Abstract
Soft-tissue sarcomas are a heterogeneous group of tumors arising from connective tissue. Recently, mutations in the neurofibromin 1 (NF1) tumor suppressor gene were identified in multiple subtypes of human soft-tissue sarcomas. To study the effect of NF1 inactivation in the initiation and progression of distinct sarcoma subtypes, we have developed a novel mouse model of temporally and spatially restricted NF1-deleted sarcoma.
To generate primary sarcomas, we inject adenovirus containing Cre recombinase into NF1flox/flox; Ink4a/Arf flox/flox mice at two distinct orthotopic sites: intramuscularly or in the sciatic nerve. The mice develop either high-grade myogenic sarcomas or MPNST-like tumors, respectively. These tumors reflect the histological properties and spectrum of sarcomas found in patients. To explore the utility of this model for preclinical studies, we performed a study of MAPK pathway inhibition with the MEK inhibitor PD325901. Treatment with PD325901 delays tumor growth through decreased cyclin D1 mRNA and cell proliferation. We also examined the effects of MEK inhibition on the native tumor stroma and find that PD325901 decreases VEGFα expression in tumor cells with a corresponding decrease in microvessel density. Taken together, our results utilize a primary tumor model to demonstrate that sarcomas can be generated by loss of NF1 and Ink4a/Arf, and that these tumors are sensitive to MEK inhibition by direct effects on tumor cells and the surrounding microenvironment. These studies suggest that MEK inhibitors should be further explored as potential sarcoma therapies in patients with tumors containing NF1 deletion.
Keywords: NF1, sarcoma, MAPK inhibition, preclinical study, mouse model
INTRODUCTION
Soft-tissue sarcomas are an aggressive and heterogeneous group of tumors that arise from connective tissue, including the muscle, nerve sheath, blood vessel, fat, and fibrous tissue. These tumors are fatal in thirty percent of patients (1), and the standard treatment of wide resection and radiation carries considerable morbidity among survivors. Recent genomic studies have identified novel mutations in a diverse spectrum of human soft tissue sarcomas that may reveal new molecular targets for therapy (2). One of the most frequently mutated genes described is neurofibromin 1 (NF1), a tumor suppressor that functions as a negative regulator of the Ras pathway. Although the loss of NF1 is well-characterized in malignant peripheral nerve sheath tumors (MPNSTs), NF1 mutations in other soft-tissue sarcomas have only been recently identified. Using genomic sequencing and copy number analysis, NF1 mutations were identified in myxofibrosarcoma (10.5%, n = 4/38) and pleomorphic liposarcoma (8%, n = 2/24) (3). Similar studies have identified genomic loss of the NF1 locus in 15% of pediatric embryonal rhabdomyosarcomas (n = 4/26) (4). This deletion was mutually exclusive with Ras gene mutations. The identification of novel mutations in sarcomas with complex karyotypes provides new opportunities to model human sarcomas and to investigate new therapies for this difficult to treat disease.
In addition to identifying NF1 mutations in spontaneous sarcomas in the general population, patients with Neurofibromatosis Type 1 (NF1) are at increased risk for developing aggressive soft-tissue sarcomas. Neurofibromatosis Type 1 effects 1 in 3000 live births and is caused by germline inactivation of a single allele of the NF1 gene (5). The most common sarcoma afflicting NF1 patients is malignant peripheral nerve sheath tumor (MPNST), with a lifetime risk of 8–13% (5). NF1 patients are also at risk for developing non-neurogenic sarcomas, including rhabdomyosarcoma (RMS) and undifferentiated pleomorphic sarcoma (UPS), with a 3–6% lifetime risk (6–8). Most malignant NF1-deleted tumors have inactivation of either p53 or Ink4a/Arf tumor suppressor genes, with over 50% of MPNSTs expressing wild-type p53 and homozygous deletion of Ink4a/Arf (9). Deletion of NF1 alone is not sufficient to cause high-grade sarcomas, as tissue-specific deletion of NF1 with either Krox20-Cre or Dhh-Cre results in neurofibromas (10, 11). Similarly, deletion of either Ink4a or Arf alone is not sufficient for formation of neurofibromas or MPNSTs on an NF1 heterozygous background from either NF1 +/−; Ink4a −/− mice (12) or NF1 +/−; Arf −/− mice (13). Additionally, we have previously reported that conditional deletion of Ink4a/Arf alone by Ad-Cre injection is insufficient for sarcoma initiation (14). Deletion of both Ink4a and Arf in an NF1 heterozygous background (NF1 +/−; Ink4a/Arf −/) generated sporadic MPNSTs in 26% of mice (12). The most widely-used NF1-dependent mouse sarcoma model is the NF1+/− ; p53+/− cis mouse, which develops NF1- and p53-null sarcomas throughout the body following loss-of-heterozygosity (15, 16). Despite the prevalence of NF1 mutations in soft-tissue sarcomas, the role of NF1 in the development of soft-tissue sarcomas, particularly in the background of wild-type p53 alleles, has not been fully explored in a primary mouse model of soft-tissue sarcoma. Therefore, we designed a study to test if deletion of both NF1 and Ink4a/Arf could generate high-grade soft-tissue sarcomas in an NF1 wild-type background.
Genetically-engineered mouse models have greatly advanced our understanding of sarcomas (14, 15, 17–19) and have served as a preclinical platform for therapies (20–25). However, for many models, mice develop sarcomas at multiple sites, which can make tumor detection and assessment of treatment response challenging. Here, we developed a novel mouse model of temporally and spatially restricted NF1-deleted sarcoma to increase our understanding of NF1-mutant tumors. Using Cre-loxP technology, tumors are initiated through Cre-mediated deletion of conditional NF1 and Ink4a/Arf floxed alleles (NF1flox/flox; Ink4a/Arf flox/flox). These mice develop both high-grade myogenic sarcomas and MPNST-like tumors, based on the site of Cre injection. These tumors reflect the histological properties and spectrum of sarcomas found in patients. Because sarcomas in this model develop in a spatially and temporally restricted manner, this model is particularly useful for testing therapeutic drug responses, and we have used this model as a preclinical platform to assess new therapies for patients with NF1-deleted sarcomas. We have identified MAPK inhibition as a promising therapy that slows the growth of primary mouse sarcomas through effects on tumor cell growth and the surrounding vasculature. Importantly, this model is able to directly assess the effects of MEK inhibition on native tumor stroma, as these primary tumors develop in immunocompetent mice with native vasculature. By demonstrating the ability of these tumors to respond to MEK inhibition, we lay a foundation for future studies using this model to test the efficacy of other drugs for NF1-deficient sarcomas in the pre-clinical setting.
MATERIALS AND METHODS
Mouse Sarcoma Model
All mouse work was performed in accordance with Duke University Institutional Animal Care and Use Committee approved protocols. The NF1flox/flox mice (11) and Ink4a/Arfflox/flox mice (26) have been previously reported. Primers sequences used to demonstrate recombination of the appropriate alleles are found in Supplemental Table 1 or have been previously reported (11). Tumors were generated by injection of Adenovirus–expressing Cre recombinase (Ad-Cre; University of Iowa, Vector core) into the lower left leg of NF1flox/flox; Ink4a/Arfflox/flox compound mutant mice as previously described (14). For sciatic nerve injections, the nerve was exposed by an incision in the surrounding muscle, followed by injection of 25 μl Ad-Cre into the nerve. Tumor volumes were calculated using the formula V = (π × L × W × H)/6, with L, W, and H representing the length, width, and height of the tumor in mm, respectively.
Histology and Immunostaining
Antibodies for immunohistochemistry included: S100 (Dako Cytomation), MyoD1 (Dako Cytomation), pERK (Cell Signaling), pS6 (Cell Signaling), Ki67 (BD Pharmagen), CD31 (BD Pharmagen), pVEGFR-2 (Cell Signaling), and MECA-32 (BD Pharmagen). All immunostaining was perfomed with citrate-based antigen retrieval, with the exception of MECA-32 and pVEGFR2, which were performed with EDTA-based retrieval. TUNEL staining used the In Situ Cell Death Detection Kit, TMR Red (Sigma), per manufacturer’s instructions. To visualize mast cells, slides were stained with Toluidine Blue solution (0.02% Toluidine Blue in 1% NaCl, pH=2.2) for 2 minutes, followed by 2 washes in distilled water and 3 washes in 100% ethanol. To quantify
PD325901 Inhibitor Treatment
Treatment started when tumors were between 200–300 mm3. Mice receiving vehicle alone were treated with NMP 10% (1-methyl-2-pyrrolidone)/PEG300 90% (Fluka) orally 5 days per week. PD325901(27) (Sigma) was dissolved in NMP 10% (1-methyl-2-pyrrolidone)/PEG300 90%. Mice were treated with 5 mg/kg of PD325901 orally 5 days per week from the first day of the treatment. Treatment for all mice continued until tumors regressed or it was necessary to sacrifice the mouse for animal welfare concerns (ie: animal was moribund; tumor volume reached 2000 mm3).
Tumor growth analysis
Criteria for defining treatment response includes both volumetric analysis and a delay in tumor growth kinetics. A partial response (PR) is classified as a tumor that: 1) demonstrates a decrease in tumor volume from baseline measurements and 2) shows delayed growth kinetics (defined as requiring more than 10 days to increase in volume 150%). For waterfall plot analyses, percent maximal change is reported as the greatest percent loss in tumor volume from baseline for tumors responding to treatment. Tumors that did not respond to treatment are shown as greatest percent gain in tumor volume from baseline over the course of the treatment. In this presentation of the data, change in tumor volume is capped at +100%.
Statistical Analysis
Graphs and statistics were performed in Graph Pad 4.0. A non-parametric student’s t-test was performed to determine differences between treatment groups. A p-value of <0.05 was considered statistically significant.
In vitro studies
The mouse sarcoma cell lines 1863 and 3017 were derived from primary sarcomas in NF1flox/flox; Ink4a/Arfflox/flox mice that developed after intra-muscular Ad-Cre injection. These cell lines were authenticated by PCR genotyping. The cells were cultured in DMEM + 10% FBS. For colony formation studies, cells were seeded at 500/well in a 6-well dish and treated with DMSO or 50 nM PD325901 (Sigma) for 3–7 days. The number of existing colonies was determined by crystal violet staining. Each experiment was conducted in triplicate, and the experiment was repeated three times. Data shown is representative of one independent experiment. For colony formation studies with prior PD325901 treatment, cells were cultured overnight in 50nM PD325901, followed by splitting the cells into DMSO control or PD325901 for colony formation. Experiments were analyzed as described above. Cell proiferation assays were performed with cells cultured in either DMSO or 50nM PD325901, with the number of cell doublings calculated daily. For quantitative RT-PCR analyses, cells were treated with 50 nM PD325901 for 4 hours, followed by RNA harvest into TRIZOL. Data shown represents the average of 4 independent experiments.
Western blot analysis
1863 cells were treated with 50 nM PD325901 for either 1 or 4 hours prior to harvest. Cells were washed once with cold PBS (Sigma) and lysed for 10 minutes on ice with RIPA buffer (Sigma), supplemented with phosphatase inhibitors (Sigma, P5726 and P0044). Protein concentration was performed with BCA Protein Concentration Assay (Thermo Scientific). MiniProtean TGX gels (BioRad) were transferred to PVDF by wet transfer.
Quantitative RT-PCR
RNA was isolated from tumors and cells by TRIZOL. cDNA synthesis was performed with iScript (BioRad). Quantitative RT-PCR was performed on an iQ5 instrument (BioRad) using the delta-delta Ct method. Primer sequences can be found in Supplementary Table 2.
RESULTS
Generating inducible NF1-deleted sarcomas in the mouse
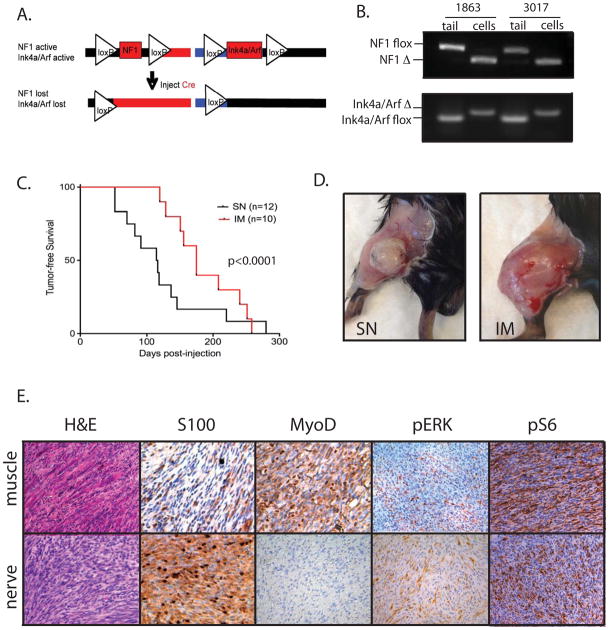
As NF1 mutations have been recently detected in a variety of human soft-tissue sarcoma subtypes (3, 4), we sought to determine if loss of NF1 and the tumor suppressor Ink4a/Arf was sufficient to generate soft-tissue sarcomas in a p53- and NF1-wild type background. Furthermore, we wished to generate an inducible mouse model of NF1-deleted sarcoma that could reflect the diverse spectrum of NF1-associated sarcomas found in patients. Therefore, we generated mice with conditional mutations in both NF1 and Ink4a/Arf (NF1flox/flox; Ink4a/Arfflox/flox). These mice were injected with an adenovirus expressing Cre recombinase (Ad-Cre) at one of two sites: intramuscular (IM) injections to model tumors arising in the skeletal muscle, and sciatic nerve (SN) injections to model tumors arising in the nerve sheath. Mice were injected into either the hind limb muscle (IM, n=10) or sciatic nerve (SN, n=12) of the lower left leg. Following injection, Cre recombinase can delete both alleles of NF1 and Ink4a/Arf (Figure 1A). Mice developed tumors 3–12 months after Cre exposure at the site of injection. We successfully derived two cell lines (1863 and 3017) from sarcomas generated after IM-injection of Ad-Cre. Using these sarcoma cell lines, we have confirmed recombination of both alleles of NF1 and Ink4a/Arf by PCR (Figure 1B), in addition to loss of both mRNA and protein for p16, p19, and NF1 (Supplemental Figure 1A–D). For clarity, we refer to sarcomas and cell lines that are deleted for NF1, p16, and p19 as “NF1-deleted”. These cell lines maintained an intact p53-dependent DNA damage response, as shown by elevation of p21 in response to doxorubicin treatment (Supplemental Figure 1E). We observed that the site of injection influences tumor initiation because sciatic nerve-derived tumors developed more quickly than intramuscularly-derived tumors (mean time to tumor, 4.1 vs 6.2 months, p<0.0001) (Figure 1C). Injection of Ad-Cre at both sites resulted in large, ovid tumors that appear well-vascularized (Figure 1C).
Figure 1. Characterization of a mouse model of NF1-deleted soft-tissue sarcoma.
A) Schematic of NF1 and Ink4a/Arf deletion following injection of an adenovirus expressing Cre recombinase. B) PCR demonstrating recombination of both NF1 and Ink4a/Arf floxed alleles in samples from paired tails and NF1-deleted sarcoma cell lines, derived from tumors generated by intramuscular injection of Ad-Cre. C) Kaplain-Meyer curve of tumor development based on site of orthotopic injection. The average time for tumor development is 4.1 months for mice injected in the sciatic nerve (SN) and 6.2 months for mice injected intramuscularly (IM). On right, photographs of NF1-deleted tumors generated from injections into the sciatic nerve (left) or muscle (right). D) Histopathology of IM and SN tumors. IM tumors resemble high-grade myogenic sarcomas and stain focally for MyoD1, but not S100. SN tumors resemble MPNSTs and stain focally for S100, but not for MyoD1. Both tumors are positive for pERK and pS6, indicating activity of the MAPK and mTOR pathways, respectively.
Histopathological analysis determined that these tumors reflect the histological properties and spectrum of disease seen in human NF1-associated sarcomas (Figure 1D). Sciatic nerve injections result in tumors with a fascicular pattern of tightly packed spindle cells surrounding the nerve (Supplemental Figure 2A). As seen in other mouse models of nerve sheath tumors (10, 22), these sarcomas demonstrate focal positivity for S100β, but do not show nuclear expression for the myogenic-lineage marker MyoD1. In contrast, IM injections generate a spectrum of high-grade spindle cell sarcomas, including rhabdomyosarcoma (RMS, n=5/10) and Undifferentiated Pleomorphic Sarcoma (UPS, n=5/10), the most common and aggressive form of soft tissue sarcoma in adults (Supplemental Figure 2B). Both RMS and UPS occur in patients with neurofibromatosis type 1 (6–8), and NF1 mutations have been identified in these sarcoma subtypes. Similar to other mouse models of myogenic sarcoma and their human tumor subtypes (14, 28), the IM tumors express the myogenic marker MyoD1, but do not stain focally for S100 β. NF1-associated sarcomas in patients have active RAS/MAPK/ERK and PI3K/AKT/mTOR signaling (29, 30). Therefore, we hypothesized that the MEK target ERK and the mTOR target S6 would be phosphorylated in these primary NF1-deleted mouse sarcomas. Immunohistochemical analysis for pERK and pS6 reveals that MAPK and mTOR pathways are active in both SN and IM primary tumors from NF1flox/flox; Ink4aflox/flox mice (Figure 1D). Taken together, these data suggest that this mouse model reflects the spectrum of NF1-deleted sarcomas found in the human population, and that distinct sarcoma subtypes can be generated from the same mouse strain by Ad-Cre injection into different orthotopic sites.
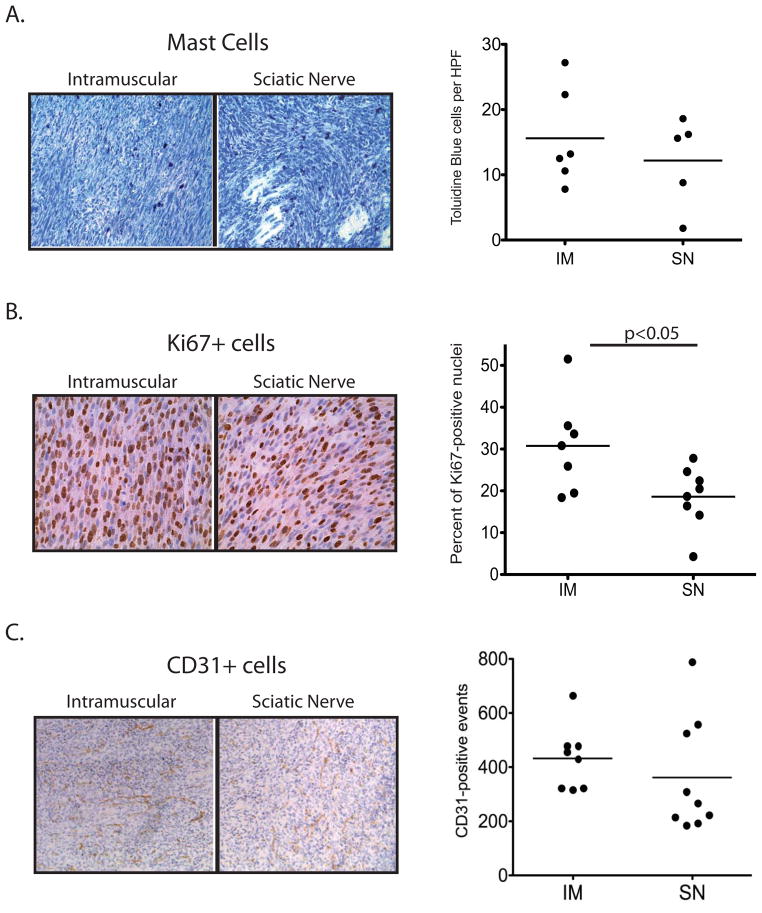
Importantly, these primary tumors develop from a small population of initiating tumor cells within the native tumor stroma, facilitating the study of endogenous immune cells, cellular growth, and native vasculature in a primary tumor context. Thus, we further characterized the similarities and differences between the sarcoma subtypes based on the site of Ad-Cre injection using histochemical analyses to examine mast cell infiltration, cell proliferation, and vascularization. First we examined mast cells, granule-rich inflammatory cells that reside within the connective tissue and are enriched in NF1-deleted neurofibromas and MPNSTs in patients (31). Infiltrating mast cells are important stromal factors that contribute to tumor formation in mouse models of NF1-deleted neurofibromas (22). In sarcomas from NF1flox/flox; Ink4aflox/flox mice, toludine blue staining shows significant infiltration of mast cells into both the sciatic nerve and intramuscular tumors. Mast cells were present at similar levels in both orthotopic injection models (Figure 2A). Ki67 staining shows a high level of proliferation in both intramuscular and sciatic nerve tumors. Quantification of Ki67-positive nuclei demonstrates that intramuscular tumors have a higher proliferative index than sciatic nerve tumors (p=0.04), although there is a wide range of proliferative index between individual tumors within each subtype (Figure 2B). In both tumor types, average CD31+ microvessel density is similar, although vessel distribution between individual tumors varies considerably (Figure 2C). Taken together, these data suggest that while this mouse model consistently develops tumors with distinct histological properties, there is individual tumor-to-tumor heterogeneity in phenotypes such as mast cell infiltration, proliferation and vascularity, with intramuscular tumors showing higher Ki67 levels. As human NF1-associated sarcomas also display heterogeneity by histological analysis (7), we hypothesize that the heterogeneity observed in this mouse model will be useful for testing therapies directed against NF1-deleted sarcoma in the preclinical setting.
Figure 2. Comparison of NF1-deleted sarcomas initiated by intramuscular (IM) or intra-sciatic nerve (SN) injection of Ad-Cre.
A) Mast cells, visualized by toludine blue, infiltrate both IM and SN tumors at similar levels. Quantification represents the average number of cells per 20x field, with 10 fields counted per tumor. B) Ki67+ proliferative index is higher in IM tumors compared to SN tumors (p<0.05). Quantification represents the percentage of Ki67+ cells normalized to total nuclei per 40x field, with 6 fields counted per tumor. C) Microvessel density, visualized by CD31+ cells, is similar between IM and SN tumors. Quantification represents the average number of cells per 20x field, with 6 fields counted per tumor.
Efficacy of MEK inhibition in NF1-deleted sarcomas
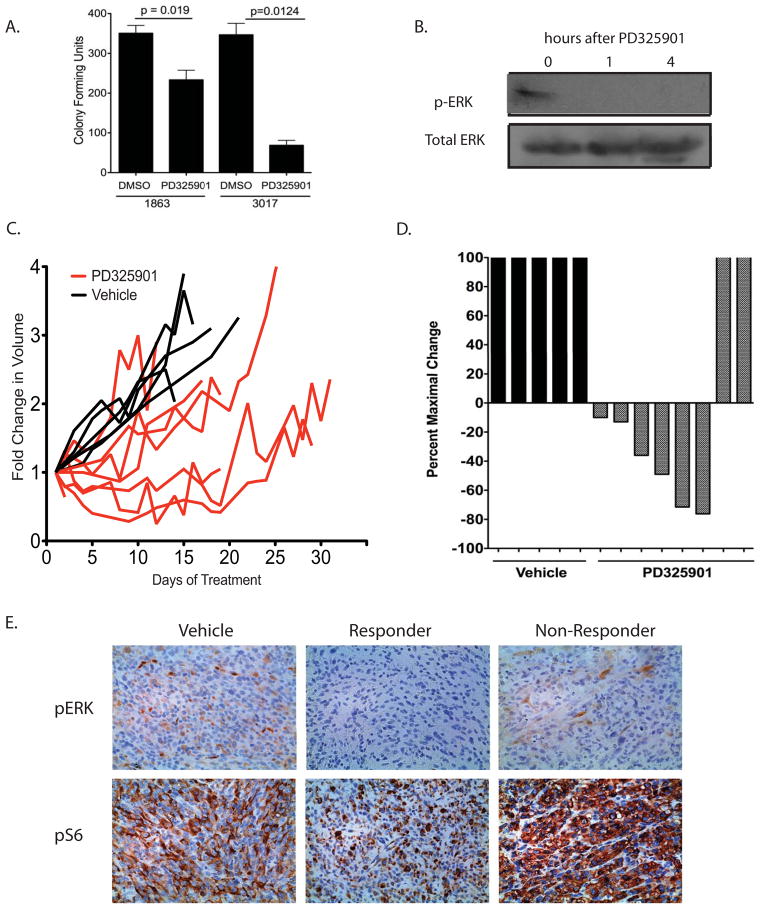
The identification of NF1 mutations in myogenic sarcoma (3, 4) provides a new foundation for preclinical studies examining targeted therapies for soft-tissue sarcomas. To determine the utility of this novel mouse model in examining therapeutic response, we hypothesized that NF1-deleted myogenic sarcomas would respond to MAPK inhibition based on two lines of evidence: (1) the role of NF1 as a negative regulator of Ras/MAPK/ERK signaling and (2) the elevation of pERK in myogenic sarcomas generated from the NF1flox/flox; Ink4aflox/flox mice. To determine the effect of MEK inhibition on cell proliferation in vitro, we generated cell lines (1863 and 3017) from NF1-deleted sarcomas that developed following IM injection of Adeno-Cre into NF1flox/flox; Ink4aflox/flox mice. We treated the cells with the MEK inhibitor PD325901 and observed decreases in colony-forming units and cell proliferation following exposure to the drug (Figure 3A and Supplemental Figure 3A). Similar results were seen when cells were treated with PD325901 prior to seeding for colony formation to account for cell-cell interaction signals and growth factors that are usually present in sub-confluent conditions and in the tumor microenvironment (Supplemental Figure 3B). Western blot analysis of 1863 cells treated with PD325901 for 4 hours showed a decrease in phosphorylation of ERK (Figure 3B), with corresponding changes in phospho-S6 and phospho-AKT levels (Supplemental Figure 4).
Figure 3. Activity of a MEK inhibitor in NF1-deleted sarcomas.
A) NF1-deleted cell lines (1863 and 3017) were exposed to either DMSO-alone or the MEK inhibitor PD325901 (50nM) for 7 days in culture. The number of colonies formed after 7 days was determined by crystal violet staining. B) Western blot showing inhibition of pERK levels following 4 hour exposure of 1863 cells to MEK inhibitor PD325901 (50nM). C) Primary high-grade myogenic sarcomas generated from intramuscular injection of Ad-Cre into NF1flox/flox; Ink4a/Arf flox/flox mice were treated with vehicle alone or PD325901. Exposure to PD325901 slowed tumor growth in a majority of tumors, as showed on the left by the fold change in tumor volume following initiation of treatment. On right, a waterfall plot shows the absolute change in tumor volume for primary NF1-deleted myogenic sarcomas following vehicle or PD325901 treatment. For tumors that showed a partial response to treatment, the maximal percent loss in tumor volume is reported. For tumors that did not respond to treatment, the maximal percent gain in tumor volume is reported. D) Immunohistochemistry of pERK and pS6 levels in primary myogenic sarcomas that received vehicle-alone or PD325901. Tumors that responded to treatment show a decrease in pERK and pS6 levels compared to vehicle-alone tumors, whereas tumors that did not respond to treatment show a corresponding upregulation in both pERK and pS6 levels.
To extend these studies to the primary mouse model of NF1-deleted myogenic sarcoma, we compared mice receiving PD325901 treatment (n=8) to mice receiving vehicle alone (n=5). Treatment began when tumors reached 200–300 mm3, with mice receiving treatment 5 times per week. Data are presented as both absolute tumor volume and in a waterfall plot that displays maximal percent change in volume for each tumor following treatment (Figure 3C). For the waterfall plots, tumors that partially responded to treatment are shown as greatest percent loss in tumor volume. Tumors that did not respond to treatment are shown as greatest percent gain in tumor volume during treatment. As predicted, treatment with PD325901 delayed tumor growth in a majority of the mice, with a partial response rate of 75% (Figure 3C). Among the tumors that responded to treatment, the degree of response varied, which has been observed in other primary sarcoma models (20, 32). The range of treatment response for individual tumors during PD325901 exposure is similar to the individual tumor-to-tumor heterogeneity observed in Ki67 and CD31 staining (Figure 2). Immunohistochemical analysis reveals that tumors responding to treatment have lower levels of MAPK and mTOR signaling compared to vehicle-treated tumors, as determined by a decrease in pERK and pS6 levels (Figure 3D). In contrast, tumors that did not respond to treatment showed hyper-elevated levels of both pERK and pS6, suggesting that these tumors escaped from treatment by alternative activation of these pathways. These results show that MEK inhibition slows growth of NF1-deleted rhabdomyosarcoma and undifferentiated pleomorphic sarcoma, as the histologies for each of these high-grade sarcoma subtype were evenly distributed between vehicle treated, responders, and non-responding tumors.
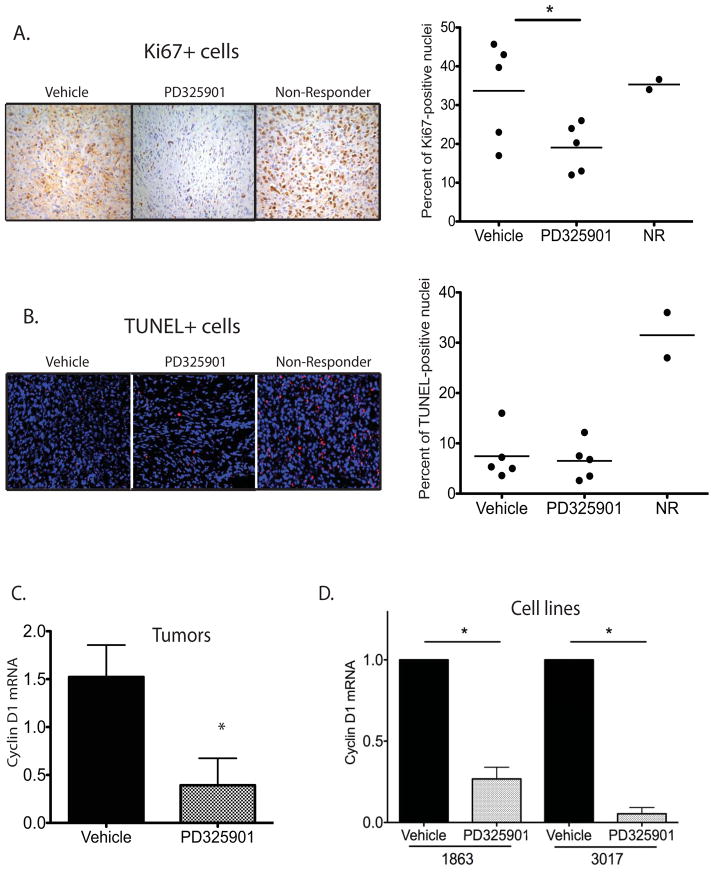
To determine the mechanism of tumor control produced by MEK inhibition, we examined the Ki67 index between vehicle and PD325901-treated tumors (Figure 4A). Tumors that responded to treatment show a decrease in proliferative index determined by Ki67 staining (p<0.05). Of note, the two non-responding (NR) tumors have Ki67 levels similar to vehicle-treated tumors, despite exposure to the inhibitor. TUNEL staining demonstrates that PD325901 treatment did not significantly increase apoptosis, although non-responders paradoxically show a high number of TUNEL-positive cells (Figure 4B). Tumors that responded to PD325901 treatment exhibit a significant decrease in levels of cyclin D1 mRNA (p<0.05), supporting previous reports that suggest inhibition of the cell cycle contributes to the decrease in tumor cell proliferation observed with this agent (33) (Figure 4C). Similar downregulation of cyclin D1 mRNA is observed during in vitro exposure of NF1-deleted sarcoma cells to PD325901, demonstrating that the cell cycle effects are cell autonomous (Figure 4D and Supplemental Figure 5). Taken together, these data suggest that PD325901 inhibits NF1-deleted sarcomas through cytostatic, but not cytotoxic, mechanisms. Very recently, others have used PD325901 to treat neurofibromatosis-associated diseases (34, 35), and treatment of a primary neurofibroma model with PD325901 resulted in decreased proliferation as measured by Ki67 staining, but not an increase in apoptosis (36). However, it is important to note that both the cellular context of the treated cancer and the specific target that is inhibited determine the effect of an agent to work in cytotoxic or cytostatic contexts.
Figure 4. NF1-deficient sarcomas respond to MEK inhibition by cytostatic mechanisms.
A) Ki67+ proliferative index is lower in tumors responding to PD325901 treatment than in vehicle-alone treated tumors (p<0.05). Tumors that did not respond to treatment (non-responders, NR) show Ki67+ levels similar to vehicle-alone treated tumors. Quantification represents the percentage of Ki67+ cells normalized to total nuclei per 40x field, with 6 fields counted per tumor. B) TUNEL staining determined that the number of apoptotic nuclei is not statistically different between vehicle-alone and PD325901-treated tumors. Tumors that did not respond to treatment (non-responders, NR) show an upregulation in TUNEL-positive cells. Quantification represents the percentage of TUNEL+ cells normalized to total nuclei per 40x field, with 6 fields counted per tumor. C) Quantitative RT-PCR demonstrates downregulation of cyclin D1 mRNA in NF1-deleted myogenic sarcomas responding to PD325901 treatment (n=4) in comparison to vehicle-alone treated tumors (n=4), (p<0.05). D) Quantitative RT-PCR demonstrates downregulation of cyclin D1 mRNA in sarcoma cell lines following a 4 hour treatment with 50nm PD325901 (p<0.05). Data represents an average of 4 independent experiments.
MEK inhibition alters the tumor microenvironment
The effect of targeted therapies on the surrounding tumor stroma is an important component of therapeutic efficacy. Recently, the MEK inhibitor PD325901 was shown to inhibit angiogenesis and VEGF signaling in xenografted tumor models of RCC and lung cancer (37, 38). While these xenograft models provide valuable information about tumor response, we sought to extend these findings to our NF1-deleted sarcoma model to determine the effect of MEK inhibition on the tumor stroma of primary immunocompetent tumors with native vasculature. First, we examined the effect of MEK inhibition on mast cell infiltration. Elevated levels of mast cells correlate with tumor aggressiveness in NF1-mutant neurofibromas (22). The role of mast cells following targeted therapy has not been examined in a primary sarcoma model, and reports of their prognostic or supportive role in other cancer types are conflicting (39–42). In our NF1-deleted sarcomas, toluidine blue staining demonstrates that tumors treated with either vehicle-alone or PD325901 have similar levels of mast cell infiltration (Figure 5A). Thus, mast cell levels were not altered by MEK inhibition in this model.
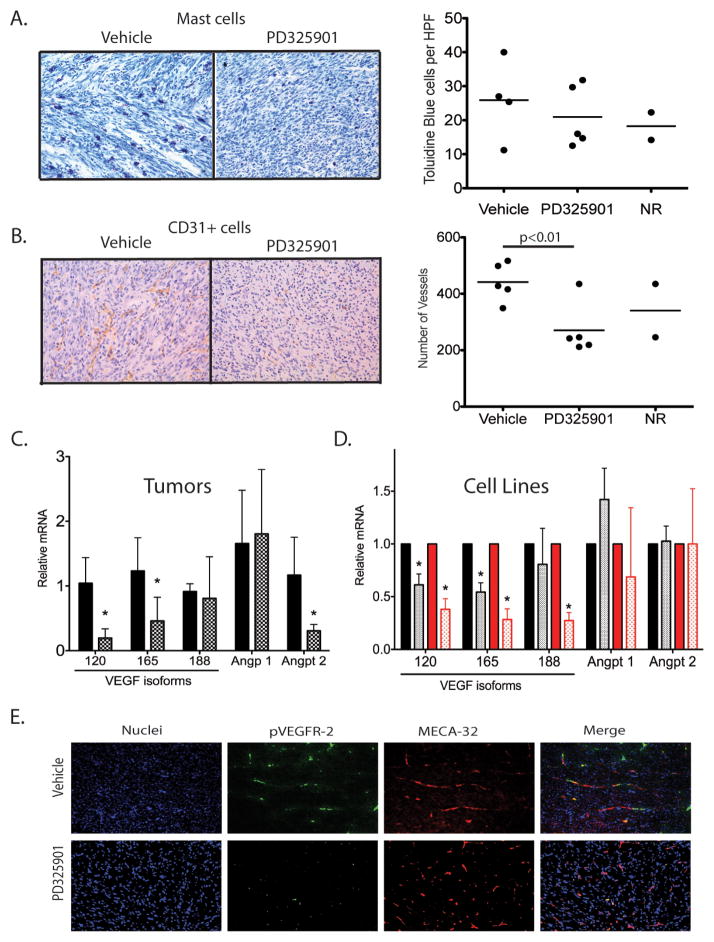
Figure 5. PD325901 alters vessel number and expression of angiogenic genes.
A) Mast cells, visualized by toludine blue, infiltrate both vehicle-treated and PD325901-treated NF1-deleted sarcomas at similar levels. Quantification represents the average number of cells per 20x field, with 10 fields counted per tumor. B) Microvessel density, visualized by CD31+ cells, is significantly decreased in tumors responding to PD325901 treatment (p<0.010). Quantification represents the average number of cells per 20x field, with 6 fields counted per tumor. C) On left, quantitative RT-PCR for angiogenic genes in primary NF1-deleted myogenic sarcomas treated with vehicle-alone (black bars) or PD325901 (shaded bars). Data represents an average of 4 tumors per group. Levels of VEGF 120, VEGF 165, and Angpt2 mRNA were significantly decreased in tumors responding to PD325901 treatment. On right, quantitative RT-PCR for angiogenic genes in sarcoma cell lines (1863-black, 3017-red) following a 4 hour treatment with either vehicle-alone (solid bars) or 50nM PD325901 (shaded bars). Data represents an average of 4 independent experiments. Levels of VEGF isoforms [120 and 165 (1863 and 3017), and 188 (3017 only)], but not Angpt2 mRNA, were significantly decreased in cells exposed to PD325901. D) Co-immunofluoresence for pVEGFR-2-positive endothelial cells in NF1-deleted myogenic sarcomas. In vehicle-treated tumors, pVEGFR-2 foci (green) are abundant and found in association with multiple endothelial cells (red, MECA-32). However, in PD325901-treated tumors, pVEGFR-2 foci are less abundant and the endothelial cell network is less extensive.
In contrast, MEK inhibition results in a significant decrease in CD31+ microvessel density (p=0.010, Figure 5B). As similar effects on the endothelium have been observed with MEK inhibition in a primary model of neurofibroma (36), we further characterized the molecular reprogramming responsible for vascular changes in our primary high-grade sarcomas. To determine the changes in several angiogenic markers that may correlate with the decreased endothelial cell content in the tumors, we performed quantitative RT-PCR on vehicle-treated and PD325901-treated tumors. Expression of the mRNA for the VEGFα isoforms VEGF 120 and VEGF 165 was decreased in treated tumors, while message levels for the VEGF 188 isoform were not significantly altered (Figure 5C). Levels of Angiopoetin 2 (Angpt 2) mRNA were lower in treated tumors, but levels of other endothelial cell markers were unaffected, including Angiopoetin 1. Additionally, mRNA levels of other angiogenesis-related genes were unchanged, including Tie2, VegfR-1, VegfR-2, Pdgf-β, PdgfR, Neuropilin, Hif1α, and Hif2α (Supplemental Figure 6). Angiopoeitin 2 can have pro-angiogenic properties when combined with increased VEGFα expression (43). Therefore, decreased levels of Angpt2, VEGF 120, and VEGF 165 mRNA suggest a potential mechanism for the decreased vascularity seen with PD325901 exposure. To determine if the change in angiogenic genes following PD325901 treatment was primarily through action on the tumor cells or on tumor stroma, we treated NF1-deficient sarcoma cells with PD325901. Following a 4 hour treatment with the inhibitor, both cell lines exhibited decreased levels of VEGF 120 and VEGF 165 mRNA, similar to the primary tumors (Figure 5C). However, levels of Angiopoeitin 2 remain unchanged in the PD325901-treated tumor cells. These data suggest that some of the decrease in VEGF mRNA levels following PD325901 treatment occurs in the sarcoma cells, but the changes in Angiopoeitin 2 levels observed in intact tumors reflect changes in the surrounding tumor stroma, perhaps in response to altered VEGF levels in the tumor cells (44).
To determine if the decreased levels in VEGF 120, VEGF 165, and Angiopoeitin 2 mRNA in the primary tumors correspond to decreased VEGFα signaling in vivo, we performed immunofluoresence for pVEGFR-2 in the primary tumors treated with PD325901 (Figure 5D). Upon VEGF ligand binding, the VEGFR-2 receptor on endothelial cells is phosporylated, activating the VEGF angiogenesis signaling cascade (45). In vehicle-treated tumors, pVEGFR-2 foci are abundant and colocalize with endothelial cells, visualized by MECA-32 staining. In contrast, PD325901-treated tumors show marked decrease in pVEGFR-2 foci, along with decay of endothelial cell architecture. These data suggest that in addition to decreasing proliferation of tumor cells, MEK inhibition has important effects on the surrounding tumor microenvironment by blocking angiogenesis and decreasing tumor microvessel density. Taken together, these preclinical data suggest that the MEK inhibitor PD325901 may be a useful therapy for patients with NF1-deficient sarcomas.
DISCUSSION
We have described a novel genetically engineered inducible mouse model of NF1-deficient sarcomas. This primary mouse model can be used to study the tumor biology and therapeutic response of sarcomas with NF1 mutation. Using temporally and spatially controlled conditional gene deletion, we have determined that inactivation of NF1 and Ink4a/Arf in the tumor cells is sufficient to initiate both myogenic and MPNST-like sarcomas in mice with a wild-type stroma. High-grade myogenic sarcomas develop after intramuscular injection of adenovirus expressing Cre recombinase, and MPNST-like sarcomas develop after sciatic nerve injection. Histopathological analysis shows the spectrum of sarcomas that develop in this model are similar to sarcomas with NF1 deletion in the patient population. By initiating sarcomas through deletion of NF1 and Ink4a/Arf, these mice model the ~50% of NF1-deleted sarcomas with wild-type p53 and complement the existing models of p53-deleted NF1 mutant sarcomas for therapeutic studies (21). In this model, tumor growth is delayed by the MEK inhibitor PD325901, which is accompanied by a decrease in tumor cell proliferation but does not increase cellular apoptosis. Importantly, in this model tumors develop within their native microenvironment, facilitating the study of stromal contribution to therapeutic response, which may be more challenging in cell transplant or xenograft studies. PD325901 treatment decreases microvessel density, at least partially through a decrease in expression of proangiogenic genes, such as VEGF 120, VEGF 165 and Angiopoeitin 2. Therefore, this study demonstrates the ability of this system to model NF1-deleted sarcomas and shows its utility in characterizing targeted therapies for NF1-deficient sarcomas. Several recent studies have also reported encouraging results using MEK inhibition to treat other neurofibromin-deficient disorders in mice (34–36).
To our knowledge, this is the first study to examine the biology of NF1-deleted high-grade myogenic sarcomas in genetically engineered mouse models, despite the prevalence of these tumors in the general population and in NF1 patients. Histological analysis shows that neither tumor location nor sarcoma subtype influences the average level of mast cell infiltration or the extent of vascularization. However, these properties are fairly heterogeneous between individual tumors, despite the fact that they all have identical initiating mutations and represent similar sarcoma subtypes. We observed a difference in Ki67 proliferative index between IM and SN subtypes, although individual tumors display a broad range of proliferative capacity within each group. This heterogeneity is a salient feature of primary tumor systems (46, 47) that permit modeling of diverse biology and may reflect differences in additional acquired somatic mutations and/or the influence of the complex microenvironment.
Indeed, the heterogeneity observed across primary tumors make them excellent models for examining differing tumor responses to therapy, and this model facilitated examination of tumors that were resistant to PD325901 treatment. Alternatively, these results can be viewed from the perspective that tumors may have sufficient phenotypic variation to account for the failure of treatment in non-responders. Although this diversity of response could be viewed as a limitation, we view it as a strength of the model. Although we are not able to determine if these tumors possessed a primary resistance to MEK inhibition or acquired resistance rapidly following the initial treatment, examining tumors that did not respond to PD325901 provides potential mechanisms to circumvent MEK inhibition. First, the resistant tumors have hyper-elevated Ki67 staining, suggesting they were able to grow through the treatment with increased cellular proliferation—even when accompanied by increased cellular apoptosis—resulting in a net gain in tumor growth. Alternatively, the increased apoptotic rate in non-responding tumors may be a reflection of a relatively low apoptotic rate of the responding tumors at this time point assayed. It is conceivable tumors that initially responded to therapies have elevated apoptosis during tumor response but have decreased apoptosis during tumor progression. Secondly, although all six tumors that responded to treatment showed low pERK and pS6 levels, the two tumors that did not respond to PD325901 treatment showed high levels of pERK and pS6, even above levels observed in vehicle-treated tumors. Thus, treatment may have been ineffective due to re-activation of these pathways. These results imply that blocking mTOR in addition to ERK would be a promising therapeutic strategy for future studies. Indeed, others have demonstrated that mTOR inhibition is effective in slowing growth of NF1-deficient MPNSTs in mouse models (21, 25, 48). However, we recognize that it can be challenging to define therapeutic windows for combination treatment of molecularly-targeted inhibitors in clinical trials. For example, recent reports have demonstrated that combination of the MEK inhibitor selumetinib with the AKT inhibitor MK-2206 results in increased toxicity when used at single-agent doses (49). Finally, the effect of PD325901 on the tumor stroma of non-responders was not well-defined, as changes in the levels of mast cells and microvessel density were varied. Although future studies will be needed to fully explore the mechanisms of resistance discussed here, this study demonstrates the utility of this mouse model to study the diversity of therapeutic responses found in NF1-deleted myogenic sarcoma.
This study examined the effects of MEK inhibition on the tumor cells and native surrounding microenvironment in a primary tumor model. Our results support a model in which PD325901 targets both the tumor cell itself and results in secondary effects on the tumor vasculature. Following MEK inhibition, the sarcoma cell decreases both cyclin D1 and VEGFα mRNA levels. Lower cyclin D1 mRNA levels decrease cell proliferation, as demonstrated by lower Ki67 levels. In parallel, lower VEGFα mRNA levels prevent activation of pVEGFR-2 on surrounding endothelial cells, ultimately lowering Angiopoeitin 2 mRNA levels in the vasculature. Together with decreased VEGFα, this results in decreased microvessel density. This model is consistent with data from previous in vitro and xenograft studies using MEK inhibitors in other tumors types (33, 37, 38). Alternatively, it is also possible that PD325901 could have direct effects on the tumor endothelial cells, which has been reported in vitro (50, 51).
In summary, we have developed a novel mouse model of NF1 deleted sarcoma with wild-type p53 status. The tumor spectrum observed after IM and SN injections of Ad-Cre is similar to the clinical spectrum of NF1-deleted sarcoma. We have used this model to assess the efficacy of PD325901 and to investigate its mechanism of action in a primary tumor model. We anticipate that this model will be useful as a pre-clinical platform for other novel therapies and for further studies to examine the interplay between the microenvironment and NF1-deficent sarcoma cells during therapeutic response.
Supplementary Material
Acknowledgments
GRANT SUPPORT
This work was supported by a Children’s Tumor Foundation Young Investigator Award (R.D. Dodd), a Liddy Shriver Sarcoma Initiative Grant (D.G. Kirsch and R.D. Dodd), and K02-AI093866 (D.G. Kirsch).
We thank Ron Dephino (MD Anderson) for providing the Ink4a/Arf flox mice. We thank Laura Jeffords, Rafaela Rodrigues, and Loretta Woodleif for assistance with mouse work. RDD is supported by postdoctoral fellowships from the American Cancer Society/Canary Foundation and the Children’s Tumor Foundation. This work is supported by RO1 CA 138265 (DGK).
Footnotes
The authors have no conflicts of interest to disclose.
References
- 1.Borden EC, Baker LH, Bell RS, Bramwell V, Demetri GD, Eisenberg BL, et al. Soft tissue sarcomas of adults: state of the translational science. Clin Cancer Res. 2003:1941–56. [PubMed] [Google Scholar]
- 2.Taylor BS, Barretina J, Maki RG, Antonescu CR, Singer S, Ladanyi M. Advances in sarcoma genomics and new therapeutic targets. Nat Rev Cancer. 2011:541–57. doi: 10.1038/nrc3087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barretina J, Taylor BS, Banerji S, Ramos AH, Lagos-Quintana M, Decarolis PL, et al. Subtype-specific genomic alterations define new targets for soft-tissue sarcoma therapy. Nature genetics. 2010:715–21. doi: 10.1038/ng.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Paulson V, Chandler G, Rakheja D, Galindo RL, Wilson K, Amatruda JF, et al. High-resolution array CGH identifies common mechanisms that drive embryonal rhabdomyosarcoma pathogenesis. Genes, chromosomes & cancer. 2011:397–408. doi: 10.1002/gcc.20864. [DOI] [PubMed] [Google Scholar]
- 5.Evans DG, Baser ME, McGaughran J, Sharif S, Howard E, Moran A. Malignant peripheral nerve sheath tumours in neurofibromatosis 1. J Med Genet. 2002:311–4. doi: 10.1136/jmg.39.5.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ferrari A, Bisogno G, Macaluso A, Casanova M, D’Angelo P, Pierani P, et al. Soft-tissue sarcomas in children and adolescents with neurofibromatosis type 1. Cancer. 2007:1406–12. doi: 10.1002/cncr.22533. [DOI] [PubMed] [Google Scholar]
- 7.Sorensen SA, Mulvihill JJ, Nielsen A. Long-term follow-up of von Recklinghausen neurofibromatosis. Survival and malignant neoplasms. N Engl J Med. 1986:1010–5. doi: 10.1056/NEJM198604173141603. [DOI] [PubMed] [Google Scholar]
- 8.McKeen EA, Bodurtha J, Meadows AT, Douglass EC, Mulvihill JJ. Rhabdomyosarcoma complicating multiple neurofibromatosis. J Pediatr. 1978:992–3. doi: 10.1016/s0022-3476(78)81233-5. [DOI] [PubMed] [Google Scholar]
- 9.Birindelli S, Perrone F, Oggionni M, Lavarino C, Pasini B, Vergani B, et al. Rb and TP53 pathway alterations in sporadic and NF1-related malignant peripheral nerve sheath tumors. Lab Invest. 2001:833–44. doi: 10.1038/labinvest.3780293. [DOI] [PubMed] [Google Scholar]
- 10.Wu J, Williams JP, Rizvi TA, Kordich JJ, Witte D, Meijer D, et al. Plexiform and dermal neurofibromas and pigmentation are caused by Nf1 loss in desert hedgehog-expressing cells. Cancer Cell. 2008:105–16. doi: 10.1016/j.ccr.2007.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhu Y, Romero MI, Ghosh P, Ye Z, Charnay P, Rushing EJ, et al. Ablation of NF1 function in neurons induces abnormal development of cerebral cortex and reactive gliosis in the brain. Genes Dev. 2001:859–76. doi: 10.1101/gad.862101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Joseph NM, Mosher JT, Buchstaller J, Snider P, McKeever PE, Lim M, et al. The loss of Nf1 transiently promotes self-renewal but not tumorigenesis by neural crest stem cells. Cancer Cell. 2008:129–40. doi: 10.1016/j.ccr.2008.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.King D, Yang G, Thompson MA, Hiebert SW. Loss of neurofibromatosis-1 and p19(ARF) cooperate to induce a multiple tumor phenotype. Oncogene. 2002:4978–82. doi: 10.1038/sj.onc.1205632. [DOI] [PubMed] [Google Scholar]
- 14.Kirsch DG, Dinulescu DM, Miller JB, Grimm J, Santiago PM, Young NP, et al. A spatially and temporally restricted mouse model of soft tissue sarcoma. Nature medicine. 2007:992–7. doi: 10.1038/nm1602. [DOI] [PubMed] [Google Scholar]
- 15.Cichowski K, Shih TS, Schmitt E, Santiago S, Reilly K, McLaughlin ME, et al. Mouse models of tumor development in neurofibromatosis type 1. Science. 1999:2172–6. doi: 10.1126/science.286.5447.2172. [DOI] [PubMed] [Google Scholar]
- 16.Vogel KS, Klesse LJ, Velasco-Miguel S, Meyers K, Rushing EJ, Parada LF. Mouse tumor model for neurofibromatosis type 1. Science. 1999:2176–9. doi: 10.1126/science.286.5447.2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nishijo K, Chen QR, Zhang L, McCleish AT, Rodriguez A, Cho MJ, et al. Credentialing a preclinical mouse model of alveolar rhabdomyosarcoma. Cancer research. 2009:2902–11. doi: 10.1158/0008-5472.CAN-08-3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haldar M, Hancock JD, Coffin CM, Lessnick SL, Capecchi MR. A conditional mouse model of synovial sarcoma: insights into a myogenic origin. Cancer Cell. 2007:375–88. doi: 10.1016/j.ccr.2007.01.016. [DOI] [PubMed] [Google Scholar]
- 19.Dodd RD, Mito J, Kirsch DG. Animal models of soft-tissue sarcoma. Disease Models and Mechanisms. 2010:557–66. doi: 10.1242/dmm.005223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim S, Dodd RD, Mito JK, Ma Y, Kim Y, Riedel RF, et al. Efficacy of phosphatidylinositol-3 kinase (PI3K) inhibitors in a primary mouse model of undifferentiated pleomorphic sarcoma (UPS) Sarcoma. 2012 doi: 10.1155/2012/680708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johannessen CM, Johnson BW, Williams SM, Chan AW, Reczek EE, Lynch RC, et al. TORC1 is essential for NF1-associated malignancies. Curr Biol. 2008:56–62. doi: 10.1016/j.cub.2007.11.066. [DOI] [PubMed] [Google Scholar]
- 22.Yang FC, Ingram DA, Chen S, Zhu Y, Yuan J, Li X, et al. Nf1-dependent tumors require a microenvironment containing Nf1+/− and c-kit-dependent bone marrow. Cell. 2008:437–48. doi: 10.1016/j.cell.2008.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abraham J, Nelon LD, Kubicek CB, Kilcoyne A, Hampton ST, Zarzabal LA, et al. Preclinical testing of erlotinib in a transgenic alveolar rhabdomyosarcoma mouse model. Sarcoma. 2011:130484. doi: 10.1155/2011/130484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu J, Dombi E, Jousma E, Scott Dunn R, Lindquist D, Schnell BM, et al. Preclincial testing of sorafenib and RAD001 in the Nf(flox/flox) ;DhhCre mouse model of plexiform neurofibroma using magnetic resonance imaging. Pediatr Blood Cancer. 2012:173–80. doi: 10.1002/pbc.23015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ghadimi MP, Lopez G, Torres KE, Belousov R, Young ED, Liu J, et al. Targeting the PI3K/mTOR Axis, Alone and in Combination with Autophagy Blockade, for the Treatment of Malignant Peripheral Nerve Sheath Tumors. Mol Cancer Ther. 2012:1758–69. doi: 10.1158/1535-7163.MCT-12-0015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aguirre AJ, Bardeesy N, Sinha M, Lopez L, Tuveson DA, Horner J, et al. Activated Kras and Ink4a/Arf deficiency cooperate to produce metastatic pancreatic ductal adenocarcinoma. Genes Dev. 2003:3112–26. doi: 10.1101/gad.1158703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brown AP, Carlson TC, Loi CM, Graziano MJ. Pharmacodynamic and toxicokinetic evaluation of the novel MEK inhibitor, PD0325901, in the rat following oral and intravenous administration. Cancer Chemother Pharmacol. 2007:671–9. doi: 10.1007/s00280-006-0323-5. [DOI] [PubMed] [Google Scholar]
- 28.Rubin BP, Nishijo K, Chen HI, Yi X, Schuetze DP, Pal R, et al. Evidence for an unanticipated relationship between undifferentiated pleomorphic sarcoma and embryonal rhabdomyosarcoma. Cancer Cell. 2011:177–91. doi: 10.1016/j.ccr.2010.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lau N, Feldkamp MM, Roncari L, Loehr AH, Shannon P, Gutmann DH, et al. Loss of neurofibromin is associated with activation of RAS/MAPK and PI3-K/AKT signaling in a neurofibromatosis 1 astrocytoma. J Neuropathol Exp Neurol. 2000:759–67. doi: 10.1093/jnen/59.9.759. [DOI] [PubMed] [Google Scholar]
- 30.Gregorian C, Nakashima J, Dry SM, Nghiemphu PL, Smith KB, Ao Y, et al. PTEN dosage is essential for neurofibroma development and malignant transformation. Proceedings of the National Academy of Sciences of the United States of America. 2009:19479–84. doi: 10.1073/pnas.0910398106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Donhuijsen K, Sastry M, Volker B, Leder LD. Mast cell frequency in soft tissue tumors. Relation to type and grade of malignancy. Pathol Res Pract. 1992:61–6. doi: 10.1016/s0344-0338(11)81157-x. [DOI] [PubMed] [Google Scholar]
- 32.De Raedt T, Walton Z, Yecies JL, Li D, Chen Y, Malone CF, et al. Exploiting cancer cell vulnerabilities to develop a combination therapy for ras-driven tumors. Cancer Cell. 2011:400–13. doi: 10.1016/j.ccr.2011.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Halilovic E, She QB, Ye Q, Pagliarini R, Sellers WR, Solit DB, et al. PIK3CA mutation uncouples tumor growth and cyclin D1 regulation from MEK/ERK and mutant KRAS signaling. Cancer research. 2010:6804–14. doi: 10.1158/0008-5472.CAN-10-0409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Staser K, Park SJ, Rhodes SD, Zeng Y, He YZ, Shew MA, et al. Normal hematopoiesis and neurofibromin-deficient myeloproliferative disease require Erk. J Clin Invest. :329–34. doi: 10.1172/JCI66167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chang T, Krisman K, Theobald EH, Xu J, Akutagawa J, Lauchle JO, et al. Sustained MEK inhibition abrogates myeloproliferative disease in Nf1 mutant mice. J Clin Invest. :335–9. doi: 10.1172/JCI63193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jessen WJ, Miller SJ, Jousma E, Wu J, Rizvi TA, Brundage ME, et al. MEK inhibition exhibits efficacy in human and mouse neurofibromatosis tumors. J Clin Invest. :340–7. doi: 10.1172/JCI60578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Takahashi O, Komaki R, Smith PD, Jurgensmeier JM, Ryan A, Bekele BN, et al. Combined MEK and VEGFR inhibition in orthotopic human lung cancer models results in enhanced inhibition of tumor angiogenesis, growth, and metastasis. Clin Cancer Res. 2012:1641–54. doi: 10.1158/1078-0432.CCR-11-2324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang D, Ding Y, Luo WM, Bender S, Qian CN, Kort E, et al. Inhibition of MAPK kinase signaling pathways suppressed renal cell carcinoma growth and angiogenesis in vivo. Cancer research. 2008:81–8. doi: 10.1158/0008-5472.CAN-07-5311. [DOI] [PubMed] [Google Scholar]
- 39.Tanaka T, Ishikawa H. Mast cells and inflammation-associated colorectal carcinogenesis. Semin Immunopathol. 2012 Sep 20; doi: 10.1007/s00281-012-0343-7. [DOI] [PubMed] [Google Scholar]
- 40.Xia Q, Wu XJ, Zhou Q, Jing Z, Hou JH, Pan ZZ, et al. No relationship between the distribution of mast cells and the survival of stage IIIB colon cancer patients. J Transl Med. 2011;9:88. doi: 10.1186/1479-5876-9-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chichlowski M, Westwood GS, Abraham SN, Hale LP. Role of mast cells in inflammatory bowel disease and inflammation-associated colorectal neoplasia in IL-10-deficient mice. PloS one. 2010:e12220. doi: 10.1371/journal.pone.0012220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gulubova M, Vlaykova T. Prognostic significance of mast cell number and microvascular density for the survival of patients with primary colorectal cancer. J Gastroenterol Hepatol. 2009:1265–75. doi: 10.1111/j.1440-1746.2007.05009.x. [DOI] [PubMed] [Google Scholar]
- 43.Lobov IB, Brooks PC, Lang RA. Angiopoietin-2 displays VEGF-dependent modulation of capillary structure and endothelial cell survival in vivo. Proceedings of the National Academy of Sciences of the United States of America. 2002:11205–10. doi: 10.1073/pnas.172161899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Felcht M, Luck R, Schering A, Seidel P, Srivastava K, Hu J, et al. Angiopoietin-2 differentially regulates angiogenesis through TIE2 and integrin signaling. J Clin Invest. 2012:1991–2005. doi: 10.1172/JCI58832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Carmeliet P, Jain RK. Molecular mechanisms and clinical applications of angiogenesis. Nature. 2011:298–307. doi: 10.1038/nature10144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen Z, Cheng K, Walton Z, Wang Y, Ebi H, Shimamura T, et al. A murine lung cancer co-clinical trial identifies genetic modifiers of therapeutic response. Nature. 2012 Mar 29;483(7391):613–7. doi: 10.1038/nature10937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Singh M, Lima A, Molina R, Hamilton P, Clermont AC, Devasthali V, et al. Assessing therapeutic responses in Kras mutant cancers using genetically engineered mouse models. Nat Biotechnol. :585–93. doi: 10.1038/nbt.1640. [DOI] [PubMed] [Google Scholar]
- 48.Johansson G, Mahller YY, Collins MH, Kim MO, Nobukuni T, Perentesis J, et al. Effective in vivo targeting of the mammalian target of rapamycin pathway in malignant peripheral nerve sheath tumors. Mol Cancer Ther. 2008:1237–45. doi: 10.1158/1535-7163.MCT-07-2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Speranza RJK, Khin S, Weil MK, Do KT, Horneffer Y, Juwara L, Allen D, Williams PM, Lih CJ, et al. Pharmacodynamic biomarker-driven trial of MK-2206, and AKT inhibitor, with AZD6244 (selumetinib), a MEK inhibitor, in patients with advanced colorectal carcinoma. J Clin Oncol. 2012 [Google Scholar]
- 50.Vart RJ, Nikitenko LL, Lagos D, Trotter MW, Cannon M, Bourboulia D, et al. Kaposi’s sarcoma-associated herpesvirus-encoded interleukin-6 and G-protein-coupled receptor regulate angiopoietin-2 expression in lymphatic endothelial cells. Cancer research. 2007:4042–51. doi: 10.1158/0008-5472.CAN-06-3321. [DOI] [PubMed] [Google Scholar]
- 51.Mavria G, Vercoulen Y, Yeo M, Paterson H, Karasarides M, Marais R, et al. ERK-MAPK signaling opposes Rho-kinase to promote endothelial cell survival and sprouting during angiogenesis. Cancer Cell. 2006 Jan;:33–44. doi: 10.1016/j.ccr.2005.12.021. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.