Abstract
Adenocarcinoma of the lung, a leading cause of cancer death, frequently displays mutational activation of the KRAS proto-oncogene but, unlike lung cancers expressing mutated EGFR, ROS1 or ALK, there is no pathway-targeted therapy for patients with KRAS mutated lung cancer. In pre-clinical models, expression of oncogenic KRASG12D in the lung epithelium of adult mice initiates development of lung adenocarcinoma through activation of downstream signaling pathways. By contrast, mutationally activated BRAFV600E, a KRAS effector, fails to initiate lung carcinogenesis despite highly efficient induction of benign lung tumorigenesis. To test if PI3′-kinase-α (PIK3CA), another KRAS effector, might cooperate with oncogenic BRAFV600E to promote lung cancer progression, we employed mice carrying a conditional allele of Pik3ca that allows conversion of the wild-type catalytic subunit of PIK3CA to mutationally activated PIK3CAH1047R. Whereas expression of PIK3CAH1047R in the lung epithelium, either alone or in combination with PTEN silencing, was without phenotype, concomitant expression of BRAFV600E and PIK3CAH1047R led to dramatically decreased tumor latency and increased tumor burden compared to BRAFV600E alone. Most notably, co-expression of BRAFV600E and PIK3CAH1047R elicited lung adenocarcinomas in a manner reminiscent of the effects of KRASG12D. These data emphasize a role for PI3′-kinase signaling, not in lung tumor initiation per se, but in both the rate of tumor growth and the propensity of benign lung tumors to progress to a malignant phenotype. Finally, biological and biochemical analysis of BRAFV600E/PIK3CAH1047R expressing mouse lung cancer cells revealed mechanistic clues as to cooperative regulation of the cell division cycle and apoptosis by these oncogenes.
INTRODUCTION
Non-small cell lung cancer (NSCLC) is a leading cause of cancer mortality (1). Recently identified mutations in proto-oncogenes in NSCLC have provided strategies for the deployment of pathway-targeted therapies (2). However, despite the success of such agents in the treatment of genetically-defined subsets of lung cancer, most NSCLC patients do not have recourse to the use of such agents.
Of the proto-oncogenes mutated in NSCLC, KRAS remains the most common (≥25%) and the most pharmacologically intractable (2). Consequently, attention has turned to inhibiting key downstream signaling pathways required for maintenance of KRAS mutated NSCLC. Moreover, KRAS effectors such as BRAF, PI3′-kinase and AKT are also directly implicated as bona fide human lung cancer genes (2, 3, 4, 5). However, a key outstanding question is, what KRASG12D regulated pathways are essential for lung carcinogenesis in GEM models? Although KRASG12D-driven tumors are MEK1/2 dependent, expression of BRAFV600E in the lung epithelium elicits benign lung tumors that rarely progress to malignancy due to a senescence-like growth arrest (6, 7, 8). By contrast, although transgenic expression of mutationally activated PIK3CAH1047R in the lung epithelium was reported to promote lung tumorigenesis, combined inhibition of PI3′-kinase/mTorc had no effect on established KRASG12D-initiated lung tumors (9). Consequently, we tested whether mutationally activated BRAFV600E and PIK3CAH1047R might cooperate in promoting lung carcinogenesis using GEM models (6, 11).
Perhaps surprisingly, we could not substantiate reports that PIK3CAH1047R can initiate lung tumorigenesis. However, concomitant expression of both BRAFV600E and PIK3CAH1047R led to rapid onset of lung tumorigenesis with evidence of malignant progression observed six months after tumor initiation. The cooperative effects of PIK3CAH1047R in vivo were AKT dependent and were modeled in vitro. Finally, we demonstrate that both BRAFV600E and PIK3CA signaling is necessary for proliferation and survival of BRAFV600E/PIK3CAH1047R expressing lung cancer cells and demonstrate that these pathways cooperatively regulate the cell division cycle and apoptosis.
RESULTS
Expression of PIK3CAH1047R combined with PTEN silencing fails to induce lung tumors in mice
To test whether PIK3CAH1047R could initiate lung tumorigenesis, we employed Pik3calat-H1047R mice (Pik3calat) carrying a modified Pik3ca allele that expresses normal PIK3CA prior to Cre-mediated recombination after which PIK3CAH1047R is expressed under control of the gene’s chromosomal regulatory elements (9, 11). Cre-mediated genetic alterations were initiated using an adenovirus encoding Cre recombinase (Ad-Cre) (6). Since expression of PIK3CAH1047R from a single allele might be insufficient for lung tumorigenesis, we generated mice with every possible heterozygous or homozygous combination of Pik3calat in combination with a conditional null allele of Pten (Ptenlox) (12).
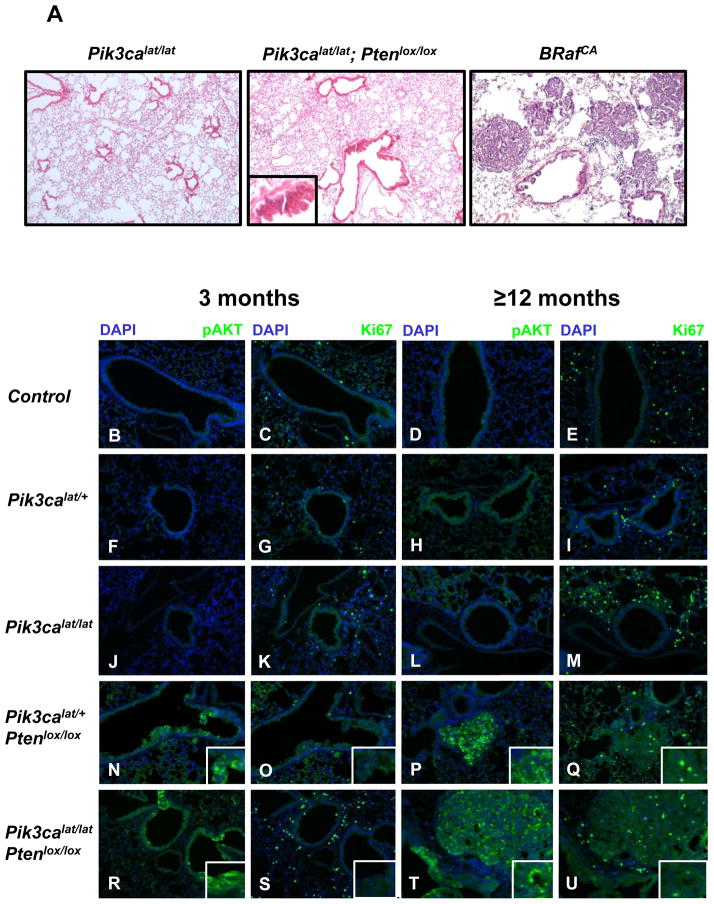
The lungs of adult mice of the various Pik3calat/Ptenlox genotypes were infected with Ad-Cre and euthanized for analysis three, six or ≥12 months post-infection. As a control for successful tumor induction, BRAFV600E expression was initiated in the lung epithelium of BRafCA mice, which developed benign lung adenomas at high multiplicity requiring euthanasia ~12 weeks post-initiation (Fig. 1A, right panel). By contrast, we failed to detect lung tumors in mice carrying conditional alleles of Pik3ca or Pten, either alone or in combination, at three or six months after Ad-Cre infection (Fig. 1A, left and middle panels). That PTEN silencing was insufficient for lung tumorigenesis was consistent with previous observations, but the lack of lung tumorigenesis in the compound Pik3calat/Ptenlox mice was surprising (13). At ≥12 months we detected benign adenomas in Pik3calat/lat; Ptenlox/lox mice however these were rare (< one tumor/mouse). Most of these tumors stained positive for PTEN expression and negative for phospho (p)-AKT suggesting that they may be spontaneously arising lung tumors unrelated to mouse genotype. However, we also detected very rare lung tumors in Pik3calat/lat; Ptenlox/lox mice that stained brightly for pAKT and may therefore due to this combination of genetic modifications (Fig. 1T). Immunofluorescence analysis of Ad-Cre infected Pik3calat or Pik3calat/lat mice failed to detect evidence of cells with pAKT or Ki67 expression above that detected in normal mouse lung (Fig. 1F–M). However, in Pik3calat mice carrying either one or two Ptenlox alleles, small numbers of airway epithelial hyperplasias were detected (Fig. 1N–U). Most prevalent in Pik3calat/lat; Ptenlox/lox mice, these lesions comprised small numbers of cells that stained negative for PTEN and positive for pAKT, with the strongest pAKT signal detected in lesions in Pik3calat/lat; Ptenlox/lox mice (Fig. 1R). These lesions were ≤50μm in diameter and not proliferative (Fig. 1S). Hence, by contrast to the effects of KRASG12D or BRAFV600E, there was no combination of Pik3calat and Ptenlox that elicited lung tumors in mice within 6 months after Ad-Cre (6, 9, 14).
Figure 1. PI3′-kinase pathway activation is insufficient to initiate tumorigenesis in the mouse lung.
A: Lungs of mice of the indicated genotypes were infected with Ad-Cre and monitored for three or ≥12 months as indicated at which time they were euthanized and their lungs processed for H&E staining. Representative H&E-stained tissue sections from Pik3calat/lat (left) or Pik3calat/lat;Ptenlox/lox (middle) animals euthanized 6 months after infection. BRafCA mice analyzed 12 weeks following Ad-Cre initiation are presented as a positive control (right).
B–U: Mouse lung sections from control (uninfected) or mice of the indicated genotypes treated as described in A were stained with DAPI, antisera against phospho-(p)-AKT or Ki67 as indicated. Insets indicate higher magnification of regions of airway hyperplasia.
PIK3CAH1047R dramatically accelerates BRAFV600E driven lung tumorigenesis
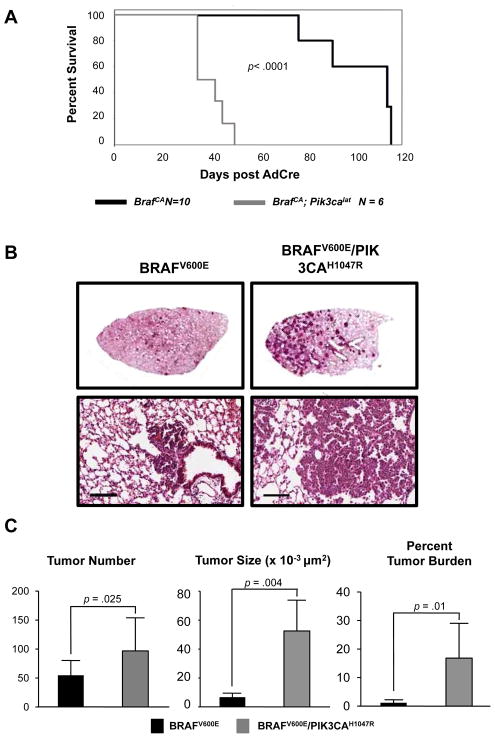
To determine if PIK3CAH1047R cooperates with BRAFV600E in lung tumorigenesis, we infected adult BRafCA or compound BRafCA; Pik3calat mice with either 5x106 or 107 pfu Ad-Cre. Kaplan-Meier survival analysis of mice infected with 5x106 pfu Ad-Cre indicated that all BRafCA; Pik3calat mice reached end-stage by ~50 days based on their body conditioning score, a time at which all BRafCA mice remained healthy (Fig. 2A) (15). Overall, BRafCA mice lived twice as long as BRafCA; Pik3calat littermates after initiation (median survival 100 vs. 39 days, p = 1.25 × 10−5, Fig. 2B). Furthermore, three weeks after infection with 107 pfu Ad-Cre, many BRafCA; Pik3calat mice displayed labored breathing and reduced body weight indicating onset of lethal lung tumorigenesis at which time representative mice were euthanized for necropsy (Fig. 2B). Compared to control BRafCA mice, BRafCA; Pik3calat mice presented with more tumors (96 vs. 54, p=0.025) and BRAFV600E/PIK3CAH1047R expressing tumors were larger than their BRAFV600E expressing counterparts (52,516 μm2 vs. 6,782.5μm2, p=0.004, Figs. 2B&C). Three weeks post-initiation, BRAFV600E/PIK3CAH1047R expressing lung lesions manifested as large adenomas, whereas BRafCA mice displayed only alveolar and/or airway hyperplasias (Figs. 2B&C). Overall, BRafCA; Pik3calat mice displayed a 14-fold higher tumor burden at three weeks post-initiation compared to BRafCA mice (16.8% vs. 1.2%, p=0.01, Fig. 2C).
Figure 2. PIK3CAH1047R accelerates lung tumorigenesis initiated by BRAFV600E.
A: BRafCA and BRafCA; Pik3calat mice were infected with Ad-Cre and monitored prospectively over ~120 days. Mice were euthanized at end-stage per IACUC regulations and a Kaplan-Meier survival curve was plotted.
B: BRafCA and BRafCA; Pik3calat mice were infected with 107 pfu Ad-Cre to initiate expression of BRAFV600E either alone or in combination with PIK3CAH1047R. Mice were euthanized three weeks later with lungs processed for H&E staining. Scale bar = 100μm.
C: Tumor number, size and burden were calculated as described in Materials and Methods in mice carrying either BRAFV600E or BRAFV600E/PIK3CAH1047R expressing tumors.
PTEN silencing enhances BRAFV600E-induced lung tumorigenesis
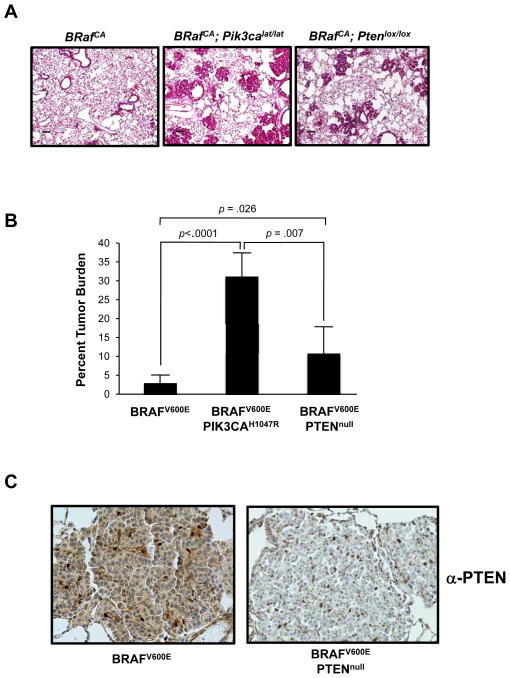
To complement analysis of the effects of PIK3CAH1047R on BRAFV600E-induced lung tumorigenesis, we tested if deregulation of PI3′-kinase signaling through PTEN silencing might have similar effects. Indeed, this is a combination of alterations commonly found in human melanoma and which cooperate in GEM models of the disease (5, 16). We infected BRafCA, BRafCA; Pik3calat/+ or compound BRafCA; Ptenlox/lox with 106 pfu Ad-Cre with mice euthanized four weeks after initiation. Compared to control BRafCA mice, BRafCA; Pik3calat and BRafCA/+; Ptenlox/lox mice displayed a ≥10-fold and a 4-fold increase in overall tumor burden respectively (2.8% vs. 31.2% vs. 10.8%, Figs. 3A & B). Whereas most lung tumor cells arising in BRafCA mice displayed readily detectable PTEN expression, the vast majority of tumor cells in BRafCA; Ptenlox/lox mice were PTEN negative (Fig 3C). These data indicate that the statistically significant difference between lung tumorigenesis in BRafCA; Pik3calat vs. BRafCA/+; Ptenlox/lox mice is not due to inefficient PTEN silencing. Overall, these data support the hypothesis that activation of PI3′-kinase signaling, either by co-expression of PIK3CAH1047R or PTEN silencing, cooperates with BRAFV600E in lung tumorigenesis (13).
Figure 3. PTEN silencing accelerates BRAFV600E-initiated lung tumorigenesis.
A: Mice of the indicated genotypes were infected Ad-Cre and euthanized four weeks later for analysis of tumor burden, which was quantified as described previously. Scale bars represent 100μM.
B: Lung tumors arising in BRafCA or BRafCA; Ptenlox/lox mice were stained with antisera against PTEN.
Pharmacological blockade of MEK or AKT prevents the growth of BRAFV600E/PIK3CAH1047R lung tumors
To assess the role of downstream signaling components on the cooperation between BRAFV600E and PIK3CAH1047R, we employed pharmacological inhibitors of either MEK1/2 (PD325901) or AKT (MK-2206) (17, 18). These agents are non-substrate competitive, allosteric inhibitors with high specificity and selectivity. BRafCA or BRafCA; Pik3calat mice were infected with 106 Ad-Cre and then separated into two groups for treatment with either vehicle control or MK-2206 (120 mg/kg, q.d., po) to inhibit AKT for an additional 4–5 weeks at which time mice were euthanized.
MK-2206 treatment had no effect on BRAFV600E-initiated lung tumor burden (vehicle - 25% vs. MK-2206 - 22%, p=0.63, Fig. S1A). By contrast, consistent with previous results, MEK1/2 inhibition largely abolished BRAFV600E-induced lung tumorigenesis (not shown) (6, 7). These data strongly suggest that BRAFV600E-induced lung tumors are AKT independent for their initial growth.
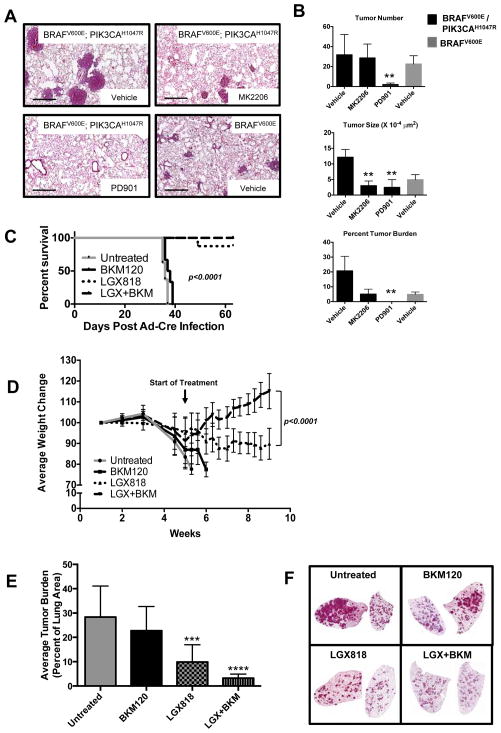
By contrast to BRAFV600E driven tumors, MK-2206 had a significant effect on BRAFV600E/PIK3CAH1047R driven lung tumorigenesis (Figs. 4A&B). Although four weeks of MK-2206 dosing had no effect on overall tumor number (vehicle = 31.8 vs. MK-2206 = 28.7; p=0.76), both tumor size (vehicle = 121,413 μm2 vs. MK-2206 = 30,083 μm2, p=<0.0001) and overall tumor burden (vehicle = 20.7%, MK-2206 = 5%, p=0.004), were significantly reduced following AKT inhibition (Figs. 4B). Indeed, tumor sizes and overall tumor burden that developed in BRafCA; Pik3calat mice in the presence of MK-2206 were roughly equivalent to that observed in BRafCA mice. These data support the hypothesis that AKT activity is required for the cooperation between BRAFV600E and PIK3CAH1047R in lung tumorigenesis but largely dispensable for the growth of BRAFV600E-induced lung tumors. Inhibition of pS473-AKT by MK-2206 was confirmed by immunoblot analysis of lysates of BRAFV600E or BRAFV600E/PIK3CAH1047R expressing lung tumors treated with either vehicle or MK-2206 two hours prior to euthanasia (Figs. S1B & S1C, respectively).
Figure 4. Pharmacological inhibition of PI3′K/AKT and MAPK signaling inhibits growth of BRAFV600E/PIK3CAH1047R tumors.
A:BRAFV600E or BRAFV600E/PIK3CAH1047R expressing lung tumors were initiated in mice of the appropriate genotype using Ad-Cre. Two weeks later mice were dosed for a further four weeks with vehicle control, MK-2206 or PD325901 as described in Materials and Methods at which time mice were euthanized and lungs prepared for H&E staining. Scale bar represents 500μM.
B: The effects of MK-2206 or PD325901 on lung tumor number, size and overall burden in mice bearing BRAFV600E or BRAFV600E/PIK3CAH1047R expressing lung tumors was estimated as described in Materials and Methods. ** represents p values less than or equal to 0.005.
C&D: Lung tumors were initiated in BRafCA; Pik3calat mice and five weeks later they were either untreated or treated with inhibitors of BRAFV600E (LGX-818) or class 1 PI3′-kinases (BKM-120) either alone or in combination as indicated. Mice were euthanized when they developed end-stage disease and this was used to calculate a Kaplan-Meier survival curve (C). In addition, the weight of every mouse in each group was measured and normalized to its starting weight, which was set at 100%, and the average normalized weight of all mice in a treatment group was used to assess their response to treatment (D).
E&F: Lung tumor burden in BRafCA; Pik3calat treated as described in C&D was assessed by H&E staining of lung tumor sections. Lung tumor specimens from untreated or BKM-120 treated mice came from mice with end-stage disease whereas the specimens from the LGX-818 or combined LGX-818 plus BKM-120 treatments were derived from mice at the end of the 4 week dosing schedule. *** represents a p value of 0.0003, **** represents a p value of less than 0.0001.
Previous studies have indicated the exquisite sensitivity of BRAFV600E driven lung tumors to MEK1/2 inhibition with PD325901 (6, 7). However, analysis of lung, pancreas and colon cancer cell lines suggest that PI3′-kinase signaling, either by mutation of PIK3CA or PTEN silencing, can render cells less sensitive to MEK inhibition possibly by sustaining the expression of D-type cyclins (19, 20). Hence, we sought to determine if BRAFV600E/PIK3CAH1047R lung tumors remained sensitive to MEK1/2 inhibition in vivo. Lung tumors were initiated in BRafCA; Pik3calat mice and two weeks later mice were dosed with vehicle or PD325901 (12.5mg/kg, q.d. po) for a further four weeks at which time mice were euthanized (Figs. 4A&B). Compared to controls, MEK1/2 inhibitor treated mice displayed a 15-fold reduction in tumor number (31.8 vs. 2.0, p=0.005). Lesions detected in PD325901 treated BRafCA; Pik3calat mice were largely small alveolar or airway hyperplasias and no fully formed adenomas were present in these mice. This resulted in a ~12-fold reduction in average tumor size (vehicle = 121,413 μm2 vs. PD325901 = 10,914μm2 for drug treated mice, p<0.0001) and a >20,000-fold reduction in overall tumor burden (vehicle = 20.7% vs. PD325901 = 0.001%, p=0.0004) compared to controls (Figure 4B). Consequently, these data indicate that BRAFV600E/PIK3CAH1047R lung tumors remain sensitive to the anti-tumor effects of MEK1/2 inhibition and highlight the central importance of BRAFV600E→MEK→ERK signaling in the growth of BRAFV600E/PIK3CAH1047R-induced lung tumors.
Enhanced therapeutic benefit of combined BRAFV600E plus PIK3CAH1047R inhibition against BRAFV600E/PIK3CAH1047R expressing lung tumors
To examine the requirement for sustained signaling in maintenance of BRAFV600E/PIK3CAH1047R expressing lung tumors, we initiated tumorigenesis in 34 BRafCA; Pik3calat mice and monitored them for five weeks until they were close to end-stage. At this time two mice were euthanized with the remaining mice divided into four cohorts for the following interventions: 1. No treatment; 2. Class 1 PI3′-kinase inhibitor (BKM-120, 60mg/kg, q.d., po); 3. BRAFV600E inhibitor (LGX-818, 30mg/kg, q.d., po) or; 4. Combination of BKM-120 and LGX-818 (60 and 30mg/kg respectively, q.d., po) (21, 22). Drug benefit was assessed by Kaplan-Meier analysis with mice euthanized when their body conditioning score (BCS) was ≤2 (15) (Fig. 4C). In addition we monitored mouse body weight, which correlates with changes in tumor burden (Fig. 4D).
There was no statistical difference in survival between untreated (n=6) or BKM-120 treated (n=8) mice, all of which lost weight requiring euthanasia within five days after dosing (Figs. 4C&D). By contrast, mice treated with LGX-818 (n=8) or combined LGX-818 plus BKM-120 (n=8) displayed increased survival (Fig. 4C) such that no mice in the combination arm required euthanasia. Although LGX-818 treated mice continued to lose weight, this was at reduced rate and only 1/8 mice developed end-stage disease. Finally, mice treated with combined LGX-818 plus BKM-120 displayed immediate signs of improvement in their BCS that continued throughout the treatment period (Fig. 4D).
Analysis of mouse lungs from each group indicated that untreated and BKM-120 treated mice displayed a substantial tumor burden (average of ~28% and ~23%) with no difference between them, consistent with the lack of benefit afforded by BKM-120 (Figs. 4E&F). By contrast, LGX-818 treated mice displayed reduced tumor burden (~10%) compared to control or BKM-120 treated mice. The therapeutic benefit of LGX-818 treated mice was further enhanced by combination with BKM-120 (~3% tumor burden) consistent with the survival and health benefits afforded these mice. Taken together, combined blockade of BRAFV600E and PIK3CAH1047R signaling in BRAFV600E/PIK3CAH1047R expressing lung tumors was superior to blockade of BRAFV600E or PIK3CAH1047R alone.
PIK3CAH1047R promotes malignant progression of BRAFV600E-induced tumors
KRASG12D-induced lung tumorigenesis in GEM models depends on RAF→MEK→ERK, PI3′-kinase, RAC1 and NF-κB signaling (7, 9, 23–26). However, to date, activation of single nodes downstream of KRAS.GTP fails to recapitulate the malignant progression displayed by KRASG12D-initiated lung tumors. Although BRAFV600E induces benign lung tumors, such lesions are low grade and fail to progress to lung cancer (6, 7). As demonstrated here, although PIK3CAH1047R is unable to initiate lung tumorigenesis, it promotes the growth BRAFV600E-initiated lung tumors. This prompted us to evaluate whether BRAFV600E and PIK3CAH1047R might cooperate for malignant lung cancer progression. To that end, tumors were initiated in either BRafCA or BRafCA; Pik3calat mice (5x105 pfu Ad-Cre) with mice euthanized 25 weeks later for evaluation (7, 27).
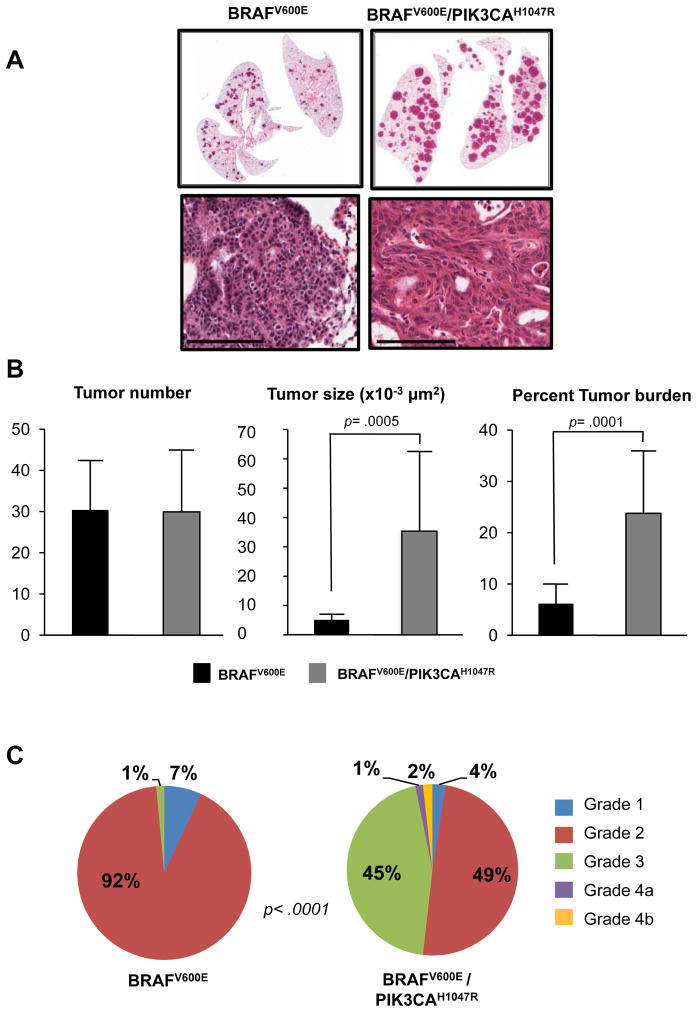
By contrast with early time points, overall tumor number was not different between BRafCA and BRafCA; Pik3calat mice with an average of 30 tumors per lobe recorded in both genotypes (Figs. 5A&B). However, individual lung tumors in BRafCA; Pik3calat mice were larger than those in BRafCA mice (35,000μm2 vs. 5,000μm2, p=0.0005, Fig. 5B). Differences in tumor size were reflected by overall increased tumor burden in BRafCA; Pik3calat mice (24% vs. 6%, p=0.0001, Fig. 5B). These data suggest that PIK3CAH1047R does not influence the frequency of BRAFV600E initiated cells in the lung epithelium, but decreases the latency period prior to tumorigenesis and also increases their overall proliferative potential.
Figure 5. PIK3CAH1047R promotes malignant progression of BRAFV600E-initiated lung tumors.
A: BRAFV600E or BRAFV600E/PIK3CAH1047R expressing lung tumors were initiated in mice of the appropriate genotype using Ad-Cre with mice monitored for 25 weeks at which time they were euthanized and their lungs processed for H&E staining. Scale bar represents 50μM. Lung tumor number, size and overall burden was assessed as described in Materials and Methods.
B: The grade of BRAFV600E or BRAFV600E/PIK3CAH1047R expressing lung tumors was assessed on a 4 point scale using the grading criteria described in Supplemental Figure S3.
One possible explanation for the larger tumors observed in BRafCA; Pik3calat mice is that each tumor arises in a polyclonal manner from more than one initiated cell. To test the clonality of BRAFV600E/PIK3CAH1047R lung tumors we generated BRafCA; Pik3calat mice containing the “Confetti” reporter (Gt(ROSA)26Sortm1(CAG-Brainbow2.1)Cle, R26Confetti hereafter) (Supp. Fig. 2) (28). The action of Cre on the R26Confetti reporter elicits a recombination event that stochastically places one of four different fluorescent proteins (FP) downstream of the CAG promoter. Hence, if BRAFV600E/PIK3CAH1047R-induced lung tumors are clonally derived, each tumor should express a single FP. However, if tumors are polyclonal in origin, they will contain a mixture of cells with different FP expression. BRafCA; Pik3calat; R26Confetti mice were infected with 107 pfu Ad-Cre with lung tumors analyzed 9 weeks later. Individual BRAFV600E/PIK3CAH1047R lung tumors were uniformly positive for a single FP, indicating that each tumor is clonally derived from a single initiated cell (Supp. Fig. 2).
To assess malignant progression, we employed a grading scheme based on the classification of lung lesions in GEM models to assess the grade of >250 tumors from mice of each genotype (Fig. 5C and Supp. Figs. 3A&B) (14, 27). Hyperplasia was designated Grade 1, benign adenomas as Grade 2, larger adenomas as Grade 3 and adeno-squamous or adenocarcinomas as Grade 4. BRAFV600E-induced lung lesions were predominantly Grade 1 or 2 with rare examples of Grade 3 detected. By contrast, BRAFV600E/PIK3CAH1047R-induced lesions were generally Grade 2 or 3 adenomas but ~3% of tumors were malignant Grade 4 adeno-squamous or adenocarcinoma lesions. These data indicate that BRAFV600E cooperates with PIK3CAH1047R to both accelerate early-stage lung tumorigenesis and to promote late-stage malignant progression (14).
To determine if the cell of origin might influence the spectrum of lung tumors initiated in these mice, we employed adenovirus vectors with lung cell specific expression of Cre recombinase (29). Ad-SPC-Cre, Ad-CCSP-Cre and Ad-CGRP-Cre direct Cre expression to Alveolar type II pneumocytes, Clara or neuroendocrine cells respectively. Of these, we noted that Ad-SPC-Cre was the most potent inducer of lung tumorigenesis in either BRafCA or BRafCA; Pik3calat mice and was equivalent in potency to Ad-CMV-Cre. Moreover, tumors emerging in mice initiated with either Ad-CCSP-Cre or Ad-CGRP-Cre, which were less potent in inducing lung tumorigenesis, displayed the cuboidal morphology and expression of SP-C characteristic of AT2 cells (data not shown).
In these experiments we assessed tumor grade at 22 weeks in groups of BRafCA or BRafCA; Pik3calat mice initiated with Ad-SPC-Cre to confirm the ability of PIK3CAH1047R to promote malignant progression of BRAFV600E-initiated lung tumors (Supp. Fig. 4). As before, the majority of BRAFV600E expressing lung tumors were small, grade 2 adenomas. By contrast, >50% of lung tumors in the BRafCA; Pik3calat mice were high-grade adenomas (Grade 3) and 8.5% were either grade 4 adeno-squamous cancers or adenocarcinomas confirming observations made with Ad-CMV-Cre (30).
TP53 constrains malignant progression of BRAFV600E/PIK3CAH1047R lung tumors
TP53 mutations frequently coexist with mutations in KRAS, BRAF or PIK3CA mutations in human lung adenocarcinomas and TP53 mutation or silencing promotes malignant progression of either KRASG12D or BRAFV600E-driven lung tumors (6, 14). Consequently, we determined if TP53 loss would enhance lung tumorigenesis initiated by combined expression of BRAFV600E and PIK3CAH1047R. Tumorigenesis was initiated in a BRafCA/CA; Pik3calat, Trp53lox/lox mouse with a BRafCA; Trp53lox/lox mouse as a littermate control. Within four weeks the former mouse displayed end-stage disease requiring euthanasia. Whereas the BRafCA; Trp53lox/lox mouse had a tumor burden of 0.2% made up of mainly small hyperplasias with a solitary benign adenoma detected (Supp. Fig. 5A), the BRafCA/CA; Pik3calat, Trp53lox/lox mouse had a tumor burden of 41% made up of high grade, anaplastic adenocarcinoma. Moreover, these high-grade tumors stained positive for Vimentin, a marker of epithelial-mesenchymal transition, lacked TTF-1/NKX2-1 expression, a marker of the distal lung epithelium and displayed a high proliferative index (Ki67, Supp. Fig. 5B). Similar results were obtained using a second BRafCA/+; Pik3calat, Trp53lox/lox mouse.
PIK3CAH1047R promotes BRAFV600E-induced oncogenic transformation in vitro
An advantage of GEM models cancer is the ability to engineer defined genetic alterations and then generate cell lines for in vitro analysis. To that end, we generated BRAFV600E/TP53Null and BRAFV600E/PIK3CAH1047R/TP53Null mouse lung cancer derived cell lines (BT and BPT cells hereafter) from suitably manipulated mice (7).
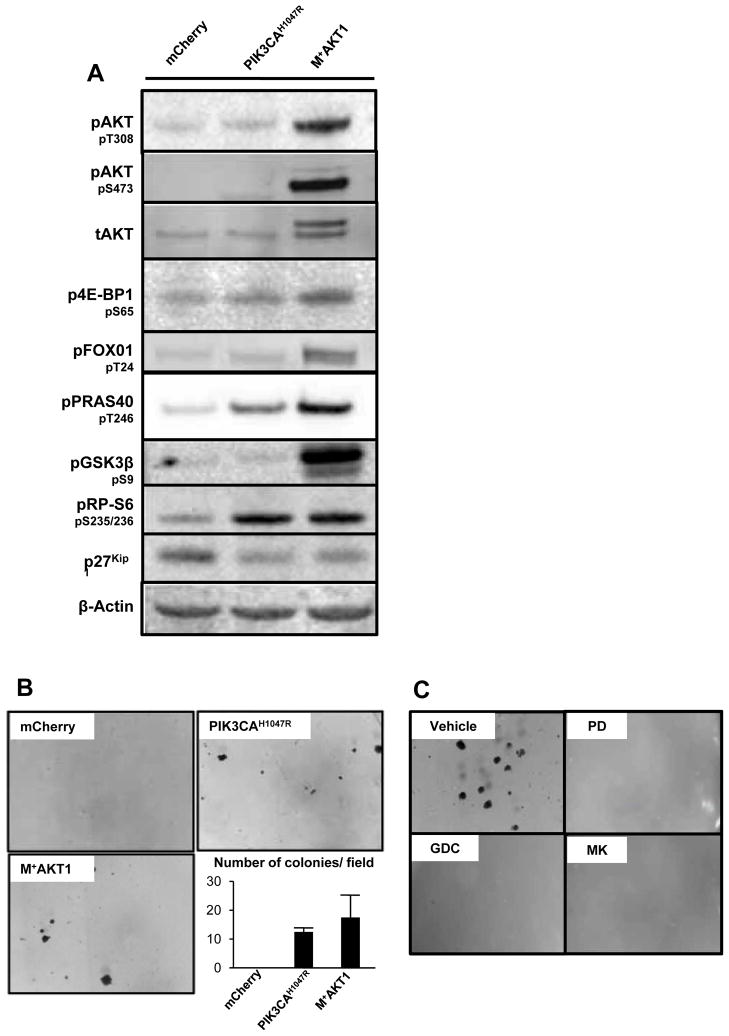
Since BT cells displayed a limited capacity for anchorage independent growth we engineered them to express either mCherry (control), PIK3CAH1047R or a myristoylated, constitutively activated AKT1 (M+AKT1) to test whether activation of PI3′-kinase signaling might cooperate with BRAFV600E to influence the behavior of these cells (Fig. 6). Cell extracts were analyzed by immunoblotting for the activation status of key nodes in the PI3′-kinase signaling pathway (Fig. 6A and Supp. Fig. 8A). Compared to control, cells expressing either PIK3CAH1047R or M+AKT1 displayed elevated pPRAS40 and pRP-S6 while M+AKT1 expressing cells displayed elevated levels of pAKT, p4E-BP1, pFOXO1 and pGSK3β. PIK3CAH1047R and M+AKT1 cells also showed a decrease in p27KIP1 expression. Consistent with the ability of PIK3CAH1047R to cooperate with BRAFV600E in lung tumorigenesis in vivo, expression of either PIK3CAH1047R or M+AKT1 led to increased anchorage-independent colony formation in vitro (Fig. 6B). Indeed, the parental and mCherry control BT lung cancer cells failed to form colonies in agarose even when cultured for ~8 weeks. By contrast, the PIK3CAH1047R or M+AKT1 expressing cells cloned with an efficiency of 1% and 1.4%, respectively, after three weeks. These data indicate that PI3′-kinase→AKT signaling cooperates with BRAFV600E in oncogenic transformation of cultured lung cancer cell lines.
Figure 6. Ectopic expression of either activated PIK3CA or AKT1 promotes anchorage independent growth to BRAFV600E; TP53null lung cancer derived cells.
A: BRAFV600E/TP53null lung cancer derived cells were infected with retroviruses as described in the text. Cell extracts were analzed by immunoblotting as indicated. Quantification of blots is included as Supp. Fig. 8A.
B: Anchorage-independent growth of BRAFV600E/TP53null lung cancer cells engineered to express mCherry, PIK3CAH1047R or M+AKT1 was quantified over three weeks.
C: BRAFV600E/PIK3CAH1047R/TP53null lung cancer cells were plated in soft agar in the absence (vehicle) or presence of inhibitors of MEK1/2 (1μM PD325901), class 1 PI3′-kinase (5μM GDC-0941) or AKT (5μM MK-2206). Anchorage-independent growth was assessed over three weeks.
The ability of BPT lung cancer cells to form colonies in agarose was assessed in the presence of various pathway-targeted inhibitors. Unlike BT cells, BPT cells are capable of anchorage-independent colony formation (Vehicle, Fig. 6C). However, when cultured in the presence of a MEK inhibitor, a class 1 PI3′-kinase inhibitor or an AKT inhibitor, colony formation was abolished even when cultured for 8 weeks (Fig. 6C) (31). These data emphasize the ability of PI3′-kinase→AKT signaling to cooperate with BRAFV600E in promoting the transformed phenotype on lung cancer cells.
BRAFV600E/PIK3CAH1047R/TP53Null lung cancer cell lines are sensitive to combined pharmacological inhibition of RAF or PI3′-kinase pathway inhibition
To assess the mechanism(s) of cooperation between BRAFV600E and PIK3CAH1047R on the behavior of BPT cells they were treated with various pathway-targeted inhibitors of BRAFV600E or PIK3CAH1047R signaling, either alone or in combination, at which time effects on cell viability or the cell division cycle were assessed (Figs. 7A–C).
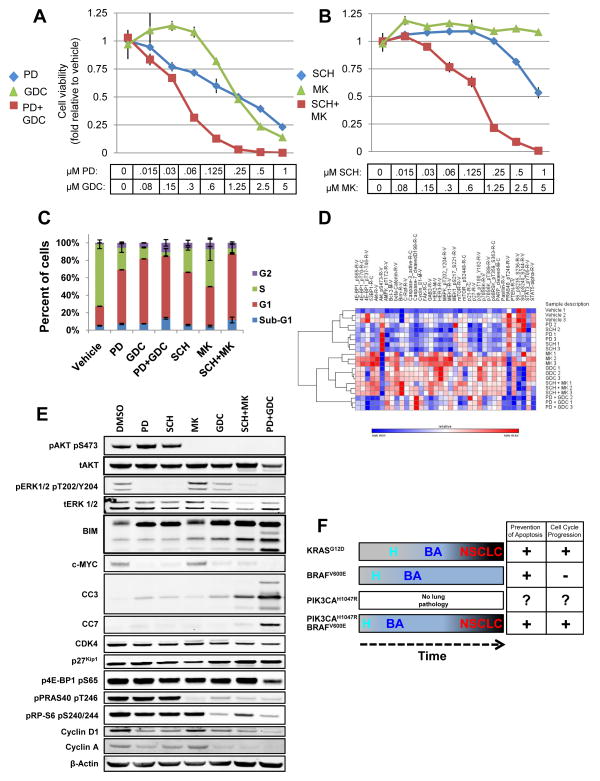
Figure 7. Cooperative effects of combined pharmacological blockade BRAFV600E and PIK3CAH1047R signaling on BRAFV600E/PIK3CAH1047R/TP53null lung cancer derived cells.
A&B: BRAFV600E/PIK3CAH1047R/TP53null lung cancer cells were treated with a range of concentrations of MEKi (PD325901), PI3′Ki (GDC-0941), ERKi (SCH772984) or AKTi (MK-2206) either alone or in the indicated combinations for 72 hours when cell viability was measured using a Cell-Titre-Glo assay (Promega) and normalized to zero concentration values.
C: BRAFV600E/PIK3CAH1047R/TP53null lung cancer cells were treated with inhibitors of MEK1/2, ERK1/2, AKT1-3 or class 1 PI3′-kinases either alone or in the indicated combinations for 24 hours at concentrations listed in Table S1. Cell cycle status was assessed by FACs analysis of cells labeled with BrdU and Propidium Iodide.
D: Unsupervised clustering of RPPA analysis of cell extracts derived from cells treated in biological triplicates with inhibitors of MEK1/2 (PD), ERK1/2 (SCH), AKT1-3 (MK) or class 1 PI3′-kinases (GDC) either alone or in the indicated combinations for 24 hours. Full dataset is included as Supp. Fig. 7 and an Excel file.
E: Cell extracts prepared from BPT lung cancer cells treated with inhibitors of MEK1/2 (PD), ERK1/2 (SCH), AKT1-3 (MK) or class 1 PI3′-kinases (GDC) either alone or in the indicated combinations for 24 hours were analyzed by immunoblotting and quantified against β-actin (Supp. Fig. 8B).
F: Mutationally activated KRASG12D promotes lung hyperplasia (H) then benign adenomas (BA), some of which progress to non-small cell lung cancers (NSCLC). Similarly initiated expression of BRAFV600E leads to more rapid and numerous hyperplasia and benign adenomas but without progression to NSCLC, most likely due to a senescence-like growth arrest. Although PIK3CAH1047R fails to initiate pathology in the lung, when combined with BRAFV600E, it promotes rapid emergence of hyperplasias and benign adenomas, some of which progress to NSCLC. At the molecular level, BRAFV600E cooperates with PIK3CAH1047R to regulate the cell division cycle and suppress apoptosis.
As expected, single agent inhibition of MEK1/2 or class 1 PI3′-kinases effectively decreased cell proliferation. Moreover, the combination of these agents had the strongest anti-proliferative effects, demonstrating at least an additive effect particularly in the mid-range of drug (Fig. 7A). Despite its potency in vivo, single agent AKT inhibition (MK-2206) was ineffective at preventing the growth of these cells (Fig. 7B) even at 5μM. Single agent ERK1/2 inhibition (SCH772984) also exerted a cytostatic effect, only decreasing cell viability at higher concentrations (>250nM) (32). However, combined ERK1/2 plus AKT inhibition displayed cooperativity such that concentrations of inhibitors that had no single agent effect had striking inhibitory effects on cell proliferation when combined (Fig. 7B), which was further confirmed by Crystal Violet staining (Supp. Fig. 6A).
To assess the effects of the various agents on the cell division cycle, BPT cells were incubated with the various agents either alone or in combination and then labeled with BrdU to assess S phase progression (Fig. 7C, see Table 1 for drug concentrations). Vehicle-treated cells displayed a high proliferative index with >75% of cells incorporating BrdU. As expected, single-agent blockade of MEK1/2, ERK1/2, PI3′-kinase or AKT led to decreased BrdU incorporation and increased the G1 population with the PI3′-kinase inhibitor most potent, the MEK and ERK inhibitors roughly equipotent and the AKT inhibitor least potent. Cells treated with combined MEK1/2 plus PI3′-kinase or ERK1/2 plus AKT inhibitors displayed the most profound effects on the cell division cycle in addition to displaying evidence of cell death as assessed by the number of non-adherent cells and cells with a sub-G1 DNA content (Fig. 7C and Table 2). Taken together, these data indicate that BRAFV600E and PIK3CAH1047R cooperatively regulate the cell division cycle and cell death in vitro.
BRAFV600E and PIK3CAH1047R likely regulate many biochemical processes in the cell to influence cell proliferation in vitro and tumorigenesis in vivo. To assess the molecular mechanism(s) of such oncogenic cooperation we employed reverse phase protein array (RPPA) analysis to quantify protein expression/modification in BPT cells treated with inhibitors (Fig. 7D). Triplicate cultures of BPT cells were treated with inhibitors of MEK1/2 (PD), ERK1/2 (SCH), class 1 PI3′-kinases (GDC) or AKT1-3 (MK) either alone or in combination as indicated (Fig. 7D). Prior to RPPA analysis, we confirmed that each of the agents had the anticipated biochemical effects (Supp. Fig. 6B). As expected, inhibition of MEK1/2 or ERK1/2 led to strongly diminished pERK1/2 and promoted a modest increase in pAKT. Inhibition of PI3′-kinases or AKT led to strongly diminished pAKT with no obvious effects on pERK. Finally, the combinations of inhibitors potently suppressed both pERK1/2 and pAKT.
The triplicate samples were subject to RPPA analysis using antisera against >150 proteins or protein modifications (33, 34). Unsupervised hierarchical clustering of the data indicated that samples clustered based on the pathway inhibited such that MEKi or ERKi treated samples clustered together as did PI3Ki and AKTi treated samples (Fig. 7D and Supp. Fig. 7). Samples from cells treated with combined pathway inhibitors clustered separately from samples from cells treated with the various single agents with the samples from cells treated with combined MEK1/2 plus class PI3′-kinase showing the greatest separation.
RPPA analysis generated predictions with regards to the consequences of inhibition of BRAFV600E or PIK3CAH1047R signaling on regulators of cell proliferation, some of which were then validated in an independent sample set by immunoblotting (Fig. 7E and Supp. Fig. 8). As predicted by RPPA analysis, inhibition of MEK1/2→ERK1/2 signaling led to increased expression of the pro-apoptotic BCL-2 family protein BIM and decreased expression of c-MYC as well as cyclins D1 and A. Inhibition of PIK3CAH1047R, but not AKT, also promoted BIM expression and inhibited c-MYC, cyclins D1 and A and led to elevated p27KIP1 expression and suppression of pRP-S6. Consistent with the lack of effect of MK-2206 against the proliferation of cultured BPT cell, we noted that inhibition of AKT had no single-agent effect on BIM, c-MYC, cyclins D1 or A, p4E-BP1 or pRP-S6 but largely extinguished phosphorylation of PRAS40, a known AKT substrate. Finally, combined inhibition of both MEK and PI3′-kinases displayed additive effects on expression of c-MYC, cyclins D1 and A and BIM, the latter correlating with increased cleaved Caspases 3 (CC3) and 7 (CC7) that were also predicted by RPPA analysis. These analyses indicate the complexity with which these pathways cooperatively regulate apoptosis and the cell division cycle, which in turn influences tumor latency, burden and malignant progression. Although these experiments do not identify a single mechanism of BRAFV600E - PIK3CAH1047R cooperation in lung carcinogenesis, they suggest numerous areas of future investigation.
DISCUSSION
The mechanism(s) by which KRASG12D promotes lung carcinogenesis in humans and mice remain incompletely understood. However, here we demonstrate that two well-credentialed KRAS.GTP effectors, BRAF and PIK3CA, cooperate to mimic the effects of oncogenic KRASG12D in GEM models of lung cancer. Whereas BRAFV600E expression elicits benign lung tumorigenesis, such tumors rarely progress to lung cancer, which contrasts with KRASG12D-initiated lung tumors (Fig. 7F). Unexpectedly, we noted that PIK3CAH1047R failed to initiate lung tumorigenesis, even when combined with PTEN silencing, a result at-odds with a previous report (9) (Fig. 7F). Indeed, no combination of heterozygous or homozygous Pik3calat and Ptenlox initiated lung tumorigenesis, contrasting with the effects of a tetracycline inducible PIK3CAH1047R transgene. It remains possible that strength of pathway activation, genetic background or the promoter system may influence these disparate results as suggested by similar observations in pancreatic cancer (24, 35). However, the lack of PIK3CAH1047R effects in lung is consistent with the effects of PIK3CAH1047R in ovarian epithelial cells (11). Furthermore, others have demonstrated that PI3′-kinase activation through PTEN silencing failed to elicit lung tumorigenesis (13). It is unclear whether the inability of PIK3CAH1047R to initiate lung tumorigenesis will be predictive of anti-tumor effects of pharmacological inhibition targeting of PI3′-kinase in the clinic (9).
Despite its inability to initiate tumorigenesis, PIK3CAH1047R cooperated with BRAFV600E in dramatic fashion to promote lethal lung tumorigenesis, which occurred with short latency and complete penetrance (Fig. 7F). That PTEN silencing also cooperated with BRAFV600E, albeit less strikingly, confirmed these observations indicating an important role for PI3′-lipid signaling. Although at early time points, BRAFV600E/PIK3CAH1047R-induced tumors were benign, six months post-initiation there was evidence of cancer progression in a manner reminiscent of KRASG12D. Finally, TP53 silencing in conjunction with combined BRAFV600E/PIK3CAH1047R expression, led to rapid lung cancer development, which showed signs of de-differentiation and epithelial-to-mesenchymal transition, hallmarks of advanced disease (36, 37, 38).
In conjunction with previous reports of the MEK1/2 and PI3′-kinase dependency of KRASG12D-induced lung tumors (39), the simplest explanation is that combined activation of RAF→MEK→ERK and PI3′-kinase→AKT signaling cooperates to accelerate benign tumorigenesis and to promote malignant lung cancer progression. Although activation of BRAF and PI3′-kinase signaling accounts for much of the phenotype associated with KRASG12D expression, a role for other KRASG12D effectors remains possible since ~10% of KRASG12D initiated tumors display malignant progression whereas only ~3–8% of BRAFV600E/PIK3CAH1047R tumors do so (7). Indeed, since ≤10% of KRASG12D- or BRAFV600E/PIK3CAH1047R-initiated lung tumors display malignant progression, there must be additional stochastic genetic/epigenetic changes in benign tumor cells that promote malignant progression as emphasized by the effects of TP53 silencing on the progression of KRASG12D- or BRAFV600E/PIK3CAH1047R-initiated lung tumors.
The absence of malignant progression of BRAFV600E-initiated lung tumors has been ascribed to a senescence-like proliferative arrest, which is due to feedback inhibition of PI3′-lipid production (10). This hypothesis is supported by our data since PIK3CAH1047R expression enhanced the proliferative capacity of BRAFV600E-initiated lung tumors. However, it remains unclear how PI3′-kinase signaling sustains the proliferation of these tumors. Previous data indicate cooperative regulation of cyclins cyclin-dependent kinase inhibitor expression by these pathways (40). Analysis of BPT cells suggests that the cell division cycle is indeed under the coordinate control of both BRAFV600E and PIK3CAH1047R. However, future experiments are required to determine which of the nodes of cell cycle regulation are essential to the observed cooperation.
BPT cells were sensitive to combined inhibition of BRAFV600E and PIK3CAH1047R with pathway-targeted agents. In general, single-agent inhibition of either pathway had inhibitory effects on the cell division cycle without promoting apoptosis. By contrast, combined pathway inhibition had stronger inhibitory effects on the cell cycle and also promoted cell death in vitro and in vivo. That AKT inhibition was without effect against lung tumors initiated by BRAFV600E but potently suppressed the cooperation between BRAFV600E and PIK3CAH1047R suggests a genotype-drug response phenotype with regards to AKT inhibition in GEM models. Whether these data will translate into the clinical utility of single or combined pathway-targeted interventions in humans with BRAF mutated lung cancer remains an open question. Whereas KRAS mutation is frequent in lung cancer, expression of BRAFV600E or PIK3CAH1047R accounts for a smaller percentage of NSCLC patients (2). Since mutated KRAS has proven an intractable pharmacological target, it will be interesting to determine whether combined pathway-targeted inhibition of RAF and PI3′-kinase signaling will promote regression of KRAS mutated lung cancers.
MATERIALS AND METHODS
Mice and Adenovirus delivery
Animal experiments were conducted in accordance with protocols approved by the UCSF Institutional Animal Care and Use Committee (IACUC). BRafCA (Braftm1Mmcm), Trp53lox (Trp53tm1Brn), PIK3CAlat-H1047R and Ptenlox mice were bred and genotyped as previously described (6, 11, 12, 14, 41). All mice were back-crossed onto the FVB/NJ genetic background for at least 10 generations. Experiments were performed using littermates as controls whenever possible. Stocks of Adenovirus encoding Cre recombinase were purchased from Viraquest (North Liberty, IA) or the University of Iowa, Gene Transfer Vector Core (29). Adenoviruses with cell specific expression of Cre recombinase were generously provided by Kate Sutherland and Anton Berns (NKI, Amsterdam) and purchased from the University of Iowa, Gene Transfer Vector Core (29). Intranasal instillation of adenovirus for infection of the mouse lung epithelium was performed as previously described (42). Tumor bearing mice were euthanized for analysis either at a pre-determined time point or when their body conditioning score (BCS) was ≤2 (15).
Histology and quantification of lung tumor burden
Lungs were removed, fixed in zinc buffered formalin (ZBS) and stored in 70%(v/v) ethanol prior to paraffin embedding. 6μm sections were stained with Hematoxylin and Eosin (H&E) on slides, scanned with an Aperio ScanScope scanner. Quantification of tumor number, size, and overall burden of individual lung sections was conducted using Aperio Spectrum ImageScope software. Overall tumor burden was calculated as (area of lung section occupied by tumor)/(total area of section) in μm2.
Treatment of mice with pathway-targeted therapeutics
PD325901 (Hansun Trading Co.) was formulated in 0.5%(w/v) Hydroxy-Propyl-Methylcellulose (HPMT, Sigma) and administered daily (q.d) by oral gavage (po) at 12.5 mg/kg. MK-2206 (Merck) was formulated in 30%(w/v) Captisol (Ligand Technology, La Jolla, Ca.) and dosed at 120 mg/kg q.d. po. LGX-818 and BKM-120 (Novartis) were formulated in 0.5% carboxymethylcellulose (Sigma) plus 0.5% Tween 80 (Sigma) and dosed q.d. po at 30 mg/kg and 60 mg/kg, respectively. Mice were dosed with drug for pre-determined time points or until their body conditioning score (BCS) was ≤2, at which point they were euthanized for analysis (15).
Immunostaining of mouse lung tissue and immunoblotting
ZBS fixed sections of mouse lungs were subject to citrate-mediated antigen retrieval and then probed with: anti-pS473-AKT, anti-Vimentin, anti-PTEN (Cell signaling technology), anti-NKX2.1, anti-Ki67, anti-SP-C and anti-p63 (Santa Cruz Biotechnology). Immunoblotting was performed on 50μg of extracted protein and probed with antisera against various phospho- or backbone specific antisera as described. Immunoblots were quantified using an Odyssey FC system and Image Studio software.
Lung tumor cell isolation, culture and analysis
Single cell suspensions were generated from BRAFV600E/TP53Null- or BRAFV600E/PIK3CAH1047R/TP53Null-induced lung cancers and cultured in Hams-F12/Glutamax media with 10%(v/v) fetal bovine serum. Following outgrowth, single cell-derived clones were isolated. Recombination of BRafCA, Trp53lox and Pik3calat alleles was verified by PCR (11). Expression of oncogenic BRAFV600E was determined by immunoblotting with a mAb (VE1) specific to BRAFV600E (43).
Cell proliferation was assessed in triplicate cultures of cells treated for 72 hours with: DMSO control, MEK1/2 inhibitor (PD325901, 15.6nM-1μM), ERK1/2 inhibitor (SCH772984, 7.8nM-5μM), AKT1-3 inhibitor (MK-2206, 80nM-5μM) or class 1 PI3′-kinase inhibitor (GDC-0941, 80nM-5μM) alone or in combination as indicated using a Cell Titre Glow assay (Promega). Cell cycle status was assessed in fixed floating and adherent cells stained with anti-BrdU-FITC (Becton-Dickinson) and propidium iodide (Sigma). Sorting of mCherry encoding ecotropic retroviruses was performed using a FACS-Calibur (Becton Dickinson) (44, 45). Anchorage-independent colony formation was performed by plating 5x103 cells in 1.4%(w/v) Sea-Plaque low melting temperature agarose (Cambrex, Rockland, ME) and culturing them for three weeks at which time colony formation was enumerated using dissecting microscope.
Reverse Phase Protein Array (RPPA) Analysis
Reverse phase protein array (RPPA) analysis (Functional Proteomics Core Facility at M.D. Anderson Cancer Center) was performed as previously described (33, 34) and in supplementary information.
Supplementary Material
Acknowledgments
We thank the members of the McMahon lab, especially David Dankort, Roch-Philippe Charles, Jillian Silva, Joseph Juan and J. Edward Van Veen for training, advice, guidance and support throughout this project. We thank Jane Gordon, Yunita Lim and Cynthia Cowdrey from the UCSF Laboratory for Cell Analysis and the Brain Tumor Research Center respectively for assistance with microscopy, FACS analysis and processing of formalin fixed samples. We thank Kate Sutherland and Anton Berns (Netherlands Cancer Institute) for access to Adenovirus-Cre vectors, Frank McCormick (UCSF) and Xiaolin Nan (OHSU) for retroviral constructs, Meredith West and Sandy DeVries from the UCSF Radiation Therapy Oncology Group for use of the Aperio Scanner, Darrin Stuart and Emmanuelle Di Tomaso (Novartis) for LGX-818 and BKM-120 respectively, Lori Friedman (Genentech) for GDC-0941, Steven Townson and Ahmed Samatar (Merck) for MK-2206 and SCH772984. We thank Dr. Michael A. Davis and the M.D. Anderson Functional Proteomics Core Facility for performing and analyzing the RPPA analysis (supported by an NCI Cancer Center Support Grant, CA-16672). CT was supported by a Genentech Foundation Graduate Student Fellowship, SG was supported by a National Science Foundation Graduate Research Fellowship, EAC was supported by K08 CA137153 and by the Pancreatic Cancer Action Network, WAP was supported by project grants from the National Health and Medical Research Council of Australia and MM was supported by the National Cancer Institute (CA131261) and by Uniting Against Lung Cancer.
LITERATURE CITED
- 1.Herbst RS, Heymach JV, Lippman SM. Lung cancer. N Engl J Med. 2008;359:1367–80. doi: 10.1056/NEJMra0802714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Heist RS, Engelman JA. SnapShot: non-small cell lung cancer. Cancer Cell. 21:448, e2. doi: 10.1016/j.ccr.2012.03.007. [DOI] [PubMed] [Google Scholar]
- 3.Network TCGAR. Diversity of Lung Adenocarcinoma Revealed by Integrative Molecular Profiling. Nature. 2013 Under Review. [Google Scholar]
- 4.Chaft JE, Arcila ME, Paik PK, et al. Coexistence of PIK3CA and other oncogene mutations in lung adenocarcinoma-rationale for comprehensive mutation profiling. Mol Cancer Ther. 2011;11:485–91. doi: 10.1158/1535-7163.MCT-11-0692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Altomare DA, Testa JR. Perturbations of the AKT signaling pathway in human cancer. Oncogene. 2005;24:7455–64. doi: 10.1038/sj.onc.1209085. [DOI] [PubMed] [Google Scholar]
- 6.Dankort D, Filenova E, Collado M, Serrano M, Jones K, McMahon M. A new mouse model to explore the initiation, progression, and therapy of BRAFV600E-induced lung tumors. Genes Dev. 2007;21:379–84. doi: 10.1101/gad.1516407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Trejo CL, Juan J, Vicent S, Sweet-Cordero A, McMahon M. MEK1/2 inhibition elicits regression of autochthonous lung tumors induced by KRASG12D or BRAFV600E. Cancer Res. 2012;72:3048–59. doi: 10.1158/0008-5472.CAN-11-3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ji H, Wang Z, Perera SA, et al. Mutations in BRAF and KRAS converge on activation of the mitogen-activated protein kinase pathway in lung cancer mouse models. Cancer Res. 2007;67:4933–9. doi: 10.1158/0008-5472.CAN-06-4592. [DOI] [PubMed] [Google Scholar]
- 9.Engelman JA, Chen L, Tan X, et al. Effective use of PI3K and MEK inhibitors to treat mutant Kras G12D and PIK3CA H1047R murine lung cancers. Nat Med. 2008;14:1351–6. doi: 10.1038/nm.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Courtois-Cox S, Genther Williams SM, Reczek EE, et al. A negative feedback signaling network underlies oncogene-induced senescence. Cancer Cell. 2006;10:459–72. doi: 10.1016/j.ccr.2006.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kinross KM, Montgomery KG, Kleinschmidt M, et al. An activating Pik3ca mutation coupled with Pten loss is sufficient to initiate ovarian tumorigenesis in mice. J Clin Invest. 2012;122:553–7. doi: 10.1172/JCI59309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Trotman LC, Niki M, Dotan ZA, et al. Pten dose dictates cancer progression in the prostate. PLoS Biol. 2003;1:E59. doi: 10.1371/journal.pbio.0000059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Iwanaga K, Yang Y, Raso MG, et al. Pten inactivation accelerates oncogenic K-ras-initiated tumorigenesis in a mouse model of lung cancer. Cancer Res. 2008;68:1119–27. doi: 10.1158/0008-5472.CAN-07-3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jackson EL, Willis N, Mercer K, et al. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes Dev. 2001;15:3243–8. doi: 10.1101/gad.943001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ullman-Cullere MH, Foltz CJ. Body condition scoring: a rapid and accurate method for assessing health status in mice. Lab Anim Sci. 1999;49:319–23. [PubMed] [Google Scholar]
- 16.Dankort D, Curley DP, Cartlidge RA, et al. Braf(V600E) cooperates with Pten loss to induce metastatic melanoma. Nat Genet. 2009;41:544–52. doi: 10.1038/ng.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ohren JF, Chen H, Pavlovsky A, et al. Structures of human MAP kinase kinase 1 (MEK1) and MEK2 describe novel noncompetitive kinase inhibition. Nat Struct Mol Biol. 2004;11:1192–7. doi: 10.1038/nsmb859. [DOI] [PubMed] [Google Scholar]
- 18.Hirai H, Sootome H, Nakatsuru Y, et al. MK-2206, an allosteric Akt inhibitor, enhances antitumor efficacy by standard chemotherapeutic agents or molecular targeted drugs in vitro and in vivo. Mol Cancer Ther. 2010;9:1956–67. doi: 10.1158/1535-7163.MCT-09-1012. [DOI] [PubMed] [Google Scholar]
- 19.Halilovic E, She QB, Ye Q, et al. PIK3CA mutation uncouples tumor growth and cyclin D1 regulation from MEK/ERK and mutant KRAS signaling. Cancer Res. 70:6804–14. doi: 10.1158/0008-5472.CAN-10-0409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wee S, Jagani Z, Xiang KX, et al. PI3K pathway activation mediates resistance to MEK inhibitors in KRAS mutant cancers. Cancer Res. 2009;69:4286–93. doi: 10.1158/0008-5472.CAN-08-4765. [DOI] [PubMed] [Google Scholar]
- 21.Drahl C. LGX818, Made To Fight Melanoma. Chemical & Engineering News. 2013;91:14. [Google Scholar]
- 22.Koul D, Fu J, Shen R, et al. Antitumor activity of NVP-BKM120--a selective pan class I PI3 kinase inhibitor showed differential forms of cell death based on p53 status of glioma cells. Clin Cancer Res. 2011;18:184–95. doi: 10.1158/1078-0432.CCR-11-1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hofmann I, Weiss A, Elain G, et al. K-RAS mutant pancreatic tumors show higher sensitivity to MEK than to PI3K inhibition in vivo. PLoS One. 2012;7:e44146. doi: 10.1371/journal.pone.0044146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Collisson EA, Trejo CL, Silva JM, et al. A central role for RAF-->MEK-->ERK signaling in the genesis of pancreatic ductal adenocarcinoma. Cancer Discov. 2012;2:685–93. doi: 10.1158/2159-8290.CD-11-0347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kissil JL, Walmsley MJ, Hanlon L, et al. Requirement for Rac1 in a K-ras induced lung cancer in the mouse. Cancer Res. 2007;67:8089–94. doi: 10.1158/0008-5472.CAN-07-2300. [DOI] [PubMed] [Google Scholar]
- 26.Meylan E, Dooley AL, Feldser DM, et al. Requirement for NF-kappaB signalling in a mouse model of lung adenocarcinoma. Nature. 2009;462:104–7. doi: 10.1038/nature08462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nikitin AY, Alcaraz A, Anver MR, et al. Classification of proliferative pulmonary lesions of the mouse: recommendations of the mouse models of human cancers consortium. Cancer Res. 2004;64:2307–16. doi: 10.1158/0008-5472.can-03-3376. [DOI] [PubMed] [Google Scholar]
- 28.Livet J, Weissman TA, Kang H, et al. Transgenic strategies for combinatorial expression of fluorescent proteins in the nervous system. Nature. 2007;450:56–62. doi: 10.1038/nature06293. [DOI] [PubMed] [Google Scholar]
- 29.Sutherland KD, Proost N, Brouns I, Adriaensen D, Song JY, Berns A. Cell of origin of small cell lung cancer: inactivation of Trp53 and Rb1 in distinct cell types of adult mouse lung. Cancer Cell. 2011;19:754–64. doi: 10.1016/j.ccr.2011.04.019. [DOI] [PubMed] [Google Scholar]
- 30.Xu X, Rock JR, Lu Y, et al. Evidence for type II cells as cells of origin of K-Ras-induced distal lung adenocarcinoma. Proc Natl Acad Sci U S A. 2012;109:4910–5. doi: 10.1073/pnas.1112499109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Raynaud FI, Eccles SA, Patel S, et al. Biological properties of potent inhibitors of class I phosphatidylinositide 3-kinases: from PI-103 through PI-540, PI-620 to the oral agent GDC-0941. Mol Cancer Ther. 2009;8:1725–38. doi: 10.1158/1535-7163.MCT-08-1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morris EJ, Jha S, Restaino CR, et al. Discovery of a novel ERK inhibitor with activity in models of acquired resistance to BRAF and MEK inhibitors. Cancer Discov. 2013 doi: 10.1158/2159-8290.CD-13-0070. [DOI] [PubMed] [Google Scholar]
- 33.Deng W, Gopal YN, Scott A, Chen G, Woodman SE, Davies MA. Role and therapeutic potential of PI3K-mTOR signaling in de novo resistance to BRAF inhibition. Pigment Cell Melanoma Res. 2011;25:248–58. doi: 10.1111/j.1755-148X.2011.00950.x. [DOI] [PubMed] [Google Scholar]
- 34.Gopal YN, Deng W, Woodman SE, et al. Basal and treatment-induced activation of AKT mediates resistance to cell death by AZD6244 (ARRY-142886) in Braf-mutant human cutaneous melanoma cells. Cancer Res. 2010;70:8736–47. doi: 10.1158/0008-5472.CAN-10-0902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eser S, Reiff N, Messer M, et al. Selective requirement of PI3K/PDK1 signaling for Kras oncogene-driven pancreatic cell plasticity and cancer. Cancer Cell. 2013;23:406–20. doi: 10.1016/j.ccr.2013.01.023. [DOI] [PubMed] [Google Scholar]
- 36.Snyder EL, Watanabe H, Magendantz M, et al. Nkx2-1 represses a latent gastric differentiation program in lung adenocarcinoma. Mol Cell. 2013;50:185–99. doi: 10.1016/j.molcel.2013.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Winslow MM, Dayton TL, Verhaak RG, et al. Suppression of lung adenocarcinoma progression by Nkx2-1. Nature. 2011;473:101–4. doi: 10.1038/nature09881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–90. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 39.Gupta S, Ramjaun AR, Haiko P, et al. Binding of ras to phosphoinositide 3-kinase p110alpha is required for ras-driven tumorigenesis in mice. Cell. 2007;129:957–68. doi: 10.1016/j.cell.2007.03.051. [DOI] [PubMed] [Google Scholar]
- 40.Mirza AM, Gysin S, Malek N, Nakayama K, Roberts JM, McMahon M. Cooperative regulation of the cell division cycle by the protein kinases RAF and AKT. Mol Cell Biol. 2004;24:10868–81. doi: 10.1128/MCB.24.24.10868-10881.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jonkers J, Meuwissen R, van der Gulden H, Peterse H, van der Valk M, Berns A. Synergistic tumor suppressor activity of BRCA2 and p53 in a conditional mouse model for breast cancer. Nat Genet. 2001;29:418–25. doi: 10.1038/ng747. [DOI] [PubMed] [Google Scholar]
- 42.Fasbender A, Lee JH, Walters RW, Moninger TO, Zabner J, Welsh MJ. Incorporation of adenovirus in calcium phosphate precipitates enhances gene transfer to airway epithelia in vitro and in vivo. J Clin Invest. 1998;102:184–93. doi: 10.1172/JCI2732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Capper D, Preusser M, Habel A, et al. Assessment of BRAF V600E mutation status by immunohistochemistry with a mutation-specific monoclonal antibody. Acta Neuropathol. 2011;122:11–9. doi: 10.1007/s00401-011-0841-z. [DOI] [PubMed] [Google Scholar]
- 44.Morita S, Kojima T, Kitamura T. Plat-E: an efficient and stable system for transient packaging of retroviruses. Gene Ther. 2000;7:1063–6. doi: 10.1038/sj.gt.3301206. [DOI] [PubMed] [Google Scholar]
- 45.Zhu J, Woods D, McMahon M, Bishop JM. Senescence of human fibroblasts induced by oncogenic Raf. Genes Dev. 1998;12:2997–3007. doi: 10.1101/gad.12.19.2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.