Summary
The benzoaxines have been developed from structurally similar chromones as specific inhibitors of the PI3K family to sensitize cancer cells to the effects of chemotherapeutic agents; most have been shown to do this through specific inhibition of DNA-PK and DNA repair mechanisms. In this study we examined the benzoxazine, 2-((3-methoxybut-3en-2-yl)amino)-8methyl-4H-benzo[1,3]oxazin-4one (LTUSI54). This compound had no DNA-PK or PI3K inhibitory activity but still sensitized HeLa cells to the effects of Etoposide. LTUSI54 works synergistically with Etoposide to inhibit growth of HeLa cells and sub G1 analysis indicates that this is not due to an increase in apoptosis. LTUSI54 neither enhances DSB formation due to Etoposide nor does it delay the repair of such damage. Cell cycle analysis shows a clear G2 block with Etoposide alone while, in combination with LTUSI54 there is an additional S phase arrest. Phospho-kinase analysis indicated that LTUSI54 engages key regulators of cell cycle progression, specifically p38α, p53 and ERK 1/2. From our results we hypothesize that LTUSI54 is promoting the cell cycle arrest through activation of p38α pathways, independent of p53 mechanisms. This results in a decrease in p53 phosphorylation and hence, restricted apoptosis. Changes in cell number appear to be the result of p38α pathways disrupting cell cycle progression, at the S and G2 checkpoints. Further investigation into the finer mechanisms by which LTUSI54 effects cell cycle progression would be of great interest in assessing this compound as a chemosensitising agent.
Keywords: Chemosensitisation, Benzoxazines, Etoposide, HeLa, Cancer, Cell cycle arrest
Introduction
Radiation and some chemotherapeutics, such as Etoposide, cause double strand DNA breaks (DSB). Etoposide is a topoisomserase II (topo II) inhibitor. Topo II is an enzyme that has multiple functions including relaxation of supercoiled DNA, cell cycle regulation and decatenation of sister chromatids [1, 2]. During the catalytic cycle of topo II, to relax the DNA helix, topo II creates a transient covalent complex (“cleavable complex”) in the DNA by cleaving the phosphodiester backbone, essentially creating a DSB for another DNA duplex to pass through before the DSB is religated [1–3]. In the presence of topo II inhibitors such as Etoposide, the normally transient cleavable complex is stabilised resulting in permanent DSBs [1–3] which are identified by the cell cycle checkpoints. In eukaryotic cells there are three major checkpoints where the cell cycle can be blocked due to DNA damage or incomplete replication; they are necessary for the maintenance of genome integrity and they occur at the transition between G1-S, within S and at the G2-M boundary during the cell cycle. Additionally there is a checkpoint at the exit of mitosis [4, 5]. Activation of cell cycle checkpoints is initiated by the protein kinase signalling molecules ATM and ATR [4, 6].
Until recently it was thought that checkpoint pathways only regulated cell cycle transition, however, it is now recognised that signal transduction cascades link DNA damage signalling, checkpoints and repair [6, 7]. Once the detection of damage occurs, the cell cycle is arrested before one of three possible outcomes is determined; damage can either be repaired in which case the cell continues to cycle, the cell can enter senescence and progresses no further or ideally the cell will be unable to repair the damage and cell death will result by apoptosis [8]. Etoposide has been shown to cause DSBs, ATM activation and apoptosis in all phases of the cell cycle [3].
Increasing the sensitivity of cancer cells to therapy has been the focus of a number of studies [9–14]. One mechanism that has been of interest is inhibiting the DNA repair pathways of cancer cells subsequent to cell cycle arrest and many inhibitors of these pathways have been developed, some of which are currently undergoing clinical trials [11–15]. The chromones, LY294002 and wortmannin, were found to a promising family of chemo- and radio-sensitisers by inhibition of DNA repair through inhibition of the PI3K family, including DNA-PK [16–18]. However, the underlying mechanisms of action have seldom been specifically elucidated and these inhibitors have consistently been shown to have broad specificity for other members of the PI3K family as well as DNA-PK. Hence it is possible that interactions of these inhibitors with enzymes other than DNA-PK are a significant contributor, or even an essential part, of their sensitizing ability. We have developed a series of benzoxazine analogues of the chromones with a view to increasing their specificity for DNA-PK, while maintaining potency. The compound used in this present study illustrates the above point in that it has minimal activity against DNA-PK or PI3K isoforms and yet, it is a potent sensitiser to Etoposide.
In this study we show that treatment of HeLa, human cervical cancer, cells with the topo II inhibitor Etoposide can be enhanced by the presence of the benzoxazine, 2-((3-methoxybut-3en-2-yl)amino)-8methyl-4H-benzo[1,3]oxazin-4one (LTUSI54) (Fig. 1). We show that the increased sensitivity to Etoposide is independent of DNA repair inhibition or apoptosis but involves cell cycle arrest.
Fig. 1.
Chemical structure of 2-((3-methoxybut-3en-2-yl)amino)-8methyl-4H-benzo[1,3]oxazin-4one (LTUSI54)
Materials and methods
Cell lines
HeLa, cervical cancer, cell line used were obtained from the American Type Culture Collection and was cultured in Dulbecco’s Modified Eagle Medium (DMEM, Invitrogen, Grand Island) containing phenol red, supplemented with 10 % foetal bovine serum (FBS, Sigma-Aldrich, St Louis).
Preparation of stock solutions
All stock solutions were diluted using DMSO (Sigma-Aldrich, St Louis); LTUSI54 was made up to 1 mM (kindly supplied by Dr. Jasim Al-Rawi and Saleh Ihmaid, La Trobe University Bendigo, Australia). Etoposide (Sigma-Aldrich, St Louis) was made up to stocks of 10 mM.
It should be noted that in accordance with Lee et al. [19] the amount of DMSO added to both test and control media never exceeded 0.5 % of the total volume, and control cultures were treated with equivalent volumes of DMSO that was used to prepare stock solutions of the drug to eliminate adverse effects of DMSO.
Inhibition assay
The PI3K and DNA-PK inhibition assay was performed by Reaction Biology Corporation USA. IC50 values greater than 100 μM were said to be not active against the corresponding enzyme.
Toxicity of LTUSI54
Sulphorhodamine blue assay
The method put forward by Freshney [20] was used to carry out the assay with slight variations. Cells were seeded on 24 well plates at a concentration of 2x103cells/well. After cells had adhered overnight, 1 ml of treatment media was added for 4 h. Treatment media was removed and cells were washed with 0.01 M PBS before 1 ml of recovery media was added to each well for 48 h. For control cells and Etoposide alone treated cells recovery media contained media with DMSO only, recovery media for LTUSI54 alone and Etoposide plus LTUSI54 treated cells contained 200nM LTUSI54. Absorbance was measured at 538 nm using a Flex Station 3 (Molecular Devices, California).
CellTox™ green cytotoxicity assay
Promega’s CellTox™ cytotoxicity assay was used to investigate the cytotoxic effect of LTUSI54 on HeLa cells. Cells were seeded in a 96 well plate at a concentration of 1x104cells/well and allowed to adhere overnight. Fifty microlitres of test media was added to each well containing 0.1 % CellTox green dye, and incubated for 24 h before plate was read using an excitation wavelength of 485 nm and emission filter of 520 nm on the Flex Station 3 (Molecular Devices, California).
Flow cytometry for γ-H2AX expression
This assay is essentially as described in MacPhail et al. 2003 [21], cells were seeded at a concentration of 2 × 105 cells per well in a 6 well plate (BD Biosciences, San Jose) and allowed to adhere overnight. Treatment was with either; 0.5 % DMSO in DMEM, 25 μM Etoposide alone, 200nM LTUSI54 or the combination of Etoposide and LTUSI54. Cells were treated for either 4 h before fixation or alternatively for 4 h in initial treatment followed by 2 days recovery in recovery media containing LTUSI54, as outlined in the SRB method.
After treatment, cells were harvested, fixed in 70 % ethanol at 20 °C overnight before rehydration and staining. Cells were stained with 100 μL anti H2AX (pSer139) rabbit polyclonal antibody (Santa Cruz Biotechnology, Santa Cruz, Cat# sc-101696) diluted 1:500 for 2 h. Following washing, cells were resuspended in 100 μL goat anti-rabbit IgG (H + L) Alexa Fluor 488 conjugate (Invitrogen, Grand Island, Cat# A-11008) diluted 1:200. Finally cells were resuspended in a 5 μg/ml propidium iodide and analysed on Accuri C6 Flow cytometer (BD Biosciences, San Jose) and gated to omit doublets.
Cell cycle analysis
Cell cycle and sub G1 analysis was achieved as outlined by Riccardi and Nicoletti [22]. After treatment, test media was collected in a centrifuge tube, washed with 0.01 M PBS, trypsinized and centrifuged at 200 g for 5 min. Cells were then washed with 0.01 M PBS before being resuspended in 70 % ethanol and stored at −20 °C generally overnight.
Fixed cells were centrifuged at 200 g for 5 min, washed with 0.01 M PBS and resuspended in 250 μL stain solution containing 25 μg/ml Propidium iodide and 100 μg/ml RNase. Cells were then incubated at 37 °C in the dark for 30 min before flow cytometry analysis on the Accuri C6 Flow cytometer (BD Biosciences, San Jose) and gated to omit doublets and debris.
Human phospho-kinase array
Small flasks of HeLa cells were treated for 4 h with either media alone, 25 μM Etoposide, 200nM LTUSI54 or a combination of Etoposide and LTUSI54. Following treatment, the R&D systems Proteome Profiler™ Human Phospho-Kinase array (R&D Systems, Minneapolis, Cat# ARY003) was used to analyse protein phosphorylation in each treatment group.
The procedure was carried out as per the manufacturer’s instructions, cells were lysed using lysis buffer provided and membranes were incubated overnight in cell lysate at 4 °C to allow for protein and antibody binding. Protein concentration of lysate was assayed by the BioRad DC Protein Assay Kit II (Cat #500-0112, Bio-Rad Laboratories, Philadelphia) and used at 150 μg/ml. Membranes were washed and exposed to the appropriate detection antibody cocktail for 2 h at room temperature. Again membranes were washed and then exposed to Streptavidin-HRP for 30 min. After a final wash membranes were exposed to the prepared Chemi reagent mix for 1 min before being read at increasing exposures using a Bio-Rad Chemi Doc station.
Statistics and analysis
Data was graphed using Graph Pad prism 5.0 and presented as mean ± SEM. Statistical significance was determined using two-tailed unpaired Students t-test, p values less than 0.05 were taken as statistically significant.
Results
Inhibition assays
LTUSI54 was found to have no inhibitory effect on any of the PI3K isoforms or DNA-PK. IC50 values greater than 100 μM were taken as not active (data not shown).
Toxicity of LTUSI54
The SRB assay was used to show changes in cell number in the presence of LTUSI54 (Fig. 2a). At low concentrations there was no significant effect of LTUSI54 on HeLa cell number. It was not until HeLa cells were exposed to 100 μM of LTUSI54 that there was an observed significant increase in cell number when compared to the control (p = 0.0003). Similarly, the Promega CellTox™ green dye assay results (Fig. 2b) indicated that LTUSI54 had no cytotoxic effect on HeLa cells at 200nM, however, as the concentration increased there was an observed increase in fluoresence indicating an increase in cytotoxicity. The greatest amount of CellTox fluoresecence was seen at 100 μM LTUSI54, 299.3 RFUs, a significant increase from the control group, 191 RFUs, generating a p value <0.0001.
Fig. 2.
Results of HeLa cells treated with varying concentrations of LTUSI54. a Results of SRB assay after 2 days treatment, graph indicates mean of 4 replicates ± SEM of four replicates. b Result obtained using the cell toxicity assay after 24 h treatment, graph indicates mean of eight replicates ± SEM. Statistical significance was determined using two-tailed t-tests, (asterisk) indicates results statistically significant to the control
Growth inhibition
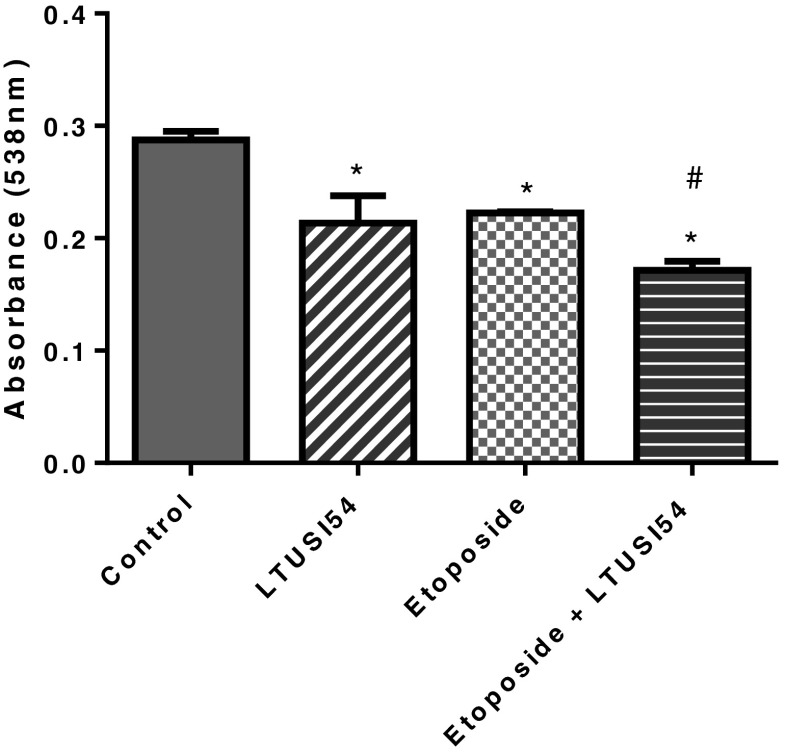
When HeLa cells were cultured in the presence of Etoposide alone there was a significant reduction in cell number compared to control (p < 0.0001). Similarly, treatment with LTUSI54 alone resulted in a significant reduction in cell numbers (p = 0.0115). The combination treatment, Etoposide and LTUSI54, further reduced HeLa cell number (p < 0.0001), enhancing the effect of Etoposide (Fig. 3).
Fig. 3.
SRB results of HeLa cells treated with 25 μM Etoposide both in the presence and absence of 200nM LTUSI54 for 4 h and allowed to recover for 2 days in the presence of LTUSI54. Graph indicates mean ± SEM of four replicates. Statistical significance was determined using two-tailed t-tests, (asterisk) indicates treatments significantly different to the control treatment group and (number sign) indicates treatments significantly different to Etoposide alone treatment group
Effect on apoptosis
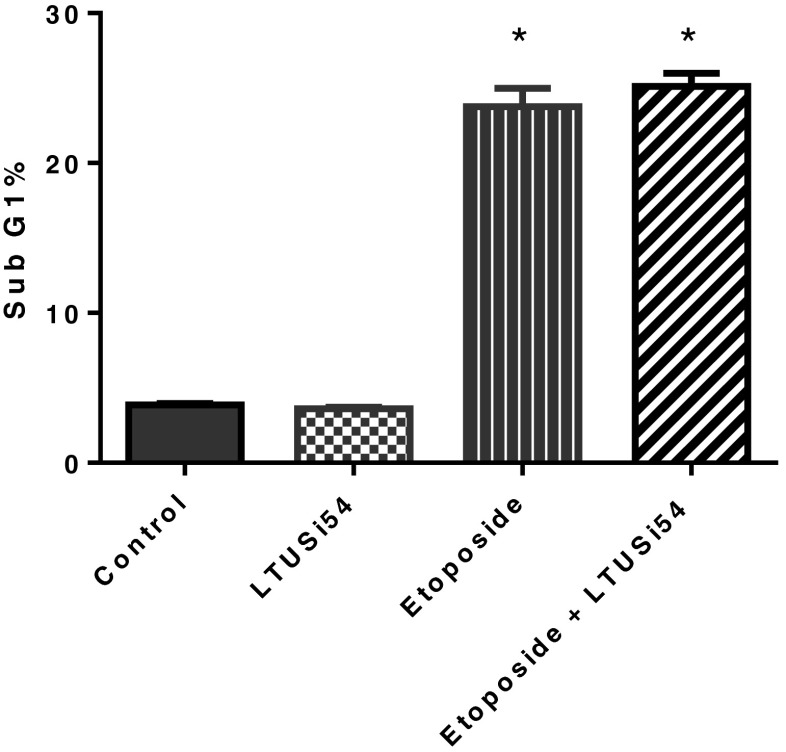
HeLa cells were culture for 4 h in the various combinations of Etoposide and LTUSI54, and then allowed to recover for 2 days. The effect on cellular apoptosis was determined by analysing cells in sub G1 phase (Fig. 4). After 2 days recovery time, Etoposide treatment caused a significant increase in apoptotic cells, p < 0.0001. However, the combined treatment with LTUSI54 neither enhanced nor inhibited this effect. Results showed that after 4 h treatment there was no significant change in sub G1 populations regardless of the treatment group (data not shown).
Fig. 4.
Sub G1 analysis of HeLa cells treated with 25 μM Etoposide both in the presence and absence of 200nM LTUSI54 for 4 h and allowed to recover for 2 days in LTUSI54 recovery media. Graph indicates mean ± SEM of three replicates. Statistical significance was determined using two-tailed t-tests, (asterisk) indicates treatments significantly different to control group. (Flow cytometry profiles shown in Fig. 6b)
Flow cytometry for γ-H2AX
γ-H2AX formation was assessed by flow cytometry (Fig. 5). After 4 h treatment, LTUSI54 alone had no effect on the number of HeLa cells that stained positive for γ-H2AX (7.3 % of cells were positive), and thus DSBs. As expected, treatment with Etoposide increased γ-H2AX expression (about 80 % of cells positive), and thus caused DSBs. Combination treatment with Etoposide and LTUSI54 resulted in no further effect on the number of HeLa cells that stained positive for γ-H2AX (about 80 %) when compared to Etoposide alone treated cells.
Fig. 5.
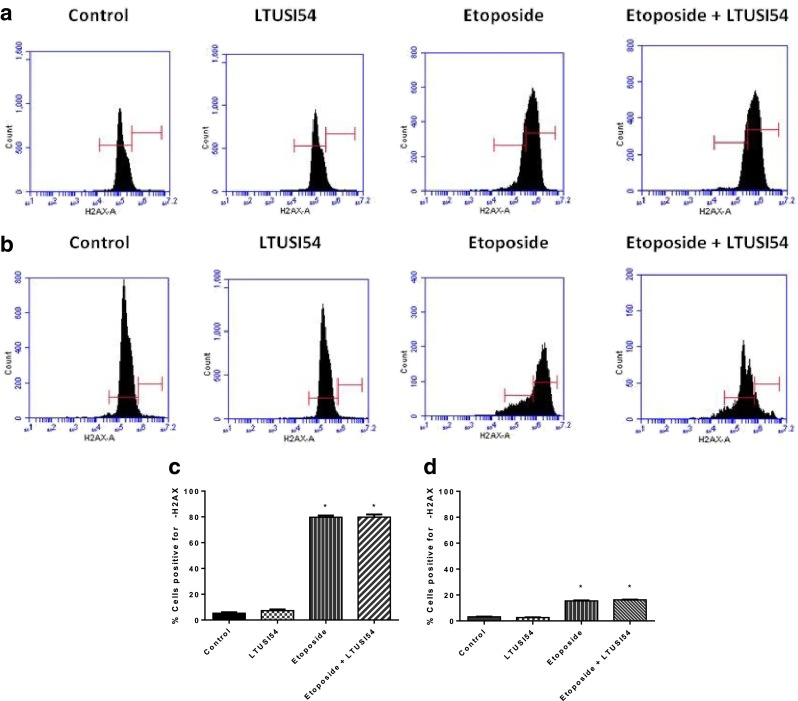
γ-H2AX results obtained for HeLa cells treated with Etoposide in presence and absence of 200nM LTUSI54. a Flow cytometry results following 4 h treatment alone with Etoposide, 200nM LTUSI54 alone and in combination or (b) 4 h treatment followed by 2 days in recovery media. Left hand gate indicated cells that are considered to have been negative for γ-H2AX antibody, while right hand gate indicates cells that were considered to have stained positive for γ-H2AX antibody. c Percent of cells stained positive for γ-H2AX following 4 h treatment alone and (d) 4 h treatment followed by 2 days recovery. Graphs indicate mean of three replicates ± SEM. Statistical significance was determined using two-tailed T tests. (Asterisk) Indicates treatments significantly different to control
When HeLa cells were allowed to recover for 2 days, a reduction in γ-H2AX indicated that cells were able to repair the DSBs. When HeLa cells were allowed to recover after treatment with Etoposide there was a decrease in γ-H2AX expression (only 15 % of cells were positive), suggesting the majority of samples had DSBs repaired. This repair occurred even in the presence of LTUSI54 (16 % of cells were positive) suggesting this compound has no effect on the repair pathways. Treatment of HeLa cells with LTUSI54 alone had no effect on cells that stained positive for γ-H2AX (2.7 % of cells were positive). The more than 5 fold decrease in γ-H2AX positive cells, and thus DSBs repair suggests that Etoposide is causing DSB, but when cells are allowed to recover, the DNA repair pathways are not inhibited by the presence of LTUSI54.
Cell cycle effects
Cell cycle analysis was used to investigate the effects of treatment on cell cycle progression, results are shown in Fig. 6a. Exposure to LTUSI54 alone for 4 h caused a significant increase in cells in S phase (p = 0.0074) and a corresponding significant reduction of cells in G2/M phase (p = 0.0036). Etoposide alone treatment also caused an increase in cells in S phase (p = 0.0059) which was further enhanced by the presence of LTUSI54 (p = 0.0006) when compared to Etoposide alone treatment. This significant S phase block evident in the combination treatment also resulted in a reduction of cells in the G2/M phase when compared to Etoposide alone treatment (p = 0.0265).
Fig. 6.
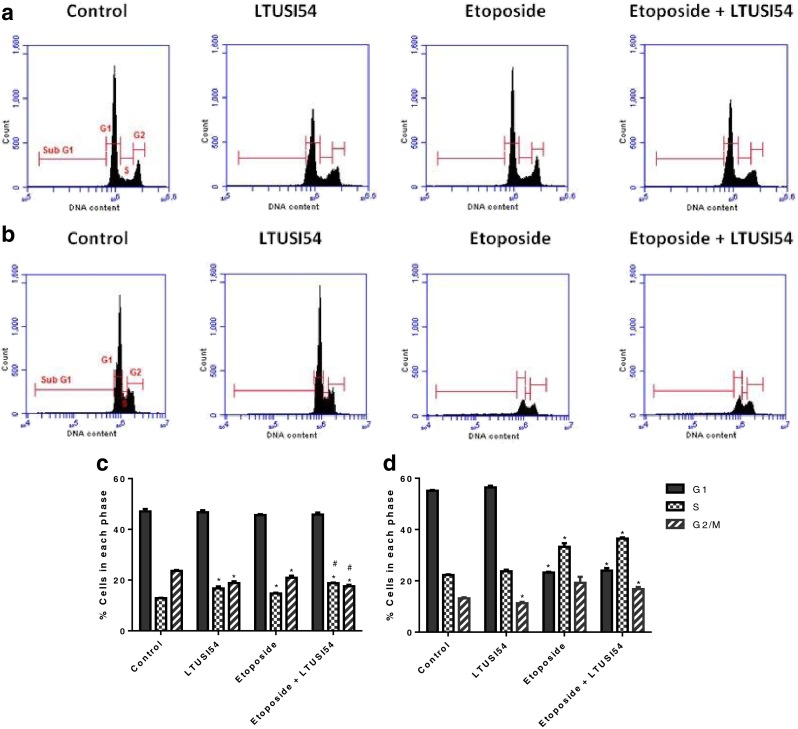
Cell cycle profiles from flow cytometry analysis of HeLa cells treated with Etoposide in the presence and absence of LTUSI54 for (a) 4 h alone and (b) when allowed to recover for 2 days. Percent of cells in each phase of the cell cycle following (c) 4 h treatment alone and (d) following 2 days recovery time in recovery media containing LTUSI54. Graphs indicate mean ± SEM of three independent experiments. (Asterisk) Indicates treatments significantly different to control, (number sign) indicates combination treatments significantly different to Etoposide alone treatment
When cells were allowed to recover for 2 days (Fig. 6b), a significant reduction in G1 cells was seen with Etoposide and combination treated cells when compared to control and LTUSI54 alone treated cells (p < 0.0001). There was also a significant increase in cells in both the S phase for Etoposide alone (p = 0.0016) and the combination treatment (p < 0.0001) when compared to the control group. A reduction of cells in G2/M phase was seen when cells were exposed to the LTUSI54 alone (p = 0.0417), conversely combination treatment resulted in an increase of cells in this phase (p = 0.0160). However, there was no significant difference in the number of cells in the G2/M phase between the Etoposide alone treated cells and the combination treated cells.
Human phospho-kinase array
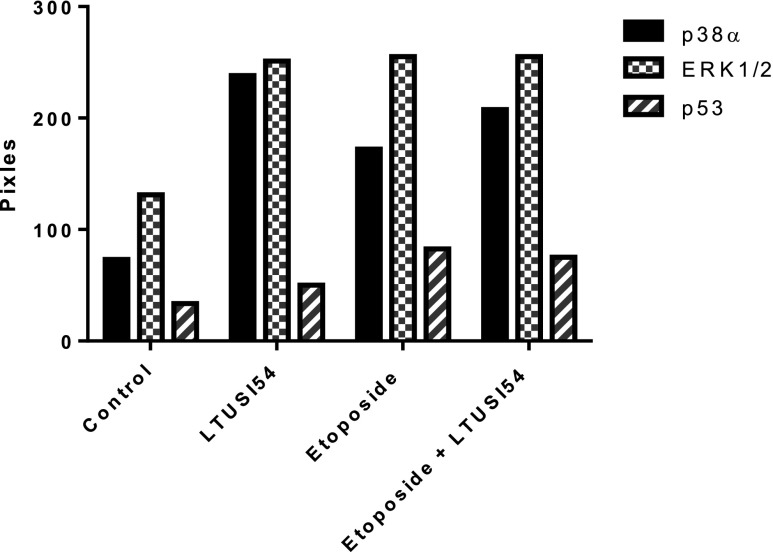
To determine which proteins are involved in the signalling pathway, phosphorylation of key protein kinases was analysed after 4 h treatment using the R&D systems proteome profiler human phospho-kinase array (Fig. 7). Analysis revealed that cells treated with LTUSI54 alone had a 3.645 fold increase in the activity of p38α when compared to control levels, this effect was more significant than Etoposide alone, which only had a 2.64 fold increase in p38α activity, compared to control levels. Combination treatment resulted in a 3 fold increase in the activity of p38α when compared with control. All three treatment groups showed about a 2 fold increase in activity of Extracellular signal-regulated protein kinases 1 and 2 (ERK1/2) when compared to the control. The activation of p53, a key checkpoint protein, was most significantly increased following treatment with Etoposide (3.5 fold increase). This increase was somewhat inhibited in the combination treatment with LTUSI54 showing only 2.35 fold increase when compared to control. LTUSI54 alone also increased the activity of p53 1.5 fold when compared to the control group, much less than Etoposide alone treated cells.
Fig. 7.
Example results of Human phospho-kinase of HeLa cells following 4 h treatment alone with 25 μM Etoposide, 200nM LTUSI54 or in combination. Experiment was repeated with both runs containing duplicate antibody spots, a similar trend as shown was observed
Discussion
The benzoaxines have been developed from structurally similar chromones as specific inhibitors of the PI3K family to sensitize cancer cells to the effects of chemotherapeutic agents; most have been shown to do this through specific inhibition of DNA-PK and DNA repair mechanisms [23, 24]. Structure activity relationship studies (SAR) have identified key substitutions leading to enhancement of this DNA-PK inhibition, specifically at position 8 [25]. In this study we examined a benzoxazine with an alanine substituted at position 2. This compound, LTUSI54, had no DNA-PK or PI3K inhibitory activity but still sensitized HeLa cells to the effects of Etoposide.
The chemotherapeutic effect of Etoposide is characterized by a reduction in cell number as a result of initiating apoptosis and mitotic catastrophy following an induction of G2 cell cycle arrest [26]. Consistent with previous studies [26], treatment of HeLa cells cells with Etoposide resulted in a significant reduction in cell number and an increase in apoptosis. In the presence of LTUSI54, cell numbers were further reduced following treatment with Etoposide although this was not found to be due to an increase in apoptosis. However, we did find that LTUSI54 enhanced the Etoposide induced G2 block.
Consistent with the observation that LTUSI54 had no effect on the amount of apoptosis induced by Etoposide we also found no change in the number of DSBs induced by Etoposide. The phosphorylation of histone H2AX (to γ-H2AX) following treatment was explored, as an initial signalling event for DNA damage [3, 21, 27–29]. It was evident that HeLa cells treated with Etoposide resulted in DSBs within the 4 h treatment, consistent with previous studies [3]. When HeLa cells were allowed 2 days to recover from this damage, γ-H2AX expression levels had decreased indicating that DNA repair had taken place. The presence of LTUSI54 during this recovery time had no effect on the repair of this damage. The inability of LTUSI54 to inhibit or prolong the repair of DSBs caused by Etoposide again reinforces the fact that LTUSI54 has no effect on the activity of DNA-PK or any other elements involved in DSB damage repair pathways.
The function of many chemotherapy treatments is to cause sufficient DNA damage that the cell is unable to repair the damage and goes into apoptosis. In order for this to occur, mechanisms within the cell must first detect the DNA damage, institute cell cycle arrest and initiate DNA repair pathways. Aside from inhibiting DNA repair another mechanism to enhance the cytotoxic effect of chemotherapy agents is through disrupting cell cycle progression [5, 30–34]. As the cell moves through the cell cycle, there are several cell cycle checkpoints that monitor DNA integrity and inhibit cell cycle progression if damage is detected. These check points have been identified between each phase: between G1 and S phase, S phase and the G2 phase and before cells enter the M phase [4, 5]. LTUSI54 alone was found to cause cell cycle arrest at the S checkpoint following initial treatment. In combination, the arrest of cells in the S phase of the cell cycle induced by Etoposide was found to be enhanced by the presence of LTUSI54, which resulted in a corresponding decrease of cells in the G2/M phase. Following 2 days recovery, LTUSI54 alone treated cells showed a similar cell cycle profile to that of the control group, with the exception of a decrease of cell in the G2/M phase. Similarly, combination treatment showed no significant difference to Etoposide alone treated cells. This return to normal and no further enhancement of Etoposide treatment may be due to the stability of LTUSI54 in the cellular environment. LTUSI54 may be broken down over this period of time eliminating any further effect on the cell cycle.
These results suggest that the enhanced reduction in cell numbers seen with the treatment of Etoposide and LTUSI54, which was found to be independent of an increase in apoptosis and prolonged DSB repair, is due to early disruption of cell progression through the cell cycle. In recent years cell cycle progression has become a major target for improving anticancer therapy. The majority of these studies looked at eliminating the cell cycle checkpoints [5, 30–32] while others have found benefit in inducing and enhancing cell cycle arrest [33, 34] and many current chemotherapeutic agents elicit their therapeutic benefit by halting the cell cycle.
To determine the pathway, and possible target molecules for LTUSI54, the effects on cellular kinases was assessed in both treated and untreated cells. The kinase array results indicated a change in phosphorylation levels of three key kinases, p53, p38α and ERK1/2. p53 is a well-known tumour suppressor, the protein levels and functional activity of p53 are commonly up-regulated during chemotherapy treatment of cancers [35]. Treatment with Etoposide resulted in increased phosphorylation of p53, and increased levels of apoptosis, suggesting Etoposide is stimulating apoptosis through a p53 dependant mechanisms as has been demonstrated before [3]. In response to DNA damage, activated ATM family kinases phosphorylate p53 at Ser 15. Phosphorylation at this site disrupts the interaction of p53 with HDM2 (the human homolog of the murine double minute 2 oncogene), leading to the stabilization of p53 protein [36, 37]. p53 is normally rapidly degraded by HDM2-mediated ubiquitin-dependent proteolysis [38–40]. p53 has two functional roles, it can induce cell cycle arrest, through the transactivation of the Cdk inhibitor p21, or apoptotic cell death through transcription-dependent and -independent mechanisms [41]. p53 mediated cell cycle arrest at the transition of cells from G2-M phase involves upregulation of cell cycle inhibitors such as the CDK inhibitor p21, GADD45a (growth arrest and DNA-damage-inducible 45 alpha) as well as 14-3-3 sigma proteins [42]. These factors all play a key role for cells to progress through the G2/M checkpoint of the cell cycle by inhibiting the activation of Cdc2/Cyclin B complex by Cdc25that is needed for entry into mitosis, hence resulting in a G2 arrest [40, 43].
p53 mediated apoptosis occurs through the intrinsic pathway, which is regulated by the pro- and anti-apoptotic Bcl-2 family of proteins that directly affect mitochondrial outer-membrane permeabilization (MOMP) [38, 39]. The two main Bcl-2 family members that influence p53 mediated MOMP are Bax and Bak. Oligomerization of these proteins occurs by the direct or indirect activation of Bcl-2-homology domain-3 (BH3)-only pro-apoptotic Bcl-2 family members [38, 39, 44]. Bax and Bak p53-induced activation triggers the release of mitochondrial cytochrome c and APAF which activate the caspase cascade of apoptosis leading to cell death [38, 39].
Combination treatment of Etoposide with LTUSI54, showed an inhibitory effect on p53 phosphorylation when compared to Etoposide alone treated cells. However, LTUSI54 alone only showed a 1.5 fold increased in p53 phosphorylation much less than Etoposide, with no corresponding increase in apoptosis, suggesting that LTUSI54 has a positive effect on the p53 dependent role in cell cycle arrest rather than apoptosis.
In response to DSBs, such as those caused by Etoposide, activation of p38α occurs and leads to the establishment of a G2/M cell cycle arrest [45]. p38α is a member of the mitogen activated protein kinases (MAPK) [45], it is a stress activated protein kinase that inhibits cell cycle progression at both the G1/S and the G2/M checkpoints [46]. Treatment of HeLa cells with LTUSI54 alone showed a much greater increase in p38α phosphorylation than Etoposide alone treatment when compared to the control group (3.645 and 2.64 fold increase respectively). Treatment with Etoposide alone resulted in phosphorylation of p38α and cell cycle arrest at the G2/M checkpoint, consistent with the role of p38α in G2/M cell cycle arrest [40, 47]. DNA damage is detected by ATM and ATR kinases, ATM indirectly phosphorylate p38α via activation of the Thousand and one (Tao) kinases [45]. Once activated p38α can induce cell cycle arrest via 2 pathways, one involving p53 as previously described, the other through activation of MAPK activated kinase 2 (MK2) [40, 43]. MK2 in turn phosphorylates Cdc25B and Cdc25C, which induces their binding to 14-3-3 proteins, resulting in the prevention of Cdc25 activating the Cdc2/Cyclin B complex that is needed for entry into mitosis [40, 43]. Similarly, MK2 activation has previously been found to initiate the S phase checkpoint via phosphorylation-dependent depletion of Cdc25A [40], which could explain to the observed increase in cells in the S phase following LTUSI54 treatment.
HeLa cells treated with LTUSI54 alone, resulted in a significantly higher p38α phosphorylation but lower p53 activation, compared to Etoposide alone treatment. This can be attributed to the actions of p38α and MK2 in destabilizing p53. Initially, activation of both p38α and MK2 stabilises and activates p53, however activation of MK2 also leads to the phosphorylation of HDM2 [48]. Activation of HDM2 leads to a reduction in p53 stability and may play a role in moderating the extent and duration of a stress induced p53 response. Previous studies have shown that phosphorylation of HDM2 at Ser 157 and 166 reduces the stabilisation of p53 and promotes its degradation [48].
Consistent with previous studies treatment of HeLa cells with Etoposide resulted in increased ERK1/2 expression. ERK1/2 has been found to play a role in the progression through the cell cycle, mainly the transition of cells from G1 phase into S phase [49]. ERK1/2 is phosphorylated downstream of MEKs in the RAF-MEK-ERK1/2 pathway, it promotes G1 progression by interactions with D-type cyclins, Cdk4, stabilisation of c-Myc, regulation of p21 and p27 as well as down regulating anti-proliferative genes such as Tob1, Ddit3 and JunD [49–51]. All three treatment groups up regulated ERK1/2 indicating that they all activate the RAF-MEK-ERK pathway, as the main role for ERK phosphorylation is understood to be the progression of cells through G1 into S phase, it is no surprise that the observed results showed no arrest of cells in the G1 phase.
From our results we hypothesize that LTUSI54 is promoting the MK2 pathway of cell cycle arrest over that of p53 dependent mechanisms (summarised in Fig. 8). However, as the phosphorylation of p53 increased in the presence of LTUSI54 it appears that the p53 dependent pathway of cell cycle arrest is still activated. As the MK2 pathway is up regulated, phosphorylation of HDM2 occurs at Ser 157 and 166, promoting degradation of p53 [48]. Such degradation of p53 would account for the reduction in p53 expression levels observed when LTUSI54 was present in combination with Etoposide when compared to the Etoposide alone treated HeLa cells. Similarly, p53 mediated apoptosis would be effected by the presence of LTUSI54 and its corresponding HDM2 phosphorylation. This would account for the lack of increased apoptosis observed when LTUSI54 was present in combination with Etoposide. LTUSI54 could possibly be promoting the MK2 dependent pathway of cell cycle arrest and hence p53 degradation by activating proteins downstream of p38α. Possible targets for LTUSI54 include p38α itself, MK2 or even HDM2, possibly at the Ser 157 and 166 sites that promote the degradation of p53 and reduce p53 dependent apoptotic pathways.
Fig. 8.
Proposed mechanism of action for Etoposide and LTUSI54. LTUSI54 appears to promote the MK2 dependent (p53 independent) pathway of cell cycle arrest over p53 dependent mechanisms. As MK2 is phosphorylated, it phosphorylates HDM2, which destabilises p53, inhibiting the p53 mediated apoptosis pathways
LTUSI54 is a novel compound which has been found to enhance the cytotoxic effects of Etoposide on the HeLa cervical cancer cell line. This response is independent of increased apoptosis and appears to be the result of disrupted cell cycle progression, at the S and G2 checkpoints. Further investigation into the finer mechanisms by which LTUSI54 up regulates the p53 independent cell cycle arrest, including its effects on MK2 and HDM2, would be of great interest in assessing this compound as a chemosensitising agent.
References
- 1.Willmore E, Frank AJ, Padget K, Tilby MJ, Austin CA. Etoposide targets topoisomerase IIa and IIb in leukemic cells: isoform-specific cleavable complexes visualized and quantified in situ by a novel immunofluorescence technique. Mol Pharmacol. 1998;53:78–85. doi: 10.1124/mol.54.1.78. [DOI] [PubMed] [Google Scholar]
- 2.Wilmore E, Sd C, Sunter NJ, Tilby MJ, Jackson GH, Austin CA, Durkacz BW. A novel DNA-dependent protein kinase inhibitoe, NU7026, potentiates the cytotoxicity of topoisomerase II poisons used in the treatment of leukemia. Blood. 2004;103(12):4659–4665. doi: 10.1182/blood-2003-07-2527. [DOI] [PubMed] [Google Scholar]
- 3.Tanaka THH, Traganos F, Seiter K, Darzynkiewicz Z. Induction of ATM activation, histone H2AX phosphorylation and apoptosis by etoposide. Relation to cell cycle phase. Cell Cycle. 2007;6(3):6. doi: 10.4161/cc.6.3.3835. [DOI] [PubMed] [Google Scholar]
- 4.Karlsson-Rosenthal C, Millar JBA. Cdc25: mechanisms of checkpoint inhibition and recovery. Trends Cell Biol. 2006;16(6):285–292. doi: 10.1016/j.tcb.2006.04.002. [DOI] [PubMed] [Google Scholar]
- 5.Gabrielli B, Brooks K, Pavey S (2012) Defective cell cycle checkpoints as targets for anti-cancer therapies. Front Pharmacol 3 [DOI] [PMC free article] [PubMed]
- 6.Alexander BM, Pinnell N, Wen PY, D’Anrea A. Targeting DNA repair and the cell cycle in gliolastoma. J Neuron Oncolocy. 2012;107:463–477. doi: 10.1007/s11060-011-0765-4. [DOI] [PubMed] [Google Scholar]
- 7.Kawabe T. G2 checkpoint abrogators as anticancer drugs. Mol Cancer Ther. 2004;3(4):513–519. [PubMed] [Google Scholar]
- 8.Kung G, Konstantinidis K, Kitsis RN. Programed necrosis, not apoptosis, in the heart. Circ Res. 2011;108:1017–1036. doi: 10.1161/CIRCRESAHA.110.225730. [DOI] [PubMed] [Google Scholar]
- 9.Munck JM, Batey MA, Zhao Y, Jenkins H, Richardson CJ, Cano C, Tavecchio M, Barbeau J, Bardos J, Cornell L, Griffin RJ, Menear K, Slade A, Thommes P, Martin NM, Newell DR, Smith GC, Curtin NJ. Chemosensitization of cancer cells by KU-0060648, a dual inhibitor of DNA-PK and PI-3K. Mol Cancer Ther. 2012;11(8):1789–1798. doi: 10.1158/1535-7163.MCT-11-0535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Griffin RJ, Fontana G, Golding BT, Guiard S, Hardcastle IR, Leahy JJJ, Martin N, Richardson C, Rigoreau L, Stockley M, Smith GCM. Selective benzopyranone and pyrimido[2,1-a]isoquinolin-4-one inhibitors of DNA-dependent protein kinase: synthesis, structure-activity studies, and radiosensitization of a human tumor cell line in vitro. J Med Chem. 2005;48:569–585. doi: 10.1021/jm049526a. [DOI] [PubMed] [Google Scholar]
- 11.Ding J, Miao Z-H, Meng L-H, Geng M-Y. Emerging cancer therapeutic opportunities target DNA-repair systems. Trends Pharmacol Sci. 2006;27(6):338–344. doi: 10.1016/j.tips.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 12.Martin SA, Lord CJ, Ashworth A. DNA repair deficiency as a therapeutic target in cancer. Curr Opin Genet Dev. 2008;18(1):80–86. doi: 10.1016/j.gde.2008.01.016. [DOI] [PubMed] [Google Scholar]
- 13.Sánchez-Pérez I. DNA repair inhibitors in cancer treatment. Clin Transl Oncol. 2006;8(9):642–646. doi: 10.1007/s12094-006-0034-8. [DOI] [PubMed] [Google Scholar]
- 14.Veuger SJ, Curtin NJ, Richardson CJ, Smith GC, Durkacz BW. Radiosensitization and DNA repair inhibition by the combined use of novel inhibitors of DNA-dependent protein kinase and poly (ADP-ribose) polymerase-1. Can Res. 2003;63(18):6008–6015. [PubMed] [Google Scholar]
- 15.Hosoya N, Miyagawa K. Clinical importance of DNA repair inhibitors in cancer therapy. Memo. 2009;2(1):9–14. doi: 10.1007/s12254-008-0081-7. [DOI] [Google Scholar]
- 16.Workman P, Clarke PA, Raynaud FI, van Montfort RL. Drugging the PI3 kinome: from chemical tools to drugs in the clinic. Can Res. 2010;70(6):2146–2157. doi: 10.1158/0008-5472.CAN-09-4355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sarbassov DD, Guertin DA, Ali SM, Sabatini DM. Phosphorylation and regulation of Akt/PKB by the Rictor-mTOR complex. Science. 2005;307(5712):1098–1101. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- 18.Chiosis G, Rosen N, Sepp-Lorenzino L. LY294002-geldanamycin heterodimers as selective inhibitors of the PI3K and PI3K-related family. Bioorg Med Chem Lett. 2001;11(7):909–913. doi: 10.1016/S0960-894X(01)00099-3. [DOI] [PubMed] [Google Scholar]
- 19.Lee CM, Fuhrman CB, Planelles V, Peltier MR, Gaffney DK, Soisson AP, Dodson MK, Tolley HD, Green CL, Zempolich KA. Phosphatidylinositol 3-kinase inhibition by LY294002 radiosensitizes human cancer cell lines. Clin Cancer Res. 2006;12(1):250–256. doi: 10.1158/1078-0432.CCR-05-1084. [DOI] [PubMed] [Google Scholar]
- 20.Freshney RI (1992) Animal cell culture: a practical approach. IRL Press at Oxford University Press
- 21.MacPhail SH, Banath JP, Yu TY, Chu EHM, Lambur H, Olive PL. Expression of phosphorylated histone H2AX in culture cell lines following exposure to X-rays. Int J Radiat Biol. 2003;79(5):351–358. doi: 10.1080/0955300032000093128. [DOI] [PubMed] [Google Scholar]
- 22.Riccardi C, Nicoletti I. Analysis of apoptosis by propidium iodide staining and flow cytometry. Nat Protoc. 2006;1(3):1458–1461. doi: 10.1038/nprot.2006.238. [DOI] [PubMed] [Google Scholar]
- 23.Ihmaid S, Al-Rawi J, Bradley C, Angove MJ, Robertson MN, Clark RL. Synthesis, structural elucidation, DNA-PK inhibition, homology modelling and anti-platelet activity of morpholino-substituted-1,3-naphth-oxazines. Bioorg Med Chem. 2011;19(13):3983–3994. doi: 10.1016/j.bmc.2011.05.032. [DOI] [PubMed] [Google Scholar]
- 24.Ihmaid SK, Al-Rawi JMA, Bradley CJ, Angove MJ, Robertson MN. Synthesis, DNA-PK inhibition, anti-platelet activity studies of 2-(N-substituted-3-aminopyridine)-substituted-1,3-benzoxazines and DNA-PK and PI3K inhibition, homology modelling studies of 2-morpholino-(7,8-di and 8-substituted)-1,3-benzoxazines. Eur J Med Chem. 2012;57:85–101. doi: 10.1016/j.ejmech.2012.08.035. [DOI] [PubMed] [Google Scholar]
- 25.Clapham KM, Bardos J, Finlay MRV, Golding BT, Griffen EJ, Griffin RJ, Hardcastle IR, Menear KA, Ting A, Turner P. DNA-dependent protein kinase (DNA-PK) inhibitors: structure–activity relationships for O-alkoxyphenylchromen-4-one probes of the ATP-binding domain. Bioorg Med Chem Lett. 2011;21(3):966–970. doi: 10.1016/j.bmcl.2010.12.047. [DOI] [PubMed] [Google Scholar]
- 26.Lock RB, Stribinskiene L. Dual modes of death induced by etoposide in human epithelial tumor cells allow Bcl-2 to inhibit apoptosis without affecting clonogenic survival. Cancer Res. 1996;56:4006–4012. [PubMed] [Google Scholar]
- 27.Kao J, Lavaf A, Lan C-H, Fu S. Inhibition of γ-H2AX after ionizing radiation as a biological surrogate of impaired upstream DNA damage signaling and radiosensitivity. J Cancer Mol. 2010;5(2):49–54. [Google Scholar]
- 28.Banáth JP, Olive PL. Expression of phosphorylated histone H2AX as a surrogate of call killing by drugs that create DNA double-strand breaks. Cancer Res. 2003;63:4347–4350. [PubMed] [Google Scholar]
- 29.Celeste A, Fernandez-Capetillo O, Kruhlak MJ, Pilch DR, Staudt DW, Lee A, Bonner RF, Bonner WM, Nussenzweig A. Histone H2AX phosphyrlation is dispensable for the inital recognition of DNA breaks. Nat Cell Biol. 2003;5(7):675–679. doi: 10.1038/ncb1004. [DOI] [PubMed] [Google Scholar]
- 30.Collins I, Garrett MD. Targeting the cell division cycle in cancer: CDK and cell cycle checkpoint kinase inhibitors. Curr Opin Pharmacol. 2005;5(4):366–373. doi: 10.1016/j.coph.2005.04.009. [DOI] [PubMed] [Google Scholar]
- 31.de Klein A, Muijtjens M, van Os R, Verhoeven Y, Smit B, Carr AM, Lehmann AR, Hoeijmakers JHJ. Targeted disruption of the cell-cycle checkpoint gene ATR leads to early embryonic lethality in mice. Curr Biol. 2000;10(8):479–482. doi: 10.1016/S0960-9822(00)00447-4. [DOI] [PubMed] [Google Scholar]
- 32.Yves P. Camptothecins and topoisomerase I: a foot in the door. Targeting the genome beyond topoisomerase I with camptothecins and novel anticancer drugs: importance of DNA replication, repair and cell cycle checkpoints. Curr Med Chem Anti Cancer Agents. 2004;4(5):429–434. doi: 10.2174/1568011043352777. [DOI] [PubMed] [Google Scholar]
- 33.Schwartz GK, Shah MA. Targeting the cell cycle: a new approach to cancer therapy. J Clin Oncol. 2005;23(36):9408–9421. doi: 10.1200/JCO.2005.01.5594. [DOI] [PubMed] [Google Scholar]
- 34.Bai Y, Mao Q-Q, Qin J, Zheng X-Y, Wang Y-B, Yang K, Shen H-F, Xie L-P. Resveratrol induces apoptosis and cell cycle arrest of human T24 bladder cancer cells in vitro and inhibits tumor growth in vivo. Cancer Sci. 2010;101(2):488–493. doi: 10.1111/j.1349-7006.2009.01415.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Johnstone RW, Ruefli AA, Lowe SW. Apoptosis: a link between cancer genetics and chemotherapy. Cell. 2002;108(2):153–164. doi: 10.1016/S0092-8674(02)00625-6. [DOI] [PubMed] [Google Scholar]
- 36.Shieh S-Y, Ikeda M, Taya Y, Prives C. DNA damage-induced phosphorylation of p53 alleviates inhibition by MDM2. Cell. 1997;91(3):325–334. doi: 10.1016/S0092-8674(00)80416-X. [DOI] [PubMed] [Google Scholar]
- 37.Banin S, Moyal L, Shieh S-Y, Taya Y, Anderson CW, Chessa L, Smorodinsky NI, Prives C, Reiss Y, Shiloh Y, Ziv Y. Enhanced phosphorylation of p53 by ATM in response to DNA damage. Science. 1998;281(5383):1674–1677. doi: 10.1126/science.281.5383.1674. [DOI] [PubMed] [Google Scholar]
- 38.Schuler M, Green D. Mechanisms of p53-dependent apoptosis. Biochem Soc Trans. 2001;29(6):684–687. doi: 10.1042/bst0290684. [DOI] [PubMed] [Google Scholar]
- 39.Chipuk J, Green D. Dissecting p53-dependent apoptosis. Cell Death Differ. 2006;13(6):994–1002. doi: 10.1038/sj.cdd.4401908. [DOI] [PubMed] [Google Scholar]
- 40.Reinhardt HC, Aslanian AS, Lees JA, Yaffe MB. p53-deficient cells rely on ATM- and ATR-mediated checkpoint signaling through the p38MAPK/MK2 pathway for survival after DNA damage. Cancer Cell. 2007;11(2):175–189. doi: 10.1016/j.ccr.2006.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Skorski T (2007) DNA damage-dependent apoptosis. In: Srivastava R (ed) Apoptosis, cell signaling, and human diseases. Humana Press, pp 263–272
- 42.Kastan MB, Bartek J. Cell-cycle checkpoints and cancer. Nature. 2004;432:316–323. doi: 10.1038/nature03097. [DOI] [PubMed] [Google Scholar]
- 43.Manke IA, Nguyen A, Lim D, Stewart MQ, Elia AEH, Yaffe MB. MAPKAP kinase-2 is a cell cycle checkpoint kinase that regulates the G2/M transition and S phase progression in response to UV irradiation. Mol Cell. 2005;17(1):37–48. doi: 10.1016/j.molcel.2004.11.021. [DOI] [PubMed] [Google Scholar]
- 44.Roos WP, Kaina B. DNA damage-induced cell death by apoptosis. Trends Mol Med. 2006;12(9):440–450. doi: 10.1016/j.molmed.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 45.Thornton TM, Rincon M. Non-classical P38 map kinase functions: cell cycle checkpoints and survival. Int J Biol Sci. 2009;5(1):44–52. doi: 10.7150/ijbs.5.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dolado I, Nebreda AR. Regulation of tumorigenesis by p38α MAP kinase. Stress Activated Protein Kinases Top Curr Genet. 2008;20:99–128. doi: 10.1007/4735_2007_0245. [DOI] [Google Scholar]
- 47.Mikhailov A, Patel D, McCance DJ, Rieder CL. The G2 p38-mediated stress-activated checkpoint pathway becomes attenuated in transformed cells. Curr Biol. 2007;17(24):2162–2168. doi: 10.1016/j.cub.2007.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weber HO, Ludwig RL, Morrison D, Kotlyarov A, Gaestel M, Vousden KH. HDM2 phosphorylation by MAPKAP kinase 2. Oncogene. 2005;24(12):1965–1972. doi: 10.1038/sj.onc.1208389. [DOI] [PubMed] [Google Scholar]
- 49.Mebratu Y, Tesfaigzi Y. How ERK1/2 activation controls cell proliferation and cell death is subcellular localization the answer? Cell Cycle. 2009;8(8):1168–1175. doi: 10.4161/cc.8.8.8147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Meloche S, Pouyssegur J. The ERK1/2 mitogen-activated protein kinase pathway as a master regulator of the G1-to S-phase transition. Oncogene. 2007;26(22):3227–3239. doi: 10.1038/sj.onc.1210414. [DOI] [PubMed] [Google Scholar]
- 51.Balmanno K, Cook S. Tumour cell survival signalling by the ERK1/2 pathway. Cell Death Differ. 2009;16(3):368–377. doi: 10.1038/cdd.2008.148. [DOI] [PubMed] [Google Scholar]