Abstract
Aims/hypothesis
Candidate gene and genome-wide association studies have identified ∼60 susceptibility loci for type 2 diabetes. A majority of these loci have been discovered and tested only in European populations. The aim of this study was to assess the presence and extent of trans-ethnic effects of these loci in an East Asian population.
Methods
A total of 9,335 unrelated Chinese Han individuals, including 4,535 with type 2 diabetes and 4,800 non-diabetic ethnically matched controls, were genotyped using the Illumina 200K Metabochip. We tested 50 established loci for type 2 diabetes and related traits (fasting glucose, fasting insulin, 2 h glucose). Disease association with the additive model of inheritance was analysed with logistic regression.
Results
We found that 14 loci significantly transferred to the Chinese population, with two loci (p = 5.7 × 10−12 for KCNQ1; p = 5.0 × 10−8 for CDKN2A/B-CDKN2BAS) reaching independent genome-wide statistical significance. Five of these 14 loci had similar lead single-nucleotide polymorphisms (SNPs) as were found in the European studies while the other nine were different. Further stepwise conditional analysis identified a total of seven secondary signals and an independent novel locus at the 3′ end of CDKAL1.
Conclusions/interpretation
These results suggest that many loci associated with type 2 diabetes are commonly shared between European and Chinese populations. Identification of population-specific SNPs may increase our understanding of the genetic architecture underlying type 2 diabetes in different ethnic populations.
Electronic supplementary material
The online version of this article (doi:10.1007/s00125-013-3047-1) contains peer-reviewed but unedited supplementary material, which is available to authorised users.
Keywords: Ethnic difference, Genetic association, Type 2 diabetes
Introduction
Type 2 diabetes and its associated complications pose a major global healthcare burden. It is estimated that 552 million people worldwide will be affected by diabetes by the year 2030 and a majority of the affected will be Asians [1, 2]. Due to an exponential population growth, ageing population and increased rate of urbanisation, there is a rapidly emerging diabetes epidemic in Asia [3]. Exploring the underlying genetic architecture of type 2 diabetes in Asian populations may improve our understanding of the pathogenesis of this devastating disease and aid in the development of novel, effective and safe therapeutic alternatives to reduce its risk.
Candidate gene and genome-wide association studies (GWAS) have identified ∼60 loci associated with type 2 diabetes and related traits (fasting glucose, fasting insulin and 2 h glucose), but a majority of the heritability remains unexplained [4]. Most of these loci were initially identified from studies of European ancestry, with the exception of KCNQ1, UBE2E2, C2CD4A/B, PTPRD, SRR, SPRY2, PEPD, KCNK16, MAEA, GCC1-PAX4, PSMD6 and ZFAND3, which were first discovered in East Asian groups [5–8]. The transferability of the risk variants of type 2 diabetes genes across different populations has not been consistently observed. In some cases, this discrepancy may reflect substantial differences in the affect allele frequency between race/ethnic groups. For example, in an earlier study of TCF7L2, the strongest risk variant associated with type 2 diabetes in multiple European populations was found in the T allele of rs7903146 at the 5′ end of the gene [9]. However, the frequency of the T allele of rs7903146 in Asian individuals was rather rare (minor allele frequency 2–2.5%). Instead, rs290487, located in a linkage disequilibrium (LD) block at the 3′ end of the gene, was associated with type 2 diabetes in Chinese individuals, suggesting a distinct genetic variation of TCF7L2 in East Asians compared with that in Europeans [10]. Similarly, in another study examining the transferability of type 2 diabetes loci from European studies in 10,718 individuals of Chinese, Malay and East Asian Indian ethnicities, there was evidence of a population-specific effect, allelic heterogeneity and LD variations at CDKAL1 and HHEX/IDE/KIF11 loci in all three cohorts [11].
Recent studies suggest that there is a phenotypic distinction in the clinical presentation of type 2 diabetes between East Asians and Europeans [1, 3, 12, 13], hence the importance of delineating the specific susceptibilities in each group. Thus, examining population-specific signals may help to detect the underlying causal variant(s) that affect(s) different populations and may provide insights into the functional biology that may differ among different ethnic groups [14].
Fine mapping through dense genotyping of a locus of interest represents one approach for detecting population-specific variants. This approach has been successfully applied on a locus-by-locus basis for different diseases (e.g. SORT1 at the 1p13 locus for myocardial infarction and LDL-cholesterol [15] or ZNF365D in Crohn’s disease [16]). The Metabochip was developed to fine-map multiple metabolic and cardiovascular-related loci simultaneously in a cost-effective manner [17]. Approximately 43,292 of the 196,725 single-nucleotide polymorphisms (SNPs) on the Metabochip, including many less-common and rare variants from the 1,000 Genome Project, were selected to fine-map the previously identified type 2 diabetes and related-trait loci.
Here we report the association results for these fine-mapping SNPs on the Metabochip in a case–control study of 4,535 unrelated Chinese individuals with type 2 diabetes and 4,800 non-diabetic controls.
Methods
Ethics statement
This study was performed in accordance with the tenets of the Declaration of Helsinki and approved by the Institutional Review Boards of each participating centre in the USA and Taiwan. Informed consent was obtained from the study participants.
Cohorts
The TaiChi consortium was formed through a collaborative effort between investigators based in the USA and Taiwan. The consortium’s primary aim is to identify genetic determinants of atherosclerosis- and diabetes-related traits in East Asians and to fine-map validated loci identified in other race/ethnic groups.
The main academic sites participating in the TaiChi consortium in Taiwan include Taichung Veteran’s General Hospitals, Taipei Veterans General Hospital, National Health Research Institutes, Tri-Service General Hospital and National Taiwan University Hospital. The main US academic sites include Stanford University School of Medicine in Stanford, CA, Hudson-Alpha Biotechnology Institute in Huntsville, AL, Cedars-Sinai Medical Center (CSMC) in Los Angeles, CA and Harbor-UCLA Medical Center in Torrance, CA. TaiChi brings together seven principal cohorts formed in Taiwan, producing a bio-resource that includes a total of 11,859 individuals. Each cohort is described in more detail in the electronic supplementary material (ESM) text.
Diagnosis of type 2 diabetes and related traits
A total of 9,335 unrelated individuals, comprising 4,535 patients with type 2 diabetes and 4,800 non-diabetic ethnically matched controls, were included in this study. The diagnosis of diabetes was based on a fasting blood glucose level ≥6.99 mmol/l (126 mg/dl), a positive diabetes history or the use of diabetic medications.
Data for vital signs, fasting glucose, HbA1c and creatinine were measured in all participants as part of routine clinical and laboratory panels. Plasma glucose was measured by the glucose oxidase–peroxidase method (Wako Diagnostics, Tokyo, Japan).
Genotyping and quality control
Blood samples were obtained from participants and DNA samples were extracted from buffy coats using the QIAamp DNA mini Kit (Qiagen, Valencia, CA, USA). Genotyping with the Metabochip [17] was performed at the Hudson-Alpha Biotechnology Institute in Huntsville, AL, USA and at the Medical Genetics Institute and the Clinical and Translational Science Institute of CSMC. Infinium technology [18] was used for genotyping participants on the 200K Metabochip, following the manufacturer’s protocol (Illumina, San Diego, CA, USA). Genotypes were automatically called by GenCall, a clustering algorithm, in Genome Studio as an initial screen, and data from the two genotyping centres were combined before a trained specialist at CSMC manually reviewed the cluster plots.
SNPs with a missingness >2%, minor allele frequency (MAF) <1%, departure from the Hardy–Weinberg equilibrium (p < 10−7), located on the sex chromosomes or which were monomorphic were removed (ESM Table 1). While a total of 93,235 SNPs passed quality control (QC) measures, only those related to the 50 type 2 diabetes and related-trait loci on the Metabochip were analysed (n = 18,638, n = 9,055 after LD pruning).
Cryptic relatedness was defined at PI-HAT (PI) >0.12. Where there were family members in the cohorts, a majority of the related family members were first and second degree relatives. In these cases, only one individual from each family is represented in the current study.
Principal component analysis (PCA) using EIGENSTRAT was conducted to evaluate for potential population stratification among study participants and also to map the participants with the population panels from the International HapMap 3 dataset [19]. Any participants who did not cluster together with HapMap Chinese samples were excluded for further association analyses. Ten Eigenvalues were generated and participants greater than 10 SD from any component were also excluded from the analysis.
In total, participants with a missingness >2%, excessive heterozygosity, cryptic relatedness (n = 1,324), sex mismatch (n = 151), missing identity numbers (n = 460), ambiguous diabetes status (n = 390) or population outliers (n = 199) as defined by PCA were removed, leaving 9,335 participants for analysis.
Statistical analysis
Disease association with the additive model of inheritance was analysed with logistic regression. Sex chromosomes and mitochondrial DNA were excluded from the analysis. Age, sex and the first principal component (PC) from PCA were included as covariates in all analyses.
Of the ∼60 loci associated with type 2 diabetes that have been discovered mainly from studies of European ancestries, ∼50 loci are represented on the Metabochip (ESM Table 2). Thus, statistical significance for confirmation in TaiChi was defined at p < 10−3 after locus-specific Bonferroni correction (p = 0.05/50 = 1 × 10−3). Only those loci achieving this level of significance were evaluated further in this study. Results for the additional loci achieving a nominal level of significance of p < 0.05 (24 loci) are summarised in ESM Table 2.
A more stringent statistical analysis for association was also performed for all unlinked markers (n = 9,055), resulting in a Bonferroni correction of 5.5 × 10−6 (=0.05/9,055). All analyses were carried out using PLINK [20]. Regional association plots of the top SNPs were generated using LocusZoom [21] and the calculation of r 2 between SNPs was generated using SNAP [22]. Haplotype blocks and LD structure were generated using Haploview [23]. The expression quantitative trait loci (eQTL) browser at the University of Chicago [24] was used for correlation comparison of the expression SNPs (eSNPs) at the CDKAL1 locus in the present study. Direction of effect, minor allele, MAF and OR were also compared between top European and Asian SNPs.
Conditional analysis
Conditional analysis, in which SNPs were added as additional covariates in the regression model, were performed in two ways to indicate signals that were either replication signals or additional secondary signals. First, we ran the conditional analysis by conditioning on the previously reported European SNP. If the result was no longer significant, this SNP was a replication signal. If the result was significant, this SNP was a secondary signal. Second, we also ran the conditional analysis by conditioning on the top SNP in our sample. This approach was used to determine additional secondary signals and was performed by adding SNPs one at a time until no significance was seen. Consistent with the locus-specific Bonferroni model, statistical significance for the conditional analysis was defined at p < 0.05, which followed the αs used in an earlier trans-ethnic study of type 2 diabetes mellitus [11].
Results
Demographics
A total of 9,335 individuals, constituting 4,535 with type 2 diabetes (cases) and 4,800 non-diabetic ethnically matched controls, remained for analysis after sample QC. The demographics of this cohort are summarised in Table 1. Cases and controls were well matched for age and sex. The cases exhibited a higher BMI (25.2 kg/m2 vs 24.7 kg/m2), a worse HbA1c (8.9% vs 5.9% [74 vs 41 mmol/mol]) and a higher fasting plasma glucose level (9.4 vs 5.56 mmol/l) compared with the controls, consistent with the clinical criteria and risk factors for type 2 diabetes (Table 1).
Table 1.
Demographics of TaiChi type 2 diabetes cohort
| Characteristic | Controls | Cases | p value |
|---|---|---|---|
| n | 4,800 | 4,535 | |
| Age (years) | 65.3 ± 0.2 | 64.9 ± 0.2 | NS |
| Sex (% female) | 37.8 | 39.1 | NS |
| BMI (kg/m2) | 24.7 ± 0.1 | 25.2 ± 0.1 | <0.0001 |
| HbA1c (%) | 5.9 ± 0.04 | 8.9 ± 0.04 | <0.0001 |
| HbA1c (mmol/mol) | 41 | 74 | <0.0001 |
| FPG (mmol/l) | 5.56 ± 0.08 | 9.4 ± 0.07 | <0.0001 |
Values for continuous variables denote mean ± SEM
FPG, fasting plasma glucose
Quality control
A total of 18,638 fine-mapping SNPs (those related to type 2 diabetes and related traits) passed QC measures and were used for association testing. For data QC purposes, the quantile–quantile plot was generated with all 93,235 SNPs that passed QC (genomic inflation factor = 1.14) and LD pruned (genomic inflation factor = 1.04) (ESM Fig. 1a). The PCA plot after ancestry exclusions indicates that individuals from the TaiChi cohort clustered together with the HapMap CHB+CHD (Chinese in Beijing + Chinese in Denver) populations (ESM Fig. 1b).
Type 2 diabetes and related-trait loci with known lead SNPs
Of the ∼50 type 2 diabetes and related-trait loci represented on the Metabochip, 14 loci significantly transferred to the Chinese individuals after locus Bonferroni correction (ESM Table 2), with the lowest p values observed for the lead SNPs in KCNQ1 (rs2237897, p = 5.7 × 10−12) and CDKN2A/B-CDKN2BAS (rs10811661, p = 5.0 × 10−8), respectively (Table 2). Another 24 loci achieved a nominal significance at p < 0.05 (ESM Table 2); thus, 76% (38/50) of loci associated with type 2 diabetes and related traits appear to be commonly shared between the European and Chinese populations. In addition, if one examines the direction of effect, 35 of the available 41 loci with proxies (85.4%) significantly shared the association in the same direction as in the Europeans (p = 1.8 × 10−6, ESM Tables 2 and 3). However, 12 loci were not observed in the Chinese population even at the nominal p value. These were NOTCH2, SLC2A2, WFS1, GCK, HNF1A, MTNR1B, HMGA2, KLF14, TP53INP1, CHCHD9/TLE4, SREBF1 and ZFAND6, of which 9/9 (with available proxies, 100%) shared the association in the same direction as in the European populations (ESM Tables 2 and 3; three were not available for evaluation due to no proxies). Of the 14 loci that met the locus-specific Bonferroni correction, the lead SNP of each locus was a common variant (MAF ≥5%), and each demonstrated a small effect on the association (Table 2). We performed a more stringent statistical analysis for association by LD pruning of type 2 diabetes loci and found that five loci remained statistically significant after Bonferroni correction for unlinked markers (p < 5.5 × 10−6; Table 2). Of those five loci, only one locus had a putative novel SNP in our East Asian population. These data suggest that the loci with the strongest signals are those that are present in the European population and are trans-ethnically shared with the East Asians.
Table 2.
Top 14 loci of type 2 diabetes from European individuals and the lead SNP observed in Chinese individuals
| Locus | Chr | Lead SNP | Putative novel SNP | SNP category | A1 | A2 | MAF | OR | p value (adjusted for age, sex, PC1) |
|---|---|---|---|---|---|---|---|---|---|
| KCNQ1* | 11 | rs2237897 | No | Type 2 diabetes | T | C | 0.34 | 0.80 | 5.7 × 10−12 |
| CDKN2A/B-CDKN2BAS* | 9 | rs10811661 | No | Type 2 diabetes | C | T | 0.41 | 0.85 | 5.0 × 10−8 |
| CDC123/CAMK1D* | 10 | rs11257655 | No | Type 2 diabetes, fasting glucose | C | T | 0.43 | 0.87 | 2.3 × 10−6 |
| KCNJ11/ABCC8* | 11 | rs2074314 | Yes | Type 2 diabetes | G | A | 0.37 | 1.15 | 3.5 × 10−6 |
| IGF2BP2* | 3 | rs6769511 | No | Type 2 diabetes | C | T | 0.26 | 1.17 | 5.0 × 10−6 |
| HNF1B (TCF2) | 17 | rs8064454 | Yes | Type 2 diabetes | A | C | 0.28 | 1.16 | 1.3 × 10−5 |
| CDKAL1 | 6 | rs9356744 | Yes | Type 2 diabetes | C | T | 0.36 | 1.14 | 2.1 × 10−5 |
| IDE/KIF11/HHEX | 10 | rs10882091 | Yes | Type 2 diabetes, fasting glucose | C | T | 0.08 | 1.30 | 2.9 × 10−5 |
| UBE2E2 | 3 | rs17012800 | Yes | Type 2 diabetes | G | A | 0.19 | 0.86 | 4.3 × 10−5 |
| GCKR | 2 | rs1260326 | No | Type 2 diabetes, 2 h glucose, fasting glucose | T | C | 0.49 | 0.89 | 9.6 × 10−5 |
| IRS1-LOC646736 | 2 | rs2943632 | Yes | Type 2 diabetes | A | G | 0.08 | 0.82 | 1.8 × 10−4 |
| SLC30A8 | 8 | rs11774700 | Yes | Type 2 diabetes, 2 h glucose, fasting glucose | C | T | 0.45 | 0.90 | 2.7 × 10−4 |
| GLIS3 | 9 | rs12378556 | Yes | Fasting glucose | G | A | 0.38 | 1.11 | 4.0 × 10−4 |
| PRC1 | 15 | rs3743451 | Yes | Type 2 diabetes | T | C | 0.48 | 0.90 | 8.9 × 10−4 |
*Loci that met LD pruned Bonferroni correction of 5.5 × 10−6
A1, minor allele; A2, major allele; Chr, chromosome; PC1, first PC
Further examination of these 14 loci revealed that for five (rs2237897, KCNQ1; rs10811661, CDKN2A/B-CDKN2BAS; rs11257655, CDC123/CAMK1D; rs6769511, IGF2BP2; rs1260326, GCKR), the lead SNPs are the same lead SNPs observed in the European populations (Table 2). The regional association plots of these loci show robust signals in these regions (ESM Fig. 2). In evaluating the transferability of the same lead SNPs, we found the SNP effects were in the same direction for all five SNPs. The direction of effect for other SNPs is also summarised in ESM Table 3.
To investigate the possible existence of a secondary signal, we performed a stepwise conditional analysis using the previously reported European SNPs as covariates. Through these analyses, we identified one additional SNP, rs11024184 (KCNQ1, p = 9.1 × 10−4 and p.condition1 = 8.1 × 10−8; ESM Table 4 and ESM Fig. 3). We next evaluated the haplotype block structure of KCNQ1 and found that these two SNPs (rs11024184 and rs2237897) were not correlated (r 2 = 0.07) and were on different LD blocks, suggesting that this was an independent secondary signal (ESM Fig. 4).
Type 2 diabetes and related-trait loci with putative novel lead SNPs
Of the 14 European type 2 diabetes loci that were also significant in the Chinese population, the strongest p value found in nine loci was for a putative novel lead SNP (Table 2). The regional association plots of these loci also showed robust signals in these regions (ESM Fig. 5).
To investigate whether these putative novel SNPs were replication or secondary signals, we performed conditional analysis using the European SNP as covariates similar to the method described above. Of these nine lead SNPs, five (rs2943632, IRS1; rs9356744, CDKAL1; rs11774700, SLC30A8 [ESM Fig. 6]; rs2074314, KCNJ11/ABCC8; rs8064454, HNF1B [TCF2]) became insignificant, indicating they were replication SNPs (in LD with previously reported European SNPs). Two SNPs failed QC and had no proxies on the Metabochip; thus their independence could not be assessed. Two SNPs (rs12378556, GLIS3, p.condition1 = 2.4 × 10−3 and rs10882091, IDE/KIF11/HHEX, p.condition1 = 4.9 × 10−3, Fig. 1) remained significant, suggesting novel independent signals (ESM Table 5).
Fig. 1.
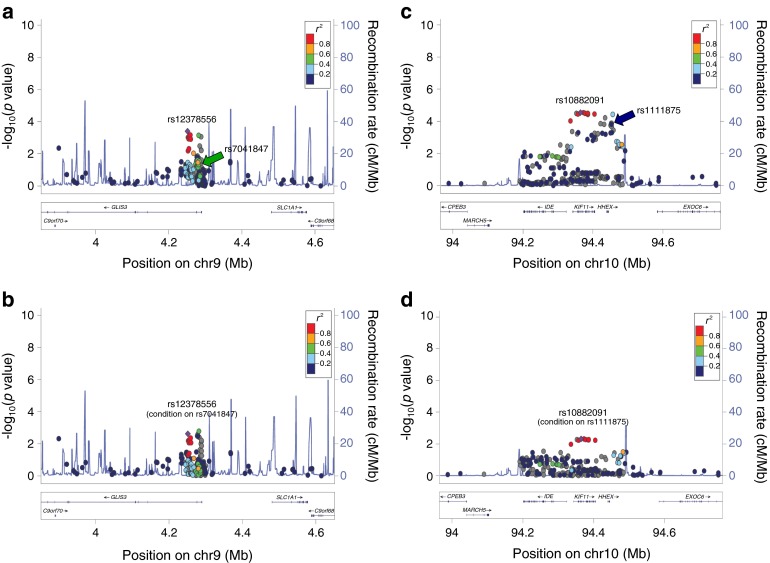
Regional association plot of unconditional (a, c) and conditional (b, d) analysis of GLIS3 (a, b) and IDE/KIF11/HHEX (c, d); examples of retaining significance after performing a conditional analysis on a previously reported European SNP. The lead East Asian SNP is represented by a purple diamond. The European SNP is indicated by an arrow, with the colour corresponding to the r 2 of the two SNPs plotted in the Asian LD pattern. Chr, chromosome
Furthermore, using a stepwise conditional analysis in which we conditioned on the top lead SNP in our dataset, we identified four additional SNPs (rs2138157, IRS1, p.condition2 = 0.03; rs7773318, CDKAL1, p.condition2 = 3.0 × 10−4; rs9465994, CDKAL1, p.condition2 = 4.1 × 10−3; rs10974438, GLIS3, p.condition2 = 0.03) that appeared to be independent signals (ESM Table 5). Of interest, two of the four independent signals were found at the 3’ end of the CDKAL1 gene.
In summary, a total of 10 SNPs replicated a previously reported European signal. Though five were more significant in the Asian population, they were in LD with previous European signals. A total of seven novel independent secondary signals were identified in this study (Table 3).
Table 3.
Summary of top SNPs, either as replication or secondary signals
| Locus | Chr | SNPs | Replication or secondary signal | Replication signals |
|---|---|---|---|---|
| GCKR | 2 | rs1260326 | Replication | Same as European |
| IRS1/LOC646736 | 2 | rs2943632 | Replication | More significant in Asians |
| rs2138157 | Secondary | |||
| UBE2E2 | 3 | rs17012800 | NA (no proxy) | |
| IGF2BP2 | 3 | rs6769511 | Replication | Same as European |
| CDKAL1 | 6 | rs9356744 | Replication | More significant in Asians |
| rs7773318 | Secondary | |||
| rs9465994 | Secondary | |||
| SLC30A8 | 8 | rs11774700 | Replication | More significant in Asians |
| GLIS3 | 9 | rs12378556 | Secondary | |
| rs10974438 | Secondary | |||
| CDKN2A/B-CDKN2BAS | 9 | rs10811661 | Replication | Same as European |
| CDC123/CAMK1D | 10 | rs11257655 | Replication | Same as European |
| IDE/KIF11/HHEX | 10 | rs10882091 | Secondary | |
| KCNQ1 | 11 | rs2237897 | Replication | Same as European |
| rs11024184 | Secondary | |||
| KCNJ11/ABCC8 | 11 | rs2074314 | Replication | More significant in Asians |
| PRC1 | 15 | rs3743451 | NA (no proxy) | |
| HNF1B (TCF2) | 17 | rs8064454 | Replication | More significant in Asians |
SNPs that are in LD with a previous European SNP are replication signals (i.e. not significant after conditional analysis). SNPs that not in LD with a previous European SNP are secondary signals (i.e. significant after conditional analysis)
Chr, chromosome
Novel locus at the 3′ end of CDKAL1
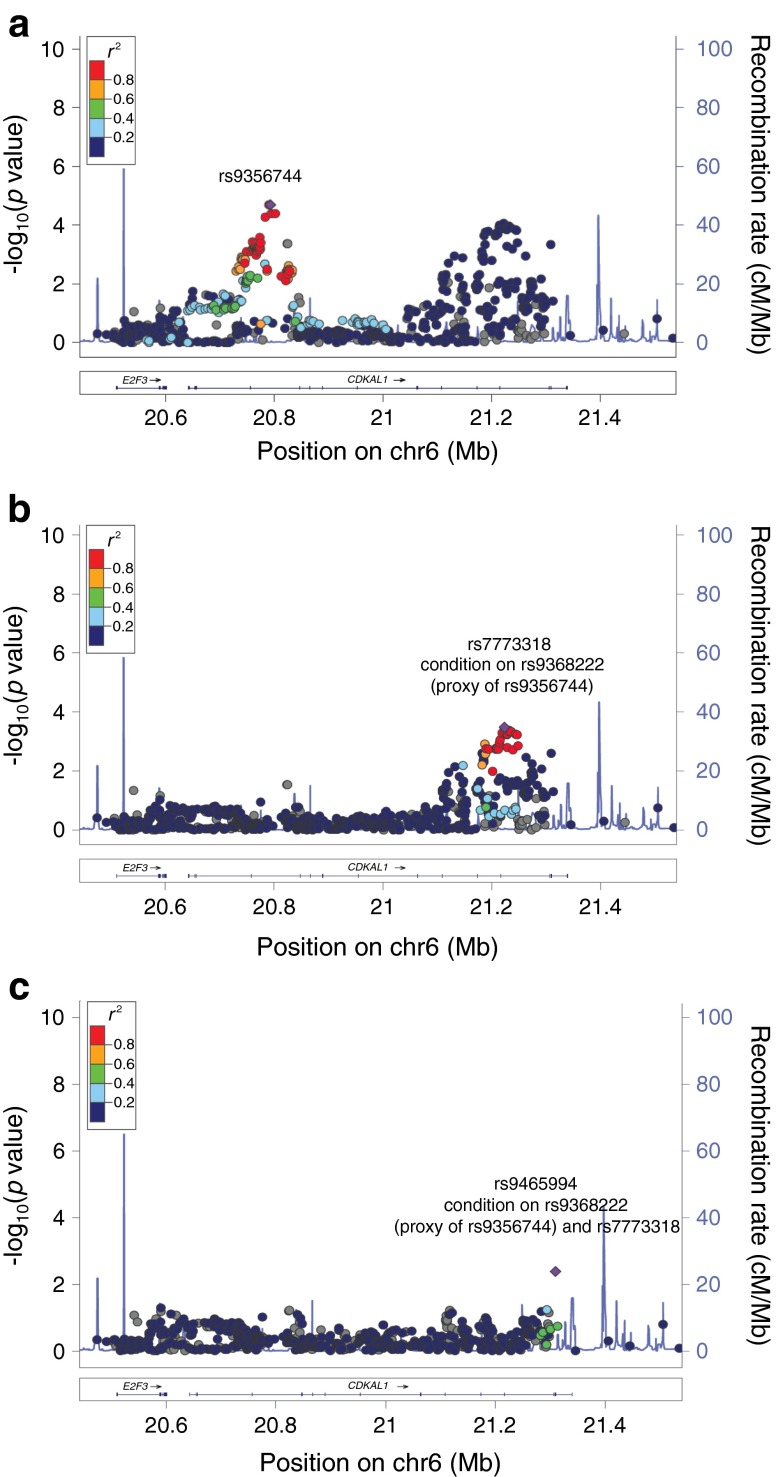
Using fine mapping, our data demonstrated that there were two peaks associated with type 2 diabetes on the CDKAL1 gene (Fig. 2a). The first peak lay at the 5′ end of the gene, closer to E2F3, with a significance of p = 2.1 × 10−5 observed by the lead SNP rs9356744. SNP rs9356744 was in tight LD with rs10440833 (proxy, rs9368222), which was previously reported in a large European type 2 diabetes study of 42,542 cases and 98,912 controls [25]. Conditional analysis on rs9368222 diminished all the signals at the 5′ end of CDKAL1 and negated the statistical significance (Fig. 2b and ESM Table 5). On the other hand, it did not affect the second peak, which lay at the 3′ end of CDKAL1 (Fig. 2b).
Fig. 2.
Regional association plot shows two peaks associated with type 2 diabetes at CDKAL1: a 5′ peak that is closer to E2F3 and another peak closer to the 3′ of CDKAL1 (a). Conditional analysis performed on rs9368222, which is a proxy for rs9356744 and rs10440833, negated the statistical significance at the 5′ peak but had no effect on the 3′ peak, indicating a novel independent locus at the 3′ end of CDKAL1 for type 2 diabetes in the Chinese population and demonstrates that rs7773318 (b) and rs9465994 (c) are novel ethnic-specific SNPs
Further stepwise conditional analysis thus identified two independent signals at the 3′ end: SNPs rs7773318 and rs9465994 (Fig. 2b, c; ESM Table 5). This result demonstrates that the 3′ end of CDKAL1 is an independent locus observed only in the Chinese population to date (Fig. 2b, c). This region was also plotted using Asian and European LD patterns to demonstrate an intrinsic LD difference between the two ethnicities (ESM Fig. 7).
Discussion
We have demonstrated that a majority of loci associated with type 2 diabetes discovered in European populations appear to also serve as susceptibility loci for the same trait in the Chinese population. Of the 50 loci tested, 14 of the loci met our locus Bonferroni criteria and another 24 were nominally significant. Furthermore, we identified a total of seven novel ethnic-specific variants for type 2 diabetes in the Chinese population using a fine-mapping approach. Of particular interest, two independent SNPs lie at the 3′ end of the CDKAL1 gene. These latter data thus split the CDKAL1 gene into two loci, the 5′ end of which is seen in both Europeans and East Asians and the 3′ end of which appears to be a novel independent locus for type 2 diabetes in Chinese individuals.
Our most important finding may well be the identification of two peaks on CDKAL1. All previously reported SNPs of CDKAL1 in type 2 diabetes (rs7756992 [26], rs7754840 [27, 28], rs4712523 [27, 29, 30], rs10946398 [31], rs9465871 [31, 32], rs4712524 [5], rs9295474 [11], and rs10440833 [25]) lie within the 5′ end of the gene, and many of these SNPs are also observed in Chinese individuals [11, 33, 34]. None of the previously reported SNPs of CDKAL1 in type 2 diabetes lie within the 3′ end of the gene. Our finding was possible because CDKAL1 was one of the five selected loci to be fine-mapped on the Metabochip [17].
CDKAL1 catalyses a methyl-thio group, which possibly causes misfolding of proinsulin [35] and inhibits pancreatic CDK5/p35 complex [26], thereby altering beta cell function and insulin production. Earlier GWAS studies found variants at the 5′ of CDKAL1 in individuals with impaired insulin secretion but the functional variant has yet to be determined. We therefore used an available database to discover whether the two novel SNPs at the 3’ end of CDKAL1 (rs7773318 and rs9465994) were eSNPs [24]. Though both rs7773318 and rs9465994 are neither eSNPs nor in LD with previously reported eSNPs (rs9460563, rs9460612, rs59633892, rs62404554, and rs10946439) on CDKAL1, we note that eSNPs located at the 5′ end of CDKAL1 are mostly trans-acting regulators, while the eSNPs located at the 3’ end of CDKAL1 are all cis-acting. This observation supports the concept that SNPs in the 3’ end of CDKAL1 regulate the expression of this gene.
In this study, although we chose a locus-specific Bonferroni correction (a less stringent statistical cut-off for association), we also performed a more stringent statistical analysis for unlinked markers. We found that five loci remained significant after correction for multiple testing; however, only one locus had a putative novel SNP in the East Asian population. This locus was later found through conditional analysis to be highly correlated with an SNP previously reported in a European population.
Of the 50 tested loci, 14 loci were significant in the Chinese population after locus Bonferroni correction. Collectively, a total of 38 loci (76%, 38/50 loci) transferred to the Chinese population with at least a nominal significance, highlighting a great deal of genetic homogeneity for type 2 diabetes between the European and Chinese populations.
In this study, 12 of the 50 loci were not observed to be significant. In comparison with earlier studies involving Chinese individuals, our result is similar (i.e. non-significant in both this study and other Chinese cohorts) for NOTCH2 [33], SLC2A2 [36], WFS1 [33, 37], GCK [38] and HNF1A [39, 40] but different (i.e. non-significant in this study but was significant in other Chinese cohorts) for MTNR1B [41–43], GCK [41, 42] and SREBF1 [44]. Comparisons could not be made for KLF14, TP53INP1, CHCHD9/TLE4, HMGA2 and ZFAND6 as these genes were not tested in other Chinese cohorts. Although our result was not significant for MTNR1B, GCK and SREBF1, the direction of effect was concordant with other Chinese [41, 42] and European [25, 45] studies for both MTNR1B and GCK, but was unavailable (no proxy) for SREBF1.
Through conditional analysis, a total of seven potential secondary signals were identified. To illustrate ethnic specificity, we give an example for SNP rs11024184 on KCNQ1. Using HapMap, the allele frequency of the A allele is seen in 9.2% of East Asians, but in as many as 53.3% of Europeans. SNP rs11024184 lies 25 kb upstream of rs2237897 (the previously reported European SNP) on KCNQ1 and the two SNPs are neither in LD with each other nor on the same LD block. Furthermore, rs11024184 does not tag any other SNP in the region at r 2 > 0.8. Collectively, these data suggest this is an independent signal found in the Chinese population.
Comparing the Chinese and European populations, among the other six potential secondary signals, the minor allele frequency is similar for GLIS3 (rs12378556; rs10974438) but different for IDE/KIF11/HHEX (rs10882091) and IRS1 (rs2138157). For CDKAL1, the minor allele frequency is similar for rs9465994 but different for rs7773318.
There are several strengths to this study. First, this is a homogenous group of Chinese individuals, recruited at seven principal sites in Taiwan, with well-defined phenotype and ethnically matched controls. Second, to our knowledge, this is the first Metabochip study using fine mapping of type 2 diabetes and related traits in East Asians. Third, and most importantly, using this fine-mapping approach allows for the redefining of the association signals at previously established loci and the identification of a novel locus at the 3′ end of CDKAL1, which to date is only observed in the Chinese population. There are also several limitations. The first is the disparity of the regions covered on the Metabochip. Some regions are more extensively fine-mapped than others, thus there is a higher probability and opportunity to uncover independent signals at these regions. Second, in the most recent report from the MAGIC (Meta-Analyses of Glucose and Insulin-related traits Consortium) and DIAGRAM (DIAbetes Genetics Replication And Meta-analysis) consortia, a number of additional type 2 diabetes loci have been identified in the European population [46, 47]. We examined, in this report, the 50 loci known to be associated with type 2 diabetes or its related traits at the time of this investigation. Last, the Metabochip is a pre-designed genotyping array of cardiovascular and metabolic traits discovered in the European population. Thus, the Metabochip is designed to test for SNPs and loci only on the platform and is not designed to discover novel SNPs and loci not previously related to cardiovascular or metabolic traits in a genome-wide fashion.
In summary, we have identified a few ethnic-specific variants and demonstrated a novel independent type 2 diabetes locus at the 3′ end of CDKAL1 in the Chinese population. These findings provide initial clues to differences in the genetic architecture underlying type 2 diabetes among various ethnic populations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Quantile-quantile plot showing deviation of association from the expected for unpruned (top) and LD pruned (bottom) (A). The principal component analysis (PCA) plot after ancestry exclusions indicates that subjects in TaiChi clustered with the HapMap CHB+CHD populations (B). (PDF 470 kb)
Regional association plots showing chromosomal position and P-value of SNPs, plotted as –log10 (P-value), in known T2D loci with prior lead SNPs. The lead SNP of each locus is plotted in purple diamond. (PDF 599 kb)
Regional association plot of KCNQ1 before (A) and after conditioning on rs2237897 (B). (PDF 753 kb)
Regional association and linkage disequilibrium (LD) plot of KCNQ1. The dotted line represents the level of association that reached statistical significance after locus Bonferroni correction. The solid circles indicate the unconditional –log10 P-value and the open circles indicate the conditional –log10 P-value for each of the SNPs in this region after conditioning on prior reported SNP (rs2237897, single asterisk). Novel SNP (rs11024184, double asterisks) remained significant. LD value is displayed in r2. (PDF 734 kb)
Regional association plots showing chromosomal position and P-value of SNPs, plotted as –log10 (P-value), in known T2D loci with putative novel lead SNPs. The lead East Asian SNP of each locus is plotted in purple diamond. The prior European SNP is indicated by a colored arrow, with the color corresponding to the r2 of the two SNPs plotted in Asian LD pattern. (PDF 912 kb)
Regional association plot of SLC30A8, an example of a loss of significance after performing a conditional analysis on a prior European SNP. The lead East Asian SNP is plotted in purple diamond. The prior European SNP is indicated by a colored arrow, with the color corresponding to the r2 of the two SNPs plotted in Asian LD pattern. (PDF 600 kb)
Regional association plots of CDKAL1 with the lead East Asian SNP plotted in purple diamond using Asian LD (A) and European LD (B) patterns. (PDF 968 kb)
(PDF 61 kb)
(PDF 81 kb)
(PDF 88 kb)
(PDF 58 kb)
(PDF 74 kb)
(PDF 70 kb)
Acknowledgements
We thank all the investigators and staff who contributed to this study by collecting the data used and presented in this manuscript. We are also grateful to the patients and their families for their participation.
Funding
This study was supported by the National Institutes of Health, USA (HL087647, HL088457, EY014684, DK079888), ARRA Supplement (EY014684-03S1, -04S1), the National Health Research Institutes, Taiwan (PH-100-SP-01, BS-094-PP-01, PH-100-PP-03), the National Science Council, Taiwan (Grant Nos NSC 98-2314-B-075A-002-MY3, NSC 96-2314-B-002-151, NSC 96-2314-B-002-152, NSC 98-2314-B-002-122-MY2, NSC 100-2314-B-002-115, NSC 101-2325-002-078, 101-2314-B-075A-006-MY3), the Taichung Veterans General Hospital, Taiwan (TCVGH-1013001C, TCVGH-1013002D), the National Taiwan University Hospital, Taiwan (NTUH 98-N1266, NTUH 100-N1775, NTUH 101-N2010, NTUH 101-N, VN101-04, NTUH 101-S1784), the UCLA/Cedars-Sinai/Harbor/Drew Clinical and Translational Science Institute (CTSI) Grant (UL1 TR000124-01), the Southern California Diabetes Research Grant (DRC) (DK063491), the Cedars-Sinai Board of Governor’s Chair in Medical Genetics and the Cedars-Sinai Winnick Clinical Scholars Award.
Duality of interest
The authors declare that there is no duality interest associated with this manuscript.
Contribution statement
All authors made substantial contributions to the concept and design of this study, acquisition of data or analysis and interpretation of data and to drafting of the article and revised it critically for intellectual content. All authors gave final approval of the version to be published. YIC and JIR are the guarantors of this work, had full access to all the data and take full responsibility for the integrity of data and the accuracy of data analysis.
Abbreviations
- CHB+CHD
Chinese in Beijing + Chinese in Denver
- CSMC
Cedars-Sinai Medical Center
- eQTL
Expression quantitative trait locus
- eSNPs
Expression SNPs
- GWAS
Genome-wide association studies
- LD
Linkage disequilibrium
- MAF
Minor allele frequency
- PC
Principal component
- PCA
Principal component analysis
- QC
Quality control
- SNPs
Single-nucleotide polymorphisms
Footnotes
Jane Z. Kuo, Wayne Huey-Herng Sheu and Themistocles L. Assimes share first authorship of this article.
Chao A. Hsiung, Thomas Quertermous, Jerome I. Rotter and Yii-Der I. Chen contributed equally to this study.
References
- 1.Chan JC, Malik V, Jia W, et al. Diabetes in Asia: epidemiology, risk factors, and pathophysiology. JAMA. 2009;301:2129–2140. doi: 10.1001/jama.2009.726. [DOI] [PubMed] [Google Scholar]
- 2.Whiting DR, Guariguata L, Weil C, Shaw J. IDF diabetes atlas: global estimates of the prevalence of diabetes for 2011 and 2030. Diabetes Res Clin Pract. 2011;94:311–321. doi: 10.1016/j.diabres.2011.10.029. [DOI] [PubMed] [Google Scholar]
- 3.Ramachandran A, Ma RC, Snehalatha C. Diabetes in Asia. Lancet. 2010;375:408–418. doi: 10.1016/S0140-6736(09)60937-5. [DOI] [PubMed] [Google Scholar]
- 4.Billings LK, Florez JC. The genetics of type 2 diabetes: what have we learned from GWAS? Ann N Y Acad Sci. 2010;1212:59–77. doi: 10.1111/j.1749-6632.2010.05838.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Unoki H, Takahashi A, Kawaguchi T, et al. SNPs in KCNQ1 are associated with susceptibility to type 2 diabetes in East Asian and European populations. Nat Genet. 2008;40:1098–1102. doi: 10.1038/ng.208. [DOI] [PubMed] [Google Scholar]
- 6.Yamauchi T, Hara K, Maeda S, et al. A genome-wide association study in the Japanese population identifies susceptibility loci for type 2 diabetes at UBE2E2 and C2CD4A-C2CD4B. Nat Genet. 2010;42:864–868. doi: 10.1038/ng.660. [DOI] [PubMed] [Google Scholar]
- 7.Tsai FJ, Yang CF, Chen CC, et al. A genome-wide association study identifies susceptibility variants for type 2 diabetes in Han Chinese. PLoS genetics. 2010;6:e1000847. doi: 10.1371/journal.pgen.1000847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cho YS, Chen CH, Hu C, et al. Meta-analysis of genome-wide association studies identifies eight new loci for type 2 diabetes in east Asians. Nat Genet. 2012;44:67–72. doi: 10.1038/ng.1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sladek R, Rocheleau G, Rung J, et al. A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature. 2007;445:881–885. doi: 10.1038/nature05616. [DOI] [PubMed] [Google Scholar]
- 10.Chang YC, Chang TJ, Jiang YD, et al. Association study of the genetic polymorphisms of the transcription factor 7-like 2 (TCF7L2) gene and type 2 diabetes in the Chinese population. Diabetes. 2007;56:2631–2637. doi: 10.2337/db07-0421. [DOI] [PubMed] [Google Scholar]
- 11.Sim X, Ong RT, Suo C, et al. Transferability of type 2 diabetes implicated loci in multi-ethnic cohorts from Southeast Asia. PLoS genetics. 2011;7:e1001363. doi: 10.1371/journal.pgen.1001363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang JS, Tu ST, Lee IT, et al. Contribution of postprandial glucose to excess hyperglycaemia in Asian type 2 diabetic patients using continuous glucose monitoring. Diabetes Metab Res Rev. 2011;27:79–84. doi: 10.1002/dmrr.1149. [DOI] [PubMed] [Google Scholar]
- 13.Carnethon MR, de Chavez PJ, Biggs ML, et al. Association of weight status with mortality in adults with incident diabetes. JAMA. 2012;308:581–590. doi: 10.1001/jama.2012.9282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goodarzi MO, Rotter JI. Testing the gene or testing a variant? The case of TCF7L2. Diabetes. 2007;56:2417–2419. doi: 10.2337/db07-0923. [DOI] [PubMed] [Google Scholar]
- 15.Musunuru K, Strong A, Frank-Kamenetsky M, et al. From noncoding variant to phenotype via SORT1 at the 1p13 cholesterol locus. Nature. 2010;466:714–719. doi: 10.1038/nature09266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haritunians T, Jones MR, McGovern DP, et al. Variants in ZNF365 isoform D are associated with Crohn’s disease. Gut. 2011;60:1060–1067. doi: 10.1136/gut.2010.227256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Voight BF, Kang HM, Ding J, et al. The metabochip, a custom genotyping array for genetic studies of metabolic, cardiovascular, and anthropometric traits. PLoS genetics. 2012;8:e1002793. doi: 10.1371/journal.pgen.1002793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gunderson KL, Steemers FJ, Ren H, et al. Whole-genome genotyping. Methods Enzymol. 2006;410:359–376. doi: 10.1016/S0076-6879(06)10017-8. [DOI] [PubMed] [Google Scholar]
- 19.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 20.Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pruim RJ, Welch RP, Sanna S, et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26:2336–2337. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johnson AD, Handsaker RE, Pulit SL, Nizzari MM, O’Donnell CJ, de Bakker PI. SNAP: a web-based tool for identification and annotation of proxy SNPs using HapMap. Bioinformatics. 2008;24:2938–2939. doi: 10.1093/bioinformatics/btn564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 24.eQTL resources @ the pritchard lab http://eqtl.uchicago.edu/cgi-bin/gbrowse/eqtl; accessed 28 August 2012
- 25.Voight BF, Scott LJ, Steinthorsdottir V, et al. Twelve type 2 diabetes susceptibility loci identified through large-scale association analysis. Nat Genet. 2010;42:579–589. doi: 10.1038/ng.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Steinthorsdottir V, Thorleifsson G, Reynisdottir I, et al. A variant in CDKAL1 influences insulin response and risk of type 2 diabetes. Nat Genet. 2007;39:770–775. doi: 10.1038/ng2043. [DOI] [PubMed] [Google Scholar]
- 27.Scott LJ, Mohlke KL, Bonnycastle LL, et al. A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science. 2007;316:1341–1345. doi: 10.1126/science.1142382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saxena R, Voight BF, Lyssenko V, et al. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science. 2007;316:1331–1336. doi: 10.1126/science.1142358. [DOI] [PubMed] [Google Scholar]
- 29.Takeuchi F, Serizawa M, Yamamoto K, et al. Confirmation of multiple risk loci and genetic impacts by a genome-wide association study of type 2 diabetes in the Japanese population. Diabetes. 2009;58:1690–1699. doi: 10.2337/db08-1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rung J, Cauchi S, Albrechtsen A, et al. Genetic variant near IRS1 is associated with type 2 diabetes, insulin resistance and hyperinsulinemia. Nat Genet. 2009;41:1110–1115. doi: 10.1038/ng.443. [DOI] [PubMed] [Google Scholar]
- 31.Zeggini E, Weedon MN, Lindgren CM, et al. Replication of genome-wide association signals in UK samples reveals risk loci for type 2 diabetes. Science. 2007;316:1336–1341. doi: 10.1126/science.1142364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wellcome Trust Case Control Consortium Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hu C, Zhang R, Wang C, et al. PPARG, KCNJ11, CDKAL1, CDKN2A-CDKN2B, IDE-KIF11-HHEX, IGF2BP2 and SLC30A8 are associated with type 2 diabetes in a Chinese population. PloS one. 2009;4:e7643. doi: 10.1371/journal.pone.0007643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu Y, Li H, Loos RJ, et al. Common variants in CDKAL1, CDKN2A/B, IGF2BP2, SLC30A8, and HHEX/IDE genes are associated with type 2 diabetes and impaired fasting glucose in a Chinese Han population. Diabetes. 2008;57:2834–2842. doi: 10.2337/db08-0047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kaufman RJ. Beta-cell failure, stress, and type 2 diabetes. N Engl J Med. 2011;365:1931–1933. doi: 10.1056/NEJMcibr1109442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hu C, Zhang R, Wang C, et al. Variants from GIPR, TCF7L2, DGKB, MADD, CRY2, GLIS3, PROX1, SLC30A8 and IGF1 are associated with glucose metabolism in the Chinese. PloS one. 2010;5:e15542. doi: 10.1371/journal.pone.0015542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Han X, Luo Y, Ren Q, et al. Implication of genetic variants near SLC30A8, HHEX, CDKAL1, CDKN2A/B, IGF2BP2, FTO, TCF2, KCNQ1, and WFS1 in type 2 diabetes in a Chinese population. BMC Med Genet. 2010;11:81. doi: 10.1186/1471-2350-11-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Qi Q, Wu Y, Li H, et al. Association of GCKR rs780094, alone or in combination with GCK rs1799884, with type 2 diabetes and related traits in a Han Chinese population. Diabetologia. 2009;52:834–843. doi: 10.1007/s00125-009-1290-2. [DOI] [PubMed] [Google Scholar]
- 39.Rissanen J, Wang H, Miettinen R, et al. Variants in the hepatocyte nuclear factor-1alpha and -4alpha genes in Finnish and Chinese subjects with late-onset type 2 diabetes. Diabetes Care. 2000;23:1533–1538. doi: 10.2337/diacare.23.10.1533. [DOI] [PubMed] [Google Scholar]
- 40.Yang Z, Wu SH, Zheng TS, Lu HJ, Xiang KS. Identification of four novel mutations in the HNF-1A gene in Chinese early-onset and/or multiplex diabetes pedigrees. Chin Med J (Engl) 2006;119:1072–1078. [PubMed] [Google Scholar]
- 41.Hu C, Zhang R, Wang C, et al. Effects of GCK, GCKR, G6PC2 and MTNR1B variants on glucose metabolism and insulin secretion. PloS one. 2010;5:e11761. doi: 10.1371/journal.pone.0011761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tam CH, Ho JS, Wang Y, et al. Common polymorphisms in MTNR1B, G6PC2 and GCK are associated with increased fasting plasma glucose and impaired beta-cell function in Chinese subjects. PloS one. 2010;5:e11428. doi: 10.1371/journal.pone.0011428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu C, Wu Y, Li H, et al. MTNR1B rs10830963 is associated with fasting plasma glucose, HbA1C and impaired beta-cell function in Chinese Hans from Shanghai. BMC Med Genet. 2010;11:59. doi: 10.1186/1471-2350-11-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu JX, Liu J, Li PQ, et al. Association of sterol regulatory element-binding protein-1c gene polymorphism with type 2 diabetes mellitus, insulin resistance and blood lipid levels in Chinese population. Diabetes Res Clin Pract. 2008;82:42–47. doi: 10.1016/j.diabres.2008.06.017. [DOI] [PubMed] [Google Scholar]
- 45.Dupuis J, Langenberg C, Prokopenko I, et al. New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat Genet. 2010;42:105–116. doi: 10.1038/ng.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Morris AP, Voight BF, Teslovich TM, et al. Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat Genet. 2012;44:981–990. doi: 10.1038/ng.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Scott RA, Lagou V, Welch RP, et al. Large-scale association analyses identify new loci influencing glycemic traits and provide insight into the underlying biological pathways. Nat Genet. 2012;44:991–1005. doi: 10.1038/ng.2385. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Quantile-quantile plot showing deviation of association from the expected for unpruned (top) and LD pruned (bottom) (A). The principal component analysis (PCA) plot after ancestry exclusions indicates that subjects in TaiChi clustered with the HapMap CHB+CHD populations (B). (PDF 470 kb)
Regional association plots showing chromosomal position and P-value of SNPs, plotted as –log10 (P-value), in known T2D loci with prior lead SNPs. The lead SNP of each locus is plotted in purple diamond. (PDF 599 kb)
Regional association plot of KCNQ1 before (A) and after conditioning on rs2237897 (B). (PDF 753 kb)
Regional association and linkage disequilibrium (LD) plot of KCNQ1. The dotted line represents the level of association that reached statistical significance after locus Bonferroni correction. The solid circles indicate the unconditional –log10 P-value and the open circles indicate the conditional –log10 P-value for each of the SNPs in this region after conditioning on prior reported SNP (rs2237897, single asterisk). Novel SNP (rs11024184, double asterisks) remained significant. LD value is displayed in r2. (PDF 734 kb)
Regional association plots showing chromosomal position and P-value of SNPs, plotted as –log10 (P-value), in known T2D loci with putative novel lead SNPs. The lead East Asian SNP of each locus is plotted in purple diamond. The prior European SNP is indicated by a colored arrow, with the color corresponding to the r2 of the two SNPs plotted in Asian LD pattern. (PDF 912 kb)
Regional association plot of SLC30A8, an example of a loss of significance after performing a conditional analysis on a prior European SNP. The lead East Asian SNP is plotted in purple diamond. The prior European SNP is indicated by a colored arrow, with the color corresponding to the r2 of the two SNPs plotted in Asian LD pattern. (PDF 600 kb)
Regional association plots of CDKAL1 with the lead East Asian SNP plotted in purple diamond using Asian LD (A) and European LD (B) patterns. (PDF 968 kb)
(PDF 61 kb)
(PDF 81 kb)
(PDF 88 kb)
(PDF 58 kb)
(PDF 74 kb)
(PDF 70 kb)