Abstract
BACKGROUND
Loss of imprinting (LOI) is an epigenetic alteration involving loss of parental origin-specific expression at normally imprinted genes. A LOI for IGF2, a paracrine growth factor, has been implicated in the development of prostate and other cancers. In the current study, we define IGF2 LOI in histologically normal prostate tissues in relationship to tumor foci and gene expression.
METHODS
Microdissected tumor associated (TA) adjacent (2 mm) and distant (10 mm) tissues surrounding tumor foci were generated. IGF2 imprinting in informative prostate tissue sets was quantitated using a fluorescent primer extension assay and expression analyzed utilizing quantitative PCR. DNA methylation analyses were performed using quantitative pyrosequencing.
RESULTS
A marked IGF2 LOI was found in adjacent TA tissues (39 ± 3.1%) and did not significantly decrease in tissues distant (38 ± 5.3%) from tumor foci (45 ± 2.9%; P = 0.21). IGF2 imprinting correlated with IGF2 expression in TA tissues, but not within the tumor foci. Hypomethylation of the IGF2 DMR0 region correlated with decreased IGF2 expression in tumors (P < 0.01). The expression of IGF2 and its adjacent imprinted gene H19 were increased in adjacent and distant tissues compared to tumors (P < 0.05) indicating the importance of factors other than LOI in driving IGF2 expression.
CONCLUSIONS
LOI of IGF2 occurs not only adjacent to prostate tumor foci, but is widely prevalent even in distant areas within the peripheral zone. These data provide evidence for a widespread epigenetic field defect in histologically normal tissues that might be employed to identify prostate cancer in patients.
Keywords: field effect, IGF2, H19, prostate cancer
INTRODUCTION
The concept of “field cancerization” or “field effect” was first proposed by Slaughter et al. [1]. These authors concluded that in oral cancer, the frequency of independent multiple tumors far exceeded that expected by chance alone. Multifocal cancer has been observed in colon, bladder, esophagus, breast, and prostate [2]. In radical prostatectomy specimens removed for prostate cancer (PCa), typically four or more independent foci of cancer are found [3]. Several distinct precancerous lesions for PCa, prostatic intra-epithelial neoplasia (PIN), and proliferative inflammatory atrophy (PIA), also arise in a multifocal pattern [4,5]. This diffuse pattern of cancer development within the peripheral prostate suggests a carcinogenic stimulus has lead to cancer promotion in multiple susceptible cells [6].
Insulin-like Growth Factor-2 (IGF2) is a major embryonic mitogen and has an important role in the adult prostate as a paracrine and autocrine regulator of cell proliferation [7]. The production of IGF2 has been demonstrated in the media of cultured prostatic stromal cells [8] and its protein levels increase with aging in the human prostate [9]. IGF2 has been implicated in the neoplastic transformation of susceptible cells. Transgenic mice engineered to overexpress IGF2 as adults develop diverse carcinomas after a long latency period [10]. In addition, the overexpression of both IGF2, and IGF-IR, in pancreatic cells results in accelerated ß-cell tumor formation, with the rapid development of lymph node metastases [11]. These studies provide in vivo evidence that IGF2 can drive the acquisition of a cancer phenotype in susceptible cells.
Genomic imprinting is an epigenetic control in which one allele is expressed and other allele silenced based on the parental (maternal or paternal) origin of DNA. IGF2 displays genomic imprinting and is a paternally imprinted in most tissues [12]. The current model for the mechanism underlying this reciprocal imprinting (enhancer competition model) proposes a critical role for CTCF, which binds to the unmethylated maternal imprint control region (ICR) [13]. Normal development requires accurate IGF2 expression, and several disorders can be attributed to an abnormally high dose of IGF2 potentially caused by LOI. LOI has been reported in colorectal carcinomas [14], Wilm’s tumor [15], esophageal carcinoma [16], childhood acute lymphoblastic leukemia [17], and prostate cancer [18]. In cells that express both parental IGF2 alleles, the increase in IGF2 production may be a mechanism for promoting cancer development. In the mouse, CTCF serves as a strategic protein that implements DNA loops and helps silence DNA transcription [13,19]. A mouse model of IGF2 LOI and overexpression supports a role for IGF2 as a tumor initiator in intestinal cancers [20]. However, recent studies have cast doubt in humans on the link between IGF2 LOI and increased IGF2 expression in tumor tissues [16].
Previous studies in our laboratory have demonstrated that IGF2 LOI occurs in prostate cancer, and surprisingly within normal tissues from the peripheral prostate [18]. In contrast, the transition zone of the prostate, which rarely develops cancer, maintains the IGF2 imprint as do virtually all other adult tissues [18]. Aging human and mice prostate tissues show a relaxation of IGF2 imprinting associated with increased IGF2 expression [21]. This LOI is more pronounced in histologically normal tissues from men with cancer compared to those without [21]. In the present study, we define whether IGF2 LOI occurs as a widespread field defect within prostate tissues containing cancer, or whether it is a response related to the adjacent tumor (i.e., field cancerization). We demonstrate LOI in tissues adjacent (2 mm) to tumors, but also in regions distant (10 mm) from tumor foci. Notably, IGF2 levels were 2–5 fold higher in adjacent and distant normal regions when compared to tumor foci. These data indicate that IGF2 LOI marks a widespread field defect within the peripheral prostate, and that elevated IGF2 levels seen in the histologically normal prostate may be important in driving the development of multifocal prostate cancers during aging.
MATERIALS AND METHODS
Tissue Samples and Identification of IGF2 Informative Specimens
Prostatectomy samples containing tumor and associated normal tissue (TA) were obtained from men diagnosed with cancer, ranging in age from 44 to 69 years under IRB approved protocol. DNA from these 18 samples was sequenced for an exon 7 IGF2 single nucleotide polymorphism (SNP; C to G) at position 1926 (genbank accession: X07868). Nine samples were informative for this IGF2 polymorphism and were used for quantitating the IGF2 imprint status. Normal prostate samples without any associated tumors (NTA) were also obtained from age-matched cystoprostatectomy cases and from men undergoing organ donation under IRB approved protocols.
Microdissection of Prostate Tumor, Adjacent and Distant Regions
To define the relationship of IGF2 LOI to tumor foci, histological sections containing both cancer and normal regions were generated. Microdissection was performed to obtain normal tissue from regions adjacent to tumor foci (2 mm) and at a greater distance (10 mm) (Fig. 1A) as described [22]. Tissue was collected in RNAlater® Solution (Invitrogen, CA) for RNA analysis and portion stored on dry ice for further studies.
Fig. 1.
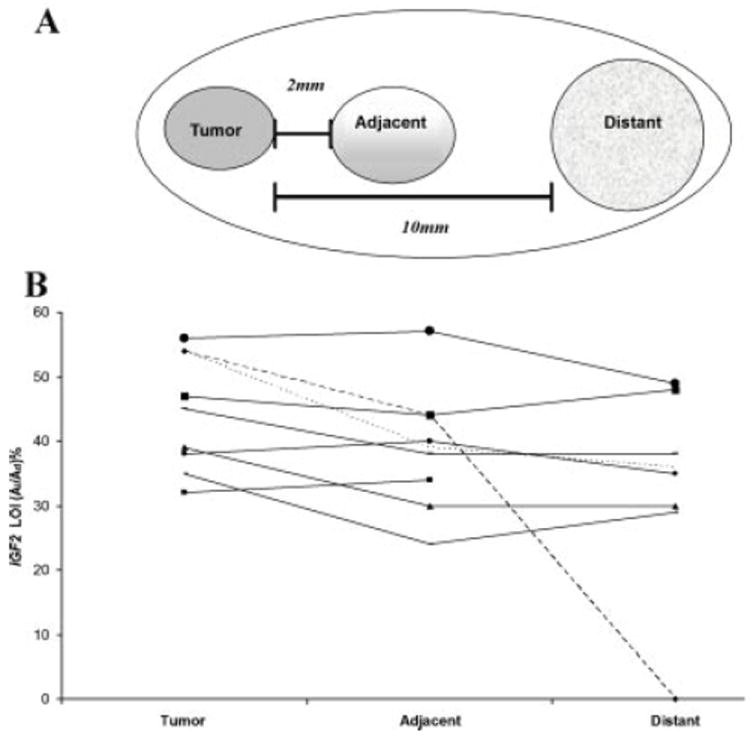
(A) Schematic showing prostate tumor, adjacent and distant normal tissue microdissection. Radical prostatectomy samples containing tumor foci were sectioned and samples microdissected for subsequent analyses. Adjacent and distant tumor-associated (TA) tissues did not contain histologic cancer. Samples were assessed in a three-dimensional pattern. (B) Fluorescent primer extension (FluPE) assay for IGF2 loss ofimprinting (LOI). Analysis of 9 informative sample sets using FluPE was performed and image analysis done using ImageJ software. LOI in prostate tumor foci ranged from 32 to 68% (mean 45%). Mean LOI in adjacent and distant TA regions was 39 and 38%, respectively. Analyses were performed in duplicate.
RNA Extraction and cDNA Synthesis
RNA was extracted from tissues using RNeasy Mini Kit (Qiagen, Valencia, CA) following manufacturer’s instructions. DNase-treated RNA was reverse-transcribed to cDNA using QuantiTect Rev. Transcription Kit (Qiagen, Valencia, CA) following manufacturer’s instructions. Briefly, 250 ng of RNA was incubated at 42°C for 2 min with genomic DNA wipe-out buffer (Qiagen, Valencia, CA) to removes genomic DNA contamination. In next step, RNA was reverse-transcribed using Quantiscript RT, buffer and poly dT primers at 42°C for 30 min. Lastly, reaction is incubated at 95°C for 3 min to inactivate Quantiscript Reverse Transcriptase.
Analysis of IGF2 LOI Using Fluorescent Primer Extension Assay
IGF2 imprinting assay was performed using a SNP located in exon 7 at 1926 position (C to G; accession: X07868) using fluorescent primer extension (FluPE) as initially described by Bennett-Baker et al. [23] and our laboratory [21]. Briefly, cDNAs were generated from DNAse treated-RNA, and a 245bp PCR product amplified from cDNA using primer sequences GCATATCTAAGCAACTACG (forward) and GTCATGGTGGAAA-CATGGAA (reverse) [18]. Products were then quantified using picogreen (Invitrogen, CA) and subject to primer extension using fluorescent primer (5′-CCAATGTTTTCATGGTCTGAGCG) and specific dNTP/ddNTPs. A 3bp or a 5bp extension for an imprinted allele or both if imprinting is no longer maintained was visualized on a sequencing gel. Quantitation was performed using ImageJ software (NIH, USA). A standard curve with variable ratios of each allele was run with each assay confirming a linear relationship with the above parameters.
Quantitative Real-time PCR
Quantitative real-time PCR (qPCR) expression profiles of IGF2, CTCF, H19, WT1, and PTEN were measured as previously described [21] using a MyiQ™ Two-Color Real-Time PCR Detection System (Bio-Rad, Hercules, CA). Primers were designed using Primer Express (Applied Biosystems, Foster City, CA) (Table I). Relative quantification of gene expression changes was recorded after normalizing for 18S expression, computed by using the 2−ΔΔCT method (user manual 2, ABI Prism 7700 SDS). In the 2−ΔΔCT analysis, the threshold cycle (CT) from tumor was used as a calibrator sample. Statistical analyses of the data were performed by comparing tumor CT with associated near, far tumor CT for each gene using a two-tailed t-test with unequal group variance.
TABLE I.
Clinical and Pathologic Characteristics of Patients with Prostate Cancer
| Specimen | Age (years) | Gleason’s grade | Tumor volume (%) | Stage (T) | Margin (+ or −)1 | Preoperative PSA (ng/ml) | Family history |
|---|---|---|---|---|---|---|---|
| 1 | 44 | 3 + 4 | 25 | T2C | + | 10 | − |
| 2 | 58 | 3 + 4 | 10 | T3A | − | 5 | + |
| 3 | 65 | 3 + 4 | 10 | T2C | − | 4.6 | + |
| 4 | 56 | 3 + 3 | 5 | T2C | − | 4.1 | − |
| 5 | 52 | 3 + 5 | 25 | T2C | − | 12.6 | N/A |
| 6 | 54 | 3 + 4 | 20 | T2C | − | 5.4 | − |
| 7 | 63 | 3 + 4 | 70 | T3B | + | 6.7 | N/A |
| 8 | 59 | 3 + 4 | 20 | T2C | − | 5.7 | + |
| 9 | 69 | 3 + 4 | 30 | T2b | − | 6.5 | − |
| 10 | 63 | 3 + 4 | 50 | T2C | + | 4.6 | + |
| 11 | 58 | 3 + 4 | 10 | T2C | − | 9.3 | + |
| 12 | 59 | 3 + 3 | 5 | T2C | − | 4.2 | + |
| 13 | 63 | 3 + 3 | 10 | T2A | − | 4 | − |
| 14 | 53 | 3 + 3 | 5 | T2C | − | 5.2 | − |
| 15 | 52 | 3 + 3 | 6 | T2C | − | 4.6 | + |
| 16 | 62 | 3 + 4 | 10 | T2C | − | 3.3 | N/A |
| 17 | 61 | 3 + 4 | 20 | T3A | + | 4.2 | − |
| 18 | 64 | 4 + 5 | 15 | T2B | − | 9.5 | − |
N/A indicates information not available.
Surgical margin status in the postoperative specimen.
Methylation Analysis in IGF2 DMR0 and H19 ICR Region
DNA methylation was analyzed in IGF2 DMR0 and IGF2-H19 imprint control region (ICR) using pyrosequencing methodology using a PSQ HS 96 and PyroMark MD System and Pyro Gold Reagent kits (Biotage, Uppsala, Sweden) as described [24]. Methylation was quantified using Pyro Q-CpG Software (Biotage, Uppsala, Sweden), which calculates the ratio of converted Cs (Ts) to unconverted Cs at each CpG and expresses this as a percentage methylation. Average methylation of sample sets across each region at each CpG was calculated. The following controls were included on each 96-well plate: prostate cell line PPC1 (0–20% methylation at IGF2 DMR0), M. SssI-treated methylated DNA from PPC1 (100% methylation), and a negative control for PCR amplification.
Statistical Analysis
Correlation analysis was done using GraphPad Prizm (San Diego, CA) between IGF2, H19, and CTCF expression in the distant region using Spearman’s correlation with significance measured using P-values less than 0.05 derived from two-tailed t-test.
RESULTS
IGF2 LOI is Widely Demonstrated Throughout Histologically Normal Prostate Tissues Associated with Cancer
The clinical and pathological characteristics at radical prostatectomy of the prostate cancer study population are presented in Table I. Average tumor volume was 19% (range 5–70%) and all pre-operative PSAs were less than 12. Genotyping revealed 9 of 18 prostate samples to be informative for exon 7 IGF2 polymorphism (50%). The FluPE assay generates some “leakage” from the upper allele (12%) when the lower allele is dominant that was corrected for in the subsequent analyses.
Tumor, adjacent and distant region DNA (Fig. 1A) from these nine informative samples was subsequently utilized for IGF2 LOI analysis using FluPE. LOI in prostate cancer samples ranged from 32 to 68% (mean 45%) (Fig. 1B). Mean LOI in adjacent and distant TA regions (39 and 38%, respectively) did not significantly differ from tumor. One sample (No. 3; Table I) demonstrated imprinting in a distant TA region. Controls including associated seminal vesicle, bladder, and kidney tissues demonstrated minimal (10–15%) but detectable expression of the silenced allele by FluPE. This low level appears to represent the normal physiologic “silenced” state of associated genitourinary tissues, and was significantly lower than that seen in all prostate tissues analyzed.
IGF2 and H19 Expression is Higher in the Normal Tissue of Prostates Containing Cancer than in Tissues without Associated Cancer or in Tumors
IGF2 changes in expression have been linked to altered imprinting in mouse models [20,25], but results in human tumors have been conflicting. Relative expression of IGF2 and its adjacent imprinted 3′ neighbor H19 were measured in adjacent and distant TA regions and were compared to tumor foci using qPCR. When all 18 prostate samples were analyzed, IGF2 mRNA expression was significantly greater in normal adjacent and distant TA tissues compared to their associated tumors (Fig. 2A; P < 0.05). However, in a subset of five TA tissues, IGF2 expression was higher in the associated tumor foci. We also compared IGF2 expression in the normal TA samples to a separate panel of age-matched peripheral prostate tissues from prostate specimens without any associated cancer (NTA). In TA tissues associated with tumors, IGF2 expression was significantly higher by 3.1 fold (dCT, 13.29 ± 0.32; N = 14) compared to NTA normal prostate tissues (dCT, 14.92 ± 0.65; N = 6; P = 0.025). In summary, the expression of IGF2 is greater in TA tissues than tissues without associated cancer or in tumors.
Fig. 2. IGF2 and H19 expression in tumor and TA tissues.
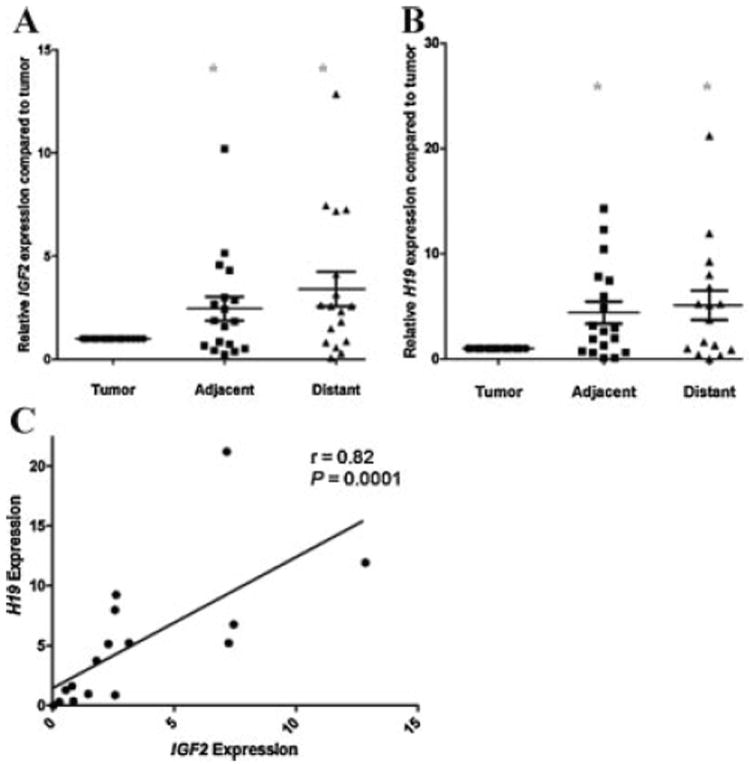
Gene expression was measured using quantitative PCR(qPCR) in adjacent and distant TA tissues relative to the tumor for 18 microdissected sample sets (A), IGF2 mRNA expression (B), H19 mRNA expression (C). In contrast to predicted results, a positive correlation between IGF2 and H19 expression in distant TA tissues was seen (r = 0.82, P = 0.0001). No correlation was found between IGF2 and H19 gene expressionin tumors (data not shown) (* = P < 0.05).
H19 is inversely linked to IGF2 expression in mouse models [26] and may play a role in human cancer progression [27]. H19 mRNA expression was significantly greater in TA adjacent and distant tissues when compared to tumors (Fig. 2B; P < 0.05). However, several samples [5] demonstrated decreased expression in TA tissues compared to tumor foci. A positive correlation was found between IGF2 and H19 expression in distant TA tissues (P = 0.0001; Fig. 2C). Notably, this correlation was not demonstrated within tumors (r = 0.41; P = 0.11). Thus, IGF2 and H19 expression is greater in adjacent and distant TA tissues than in the associated tumors. Furthermore, the inverse correlation in expression between IGF2 and H19 that genetic mouse models predict [28,29] is not seen in these human prostate tissues.
IGF2 Imprinting and Expression Correlates in TA Tissues, but not Prostate Tumors
Studies have suggested that IGF2 LOI is associated with increased IGF2 expression [30,31]. No correlation was seen between IGF2 LOI and expression in prostate tumors (r = −0.49, P = 0.09; Fig. 3A). However, in distant TA tissues LOI positively correlated with increased IGF2 expression (r = 0.58, P = 0.017; Fig. 3B). No association was seen between IGF2 LOI and H19 expression in any of the tissues tested.
Fig. 3. Correlation between IGF2 LOI and IGF2 expression.
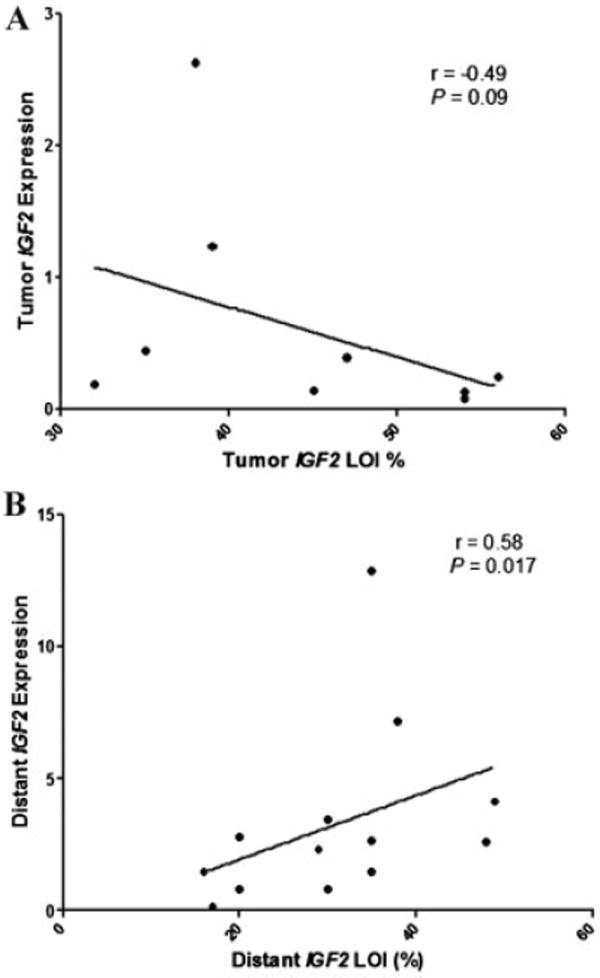
Gene expression was analyzed using qPCR and FLuPE utilized for imprinting analyses. Correlation analyses were performed using Spearman’s correlation. (A) Tumor samples (r = −0.49, P = 0.09), (B) distant TA tissues (r = 0.58, P = 0.017).
Increased IGF2 Expression is Associated with Decreased DNA Methylation at DMR0
Several genomic areas within 11p15.2 have been postulated to modulate IGF2 expression [24,31]. We examined the methylation status of two of these, DMR0 and CTCF6 (a CTCF binding site within the ICR) using pyrosequencing in prostate tumor-normal pairs showing either increased (n = 5) or decreased (n = 7) IGF2 tumor expression (Fig. 4A). In tumor samples with decreased tumor IGF2 expression, five of six CpGs in the DMR0 region showed decreased methylation compared to TA tissues with higher IGF2 levels (Fig. 4B; P < 0.05). In a correlation analysis of all 12 T–N pairs, significance was observed between lower tumor IGF2 levels and decreased methylation at the DMR0 region of tumor samples (r = 0.40 to 0.75, P < 0.05). No significant methylation changes within ICR CTCF#6 correlated with expression.
Fig. 4. Analysis of methylation status in paired prostate tumors and TA tissues.
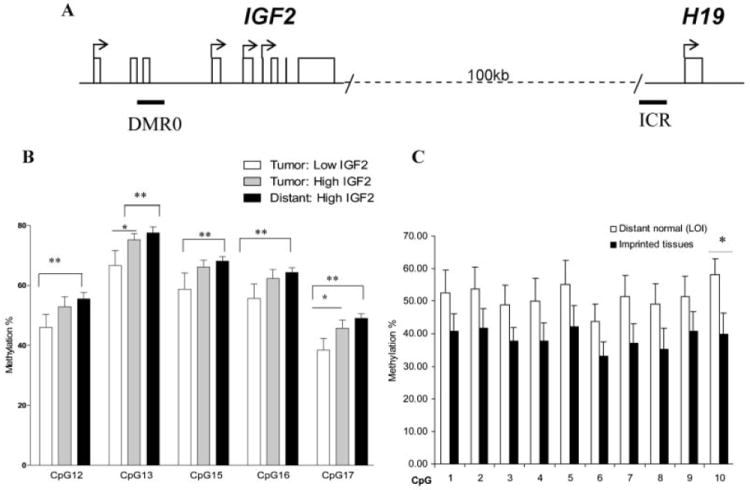
(A) Alterations in IGF2 expression have been linked to changes in methylation within DMR0 of the IGF2 promoter, as well as within CTCF#6 in the imprint control region (ICR) [24,54]. Methylation was analyzed using pyrosequencing specific primers. Tissues analyzed included prostate tumors with low or high IGF2 expression and distant TA tissues. An additional set of tissues from bladder, kidney, and prostates without cancer (NTA) that demonstrated maintained IGF2 imprinting were also analyzed for methylation. (B) At DMR0, tumors with low IGF2 expression demonstrate significantly decreased methylation compared to distant TA tissues. (C) CpG methylation analysis of CTCF6 in NTA tissues. TA prostate tissues from men with associated cancer (n = 6) demonstrating LOI is compared to imprinted NTA tissues (n = 7). A trend towards increased methylation in distant TA tissues is noted with significance seen at CpG 10 (P = 0.02).
Hypermethylation with in the Imprint Control Region (ICR) is Associated with IGF2 LOI in TA Prostate Tissues Compared to NTAT issues Displaying Imprinting
The striking finding of IGF2 LOI throughout histologically normal peripheral prostate tissues associated with cancer lead us to next examine altered DNA methylation as an underlying mechanism. The IGF2-H19 ICR contains several CTCF binding sites that can be methylated leading to IGF2 LOI in several mouse models [28,32]. We compared the methylation status at the critical CTCF6 region in TA distant tissues (n = 6) demonstrating IGF2 LOI to NTA normal tissues from imprinted urologic organs including kidney, bladder, seminal vesicles, and prostates without cancer (n = 7). Increased methylation at CTCF#6 was seen in TA prostate tissues displaying LOI (Fig. 4C). In contrast, hypermethylation at CTCF#6 was not seen in tumors (all containing LOI) when compared to imprinted normal urologic tissues (Fig. 2). Consistent with the enhancer competition model [13], DNA hypermethylation with the ICR is associated with LOI in histologically normal tissues, but this model is not valid in prostate tumors.
DISCUSSION
Defining aspects of prostate cancer include both its multifocality and the marked increase in the disease with aging. These features are consistent with the presence of a field effect that arises in the human prostate. Epigenetic factors including DNA methylation, genomic imprinting, and histone modifications have been postulated as candidates for the field defect [6]. In the present study, we define IGF2 LOI in relation to distance from tumor foci and find evidence of a widespread IGF2 LOI throughout the peripheral prostate in men with prostate cancer. Furthermore, we demonstrate that levels of IGF2 are increased in these tumor-associated tissues. LOI neither occurs in periuretheral samples of benign prostatic hyperplasia (BPH), nor is it found in other adult tissues [18]. Given its role in initiating tumor growth [10], this epigenetic modification may represent an important biologic change in the development of prostate cancer.
We examined tumors and their matched adjacent (2 mm) and distant (10 mm) non-tumor tissues for imprinting and expression using a sensitive fluorescent primer extension (FluPE) assay. Samples were analyzed in a three-dimensional fashion to avoid tumor contamination. Given the surprising finding that IGF2 LOI occurs both adjacent to tumors and at more distant sites, our data indicate that LOI is unlikely to be a local (i.e., field cancerization) effect of the tumor. In contrast, other research has identified nuclear morphology [33-35] and DNA hypermethylation [36,37] changes in normal tissues immediately adjacent to a tumor focus. In these studies, ex vivo core biopsies taken at varying distances (1–4 mm) from a PCa focus revealed increased methylation of APC, RARb2, or RASSF1A in a small subset of samples (10–20%) [36]. Hanson et al. separated the adjacent tissue compartments and found methylation of GSTpi and RARb2 to be more marked in reactive stroma [37]. In the current study, IGF2 LOI occurs with more frequency (85%), and marks a wider field effect throughout the prostate rather than a local response to the tumor. LOI in tumor tissues is only slightly more extensive (mean 45%) than that seen in the adjacent and distant peripheral prostate (mean 39 and 38%, respectively; Fig. 1B). Other studies supporting a broader field defect in the prostate include microarray-based studies [38,39] and alterations in the protein markers Mcm-2, Ki67 [40] and alpha-methylacyl-CoA racemase [41].
A second major finding is that elevated levels of IGF2 are found in adjacent and distant TA tissues when compared to associated tumors and to non-cancer containing prostates. IGF2 has important mitogenic properties in normal epithelium [42], prostate cancer cell lines, and stromal cells associated with prostate cancer [43]. Analyzing all samples, IGF2 mRNA expression was greater in adjacent and distant TA tissues (2.45 ± 0.58 and 3.40 ± 0.81 fold, respectively; P < 0.05) compared to related tumor tissues. A subset of tumors (27%) was found to elaborate higher levels of IGF2. We also compared prostate tissues from age-matched men without prostate cancer (NTA) and found IGF2 levels to be significantly higher in TA tissues (3.1 fold; P = 0.025). This paradoxical increase IGF2 expression seen within TA tissues raises intriguing questions as to the role of IGF2 in prostate tumorigenesis. High levels of IGF2 result in varied carcinomas after a long latency period [10]. More modest two-fold increases in IGF2 levels lead ApcMin mice to develop twice as many intestinal tumors as control littermates do [20]. These studies suggest that chronic IGF2 exposure has the capacity to act on selected cells as an early initiation factor in the development of neoplasia.
Current hypotheses involving the enhancer competition model of IGF2 LOI would predict an increase in IGF2 expression with LOI [44-46]. We find a correlation between the extent of LOI and increased expression in TA tissue samples (P = 0.02) consistent with this model. This correlation has been noted in normal esophageal epithelia [16]. However, within prostate tumor samples, no association of IGF2 expression with LOI is found (Fig. 3A). Our data suggest other mechanisms may be more important in regulating IGF2 expression than imprinting in prostate tumors. We also examined two negative regulators of IGF2 expression, WT1 and PTEN [47], in these samples and found no correlation with expression (data not shown).
The enhancer competition model of imprinting predicts that H19 and IGF2 expression is inversely related since they compete for a common transcriptional enhancer [12,44,45]. In contrast, when analyzing TA tissues a positive correlation exists between IGF2 and H19 (e.g., both increase expression). This would suggest other factors play a role in the regulation of these genes. H19, a maternally imprinted non-coding RNA may have tumor suppressor activity [48], although more recent studies suggest a potentiating role in tumor angiogenesis, invasion, and metastasis [27,49]. We found H19 expression to be greater in adjacent and distant normal prostate (4.4 ± 1.04 and 8.05 ± 3.12, respectively; P < 0.05) compared to tumor foci. This decrease in expression putatively supports a tumor suppressive role for H19 in prostate cancer.
We used quantitative pyrosequencing to determine if alterations in methylation were linked to increased IGF2 expression. A significant correlation was seen between decreased IGF2 expression and hypomethylation at 5 CpGs within the IGF2 DMR0 region. Human DMR0, which does not bind CTCF, may act as a silencer element that can suppress IGF2 independent of the ICR-CTCF regulatory pathway [50]. The function of DMR0 is not clear although hypomethylation has been demonstrated in Wilm’s tumors [24], colorectal (94%) and breast cancers (35%) [50,51].
Another component of the enhancer competition model is hypermethylation at several CTCF binding sites within the ICR, most notably CTCF#6, leading to an inhibition of CTCF binding and biallelic expression [28,45,46]. Consistent with this, increased methylation within CTCF6 was seen in distant TA tissues (demonstrating LOI) compared to imprinted NTA tissues from prostates and other tissues without associated cancer (Fig. 4C). This indicates a role for hypermethylation of CTCF in the regulation of imprinting in prostate tissues. In contrast, significant hypermethylation of CTCF#6 was not seen in tumor foci (Fig. 2) when compared to imprinted NTA tissues. LOI may regulated by other factors in tumors, possibly an allele-specific activation of specific IGF2 promoters [52].
CONCLUSIONS
In summary, we find that LOI of IGF2 occurs not only in prostate tumor foci, but is widespread throughout the peripheral prostate providing evidence for a field defect. This may contribute to the known multifocality of prostate cancer. This data supports a recently proposed epigenetic progenitor model of cancer that suggests LOI may play a more crucial early role in progenitor cells than in established tumors [53]. IGF2 LOI in normal colonic epithelia has been associated with an increased risk of colorectal neoplasia in some studies [20]. Furthermore, IGF2 is increased in normal prostate tissue in prostates with cancer indicating that IGF2 may be important in the early stages of prostate carcinogenesis. IGF2 LOI may be a clinically relevant molecular marker found in normal tissues for prostate cancer.
Acknowledgments
This work was supported by the National Institutes of Health (5R01CA097131). The authors would like to thank Carlos G Coriano for help with prostate tissue microdissection.
Abbreviations
- FluPE
fluorescent primer extension assay
- ICR
imprint control region
- IGF2
insulin-like growth factor 2
- LOI
loss of imprinting
- DMR0
differentially methylated region 0
- TA
tumor associated
- NTA
non-tumor associated
Footnotes
Supporting information may be found in the online version of this paper.
No financial connection between any of the authors and the subject matter.
References
- 1.Slaughter DP, Southwick HW, Smejkal W. Field cancerization in oral stratified squamous epithelium. Clinical implications of multicentric origin. Cancer. 1953;6(5):963–968. doi: 10.1002/1097-0142(195309)6:5<963::aid-cncr2820060515>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 2.Qian J, Wollan P, Bostwick DG. The extent and multicentricity of high-grade prostatic intraepithelial neoplasia in clinically localized prostatic adenocarcinoma. Hum Pathol. 1997;28(2):143. doi: 10.1016/s0046-8177(97)90097-6. [DOI] [PubMed] [Google Scholar]
- 3.Epstein JI, Carmichael MJ, Partin AW, Walsh PC. Small high grade adenocarcinoma of the prostate in radical prostatectomy specimens performed for nonpalpable disease: pathogenetic and clinical implications. J Urol. 1994;151(6):1587–1592. doi: 10.1016/s0022-5347(17)35309-0. [DOI] [PubMed] [Google Scholar]
- 4.De Marzo AM, Platz EA, Sutcliffe S, Xu J, Gronberg H, Drake CG, Nakai Y, Isaacs WB, Nelson WG. Inflammation in prostate carcinogenesis. Nat Rev Canc. 2007;7(4):256. doi: 10.1038/nrc2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vukovic B, Park PC, Al-Maghrabi J, Beheshti B, Sweet J, Evans A, Trachenberg J, Squire JA. Evidence of multifocality of telomere erosion in high-grade prostatic intraepithelial neoplasia (HPIN) and concurrent carcinoma. Oncogene. 2003;22(13):1978. doi: 10.1038/sj.onc.1206227. [DOI] [PubMed] [Google Scholar]
- 6.Larisa N, Vijayalakshmi A, Peter HG. Evidence for field cancerization of the prostate. Prostate. 2009;69(13):1470–1479. doi: 10.1002/pros.20983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yu H, Rohan T. Role of the insulin-like growth factor family in cancer development and progression. J Natl Canc Inst. 2000;92(18):1472–1489. doi: 10.1093/jnci/92.18.1472. [DOI] [PubMed] [Google Scholar]
- 8.Cohen P, Peehl DM, Baker B, Liu F, Hintz RL, Rosenfeld RG. Insulin-like growth factor axis abnormalities in prostatic stromal cells from patients with benign prostatic hyperplasia. J Clin Endocrinol Metab. 1994;79(5):1410–1415. doi: 10.1210/jcem.79.5.7525636. [DOI] [PubMed] [Google Scholar]
- 9.Li S-L, Goko H, Xu Z-D, Kimura G, Sun Y, Kawachi MH, Wilson TG, Wilczynski S, Fujita-Yamaguchi Y. Expression of insulin-like growth factor (IGF)-II in human prostate, breast, bladder, and paraganglioma tumors. Cell Tissue Res. 1998;291(3):469. doi: 10.1007/s004410051016. [DOI] [PubMed] [Google Scholar]
- 10.Rogler CE, Yang D, Rossetti L, Donohoe J, Alt E, Chang CJ, Rosenfeld R, Neely K, Hintz R. Altered body composition and increased frequency of diverse malignancies in insulin-like growth factor-II transgenic mice. J Biol Chem. 1994;269(19):13779–13784. [PubMed] [Google Scholar]
- 11.Lopez T, Hanahan D. Elevated levels of IGF-1 receptor convey invasive and metastatic capability in a mouse model of pancreatic islet tumorigenesis. Cancer cell. 2002;1(4):339. doi: 10.1016/s1535-6108(02)00055-7. [DOI] [PubMed] [Google Scholar]
- 12.Ohlsson R, Hedborg F, Holmgren L, Walsh C, Ekstrom TJ. Overlapping patterns of IGF2 and H19 expression during human development: biallelic IGF2 expression correlates with a lack of H19 expression. Development. 1994;120(2):361–368. doi: 10.1242/dev.120.2.361. [DOI] [PubMed] [Google Scholar]
- 13.Phillips JE, Corces VG. CTCF: master weaver of the genome. Cell. 2009;137(7):1194. doi: 10.1016/j.cell.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cui H, Cruz-Correa M, Giardiello FM, Hutcheon DF, Kafonek DR, Brandenburg S, Wu Y, He X, Powe NR, Feinberg AP. Loss of IGF2 imprinting: a potential marker of colorectal cancer risk. Science. 2003;299(5613):1753–1755. doi: 10.1126/science.1080902. [DOI] [PubMed] [Google Scholar]
- 15.Ogawa O, Becroft DM, Morison IM, Eccles MR, Skeen JE, Mauger DC, Reeve AE. Constitutional relaxation of insulin–like growth factor II gene imprinting associated with Wilms’ tumour and gigantism. Nat Genets. 1993;5(4):408–412. doi: 10.1038/ng1293-408. [DOI] [PubMed] [Google Scholar]
- 16.Zhao R, DeCoteau JF, Geyer CR, Gao M, Cui H, Casson AG. Loss of imprinting of the insulin-like growth factor II (IGF2) gene in esophageal normal and adenocarcinoma tissues. Carcinogenesis. 2009;30(12):2117–2122. doi: 10.1093/carcin/bgp254. [DOI] [PubMed] [Google Scholar]
- 17.Peter V, Heike W, Cornelia B, Bianka H, Uwe S, Uwe M. Loss of imprinting of IGF-II gene in children with acute lymphoblastic leukemia. Leuk Res. 2003;27(9):807. doi: 10.1016/s0145-2126(03)00014-6. [DOI] [PubMed] [Google Scholar]
- 18.Jarrard DF, Bussemakers MJ, Bova GS, Isaacs WB. Regional loss of imprinting of the insulin-like growth factor II gene occurs in human prostate tissues. Clin Canc Res. 1995;1(12):1471–1478. [PubMed] [Google Scholar]
- 19.Ling JQ, Li T, Hu JF, Vu TH, Chen HL, Qiu XW, Cherry AM, Hoffman AR. CTCF mediates interchromosomal colocalization between Igf2/H19 and Wsb1/Nf1. Science. 2006;312(5771):269–272. doi: 10.1126/science.1123191. [DOI] [PubMed] [Google Scholar]
- 20.Sakatani T, Kaneda A, Iacobuzio-Donahue CA, Carter MG, de Boom Witzel S, Okano H, Ko MSH, Ohlsson R, Longo DL, Feinberg AP. Loss of imprinting of Igf2 alters intestinal maturation and tumorigenesis in mice. Science. 2005;307(5717):1976–1978. doi: 10.1126/science.1108080. [DOI] [PubMed] [Google Scholar]
- 21.Fu VX, Dobosy JR, Desotelle JA, Almassi N, Ewald JA, Srinivasan R, Berres M, Svaren J, Weindruch R, Jarrard DF. Aging and cancer-related loss of insulin-like growth factor 2 imprinting in the mouse and human prostate. Canc Res. 2008;68(16):6797–6802. doi: 10.1158/0008-5472.CAN-08-1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schwarze SR, Luo J, Isaacs WB, Jarrard DF. Modulation of CXCL14 (BRAK) expression in prostate cancer. Prostate. 2005;64(1):67. doi: 10.1002/pros.20215. [DOI] [PubMed] [Google Scholar]
- 23.Bennett-Baker PE, Wilkowski J, Burke DT. Age-associated activation of epigenetically repressed genes in the mouse. Genetics. 2003;165(4):2055–2062. doi: 10.1093/genetics/165.4.2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Murrell A, Ito Y, Verde G, Huddleston J, Woodfine K, Silengo MC, Spreafico F, Perotti D, De Crescenzo A, Sparago A, Cerrato F, Riccio A. Distinct methylation changes at the IGF2-H19 locus in congenital growth disorders and cancer. PLoS ONE. 2008;3(3):e1849. doi: 10.1371/journal.pone.0001849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pant V, Mariano P, Kanduri C, Mattsson A, Lobanenkov V, Heuchel R, Ohlsson R. The nucleotides responsible for the direct physical contact between the chromatin insulator protein CTCF and the H19 imprinting control region manifest parent of origin-specific long-distance insulation and methylation-free domains. Gene Dev. 2003;17(5):586–590. doi: 10.1101/gad.254903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li Y-M, Franklin G, Cui H-M, Svensson K, He X-B, Adam G, Ohlsson R, Pfeifer S. The H19 transcript is associated with polysomes and may regulate IGF2 expression intrans. J Biol Chem. 1998;273(43):28247–28252. doi: 10.1074/jbc.273.43.28247. [DOI] [PubMed] [Google Scholar]
- 27.Matouk IJ, DeGroot N, Mezan S, Ayesh S, Abu-Lail R, Hochberg A, Galun E. The H19 non-coding RNA is essential for human tumor growth. PLoS One. 2007;2(9):e845. doi: 10.1371/journal.pone.0000845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bell AC, Felsenfeld G. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature. 2000;405(6785):482. doi: 10.1038/35013100. [DOI] [PubMed] [Google Scholar]
- 29.Thorvaldsen JL, Duran KL, Bartolomei MS. Deletion of the H19 differentially methylated domain results in loss of imprinted expression of H19 and IGF2. Gene Dev. 1998;12(23):3693–3702. doi: 10.1101/gad.12.23.3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ravenel JD, Broman KW, Perlman EJ, Niemitz EL, Jayawardena TM, Bell DW, Haber DA, Uejima H, Feinberg AP. Loss of imprinting of insulin-like growth factor-II (IGF2) gene in distinguishing specific biologic subtypes of Wilms tumor. J Natl Canc Inst. 2001;93(22):1698–1703. doi: 10.1093/jnci/93.22.1698. [DOI] [PubMed] [Google Scholar]
- 31.Woodson K, Flood A, Green L, Tangrea JA, Hanson J, Cash B, Schatzkin A, Schoenfeld P. Loss of insulin-like growth factor-II imprinting and the presence of screen-detected colorectal adenomas in women. J Natl Canc Inst. 2004;96(5):407–410. doi: 10.1093/jnci/djh042. [DOI] [PubMed] [Google Scholar]
- 32.Holmgren C, Kanduri C, Dell G, Ward A, Mukhopadhya R, Kanduri M, Lobanenkov V, Ohlsson R. CpG methylation regulates the Igf2/H19 insulator. Curr Biol: CB. 2001;11(14):1128. doi: 10.1016/s0960-9822(01)00314-1. [DOI] [PubMed] [Google Scholar]
- 33.Rodolfo M, Lucilla D, Roberto P, Deborah T, Peter HB. Subtle changes in benign tissue adjacent to prostate neoplasia detected with a Bayesian belief network. J Pathol. 1997;182(4):442–4449. doi: 10.1002/(SICI)1096-9896(199708)182:4<442::AID-PATH866>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 34.Mairinger T, Mikuz G, Gschwendtner A. Nuclear chromatin texture analysis of nonmalignant tissue can detect adjacent prostatic adenocarcinoma. Prostate. 1999;41(1):12–19. doi: 10.1002/(sici)1097-0045(19990915)41:1<12::aid-pros3>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- 35.Veltri RW, Khan MA, Miller MC, Epstein JI, Mangold LA, Walsh PC, Partin AW. Ability to predict metastasis based on pathology findings and alterations in nuclear structure of normal-appearing and cancer peripheral zone epithelium in the prostate. Clin Canc Res. 2004;10(10):3465–3473. doi: 10.1158/1078-0432.CCR-03-0635. [DOI] [PubMed] [Google Scholar]
- 36.Jyoti M, Shobha V, Haiying W, Hsiling C, Janet V, Karen G, Raymond BN, Jaclyn RN, Abhijit M. Quantitative, spatial resolution of the epigenetic field effect in prostate cancer. Prostate. 2008;68(2):152–160. doi: 10.1002/pros.20675. [DOI] [PubMed] [Google Scholar]
- 37.Hanson JA, Gillespie JW, Grover A, Tangrea MA, Chuaqui RF, Emmert-Buck MR, Tangrea JA, Libutti SK, Linehan WM, Woodson KG. Gene promoter methylation in prostate tumor-associated stromal cells. J Natl Canc Inst. 2006;98(4):255–261. doi: 10.1093/jnci/djj051. [DOI] [PubMed] [Google Scholar]
- 38.Yu YP, Landsittel D, Jing L, Nelson J, Ren B, Liu L, McDonald C, Thomas R, Dhir R, Finkelstein S. Gene expression alterations in prostate cancer predicting tumor aggression and preceding development of malignancy. J Clin Oncol. 2004;22(14):2790. doi: 10.1200/JCO.2004.05.158. [DOI] [PubMed] [Google Scholar]
- 39.Chandran UR, Dhir R, Ma C, Michalopoulos G, Becich M, Gilbertson J. Differences in gene expression in prostate cancer, normal appearing prostate tissue adjacent to cancer and prostate tissue from cancer free organ donors. BMC Canc. 2005;5(1):45. doi: 10.1186/1471-2407-5-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ananthanarayanan V, Deaton RJ, Yang XJ, Pins MR, Gann PH. Alteration of proliferation and apoptotic markers in normal and premalignant tissue associated with prostate cancer. BMC Canc. 2006;6(1):73. doi: 10.1186/1471-2407-6-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ananthanarayanan V, Deaton RJ, Yang XJ, Pins MR, Gann PH. Alpha-methylacyl-CoA racemase (AMACR) expression in normal prostatic glands and high-grade prostatic intraepithelial neoplasia (HGPIN): association with diagnosis of prostate cancer. Prostate. 2004;63(4):341–3346. doi: 10.1002/pros.20196. [DOI] [PubMed] [Google Scholar]
- 42.Cohen P, Peehl DM, Lamson G, Rosenfeld RG. Insulin-like growth factors (IGFs), IGF receptors, and IGF-binding proteins in primary cultures of prostate epithelial cells. J Clin Endocrinol Metab. 1991;73(2):401–407. doi: 10.1210/jcem-73-2-401. [DOI] [PubMed] [Google Scholar]
- 43.Cardillo MR, Monti S, Di Silverio F, Gentile V, Sciarra F, Toscano V. Insulin-like growth factor (IGF)-I, IGF-II and IGF type I receptor (IGFR-I) expression in prostatic cancer. Anti-canc Res. 2003;23(5):3825–3836. [PubMed] [Google Scholar]
- 44.Leighton PA, Ingram RS, Eggenschwiler J, Efstratiadis A, Tilghman SM. Disruption of imprinting caused by deletion of the H19 gene region in mice. Nature. 1995;375(6526):34. doi: 10.1038/375034a0. [DOI] [PubMed] [Google Scholar]
- 45.Ohlsson R, Renkawitz R, Lobanenkov V. CTCF is a uniquely versatile transcription regulator linked to epigenetics and disease. Trend Genet. 2001;17(9):520. doi: 10.1016/s0168-9525(01)02366-6. [DOI] [PubMed] [Google Scholar]
- 46.Reik W, Murrell A. Genomic imprinting - silence across the border. Nature. 2000;405(6785):408–409. doi: 10.1038/35013178. [DOI] [PubMed] [Google Scholar]
- 47.Samani AA, Yakar S, LeRoith D, Brodt P. The role of the IGF system in cancer growth and metastasis: overview and recent insights. Endocr Rev. 2007;28(1):20–47. doi: 10.1210/er.2006-0001. [DOI] [PubMed] [Google Scholar]
- 48.Hao Y, Crenshaw T, Moulton T, Newcomb E, Tycko B. Tumour-suppressor activity of H19 RNA. Nature. 1993;365(6448):764. doi: 10.1038/365764a0. [DOI] [PubMed] [Google Scholar]
- 49.Matouk I, Ohana P, Ayesh S, Sidi A, Czerniak A, de Groot N, Hochberg A. The oncofetal H19 RNA in human cancer, from the bench to the patient. Canc Ther. 2005;3:249–266. [Google Scholar]
- 50.Vu TH, Nguyen AH, Hoffman AR. Loss of IGF2 imprinting is associated with abrogation of long-range intrachromosomal interactions in human cancer cells. Hum Mol Genet. 2010;19(5):901–919. doi: 10.1093/hmg/ddp558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Baba Y, Nosho K, Shima K, Huttenhower C, Tanaka N, Hazra A, Giovannucci EL, Fuchs CS, Ogino S. Hypomethylation of the insulin-like growth factor 2 differentially methylated region in colorectal tumors, detected by bisulfite pyrosequencing, is associated with poor prognosis. Gastroenterology. 2010;139(6):1855–1864. doi: 10.1053/j.gastro.2010.07.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ekstrom TJ, Cui H, Li X, Ohlsson R. Promoter-specific IGF2 imprinting status and its plasticity during human liver development. Development. 1995;121(2):309–316. doi: 10.1242/dev.121.2.309. [DOI] [PubMed] [Google Scholar]
- 53.Feinberg AP, Ohlsson R, Henikoff S. The epigenetic progenitor origin of human cancer. Nat Rev Genet. 2006;7(1):21. doi: 10.1038/nrg1748. [DOI] [PubMed] [Google Scholar]
- 54.Ito Y, Koessler T, Ibrahim AEK, Rai S, Vowler SL, Abu-Amero S, Silva AL, Maia AT, Huddleston JE, Uribe-Lewis S. Somatically acquired hypomethylation of IGF2 in breast and colorectal cancer. Hum Mol Genet. 2008;17(17):2633. doi: 10.1093/hmg/ddn163. [DOI] [PMC free article] [PubMed] [Google Scholar]


