-dead Baclight viability kit (Invitrogen, kit L7012) was used. Prior to attachment, 2 µL of a 1:1 Syto 9 (green fluorescent nucleic acid stain)/Propidium iodide (red-fluorescent nuclear and chromosome counterstain) mixture at 1.5 mM were added to a drop of 50 µL bacteria suspension and mixed thoroughly. The suspension was deposited in the glass petri dish where the substrate covered with C. albicans had been previously attached, and the bacteria were let to incubate with the dyes for 15 min in the dark. 4mL of buffer were then added to the petri dish, immerging both the bacteria deposited at the bottom of the petri dish and the C. albicans substrate. The colloidal probe was then mounted into the AFM and brought into contact with an isolated bacterium. When proper attachment of the bacterium was achieved, the probe was positioned over the C. albicans surface without dewetting. Using this protocol, we never (rarely) observed floating bacteria interacting with C. albicans.
Force measurements
AFM measurements were performed at room temperature (20 °C) in Tris-NaCl buffer at pH 7.5 using a Bioscope Catalyst AFM (Bruker AXS Corporation, Santa Barbara, CA). Using the inverted optical microscope, the bacterial probe was approached towards a fungal cell. Multiple forces curves were recorded on various spots using a maximum applied force of 250 pN, a contact time of 50 ms, and constant approach and retraction speeds of 1000 nm.s−1. For each condition, the interaction forces between at least 3 pairs of bacterial-fungal cells from independent cultures were measured.
Acknowledgements
Work at the Université catholique de Louvain was supported by the National Foundation for Scientific Research (FNRS), the Université catholique de Louvain (Fonds Spéciaux de Recherche), the Région Wallonne, the Federal Office for Scientific, Technical and Cultural Affairs (Interuniversity Poles of Attraction Programme), and the Research Department of the Communauté française de Belgique (Concerted Research Action). Work at VIB, KU Leuven was supported by Flemish Science Foundation (FWO), the KU Leuven and by the Interuniversity Attraction Poles Programme, initiated by the Belgian Science Policy Office (IAP; P7/28). Work at Brooklyn College was supported by NIH grant R01 GM 098616. Y.F.D. is Senior Research Associate of the FNRS.
REFERENCES
- 1.Peleg AY, Hogan DA, Mylonakis E. Nat. Rev. Microbiol. 2010;8:340–349. doi: 10.1038/nrmicro2313. [DOI] [PubMed] [Google Scholar]
- 2.Morales DK, Hogan DA. PLoS Pathog. 2010;6:1–4. doi: 10.1371/journal.ppat.1000886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shirtliff ME, Peters BM, Jabra-Rizk MA. FEMS Microbiol. Lett. 2009;299:1–8. doi: 10.1111/j.1574-6968.2009.01668.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Douglas LJ. Trends Microbiol. 2003;11:30–36. doi: 10.1016/s0966-842x(02)00002-1. [DOI] [PubMed] [Google Scholar]
- 5.Elias S, Banin E. FEMS Microbiol. Rev. 2012;36:990–1004. doi: 10.1111/j.1574-6976.2012.00325.x. [DOI] [PubMed] [Google Scholar]
- 6.Lynch AS, Robertson GT. 2008;vol. 59:415–428. doi: 10.1146/annurev.med.59.110106.132000. [DOI] [PubMed] [Google Scholar]
- 7.Harriott MM, Noverr MC. Trends Microbiol. 2011;19:557–563. doi: 10.1016/j.tim.2011.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hogan DA, Kolter R. Science. 2002;296:2229–2232. doi: 10.1126/science.1070784. [DOI] [PubMed] [Google Scholar]
- 9.Peters BM, Jabra-Rizk MA, Scheper MA, Leid JG, Costerton JW, Shirtliff ME. FEMS Immunol. Med. Mic. 2010;59:493–503. doi: 10.1111/j.1574-695X.2010.00710.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harriott MM, Noverr MC. Antimicrob. agents ch. 2009;53:3914–3922. doi: 10.1128/AAC.00657-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harriott MM, Noverr MC. Antimicrob. agents ch. 2010;54:3746–3755. doi: 10.1128/AAC.00573-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carlson E. Infect. Immun. 1983;42:285–292. doi: 10.1128/iai.42.1.285-292.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Klotz SA, Gaur NK, De Armond R, Sheppard D, Khardori N, Edwards JE, Lipke PN, El-Azizi M. Med. Mycol. 2007;45:363–370. doi: 10.1080/13693780701299333. [DOI] [PubMed] [Google Scholar]
- 14.Silverman RJ, Nobbs AH, Vickerman MM, Barbour ME, Jenkinson HF. Infect. Immun. 2010;78:4644–4652. doi: 10.1128/IAI.00685-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peters BM, Ovchinnikova ES, Krom BP, Schlecht LM, Zhou H, Hoyer LL, Busscher HJ, van der Mei HC, Jabra-Rizk MA, Shirtliff ME. Microbiology. 2012;158:2975–2986. doi: 10.1099/mic.0.062109-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Adam B, Baillie GS, Douglas LJ. J. Med. Microbiol. 2002;51:344–349. doi: 10.1099/0022-1317-51-4-344. [DOI] [PubMed] [Google Scholar]
- 17.Lidstrom ME, Konopka MC. Nat. Chem. Biol. 2010;6:705–712. doi: 10.1038/nchembio.436. [DOI] [PubMed] [Google Scholar]
- 18.Brehm-Stecher BF, Johnson EA. Microbiol. Mol. Biol. R. 2004;68:538–559. doi: 10.1128/MMBR.68.3.538-559.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Müller DJ, Helenius J, Alsteens D, Dufrêne YF. Nat. Chem. Biol. 2009;5:383–390. doi: 10.1038/nchembio.181. [DOI] [PubMed] [Google Scholar]
- 20.Benoit M, Gabriel D, Gerisch G, Gaub HE. Nat. Cell Biol. 2000;2:313–317. doi: 10.1038/35014000. [DOI] [PubMed] [Google Scholar]
- 21.Finkel JS, Mitchell AP. Nat. Rev. Microbiol. 2011;9:109–118. doi: 10.1038/nrmicro2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Beaussart A, El-Kirat-Chatel S, Herman P, Alsteens D, Mahillon J, Hols P, Dufrêne YF. Biophys. J. 2013;104:1886–1892. doi: 10.1016/j.bpj.2013.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Beaussart A, Alsteens D, El-Kirat-Chatel S, Lipke PN, Kucharíková S, Van Dijck P, Dufrêne YF. ACS Nano. 2012;6:10950–10964. doi: 10.1021/nn304505s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alsteens D, Dupres V, Klotz SA, Gaur NK, Lipke PN, Dufrêne YF. ACS Nano. 2009;3:1677–1682. doi: 10.1021/nn900078p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alsteens D, Garcia MC, Lipke PN, Dufrêne YF. Proc. Natl. Acad. Sci. U.S.A. 2010;107:20744–20749. doi: 10.1073/pnas.1013893107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gow NAR, Van De Veerdonk FL, Brown AJP, Netea MG. Nat. Rev. Microbiol. 2012;10:112–122. doi: 10.1038/nrmicro2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ovchinnikova ES, Krom BP, Busscher HJ, Van Der Mei HC. BMC Microbiol. 2012;12 doi: 10.1186/1471-2180-12-281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Klotz SA, Gaur NK, Lake DF, Chan V, Rauceo J, Lipke PN. Infect. Immun. 2004;72:2029–2034. doi: 10.1128/IAI.72.4.2029-2034.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chatterjee B, Sahoo P. Advances in Tribology. 2012 [Google Scholar]
- 30.Kogut L, Etsion I. J. appl. Mech.-T ASME. 2002;69:657–662. [Google Scholar]
- 31.Munro CA, Bates S, Buurman ET, Hughes HB, MacCallum DM, Bertram G, Atrih A, Ferguson MAJ, Bain JM, Brand A, Hamilton S, Westwater C, Thomson LM, Brown AJP, Odds FC, Gow NAR. J. Biol. Chem. 2005;280:1051–1060. doi: 10.1074/jbc.M411413200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brand A, Barnes JD, Mackenzie KS, Odds FC, Gow NAR. FEMS Microbiol. Lett. 2008;287:48–55. doi: 10.1111/j.1574-6968.2008.01301.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hauck D, Joachim I, Frommeyer B, Varrot A, Philipp B, Möller HM, Imberty A, Exner TE, Titz A. ACS Chem. Biol. 2013 doi: 10.1021/cb400371r. [DOI] [PubMed] [Google Scholar]
- 34.Ovchinnikova ES, Van der Mei HC, Krom BP, Busscher HJ. Colloid Surf. B-Biointerfaces. 2013;110:45–50. doi: 10.1016/j.colsurfb.2013.04.015. [DOI] [PubMed] [Google Scholar]
- 35.Gillum AM, Tsay EYH, Kirsch DR. Molecular and General Genetics. 1984;198:179–182. doi: 10.1007/BF00328721. [DOI] [PubMed] [Google Scholar]
- 36.Kucharíková S, Tournu H, Lagrou K, van Dijck P, Bujdáková H. J. Med. Microbiol. 2011;60:1261–1269. doi: 10.1099/jmm.0.032037-0. [DOI] [PubMed] [Google Scholar]
- 37.Nobile CJ, Schneider HA, Nett JE, Sheppard DC, Filler SG, Andes DR, Mitchell AP. Curr. Biol. 2008;18:1017–1024. doi: 10.1016/j.cub.2008.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]