Abstract
Study Objectives:
Recent genome-wide association studies (GWAS) for Caucasians identified several allelic variants associated with increased risk of developing restless legs syndrome (RLS), also known as Willis-Ekbom disease. Although the pathogenic mechanisms of RLS are not entirely understood, it is becoming increasingly evident that many diseases such as RLS can be attributed to an epistasis. The study objectives were to evaluate whether the associations of RLS with all loci determined in previous GWAS for Caucasians can be replicated significantly for the Korean population and to elucidate whether an epistasis plays a role in the pathogenesis of RLS.
Design, Setting, and Participants:
DNA from 320 patients with RLS and 320 age- and sex-matched controls were genotyped for variants in the RLS loci.
Measurements and Results:
A significant association was found for rs3923809 and rs9296249 in BTBD9 (P < 0.0001 and P = 0.001, respectively); the odds ratio (OR) for rs3923809 was 1.61 (P < 0.0001) to 1.88 (P < 0.0001) and the OR for rs9296249 was 1.44 (P = 0.001) to 1.73 (P = 0.002), according to the model of inheritance. The OR for the interaction between rs3923809 in BTBD9 and rs4626664 in PTPRD was 2.05 (P < 0.0001) in the additive model, 1.80 (P = 0.002) in the dominant model and 2.47 (P = 0.004) in the recessive model. There was no significant association between genotypes of all tested single nucleotide polymorphisms and the mean value of serum iron parameters.
Conclusions:
Our results suggest that the role of BTBD9 in the pathogenesis of restless legs syndrome is more universal across populations than previously reported and more efforts should be focused on the role of epistasis in the genetic architecture of restless legs syndrome.
Citation:
Kim MK; Cho YW; Shin WC; Cho JW; Shon YM; Kim JH; Yang KI; Earley CJ; Allen RP. Association of restless legs syndrome variants in Korean patients with restless legs syndrome. SLEEP 2013;36(12):1787-1791.
Keywords: BTBD9, epistasis, PTPRD, restless legs syndrome
INTRODUCTION
Restless legs syndrome (RLS), also known as Willis-Ekbom disease, is a common neurological disorder that is characterized by an urge to move the legs, usually associated with unpleasant sensations within the legs and that is commonly associated with sleep disturbances and an impaired quality of life.1–3 However, the pathogenic mechanisms and genetic findings of RLS are not entirely understood.
RLS is associated with a positive family history in 45-65% of patients, and twin studies showed a high concordant rate in monozygotic twins, which suggests that genetic factors play an important role in the pathogenesis of RLS.4 To date, genome-wide linkage analyses have identified at least eight major susceptibility loci for RLS: RLS1 on chromosome 12q12-q21, RLS2 on 14q13-21, RLS3 on 9p24-p22, RLS4 on 2q33, RLS5 on 20p13, RLS6 on 19p13, RLS7 on 16p12.1, and RLS8 on 2p14.5–12 Recent genome-wide association studies (GWAS) of RLS, which were performed in populations of primarily European ancestry, identified six additional loci associated with RLS, which are represented by single nucleotide polymorphisms (SNPs) on chromosome 2p14 (MEIS1 and an intergenic region 1.3 Mb downstream of MEIS1), 6p21.2 (BTBD9), 15q23 (MAP2K5/SKOR1), 9p24.1-p23 (PTPRD), and 16q12.1 (TOX3/BC034767).12–15 Replication studies for European and US populations confirmed the association of at least one SNP in MEIS1, BTBD9, MAP2K5/SKOR1, and PTPRD.16–18 However, none of the studies on RLS has yet led to the identification of disease-causing sequence variants and has never been conducted for Asian/Korean patients.
It is becoming increasingly evident that many common human diseases such as RLS cannot be attributed to a single gene. Instead, epistasis (gene-to-gene interaction) is believed to play an important role in the genetic architecture of many common human diseases,19–22 which is not yet evident in the pathogenesis of RLS.
The aims of the current study were to evaluate whether the associations of RLS with all loci determined in previous GWAS for European/American populations can be replicated in the Korean population and to elucidate whether epistasis plays a role in the pathogenesis of RLS.
METHODS
Study Population
This study was approved by local institutional review boards on human subject research and written consent was obtained from the participants. The patients were recruited in the Center for Sleep Disorders at seven tertiary hospitals that cover five of the six largest cities in Korea. The diagnosis of all RLS cases was made according to diagnostic criteria of the International RLS Study Group,2 and through face-to-face interviews and examinations by a neurologist to exclude conditions that may mimic RLS, using the validated Korean-language version of the Johns Hopkins telephone diagnostic questionnaire.23 Familial RLS was defined as at least one affected first-degree relative. Secondary RLS cases due to uremia, dialysis, peripheral neuropathy, and pregnancy were excluded. A total of 320 patients were included [113 males, mean age (standard deviation, SD) 56.1 (12.7) y]. Positive family history was reported by 84 cases and in 76 the data were not available. Phenotype assessment for each patient was performed using a standardized Web-based form (http://ecrf.rls.co.kr; Figure S1 in supplemental material) in which demographic information, past medical history, diagnostic criteria of RLS, and clinical (for presence of periodic limb movements in sleep [PLMS] and severity of RLS symptoms) and laboratory data (serum ferritin and iron level and total iron-binding capacity of blood) were taken into consideration. The demographic data and laboratory findings of successfully genotyped samples (n = 317) are summarized in Table 1.
Table 1.
Characteristics of Korean people with restless legs syndrome
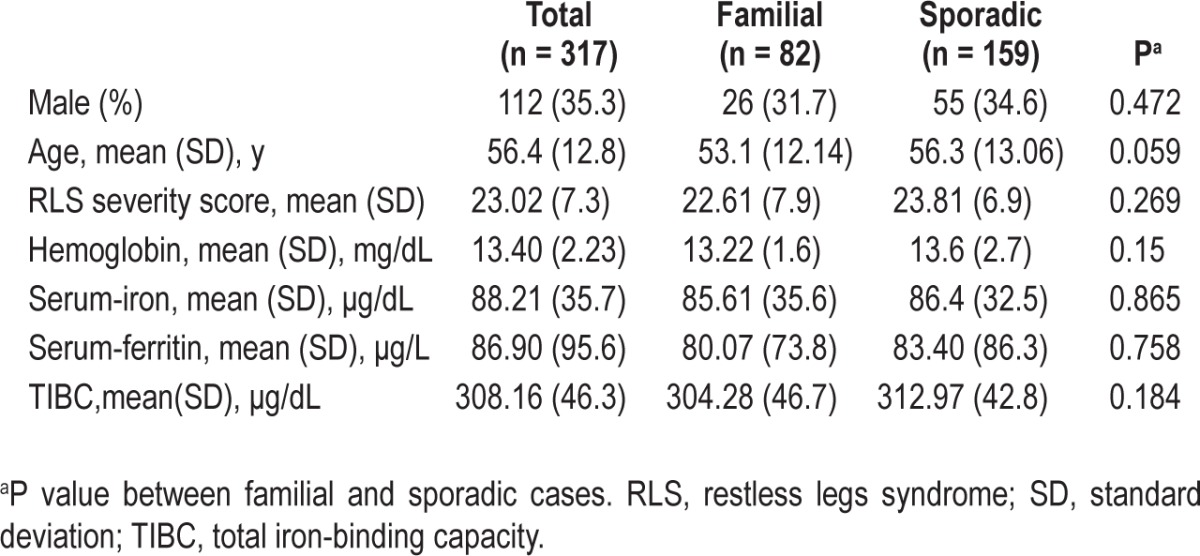
A total of 320 sex-matched controls (113 males, mean age [SD] 55.7 [9.5] y), who were screened using the diagnostic criteria of the International RLS Study Group2 for the presence of RLS, were selected from the Korean National Human Resource Bank registry of the Centers for Genome Science National Institute of Health (http://biomi.cdc.go.kr/sale_info/main.jsp).
SNP Selection
Of the seven SNPs in the four genomic regions whose association with RLS was successfully replicated by one or more GWAS with Caucasian RLS cases until the present time,14–18 six SNPs (rs2300478 in MEIS1, rs3923809, rs9296249 and rs9357271 in BTBD9, rs1026732 in MAP2K5/ SKOR1, and rs1975197 in PTPRD) were selected; another MAP2K5/SKOR1 variant, rs6494696, was excluded because it is located in the same linkage disequilibrium block as the SNP rs1026732.13 Although its association with RLS was not replicated in the follow-up GWAS,17 rs4626664 in PTPRD about which too little is known was included to extend the knowledge about its role in the pathogenesis of RLS.
Genotyping
DNA was extracted using a standard protocol and purified using a kit procedure (Qiagen, Hilden, Germany). The real-time polymerase chain reaction (PCR) using high-resolution melting (HRM) analysis was used in genotyping of the substitution of adenine for guanine or thymidine for cytosine, or vice versa, for which the accuracy of the HRM analysis was 100% according to duplicate direct sequencing analysis of, on average, 5% of the total genotype. HRM analysis was performed using Rotor-Gene 6000 (Corbett Life Science, Brisbane, Australia) according to the manufacturer's protocol.
The HRM process consists of performing the PCR in the presence of the DNA binding dye, SYTO© 9 green (Invitrogen Corp., Carlsbad, CA, USA), monitoring the progressive changes in fluorescence caused by release of the dye from a DNA duplex as it is denatured with increasing temperature, collecting a HRM curve, and identifying the samples with melting curve aberrations indicative of the presence of a sequence variant. Fluorescence intensity as a function of temperature monitored by the LightScanner© instrument (Idaho Technology, Salt Lake City, UT, USA) can reveal small changes in the melting curve shape when analyzed with the LightScanner© software using the “Scanning” mode (Idaho Technology).
PCR was performed in 20-μL reactions containing 40 ng of template DNA, 1.5 mM MgCl2, 0.5 mM deoxyribonucleotide triphosphate, 400 nM forward and reverse primers, 0.8x SYTO© 9 green, 0.5 U of AmpliTaq Polymerase Gold, and 1×PCR buffer (Applied Biosystems, Foster City, USA). The thermal cycling conditions for PCR reactions were as follows: initial denaturation at 95.0°C for 2 min with 40 cycles at 95.0°C for 5 sec and annealing/extension at 58.0 to 63.0°C for 10 sec. HRM was carried out by fluorescence acquisition during a temperature increase from a minimum of 70.0°C to a maximum of 85.0°C with increments of 0.1°C and holding steps of 10 sec.
Statistical Analysis
Genotype frequencies at each locus were tested for Hardy-Weinberg equilibrium (HWE). The threshold for the exclusion of SNP loci was P values of ≤ 0.05. The statistical evaluation of genotype data was performed with the Pearson chi-square using the IBM SPSS Statistics version 20 (SPSS Inc., Chicago, IL, USA). Fisher exact test was used if the expected cell frequencies were lower than five. The strength of the association between the mutant allele of each SNP and RLS and the effect of a biallelic interaction on susceptibility to RLS was evaluated as an odds ratio (OR), according to the mode of inheritance of a causal allele obtained with HAPSTAT 3.0 (University of North Carolina, Chapel Hill, NC, USA) that allows haplotype analysis of multiple genes as well as single- and multi-SNP analysis with missing genotypes.24 Association tests were conducted in three different settings: (1) all combined cases (familial, sporadic, and not determined; n = 317 versus all controls (n = 318); (2) familial cases (n = 82) versus controls; and (3) sporadic cases (n = 159) versus controls. The difference in the mean values of continuous variables between familial and sporadic cases was tested by two-tailed Student t-test. Analysis of variance (ANOVA) was performed to test for the existence of differences in the mean values of continuous variables among three genotypes of each SNP. The differences were considered significant for P values of ≤ 0.05.
The mode of inheritance can be additive, dominant, or recessive. Under the additive model, two copies of a causal allele have twice the effect on the trait as compared to a single copy. Under the dominant model, having one or two copies has the same effect. Under the recessive model, having only two copies of the causal allele will affect the trait. Bonferroni correction for multiple testing of seven markers and 21 possible binary interactions among the seven markers was used and the corrected P values of ≤ 0.007 and ≤ 0.0024 were considered statistically significant, respectively.
RESULTS
All SNPs tested were in HWE in both RLS cases and controls. Three hundred seventeen samples from RLS cases and 318 controls were successfully genotyped. There was no significant difference between familial (n = 82) and sporadic RLS cases (n = 159) in genotype distribution for all SNPs tested (supplemental material, Table S1), in the severity of RLS symptoms and in the mean value of serum iron, ferritin, and total iron-binding capacity (TIBC) (Table 1). Because the presence of PLMS could not be determined in more than half of the RLS cases (53.9%), the variable was excluded from analysis.
In the sample set of the combined cases versus controls, a significant association was found in genotype distribution of rs3923809 and rs9296249 in BTBD9 (P < 0.0001 and P = 0.001, respectively). The OR for rs3923809 was 1.88 (95% confidence interval [CI], 1.39-2.56; P < 0.0001) in the dominant model and 1.61 (95% CI, 1.28-2.02; P < 0.0001) in the additive model of inheritance and the OR for rs9296249 was 1.73 (95% CI, 1.23-2.44; P = 0.002) in the dominant model and 1.44 (95% CI, 1.16-1.80; P = 0.001) in the additive model of inheritance, when considering the minor allele of each SNP as a causal allele (the data for the recessive model can be found in Table 2). In the sample set of sporadic cases versus controls, the two SNPs were also associated with RLS (P < 0.0001 for rs3923809 and P = 0.007 for rs9296249) in both additive and dominant models of inheritance (data not shown). These statistical significances remained after using Bonferroni correction for multiple comparisons (supplemental material, Table S1, and Table 2).
Table 2.
Association of all tested single nucleotide polymorphisms with restless legs syndrome in a Korean population
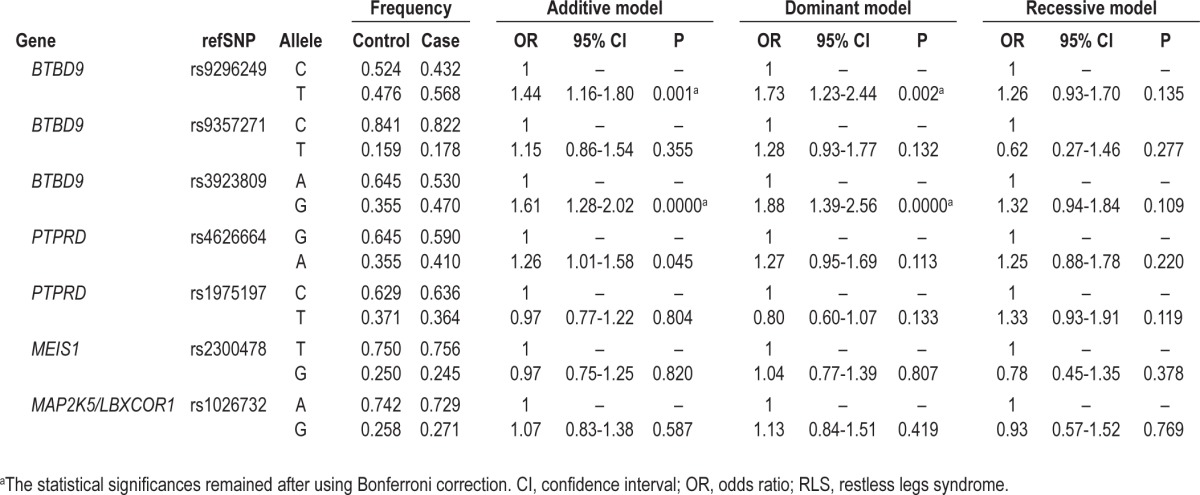
Although it seemed that rs4626664 in PTPRD in the sample set of both the combined cases versus controls and familial cases versus controls and rs3923809 and rs9296249 in BTBD9 in the sample sets of familial cases versus controls were associated with RLS, the statistical value for these associations were not significant after using Bonferroni correction for multiple comparisons (supplemental material, Table S1, and Table 2).
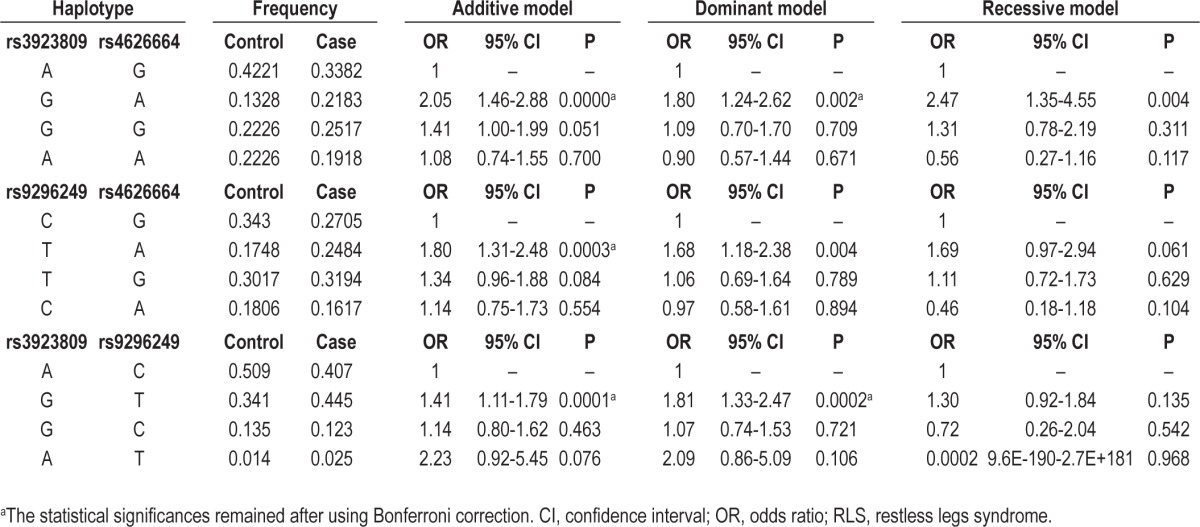
The OR for the interaction between rs3923809 in BTBD9 and rs4626664 in PTPRD was 2.05 (95% CI, 1.46-2.88; P < 0.0001) in the additive model, 1.80 (95% CI, 1.246-2.62; P = 0.002) in the dominant model, and 2.47 (95% CI, 1.35-4.55; P = 0.004) in the recessive model. The OR for the interaction between rs9296249 in BTBD9 and rs4626664 was 1.80 (95% CI, 1.31-2.48; P = 0.0003) in the additive model and 1.68 (95% CI, 1.18-2.48; P = 0.004) in the dominant model. After using Bonferroni correction for multiple testing of 21 possible binary interactions among seven markers tested in the current study, the interactions between rs3923809 and rs4626664 in the recessive model and between rs9296249 and rs4626664 in the dominant model were no longer statistically significant (Table 3). Although such a significant association with RLS was also found in the interaction between rs9296249 and rs3923809, the OR for the interaction was not higher than that for each of the SNPs; the OR was 1.41 (95% CI, 1.11-1.79; P = 0.0001) in the additive model and 1.81 (95% CI, 1.33-2.47; P = 0.0002) in the dominant model (Tables 2 and 3). No other biallelic interaction among BTBD9, MEIS1 and MAP2K5/LBXCOR1 variants was found to be associated with RLS (data not shown).
Table 3.
Effect of biallelic interaction in BTBD9 and PTPRD variants on susceptibility to restless legs syndrome
There was no significant association between genotypes of all tested SNPs and the mean value of serum iron, ferritin, and TIBC (data not shown).
DISCUSSION
The association of RLS with BTBD9 variants, rs3923809 and rs9296249, was significantly replicated in the Korean population. Furthermore, a PTPRD variant, rs4626664, seemed to increase the risk of RLS through an interaction with BTBD9 variants. To the best of our knowledge, this is the first report where the association of RLS with BTBD9 variants was successfully replicated in Korean populations and showed that an epistasis can increase the risk of RLS.
In a large-scale high-density GWAS conducted in German and Canadian populations, strong associations of RLS with variants in BTBD9, MEIS1 and MAP2K5/LBXCOR1 were found,13 which were significantly replicated for other European and US populations in subsequent studies.16–18 In the current study, a significant association was replicated only with BTBD9 variants and not with MEIS1 and MAP2K5/LBXCOR1 variants. These findings suggest that BTBD9 plays a consistent role in RLS in the Korean population and that we may need further studies with different sample sets of Korean populations to determine the exact role of MEIS1 and MAP2K5/LBXCOR1 in RLS.
It has been known that RLS is a dopamine-based disorder related to iron deficiency.25 Indeed, altered peripheral iron homeostasis, such as iron deficiency anemia and pregnancy, can produce RLS and iron concentrations are lowered in the cerebrospinal fluid and substantia nigra in patients with RLS.26–29 Although little is known about the function of BTBD9 other than it belongs to the BTB(POZ) domain-containing protein, there is some supporting evidence that BTBD9 plays a role in regulating iron homeostasis. Serum ferritin levels decreased by 13% per at-risk variant of BTBD9 rs3923809 in a GWAS in Icelandic and US populations15 and homozygosity for the at-risk variant of BTBD9 rs9296249 was associated with lower serum ferritin levels in Danish female blood donors.30 This is consistent with the suspected involvement of iron deficiency in the pathogenesis of RLS. However, an increase in serum iron levels was observed in the homozygous Btbd9 mutant mice,31 which poses a challenge to the iron deficiency hypothesis. Furthermore, in a recent larger-scale association study in a Caucasian population in which the authors asked whether known iron-related genes are candidates for association with RLS, and vice versa, whether known RLS-associated loci influence iron parameters in serum, none of the candidate SNPs at the iron-related gene loci were confirmed as significant and SNPs at the known RLS loci did not significantly affect serum iron parameters.32 This finding is consistent with results from the current study in which none of the known RLS loci tested including BTBD9 were associated with an altered iron homeostasis in this Korean population. Although it has been known that racial differences can affect serum iron parameters or risk of RLS,33 further study is needed to elucidate the exact role iron or BTBD9 plays in the etiology of RLS.
PTPRD is the fourth genome-wide significant locus for RLS. Schormair et al.14 reported that two independent SNPs in PTPRD (rs4626664 and rs1975197) were significantly associated with RLS in European and Canadian populations. The association with rs1975197 was also significantly replicated in both family-based and population-based association studies in an American Caucasian population.17 The current study showed no significant association for either rs4626664 or rs1975197 in the Korean population; however, there was a significant interaction with risk alleles in BTBD9; an increase in OR especially for the interaction between at-risk alleles of BTBD9 rs3923809 and PTPRD rs4626664 was observed compared to single SNP analysis. Epistasis is believed to play an important role in the genetic architecture of many common human diseases,19–22 but no evidence of epistasis in RLS has been reported prior to this study.14 The role of an epistasis in the pathogenesis of RLS may account for the fact that none of the studies on RLS has yet led to the identification of disease-causing sequence variants.
There are several limitations to this study. First, the sample sets recruited in the current study was relatively small in size, which makes statistical power not strong enough to detect association or interaction for risk alleles, especially those with a modest effect. Indeed, significant genotypic and allelic associations for PTPRD rs4626664 in various sample sets lost their statistical power after using Bonferroni correction for multiple testing, which may reflect lack of power due to the smaller sample size. To minimize the effect of the smaller sample size, cases and controls were matched for age and sex in the current study. Second, the association of the at-risk alleles in the RLS loci with PLMS could not be estimated in this study because PLMS were not objectively documented. A larger-scale GWAS in Icelandic and US populations showed an association of BTBD9 with PLMS but not with RLS.15 Because most RLS cases are accompanied by PLMS, further study is needed to clarify whether the RLS loci are associated with RLS or PLMS or both. Third, more studies in different sample sets are needed to replicate the evidence for epistasis in RLS proved in the current study.
In conclusion, our results suggest that the role of BTBD9 in the pathogenesis of RLS would be more universal across populations than previously expected and more efforts should be focused on the role of epistasis in the genetic architecture of RLS. Our results need to be replicated in a larger cohort of Korean descent and other large cohorts of different descent and confirmed via functional studies, but this first report of epistasis in RLS genetics opens possible new approaches to understanding the pathogenesis of RLS.
DISCLOSURE STATEMENT
This was not an industry supported study. This research was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology (2011-0009691). The authors have indicated no financial conflicts of interest.
SUPPLEMENTAL MATERIAL
Questionnaire for restless legs syndrome.
Genotype distribution of all tested single nucleotide polymorphisms by case-control status
REFERENCES
- 1.Ekbom K, Ulfberg J. Restless legs syndrome. J Intern Med. 2009;266:419–31. doi: 10.1111/j.1365-2796.2009.02159.x. [DOI] [PubMed] [Google Scholar]
- 2.Allen RP, Picchietti D, Hening WA, et al. Restless legs syndrome: diagnostic criteria, special considerations, and epidemiology. A report from the restless legs syndrome diagnosis and epidemiology workshop at the National Institutes of Health. Sleep Med. 2003;4:101–19. doi: 10.1016/s1389-9457(03)00010-8. [DOI] [PubMed] [Google Scholar]
- 3.Abetz L, Allen R, Follet A, et al. Evaluating the quality of life of patients with restless legs syndrome. Clin Ther. 2004;26:925–35. doi: 10.1016/s0149-2918(04)90136-1. [DOI] [PubMed] [Google Scholar]
- 4.Chahine LM, Chemali ZN. Restless legs syndrome: a review. CNS Spectr. 2006;11:511–20. doi: 10.1017/s1092852900013547. [DOI] [PubMed] [Google Scholar]
- 5.Desautels A, Turecki G, Montplaisir J, Sequeira A, Verner A, Rouleau GA. Identification of a major susceptibility locus for restless legs syndrome on chromosome 12q. Am J Hum Genet. 2001;69:1266–70. doi: 10.1086/324649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bonati MT, Ferini-Strambi L, Aridon P, Oldani A, Zucconi M, Casari G. Autosomal dominant restless legs syndrome maps on chromosome 14q. Brain. 2003;126:1485–92. doi: 10.1093/brain/awg137. [DOI] [PubMed] [Google Scholar]
- 7.Chen S, Ondo WG, Rao S, Li L, Chen Q, Wang Q. Genomewide linkage scan identifies a novel susceptibility locus for restless legs syndrome on chromosome 9p. Am J Hum Genet. 2004;74:876–85. doi: 10.1086/420772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Levchenko A, Provost S, Montplaisir JY, et al. A novel autosomal dominant restless legs syndrome locus maps to chromosome 20p13. Neurology. 2006;67:900–1. doi: 10.1212/01.wnl.0000233991.20410.b6. [DOI] [PubMed] [Google Scholar]
- 9.Pichler I, Marroni F, Volpato CB, et al. Linkage analysis identifies a novel locus for restless legs syndrome on chromosome 2q in a South Tyrolean population isolate. Am J Hum Genet. 2006;79:716–23. doi: 10.1086/507875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kemlink D, Plazzi G, Vetrugno R, et al. Suggestive evidence for linkage for restless legs syndrome on chromosome 19p13. Neurogenetics. 2008;9:75–82. doi: 10.1007/s10048-007-0113-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Levchenko A, Montplaisir JY, Asselin G, et al. Autosomal-dominant locus for restless legs syndrome in French-Canadians on chromosome 16p12.1. Mov Disord. 2009;24:40–50. doi: 10.1002/mds.22263. [DOI] [PubMed] [Google Scholar]
- 12.Winkelmann J, Czamara D, Schormair B, et al. Genome-wide association study identifies novel restless legs syndrome susceptibility loci on 2p14 and 16q12.1. PLoS Genet. 2011;7:e1002171. doi: 10.1371/journal.pgen.1002171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Winkelmann J, Schormair B, Lichtner P, et al. Genome-wide association study of restless legs syndrome identifies common variants in three genomic regions. Nat Genet. 2007;39:1000–6. doi: 10.1038/ng2099. [DOI] [PubMed] [Google Scholar]
- 14.Schormair B, Kemlink D, Roeske D, et al. PTPRD (protein tyrosine phosphatase receptor type delta) is associated with restless legs syndrome. Nat Genet. 2008;40:946–8. doi: 10.1038/ng.190. [DOI] [PubMed] [Google Scholar]
- 15.Stefansson H, Rye DB, Hicks A, et al. A genetic risk factor for periodic limb movements in sleep. N Engl J Med. 2007;357:639–47. doi: 10.1056/NEJMoa072743. [DOI] [PubMed] [Google Scholar]
- 16.Kemlink D, Polo O, Frauscher B, et al. Replication of restless legs syndrome loci in three European populations. J Med Genet. 2009;46:315–8. doi: 10.1136/jmg.2008.062992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang Q, Li L, Yang R, et al. Family-based and population-based association studies validate PTPRD as a risk factor for restless legs syndrome. Mov Disord. 2011;26:516–9. doi: 10.1002/mds.23459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang Q, Li L, Chen Q, Foldvary-Schaefer N, Ondo WG, Wang QK. Association studies of variants in MEIS1, BTBD9, and MAP2K5/ SKOR1 with restless legs syndrome in a US population. Sleep Med. 2011;12:800–4. doi: 10.1016/j.sleep.2011.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pharoah PD, Dunning AM, Ponder BA, Easton DF. Association studies for finding cancer-susceptibility genetic variants. Nat Rev Cancer. 2004;4:850–60. doi: 10.1038/nrc1476. [DOI] [PubMed] [Google Scholar]
- 20.Thornton-Wells TA, Moore JH, Haines JL. Genetics, statistics and human disease: analytical retooling for complexity. Trends Genet. 2004;20:640–7. doi: 10.1016/j.tig.2004.09.007. [DOI] [PubMed] [Google Scholar]
- 21.Cordell HJ. Epistasis: what it means, what it doesn't mean, and statistical methods to detect it in humans. Hum Mol Genet. 2002;11:2463–8. doi: 10.1093/hmg/11.20.2463. [DOI] [PubMed] [Google Scholar]
- 22.Moore JH. The ubiquitous nature of epistasis in determining susceptibility to common human diseases. Hum Hered. 2003;56:73–82. doi: 10.1159/000073735. [DOI] [PubMed] [Google Scholar]
- 23.Cho YW, Lee MY, Yun CH, Shin WC, Hong SB, Kim JH. The reliability and validity of the Korean version of paradigm of questions for epidemiology studies of restless legs syndrome and the Johns Hopkins telephone diagnostic interview form for the restless legs syndrome. J Korean Neurol Assoc. 2007;25:494–9. [Google Scholar]
- 24.Lin DY, Hu Y, Huang BE. Simple and efficient analysis of disease association with missing genotype data. Am J Hum Genet. 2008;82:444–52. doi: 10.1016/j.ajhg.2007.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barrière G, Cazalets JR, Bioulac B, Tison F, Ghorayeb I. The restless legs syndrome. Prog Neurobiol. 2005;77:139–65. doi: 10.1016/j.pneurobio.2005.10.007. [DOI] [PubMed] [Google Scholar]
- 26.Kavanagh D, Siddiqui S, Geddes CC. Restless legs syndrome in patients on dialysis. Am J Kidney Dis. 2004;43:763–71. doi: 10.1053/j.ajkd.2004.01.007. [DOI] [PubMed] [Google Scholar]
- 27.Manconi M, Govoni V, De Vito A, et al. Pregnancy as a risk factor for restless legs syndrome. Sleep Med. 2004;5:305–8. doi: 10.1016/j.sleep.2004.01.013. [DOI] [PubMed] [Google Scholar]
- 28.Earley CJ, B Barker P, Horská A, Allen RP. MRI-determined regional brain iron concentrations in early- and late-onset restless legs syndrome. Sleep Med. 2006;7:458–61. doi: 10.1016/j.sleep.2005.11.009. [DOI] [PubMed] [Google Scholar]
- 29.Connor JR, Boyer PJ, Menzies SL, et al. Neuropathological examination suggests impaired brain iron acquisition in restless legs syndrome. Neurology. 2003;61:304–9. doi: 10.1212/01.wnl.0000078887.16593.12. [DOI] [PubMed] [Google Scholar]
- 30.Sørensen E, Grau K, Berg T, et al. A genetic risk factor for low serum ferritin levels in Danish blood donors. Transfusion. 2012;52:2585–9. doi: 10.1111/j.1537-2995.2012.03629.x. [DOI] [PubMed] [Google Scholar]
- 31.DeAndrade MP, Johnson RL, Jr, Unger EL, et al. Motor restlessness, sleep disturbances, thermal sensory alterations and elevated serum iron levels in Btbd9 mutant mice. Hum Mol Genet. 2012;21:3984–92. doi: 10.1093/hmg/dds221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oexle K, Schormair B, Ried JS, et al. Dilution of candidates: the case of iron-related genes in restless legs syndrome. Eur J Human Genet. 2013;21:410–4. doi: 10.1038/ejhg.2012.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kutner NG, Zhang R, Huang Y, Bliwise DL. Racial differences in restless legs symptoms and serum ferritin in an incident dialysis patient cohort. Int Urol Nephrol. 2012;44:1825–31. doi: 10.1007/s11255-011-0108-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Questionnaire for restless legs syndrome.
Genotype distribution of all tested single nucleotide polymorphisms by case-control status


