Abstract
Background
A great deal has been learned over the last several decades about the function of Ras proteins in solution and membrane environments. While much of this knowledge has been derived from a plethora of experimental techniques, computer simulations have also played a substantial role.
Scope of Review
Our goal here is to summarize the contribution of molecular simulations to our current understanding of normal and aberrant Ras function. We focus on lessons from molecular dynamics simulations in aqueous and membrane environments.
Major Conclusions
The central message is that a close interaction between theory and simulation on the one hand and cell-biological, spectroscopic and other experimental approaches on the other has played, and will likely continue to play, a vital role in Ras research.
General Significance
Atomistic insights emerging from detailed simulations of Ras in solution and in bilayers may be the key to unlock the secret that to date prevented development of selective anti-Ras inhibitors for cancer therapy.
Keywords: Molecular dynamics, advanced simulations, protein motion, membrane binding, clustering, oncogenic Ras
Introduction
Ras (Rat Sarcoma) protein was discovered more than four decades ago as the first oncogene product [1, 2]. Subsequent discoveries of many other related genes gave rise to the Ras family of proteins, a group of lipid-modified and membrane-associated intracellular switches that regulate cell growth, proliferation and differentiation [3, 4]. The switching function of Ras involves cycling between a GDP-bound `off' and GTP-bound `on' conformational states [5–8]. However, this binary on/off picture is being challenged by the discovery of other (intermediate) conformational states in recent years [9–14]. Efficient cycling between the on/off states of Ras requires GDP release and GTP hydrolysis facilitated by guanine nucleotide exchange factors (GEFs) and GTPase activating proteins (GAPs) [7, 15], respectively. For instance, GAP increases the very slow intrinsic ability of Ras to hydrolyze GTP (kcat = 0.028 min−1 [16]) by about 105-fold [17]. Therefore, interaction with GAPs is crucial for turning off signal transmission. Somatic or germline Ras mutations that interfere with its intrinsic and/or GAP-assisted ability to hydrolyze GTP can result in uncontrolled cell growth or cancer [18]. In fact, Ras is found mutated in about 15% of all human tumors and in up to 90% of cases in specific tumor types [19], as well as in a number of developmental disorders [20, 21]. Therefore, a staggering number of biochemical (e.g., [22–24]), structural [25–34], spectroscopic [14, 35–38] and theoretical [39–56] studies have been devoted to investigating the mechanistic aspects of the Ras GTPase function.
Our goal here is to summarize the contribution of molecular simulations to our current understanding of normal and aberrant Ras function. We focus on lessons from molecular mechanical simulations in aqueous and membrane environments. Though ab initio simulations with various flavors of quantum mechanics continue to play a central role in studying the catalytic process within the active site of Ras [57–70], they are beyond the scope of the current review. The review is organized as follows. As a background, we first provide a general overview of the accumulated knowledge on Ras. We then turn to simulations of the soluble catalytic domain in aqueous media, followed by the isolated lipid anchor and the full-length protein in lipid bilayers. Given the large number of reports in the field, we could not cite them all and apologize to those authors whose work was left out due to space limitation.
Overview of Ras biology, biochemistry and structure
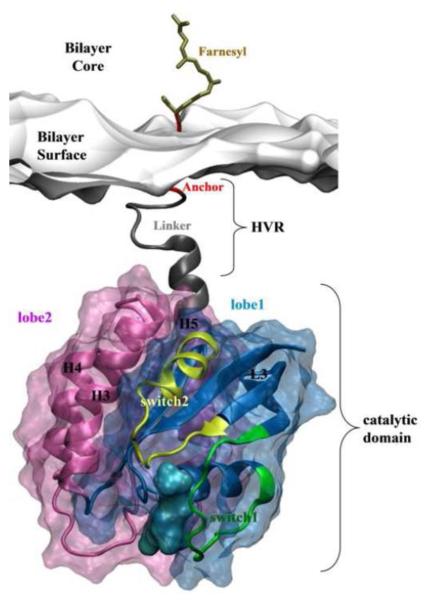
There are three major Ras isoforms in humans: N-, H- and K-Ras. These isoforms share a nearly identical water-soluble catalytic domain [7, 71] comprising the N-terminal residues 1–166. The catalytic domain can be subdivided into two lobes [42]. Lobel (residues 1–86) is strictly conserved across the Ras family and contains the functionally critical switch regions (switch 1: residues 25–40 and switch 2: residues 57–75), as well as the phosphate binding P-loop (residues 10–17) (Fig 1, also see Fig 2). Lobe2 (residues 87–166) is also structurally conserved among the Ras isoforms despite sequence variations at several positions (Fig 1), and is attached to the C-terminal hypervariable region (HVR). The HVR contains the linker segment (residues 167–179) followed by a lipid-modified membrane-binding motif (or the lipid anchor, residues 180–185/6). The HVR is believed to be responsible for the distinct sub-cellular and plasma membrane distribution and therefore diverse biological activity of Ras isoforms [39, 72–78].
Figure 1.
Structure of Ras. Lobe1 (residues 1–86) and lobe2 (residues 87–166) of the catalytic domain as well as the surface of a model monolayer to which Ras is bound are shown in blue, pink and grey surfaces, respectively. Different regions of Ras are labeled and colored in green (switch1 residues 25–40), yellow (switch2 residues 57–75), grey (linker residues 167–179) and red (lipid anchor residues 180–185). The linker and lipid anchor together constitute the hypervariable region (HVR). The structure shown here is K-Ras, which is posttranslationally modified by a single farnesyl lipid (ochre). N-Ras and H-Ras share a nearly identical catalytic domain with K-Ras, but differ in sequence at the HVR and are further lipid-modified by one and two palmitoyls, respectively. Also, their lipid anchor is longer by 1 amino acid. The GTP nucleotide is shown in a cyan surface representation.
Figure 2.
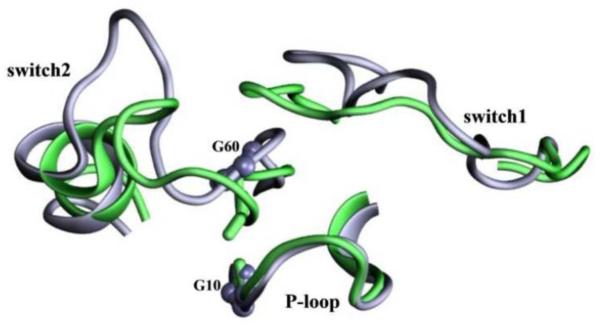
An example of large conformational changes at the switch regions of K-Ras sampled during classical molecular dynamics simulations. Also highlighted are residues Gly10 and Gly60 used to define two reaction coordinates (the N-Cα-C-O dihedral of Gly60 and distance between Gly10-Gly60 Cα atoms) that helped classify Ras structures into active, inactive, intermediate and nucleotide-free [see ref. 52 for details].
To date, over fifty-five H- and eleven K-Ras crystal structures have been deposited in the Protein Data Bank (PDB). Most of these structures were solved in the presence of GTP analogues or GDP and typically encompass residues of the catalytic domain only, with the exception of the recently reported ligand-bound K-Ras structures [79] that include the linker. Since the isolated catalytic domain of different Ras isoforms exhibits similar in vitro biochemical activity, much of the earlier structural work has focused only on H-Ras [27, 28, 31, 80–83]. Even with the recent addition of K-Ras structures, however, the non-redundant [84–87] biological activity of Ras proteins could not be fully explained in structural terms. Recent reports on the mechanism by which the HVR might modulate function has begun to shed light on this issue [72, 76–78].
Membrane binding is required for the biological function of Ras, and there is evidence that signaling specificity among Ras isoforms may be due to their distinct membrane localization and therefore differential accessibility to effectors and modulators [88–91]. A combined analysis of data from molecular dynamics (MD) simulations. Fluorescence Resonance Energy Transfer (FRET), Fluorescent Lifetime Imaging (FLIM) and functional assays has shown that GTP-H-Ras undergoes a global conformational change so that the catalytic domain directly contacts the membrane surface [43, 92, 93]. Moreover, a fraction of membrane-bound Ras proteins form dynamic clusters that are cholesterol-dependent (GDP-H-Ras and GTP-N-Ras) or -independent (GTP-H-Ras, GDP-N-Ras and K-Ras); i.e., different Ras isoforms form distinct, non-overlapping nanoclusters in an activation state-dependent manner [73, 94–96]. This differential membrane-organization explains, at least in part, the functional diversity among Ras proteins [93, 97]. However, our understanding of the molecular/atomic interactions underlying membrane binding and clustering of Ras is limited. To fill some of this gap, molecular simulations of various resolutions have been used to probe the dynamics, allosteric character and role of each structural component of Ras for isoform-specific membrane binding and assembly.
Classical MD simulations of Ras in solution
The earliest unbiased MD simulations of Ras [98–102] were very short (typically 100–500 ps) by current standards but they were able to elucidate the flexible nature of the nucleotide-binding loops (for illustration, see Fig 2 from more recent simulations). These initial studies thus helped explain why the conformation of these loops, if observed at all, differ between X-ray structures of Ras solved with GDP and GTP analogues [25, 27, 82, 83]. Another slightly longer (1 ns) simulation suggested that the active site of the oncogenic G12V mutant of H-Ras is more dynamic than the wild type [103]. Displacement of the catalytic water molecule during the simulation of another oncogenic mutant (G12D) was suggested to be one of the reasons that this variant is unable to hydrolyze GTP [104]. Comparison of multiple 10–20ns long MD trajectories on homology models of the N- and K-Ras catalytic domain with that of H-Ras suggested that the three isoforms, as well as mutant and wild type H-Ras, may differ in dynamics [42]. For instance, the amplitude of fluctuations in the atomic position of residues at the canonical switch loops, helix3 and loop7 of K-Ras were found to be larger and more similar to A59G H-Ras than wild type H-Ras [105]. Interestingly, crystallographic studies have also suggested that the flexible loop7 is part of an allosteric site [33].
Based on principal component analysis of all the crystal structures available at the time, Gorfe et al [42] has shown that the structure of G12V-H-Ras lies outside (and roughly in-between) two major groupings that are populated by structures of GDP- and GTP (or analogue)-bound Ras. X-ray structures of other Ras variants harboring mutation at the switch loops were also found to be intermediate between the canonical active and inactive states [9, 13], consistent with other observations by spectroscopic methods [12, 14, 38]. It is tempting to speculate that the energy barrier between the active and inactive states of some oncogenic variants of Ras may be lower than that in the wild type.
The simulations in ref. [42] also predicted an isoform-specific communication pathway connecting the nucleotide-binding lobe1 with the C-terminal lobe2. Subsequent simulations by another group found similar pathways and showed that these inter-lobe communications vanish in the absence of a nucleotide [53–55]. The same group also showed that transition between the inactive and active states (or vice versa) is facilitated by the higher flexibility of an intermediate state, and that this phenomenon is conserved in the entire Ras superfamily [54]. An evolutionarily conserved intrinsic flexibility of lobe1, and the nucleotide-binding site in particular, was suggested to be important for function conservation. In contrast, inter-lobe communication (i.e. lobe1-lobe2 crosstalk) was associated with functional specialization in members of a GTPase family [54]. In general, inter-lobe dynamics is a function of the bound nucleotide, with the inactive GDP-Ras being more flexible than the active GTP-Ras. This suggests that enhanced flexibility may facilitate GEF-binding and/or transition between the active and inactive states or vice versa [55].
In addition to backbone conformational changes in switches 1 and 2, transition from the active to the inactive state of Ras also involves loss of interaction between the backbone amide nitrogen of Gly60 and the y-phosphate of GTP, as well as the side chain hydroxyl of Thr35 and the Mg2+ ion [106–108]. GTP-bound Ras itself can also adopt two distinct forms: state1 and state2 [12, 106–110]. State1 differs from state2 primarily by the lack of coordination between Mg2+ and Thr35 [106–108]. Side chains of Tyr32 and Tyr64 also adopt a somewhat intermediate orientation in state1 GTP-Ras relative to their orientation in GDP-Ras and state2 GTP-Ras. Due likely to these reorganizations and the flexibility of the effector-binding loop, state1 Ras has weak affinity for effectors [12]. In fact, state1 may be an intermediate between the inactive GDP-bound and the active GTP-bound state2 [106].
Tightly bound water molecules appear to play an important role in the relative stability of the active, inactive and intermediate states of Ras. This was the conclusion of a recent study based on a series of simulations on Q61H K-Ras, with and without selected structural water molecules [52]. Analysis of the trajectories using two previously uncharacterized reaction coordinates (the distance between the Ca atoms of Gly60 and Gly10 and the N-Ca-C-O dihedral of Gly60 (see Fig 2)) revealed that a few strategically placed water molecules act as allosteric ligands that stabilize the state2 conformation of GTP-bound K-Ras [52]. The absence of some of these water molecules, particularly a single water molecule that bridges a hydrogen bond network between residues at the two lobes, induces a shift toward an inactive conformation [52].
A number of simulations have also been carried out on effector- [111–113] and GAP-bound [114, 115] Ras. For example, it was suggested that GAP-induced structural reorganization within the active site of Ras enhances GTP hydrolysis [114], even though the simulations were too short (~1ns) to draw an unambiguous conclusion. Furthermore, a combined electron spin-echo envelope modulation (ESEEM) spectroscopy and a short (~50ps) MD simulation showed movement of Gly residues, particularly Gly13 and Gly60, towards Mg2+ in the presence of GAP [116]. Taken together, these results underscore the key role of dynamics in the GTPase function of Ras. Similarly, protein motion plays an important role in the interaction of Ras with effectors [113].
Enhanced and biased simulations of Ras in solution
The atomically detailed classical MD (cMD) simulations described in the previous section provided invaluable insights into the dynamics of wild type and mutant Ras. However, computational cost rarely allowed for running cMD simulations long enough to sample large timescale global motions. Such motions have been probed by enhanced or biased simulation approaches such as accelerated MD (aMD) [117] and targeted MD (tMD) [118, 119]. For instance, nucleotide-dependent spontaneous transition between the active and inactive states of wild-type Ras has been achieved by aMD [117], since aMD allows for crossing large energy-barriers inaccessible to cMD [120]. Analysis of the aMD trajectories enabled the identification of several intermediate conformations that differ from the canonical GDP- and GTP-bound structures [117]. Moreover, aMD simulations of nucleotide-free Ras sampled a variety of conformational states including the canonical active and inactive states [121]. Based on this and other observations, it was proposed that conformational selection and population shift, and therefore allostery, might play an important role in the GTPase function of Ras and related G-proteins [121]. Though further study is required to fully establish the relative role of conformational selection and induced-fit [122], the allosteric nature of Ras is now fairly well established [33, 34, 123–125].
More than a decade ago, two groups used tMD to probe the transition path between the active and inactive states of H-Ras [118, 119]. A number of important predictions, some of which subsequently validated by experiments [9, 11], were made. These include the substantial reorientation of Tyr32, Arg68 and Tyr71 [119], and the involvement of the P-loop, the P2-P3 turn (or loop 3) and the helix3-loop7 region in state-to-state transitions [118]. These studies thus suggested that collective motions in the nucleotide-binding region could be felt by distal regions on lobe2, in agreement with subsequent findings by cMD and aMD [42, 117] as well as experiments [126, 127]. Most importantly, the tMD studies revealed that state-to-state transition involves multiple energy-barriers and therefore more than one transition state. Coarse-grain simulation approaches, such as Normal Modes Analysis (NMA), have also been used to study large-scale collective motions in Ras [128]. Among other things, NMA has shown that GTP-Ras is more rigid than GDP-Ras and that phosphate release is coupled with helix3 motion.
Atomistic simulations of Ras in membranes
In addition to characterizing the dynamics of the Ras catalytic domain in solution, cMD has played a central role in providing structural insights into bilayer-bound Ras [40, 41, 43–45, 47, 129, 130]. The simulation results were generally consistent with the available, albeit limited, experimental data from solid state NMR and other spectroscopic techniques [130–133]. One of the important observations from simulations of N- [45, 129, 130], H- [40, 41] and K-Ras [47] lipid anchors was that there is a delicate balance between hydrophobic and hydrophilic interactions that govern the positioning of the peptides on bilayer surfaces (Fig 3). In the case of the N-Ras lipid anchor, the backbone interacts with the lipid head group while the hydrophobic palmitoyl/farnesyl chains as well as side chains of Met182 and Leu184 interact with the hydrophobic interior of the bilayer [45, 129–133] (Fig 3A). This organization appears to be independent of the bilayer thickness [134]. Similarly, the polar residues of the H-Ras lipid anchor, Ser183 and Lys185, interact with the head group whereas the acyl chains of the lipid modified moieties and Met182 insert deep into the hydrophobic core of the bilayer [40, 41]. Note that the second palmitoyl in H-Ras is replaced by a Leu in N-Ras, so that the total number of the hydrophobic side chains in the two lipid anchors is the same. It is possible that this distribution of polar and apolar side chains ensures a parallel alignment of the backbone onto the membrane interface. Simulations of the K-Ras lipid anchor in a bilayer of zwitterionic POPC and negatively charged POPG lipids showed that the lysine residues immediately preceding the farnesylated cystein electrostatically interact with the POPG lipids while the farnesyl tail inserts into the hydrophobic core [47]. As a result, POPG lipids clustered around the peptides, as evidenced by analyzing time-dependent peptide-POPG hydrogen bonding and vdW interactions [47]. Insertion of the K-Ras lipid anchor brought about local changes in the bilayer structure, with the bilayer thickness near the peptides being shorter than in the rest of the bilayer or a peptide-free bilayer. A similar local bilayer perturbation was observed during MD simulations of N- and H-Ras lipid anchors in a DMPC bilayer [41, 45]. Unlike N- and H-Ras lipid anchors, however, the backbone of the K-Ras lipid anchor adopted a somewhat extended conformation with a pseudo-helical turn in the middle, and lies at an angle from the bilayer surface [47].
Figure 3.
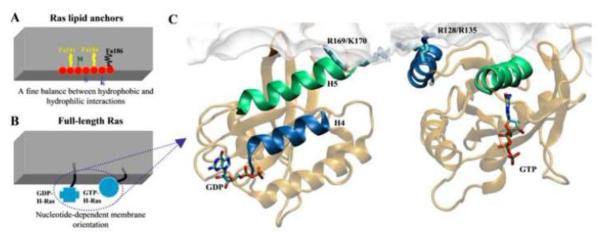
Schematic summary of the main results from atomistic MD simulations of Ras lipid anchors (A) and the full-length H-Ras (B & C). Fig A highlights the different organization of the backbone and side chains in the H-Ras lipid anchor (which has two palmitoyl (Pa) and a farnesyl (Fa) modifications). The schematic in Fig B summarizes the two different membrane-orientations of the catalytic domain observed during the simulations. Fig C shows the molecular details of GDP- and GTP-bound H-ras in a DMPC bilayer, highlighting the close contact of residues Arg169 and Lys170 of the linker and Arg128 and Arg135 of helix-4 with the bilayer, respectively. Note the different orientation of helix-4 (dark blue) and helix-5 (green) with respect to the membrane plane. Selected side chains and the nucleotide are shown in licorice with nitrogen in blue, oxygen in red, carbon in cyan and phosphate in ochre. A hypothetical bilayer surface (A and B) and a monolayer from simulations (C) are shown in grey.
Potential of mean force (PMF) calculations for the transfer of the H-Ras lipid anchor from water to bilayer yielded a large free energy gain of up to –30 kcal/mol [40, 44]. As expected, most of the contribution to the free energy of transfer came from vdW interactions between the acyl tails of the lipid anchor and those of the bilayer lipids. In fact, insertion of about half of the acyl tails of the lipid anchor into the hydrophobic core of the bilayer appears to suffice for crossing the barrier at the lipid-water interface [40, 41]. However, the interaction of polar side chains and the backbone with the head group of the bilayer lipids also contributes to binding [41]. Decomposition of the insertion free energy into enthalpic and entropic contributions led to a surprising conclusion: an enthalpy-dominated hydrophobic effect may underlie membrane binding of Ras proteins [135]. The hydrophobic effect is often associated with entropy though there are a few examples, discussed in the same paper, in which enthalpy plays a demonstrate role. Moreover, a similar insertion free energy was estimated from PMF calculations on monoand dually-palmitoylated H-Ras anchors [44], suggesting a non-additive contribution of the two palmitoyls for membrane binding. The implication of these and other PMF calculations on variants of the H-Ras lipid anchor, in which individual lipidations were systematically removed [44], was that palmitoylation at Cys184 may not be needed for affinity but rather for lateral segregation, as previously suggested [136].
Whilst the simulations on the isolated lipid anchors yielded useful insights into specific peptide-lipid atomic interactions, cMD simulations on full-length H-Ras in a DMPC bilayer indicated that the catalytic domain and the linker also play a role in membrane binding [43]. Membrane insertion and the overall organization of the anchor were similar among the isolated lipid anchor, the HVR and the full-length protein [43]. However, there were some structural differences. One of these was that the linker did not attain any specific conformation in the absence of the catalytic domain, but it adopted a coil-bend-coil geometry in the full-length protein [43]. Secondly, the linker interacts with the DMPC head group during simulations of GDP-H-Ras but it wrapped around the catalytic domain during GTP-H-Ras simulations. As a result, the backbone atoms of the lipid anchor were able to insert deeper in the former than in the latter [43]. Perhaps more significantly, the simulations of GDP-H-Ras and GTP-H-Ras in DMPC yielded two major modes of bilayer interaction (Fig 3). In one case, the catalytic domain directly interacts with and is roughly parallel to the bilayer surface. In the other case, the catalytic domain is oriented approximately perpendicular to the bilayer surface. In terms of population, GTP-bound H-Ras preferred the parallel orientation while perpendicular orientation was dominant in GDP-bound H-Ras (Fig 3B&C). Helices 4 and 5 are near the DMPC phosphates in the former whereas the N-terminal part of the linker (residues 167–172) and the β2-β3 turn make contact with the bilayer in the latter. Of special note may be the direct interaction of Arg128 and Arg135 with the phosphate head group in GTP-H-Ras; a similar interaction involving Arg169 and Lys170 exists in GDP-H-Ras (Fig 3C). These two pairs of positively charged residues thus seemed to be the key determinants of the nucleotide-dependent membrane binding of H-Ras [43]. This was found to be the case in subsequent FRET, FLIM/FRET and functional assays in cells expressing mutant Ras in which these pair of residues were separately mutated to Ala [92, 137, 138]. Recent spectroscopic studies of N-Ras in a synthetic lipid membrane [139] and the Ras-related Rheb in lipid nanodiscs [140] have observed similar nucleotide dependent membrane reorientation. Together, these results provided crucial insights into how the catalytic domain of Ras and related GTPases modulates function and how dynamics plays a critical role in the functional diversity of Ras isoforms. However, there is much more to learn about the (possibly many) different ways in which Ras proteins may interact with membrane lipids. A case in point is a recent report based on a combined cMD and spectroscopic analyses that proposed dimer formation by bilayer-bound N-Ras [141].
Coarse-grained simulations of Ras in membrane
Experiments in intact plasma membrane sheets [75] and synthetic bilayers [142, 143] have shown that Ras proteins assemble into dynamic clusters on membrane surfaces [72, 74, 75]. These nano-sized subdomains, or nanoclusters, are small (6–20nm radius) and contain about 7 proteins per cluster [72]. Different Ras isoforms form distinct and non-overlapping nanoclusters [94], with clusters of active H-Ras and K-Ras, as well as the isolated lipid anchor of K-Ras (tK), being localized at disordered membrane domains whereas clusters of inactive H-Ras and the lipid anchor of H-Ras (tH) segregate to raft-like membrane domains [142, 143]. That tK and tH form nancolusters with different domain preferences suggests the key role of the lipid-modified moiety in the formation and localization of Ras nanoclusters. However, the physical driving forces underlying Ras clustering are difficult to access by current experimental techniques or atomistic simulations. This is because nanoclusters are too dynamic to be captured by high-resolution experimental techniques and their assembly/disassembly span large spatiotemporal scales that are hard to sample by atomistic simulations. One alternative is to use coarse-grained MD (CGMD), which has been proven useful to study lipid domain formation at near atomic resolution (e.g. [144]).
CGMD has been applied on tH embedded in a phase-separating lipid mixture of DPPC, DLiPC and cholesterol (CHOL) [48]. This mixture spontaneously forms CHOL/DPPC-enriched liquid ordered (Lo) and DLiPC-enriched liquid disordered (Ld) domains that capture some of the key features of raft and non-raft-like domains, respectively [144–146]. A key result from these simulations was that, for a tH/lipid ratio of about 0.01 or higher, ~40% of the tH molecules form dynamic clusters of size 4–11 molecules [48, 50], which agrees remarkably well with experimental results [75]. The clusters accumulated at the Lo/Ld domain boundary, and their stability was found to be a function of the extent of lipid phase separation and hence domain stability [48]. As a result, decreasing the simulation temperature or increasing the cholesterol content of the bilayer led to slightly larger but significantly more stable clusters [48, 50]. Simulations in which individual lipid-modifications were systematically removed demonstrated that the segregation of the clusters to the domain boundary is a consequence of the opposite preference of the palmitoyl and farnesyl tails for the Lo and Ld domains, respectively [48] (Fig 4). Clusters of tH variants with only farnesyl modification segregated to the Ld domain whereas those with only palmitoyl modification preferred the Lo domain. It was also noted that the organization of tH within clusters allows for maximum inter-tH and tH-lipid interactions [48, 50].
Figure 4.
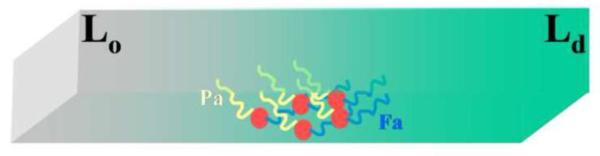
Schematic summary of microsecond scale CGMD showing clustering and localization of H-Ras lipid anchors between lipid domains. A hypothetical two-domain bilayer is shown in a gradient color with the greyish region representing the liquid order (Lo) domain and the greenish region representing the liquid disordered (Ld) domain. The H-ras lipid anchor is shown in red circle with a single palmitoyl tail in yellow and farnesyl in blue.
The effect of tH binding and clustering on the mechanical properties of the host bilayer was evaluated by computing line tension, lipid tilt and pressure profiles in the tH-bound and tH-free bilayers [48]. It was found that the line tension in the leaflet containing tH clusters was reduced relative to the corresponding monolayer of a tH-free bilayer [48]. Moreover, the asymmetric tH binding and interfacial localization caused a significant tilting of the boundary lipids and an asymmetric pressure profile, suggesting bilayer deformation.
The domain-preference of full-length H- and N-Ras proteins has also been studied by CGMD [147], in which GTP-H-Ras, GTP-N-Ras and depalmitoylated GTP-H-Ras were simulated in a bilayer made up of a similar lipid composition as that used in the tH simulations [48]. The conclusion was that GTP-H-Ras partitions into the Lo phase whereas GTP-N-Ras prefers the domain boundary. The latter is consistent with experiments [142] and simulations of tH variants containing a single palmitoyl and a farensyl [48]. It would be interesting to see if the domain preference of GDP-bound Ras would be different from that of GTP-Ras, as suggested by earlier experiments [148,149].
Perspective: Simulations can aid in anti-Ras inhibitor design
Decades of efforts by academia and industry have failed to yield selective Ras inhibitors. Complicating factors to directly targeting Ras include the conservation of the active site in a large number of small G-proteins and the high concentration and affinity of cellular GTP for Ras. Another reason could be lack of molecular-level insight into the protein-lipid interactions underlying the distinct spatiotemporal membrane-organization of different Ras proteins. The ultimate goal of the simulation approaches discussed in this review is to achieve a better understanding of Ras dynamics in solution and membrane environments, which is crucial for identifying and targeting novel pockets that transiently open during protein motion. In particular, conformers from MD trajectories can be used to find ligand binding hotspots that have distinct features in ensembles of active, inactive and intermediate states of Ras. Along this line, analysis of representative structures from MD simulations and screening of drug libraries against these conformers identified four putative ligand binding sites: two on either side of helix-2, one behind the flexible effector-binding loop and another site distal from the nucleotide-binding region [150]. Some of the small molecule ligands predicted to be targeting these sites were found to inhibit Ras signaling in cells expressing oncogenic mutant Ras [150]. While the ability of these ligands to bind Ras at the predicted sites has yet to be confirmed, a number of NMR and X-ray structures of Ras bound to several different small molecules or molecular fragments were recently reported [79, 151–153]. In some of these studies, initial hits were derived from screening of ligand libraries against less populated Ras conformations [152, 153]. Moreover, a common feature of all of the reported ligand-bound Ras structures is that a significant backbone and side chain reorganization underpins ligand binding [79, 151–153]. The key question is whether the structural changes were induced by the ligands or if the ligands selectively target specific preexisting conformations with transiently open allosteric sites. The majority of the simulation results discussed in this review and a recent report from our laboratory [46] support the latter. We are of the opinion that simulations have more to offer in future efforts to anti-cancer drug discovery. One example would be utilizing MD-derived structural ensembles of membrane-bound Ras as an initial step toward the development of isoform-selective Ras inhibitors. The inhibitory potential of ligands that interfere with the proper localization of Ras to the plasma membrane has been demonstrated in several recent reports [154–156].
Highlights
Contribution of molecular simulations to the study of Ras GTPases
Multi-scale molecular dynamics simulations of Ras in solution and in membrane
Dynamics plays an important role in the biological activity of Ras proteins
Implications of simulations and protein motion for anti-cancer Ras inhibitors
Acknowledgement
P.P. is supported by a postdoctoral training fellowship funded by the CPRIT Computational Cancer Biology Training Program (CCBTP) from the Cancer Prevention and Research Institute of Texas (CPRIT) (Grant No: RP101489). We thank the Texas Advanced Computing Center (TACC) for computational resources. This work is supported in part by grant from the National Institutes of Health General Medical Sciences (grant number R01GM100078).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Malumbres M, Barbacid M. RAS oncogenes: the first 30 years. Nat Rev Cancer. 2003;3:459–465. doi: 10.1038/nrc1097. [DOI] [PubMed] [Google Scholar]
- [2].Karnoub AE, Weinberg RA. Ras oncogenes: split personalities. Nat Rev Mol Cell Biol. 2008;9:517–531. doi: 10.1038/nrm2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Barbacid M. Ras Genes. Annu Rev Biochem. 1987;56:779–827. doi: 10.1146/annurev.bi.56.070187.004023. [DOI] [PubMed] [Google Scholar]
- [4].Bos JL. Ras oncogenes in human cancer: a review. Cancer Res. 1989;49:4682–4689. [PubMed] [Google Scholar]
- [5].Bourne HR, Sanders DA, McCormick F. The GTPase superfamily: a conserved switch for diverse cell functions. Nature. 1990;348:125–132. doi: 10.1038/348125a0. [DOI] [PubMed] [Google Scholar]
- [6].Sprang SR. G proteins, effectors and GAPs: structure and mechanism. Curr Opin Struct Biol. 1997;7:849–856. doi: 10.1016/s0959-440x(97)80157-1. [DOI] [PubMed] [Google Scholar]
- [7].Vetter IR, Wittinghofer A. The Guanine Nucleotide-Binding Switch in Three Dimensions. Science. 2001;294:1299–1304. doi: 10.1126/science.1062023. [DOI] [PubMed] [Google Scholar]
- [8].Wittinghofer A, Vetter IR. Structure-function relationships of the G domain, a canonical switch motif. Annu Rev Biochem. 2011;80:943–971. doi: 10.1146/annurev-biochem-062708-134043. [DOI] [PubMed] [Google Scholar]
- [9].Hall BE, Bar-Sagi D, Nassar N. The structural basis for the transition from Ras-GTP to Ras-GDP. Proc Natl Acad Sc. 2002;99:12138–12142. doi: 10.1073/pnas.192453199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Spoerner M, Wittinghofer A, Kalbitzer HR. Perturbation of the conformational equilibria in Ras by selective mutations as studied by 31P NMR spectroscopy. FEBS Lett. 2004;578:305–310. doi: 10.1016/j.febslet.2004.11.020. [DOI] [PubMed] [Google Scholar]
- [11].Ford B, Skowronek K, Boykevisch S, Bar-Sagi D, Nassar N. Structure of the G60A Mutant of Ras. J Biol Chem. 2005;280:25697–25705. doi: 10.1074/jbc.M502240200. [DOI] [PubMed] [Google Scholar]
- [12].Spoerner M, Nuehs A, Ganser P, Herrmann C, Wittinghofer A, Kalbitzer HR. Conformational states of Ras complexed with the GTP analogue GppNHp or GppCH2p: implications for the interaction with effector proteins. Biochemistry. 2005;44:2225–2236. doi: 10.1021/bi0488000. [DOI] [PubMed] [Google Scholar]
- [13].Ford B, Hornak V, Kleinman H, Nassar N. Structure of a Transient Intermediate for GTP Hydrolysis by Ras. Structure. 2006;14:427–436. doi: 10.1016/j.str.2005.12.010. [DOI] [PubMed] [Google Scholar]
- [14].Kalbitzer HR, Spoerner M, Ganser P, Hozsa C, Kremer W. Fundamental link between folding states and functional states of proteins. J Am Chem Soc. 2009;131:16714–16719. doi: 10.1021/ja904314q. [DOI] [PubMed] [Google Scholar]
- [15].Bos JL, Rehmann H, Wittinghofer A. GEFs and GAPs: critical elements in the control of small G proteins. Cell. 2007;129:865–877. doi: 10.1016/j.cell.2007.05.018. [DOI] [PubMed] [Google Scholar]
- [16].Temeles GL, Gibbs JB, D'Alonzo JS, Sigal IS, Scolnick EM. Yeast and mammalian ras proteins have conserved biochemical properties. Nature. 1985;313:700–703. doi: 10.1038/313700a0. [DOI] [PubMed] [Google Scholar]
- [17].Gideon P, John J, Freeh M, Lautwein A, Clark R, Scheffler JE, Wittinghofer A. Mutational and kinetic analyses of the GTPase-activating protein (GAP)-p21 interaction: the C-terminal domain of GAP is not sufficient for full activity. Mol Cell Biol. 1992;12:2050–2056. doi: 10.1128/mcb.12.5.2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Cox AD, Der CJ. Ras history: The saga continues. Small GTPases. 2010;1:2–27. doi: 10.4161/sgtp.1.1.12178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Prior IA, Lewis PD, Mattos C. A comprehensive survey of Ras mutations in cancer. Cancer Res. 2012;72:2457–2467. doi: 10.1158/0008-5472.CAN-11-2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Schubbert S, Shannon K, Bollag G. Hyperactive Ras in developmental disorders and cancer. Nat Rev Cancer. 2007;7:295–308. doi: 10.1038/nrc2109. [DOI] [PubMed] [Google Scholar]
- [21].Gripp KW, Lin AE. Costello syndrome: a Ras/mitogen activated protein kinase pathway syndrome (rasopathy) resulting from HRAS germline mutations. Genet Med. 2012;14:285–292. doi: 10.1038/gim.0b013e31822dd91f. [DOI] [PubMed] [Google Scholar]
- [22].Der CJ, Finkel T, Cooper GM. Biological and biochemical properties of human rasH genes mutated at codon 61. Cell. 1986;44:167–176. doi: 10.1016/0092-8674(86)90495-2. [DOI] [PubMed] [Google Scholar]
- [23].Schweins T, Geyer M, Scheffzek K, Warshel A, Kalbitzer HR, Wittinghofer A. Substrate-assisted catalysis as a mechanism for GTP hydrolysis of p21ras and other GTP-binding proteins. Nat Struct Biol. 1995;2:36–44. doi: 10.1038/nsb0195-36. [DOI] [PubMed] [Google Scholar]
- [24].Du X, Black GE, Lecchi P, Abramson FP, Sprang SR. Kinetic isotope effects in Ras-catalyzed GTP hydrolysis: evidence for a loose transition state. Proc Natl Acad Sci. 2004;101:8858–8863. doi: 10.1073/pnas.0401675101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Krengel U, Schlichting I, Scherer A, Schumann R, Freeh M, John J, Kabsch W, Pai EF, Wittinghofer A. Three-dimensional structures of H-ras p21 mutants: molecular basis for their inability to function as signal switch molecules. Cell. 1990;62:539–548. doi: 10.1016/0092-8674(90)90018-a. [DOI] [PubMed] [Google Scholar]
- [26].Milburn MV, Tong L, deVos AM, Brunger A, Yamaizumi Z, Nishimura S, Kim SH. Molecular switch for signal transduction: structural differences between active and inactive forms of protooncogenic ras proteins. Science. 1990;247:939–945. doi: 10.1126/science.2406906. [DOI] [PubMed] [Google Scholar]
- [27].Pai EF, Krengel U, Petsko GA, Goody RS, Kabsch W, Wittinghofer A. Refined crystal structure of the triphosphate conformation of H-ras p21 at 1.35 A resolution: implications for the mechanism of GTP hydrolysis. EMBO J. 1990;9:2351–2359. doi: 10.1002/j.1460-2075.1990.tb07409.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Prive GG, Milburn MV, Tong L, de Vos AM, Yamaizumi Z, Nishimura S, Kim SH. X-ray crystal structures of transforming p21 ras mutants suggest a transition-state stabilization mechanism for GTP hydrolysis. Proc Natl Acad Sci. 1992;89:3649–653. doi: 10.1073/pnas.89.8.3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Kraulis PJ, Domaille PJ, Campbell-Burk SL, Van Aken T, Laue ED. Solution Structure and Dynamics of Ras p21.cntdot.GDP Determined by Heteronuclear Three- and Four-Dimensional NMR Spectroscopy. Biochemistry. 1994;33:3515–3531. doi: 10.1021/bi00178a008. [DOI] [PubMed] [Google Scholar]
- [30].Ahmadian MR, Stege P, Scheffzek K, Wittinghofer A. Confirmation of the arginine-finger hypothesis for the GAP-stimulated GTP-hydrolysis reaction of Ras. Nat Struct Mol Biol. 1997;4:686–689. doi: 10.1038/nsb0997-686. [DOI] [PubMed] [Google Scholar]
- [31].Scheffzek K, Ahmadian MR, Kabsch W, Wiesmüller L, Lautwein A, Schmitz F, Wittinghofer A. The Ras-RasGAP Complex: Structural Basis for GTPase Activation and Its Loss in Oncogenic Ras Mutants. Science. 1997;277:333–339. doi: 10.1126/science.277.5324.333. [DOI] [PubMed] [Google Scholar]
- [32].Pasqualato S, Cherfils J. Crystallographic evidence for substrate-assisted GTP hydrolysis by a small GTP binding protein. Structure. 2005;13:533–540. doi: 10.1016/j.str.2005.01.014. [DOI] [PubMed] [Google Scholar]
- [33].Buhrman G, Holzapfel G, Fetics S, Mattos C. Allosteric modulation of Ras positions Q61 for a direct role in catalysis. Proc Natl Acad Sci. 2010;107:4931–4936. doi: 10.1073/pnas.0912226107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Buhrman G, Kumar VS, Cirit M, Haugh JM, Mattos C. Allosteric modulation of Ras-GTP is linked to signal transduction through RAF kinase. J Biol Chem. 2011;286 doi: 10.1074/jbc.M110.193854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Allin C, Ahmadian MR, Wittinghofer A, Gerwert K. Monitoring the GAP catalyzed H-Ras GTPase reaction at atomic resolution in real time. Proc Natl Acad Sci. 2001;98:7754–7759. doi: 10.1073/pnas.131549798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Rohrer M, Prisner TF, Brugmann O, Kass H, Spoerner M, Wittinghofer A, Kalbitzer HR. Structure of the metal-water complex in Ras x GDP studied by high-field EPR spectroscopy and 31P NMR spectroscopy. Biochemistry. 2001;40:1884–1889. doi: 10.1021/bi002164y. [DOI] [PubMed] [Google Scholar]
- [37].Iuga A, Spoerner M, Kalbitzer HR, Brunner E. Solid-state 3IP NMR spectroscopy of microcrystals of the Ras protein and its effector loop mutants: comparison between crystalline and solution state. J Mol Biol. 2004;342:1033–1040. doi: 10.1016/j.jmb.2004.07.077. [DOI] [PubMed] [Google Scholar]
- [38].Liao J, Shima F, Araki M, Ye M, Muraoka S, Sugimoto T, Kawamura M, Yamamoto N, Tamura A, Kataoka T. Two conformational states of Ras GTPase exhibit differential GTP-binding kinetics. Biochem Biophys Res Commun. 2008;369:327–332. doi: 10.1016/j.bbrc.2008.01.169. [DOI] [PubMed] [Google Scholar]
- [39].Gorfe AA. Mechanisms of allostery and membrane attachment in Ras GTPases: implications for anti-cancer drug discovery. Curr Med Chem. 2010;17:1–9. doi: 10.2174/092986710789957832. [DOI] [PubMed] [Google Scholar]
- [40].Gorfe AA, Babakhani A, McCammon JA. Free Energy Profile of H-ras Membrane Anchor upon Membrane Insertion. Angew Chem Int Ed Engl. 2007;46:8234–8237. doi: 10.1002/anie.200702379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Gorfe AA, Babakhani A, McCammon JA. H-ras protein in a bilayer: interaction and structure perturbation. J Am Chem Soc. 2007;129:12280–12286. doi: 10.1021/ja073949v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Gorfe AA, Grant BJ, McCammon JA. Mapping the nucleotide and isoform-dependent structural and dynamical features of Ras proteins. Structure. 2008;16:885–896. doi: 10.1016/j.str.2008.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Gorfe AA, Hanzal-Bayer M, Abankwa D, Hancock JF, McCammon JA. Structure and dynamics of the full-length lipid-modified H-Ras protein in a 1,2-dimyristoylglycero-3-phosphocholine bilayer. J Med Chem. 2007;50:674–684. doi: 10.1021/jm061053f. [DOI] [PubMed] [Google Scholar]
- [44].Gorfe AA, McCammon JA. Similar membrane affinity of mono- and Di-S-acylated ras membrane anchors: a new twist in the role of protein lipidation. J Am Chem Soc. 2008;130:12624–12625. doi: 10.1021/ja805110q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Gorfe AA, Pellarin R, Caflisch A. Membrane localization and flexibility of a lipidated ras peptide studied by molecular dynamics simulations. J Am Chem Soc. 2004;126:15277–15286. doi: 10.1021/ja046607n. [DOI] [PubMed] [Google Scholar]
- [46].Hocker HJ, Cho K-J, Chen; N. Rambahal C-YK, Sagineedu HC, Shaari K, Stanslas J, Hancock JF, Gorfe AA. Andrographolide derivative inhibit guanine nucleotide exchange and abrogate oncogenic Ras function. Proc Natl Acad Sei. 2013 doi: 10.1073/pnas.1300016110. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Janosi L, Gorfe AA. Segregation of negatively charged phospholipids by the polycationic and famesylated membrane anchor of Kras. Biophys J. 2010;99:3666–3674. doi: 10.1016/j.bpj.2010.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Janosi L, Li Z, Hancock JF, Gorfe AA. Organization, dynamics, and segregation of Ras nanoclusters in membrane domains. Proc Natl Acad Sei. 2012;109:8097–8102. doi: 10.1073/pnas.1200773109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Kosztin I, Bruinsma R, O'Lague P, Schulten K. Mechanical force generation by G proteins. Proc Natl Acad Sei. 2002;99:3575–3580. doi: 10.1073/pnas.052209199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Li Z, Janosi L, Gorfe AA. Formation and domain partitioning of H-ras peptide nanoclusters: effects of peptide concentration and lipid composition. J Am Chem Soc. 2012;134:17278–17285. doi: 10.1021/ja307716z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Noe F, Ille F, Smith JC, Fischer S. Automated computation of low-energy pathways for complex rearrangements in proteins: application to the conformational switch of Ras p21. Proteins. 2005;59:534–544. doi: 10.1002/prot.20422. [DOI] [PubMed] [Google Scholar]
- [52].Prakash P, Sayyed-Ahmad A, Gorfe AA. The role of conserved waters in conformational transitions of Q61H K-ras. PLoS Comput Biol. 2012;8:el002394. doi: 10.1371/journal.pcbi.1002394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Raimondi F, Felline A, Portella G, Orozco M, Fanelli F. Light on the structural communication in Ras GTPases. J Biomol Struct Dyn. 2013;31:142–157. doi: 10.1080/07391102.2012.698379. [DOI] [PubMed] [Google Scholar]
- [54].Raimondi F, Orozco M, Fanelli F. Deciphering the deformation modes associated with function retention and specialization in members of the Ras superfamily. Structure. 2010;18:402–414. doi: 10.1016/j.str.2009.12.015. [DOI] [PubMed] [Google Scholar]
- [55].Raimondi F, Portella G, Orozco M, Fanelli F. Nucleotide binding switches the information flow in ras GTPases. PLoS Comput Biol. 2011;7:e1001098. doi: 10.1371/journal.pcbi.1001098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Zeng J, Treutlein HR, Simonson T. Conformation of the Ras-binding domain of Raf studied by molecular dynamics and free energy simulations. Proteins. 1998;31:186–200. doi: 10.1002/(sici)1097-0134(19980501)31:2<186::aid-prot8>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- [57].Futatsugi N, Hata M, Hoshino T, Tsuda M. Ab Initio Study of the Role of Lysine 16 for the Molecular Switching Mechanism of Ras Protein p21. Biophys J. 1999;77:3287–3292. doi: 10.1016/S0006-3495(99)77159-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Glennon TM, Villa J, Warshel A. How does GAP catalyze the GTPase reaction of Ras? A computer simulation study. Biochemistry. 2000;39:9641–9651. doi: 10.1021/bi000640e. [DOI] [PubMed] [Google Scholar]
- [59].Grigorenko BL, Nemukhin AV, Shadrina MS, Topol IA, Burt SK. Mechanisms of guanosine triphosphate hydrolysis by Ras and Ras-GAP proteins as rationalized by ab initio QM/MM simulations. Proteins. 2007;66:456–466. doi: 10.1002/prot.21228. [DOI] [PubMed] [Google Scholar]
- [60].Grigorenko BL, Nemukhin AV, Topol IA, Cachau RE, Burt SK. QM/MM modeling the Ras-GAP catalyzed hydrolysis of guanosine triphosphate. Proteins. 2005;60:495–503. doi: 10.1002/prot.20472. [DOI] [PubMed] [Google Scholar]
- [61].Grigorenko BL, Rogov AV, Nemukhin AV. Mechanism of Triphosphate Hydrolysis in Aqueous Solution: QM/MM Simulations in Water Clusters. J Phys Chem B. 2006;110:4407–4412. doi: 10.1021/jp056395w. [DOI] [PubMed] [Google Scholar]
- [62].Klahn M, Schlitter J, Gerwert K. Theoretical IR spectroscopy based on QM/MM calculations provides changes in charge distribution, bond lengths, and bond angles of the GTP ligand induced by the Ras-protein. Biophys J. 2005;88:3829–3844. doi: 10.1529/biophysj.104.058644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Langen R, Schweins T, Warshel A. On the mechanism of guanosine triphosphate hydrolysis in ras p21 proteins. Biochemistry. 1992;31:8691–8696. doi: 10.1021/bi00152a002. [DOI] [PubMed] [Google Scholar]
- [64].Martin-Garcia F, Mendieta-Moreno JI, Lopez-Vinas E, Gomez-Puertas P, Mendieta J. The Role of Gln61 in HRas GTP hydrolysis: a quantum mechanics/molecular mechanics study. Biophys J. 2012;102:152–157. doi: 10.1016/j.bpj.2011.11.4005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Rudack T, Xia F, Schlitter J, Kotting C, Gerwert K. The role of magnesium for geometry and charge in GTP hydrolysis, revealed by quantum mechanics/molecular mechanics simulations. Biophys J. 2012;103:293–302. doi: 10.1016/j.bpj.2012.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Schweins T, Langen R, Warshel A. Why have mutagenesis studies not located the general base in ras p21. Nat Struct Biol. 1994;1:476–484. doi: 10.1038/nsb0794-476. [DOI] [PubMed] [Google Scholar]
- [67].te Heesen H, Gerwert K, Schlitter J. Role of the arginine finger in Ras.RasGAP revealed by QM/MM calculations. FEBS Lett. 2007;581:5677–5684. doi: 10.1016/j.febslet.2007.11.026. [DOI] [PubMed] [Google Scholar]
- [68].Topol IA, Cachau RE, Nemukhin AV, Grigorenko BL, Burt SK. Quantum chemical modeling of the GTP hydrolysis by the RAS-GAP protein complex. Biochim Biophys Acta. 2004;1700:125–136. doi: 10.1016/j.bbapap.2004.04.007. [DOI] [PubMed] [Google Scholar]
- [69].Shurkie A, Warshel A. Why does the Ras siwtch “break” by oncogenic mutations. Proteins. 1991;55:1–10. doi: 10.1002/prot.20004. [DOI] [PubMed] [Google Scholar]
- [70].Xia F, Rudack T, Cui Q, Kotting C, Gerwert K. Detailed structure of the H2P04(-)-guanosine diphosphate intermediate in Ras-GAP decoded from FTIR experiments by biomolecular simulations. J Am Chem Soc. 2012;134:20041–20044. doi: 10.1021/ja310496e. [DOI] [PubMed] [Google Scholar]
- [71].Valencia A, Chardin P, Wittinghofer A, Sander C. The ras protein family: evolutionary tree and role of conserved amino acids. Biochemistry. 1991;30:4637–4648. doi: 10.1021/bi00233a001. [DOI] [PubMed] [Google Scholar]
- [72].Hancock JF. Lipid rafts: contentious only from simplistic standpoints. Nat Rev Mol Cell Biol. 2006;7:456–462. doi: 10.1038/nrm1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Inder K, Harding A, Plowman SJ, Philips MR, Parton RG, Hancock JF. Activation of the MAPK module from different spatial locations generates distinct system outputs. Mol Biol Cell. 2008;19:4776–4784. doi: 10.1091/mbc.E08-04-0407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Parton RG, Hancock JF. Lipid rafts and plasma membrane microorganization: insights from Ras. Trends Cell Biol. 2004;14:141–147. doi: 10.1016/j.tcb.2004.02.001. [DOI] [PubMed] [Google Scholar]
- [75].Plowman SJ, Muncke C, Parton RG, Hancock JF. H-ras, K-ras, and inner plasma membrane raft proteins operate in nanoclusters with differential dependence on the actin cytoskeleton. Proc Natl Acad Sei. 2005;102:15500–15505. doi: 10.1073/pnas.0504114102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Wolfman A. Ras Isoform-Specific Signaling: Location, Location, Location. Science Signaling. 2001;2001:pe2. doi: 10.1126/stke.2001.96.pe2. [DOI] [PubMed] [Google Scholar]
- [77].Jaumot M, Yan J, Clyde-Smith J, Sluimer J, Hancock JF. The linker domain of the Ha-Ras hypervariable region regulates interactions with exchange factors, Raf-1 and phosphoinositide 3-kinase. J Biol Chem. 2002;277:272–278. doi: 10.1074/jbc.M108423200. [DOI] [PubMed] [Google Scholar]
- [78].Rotblat B, Prior IA, Muncke C, Parton RG, Kloog Y, Henis YI, Hancock JF. Three separable domains regulate GTP-dependent association of H-ras with the plasma membrane. Mol Cell Biol. 2004;24:6799–6810. doi: 10.1128/MCB.24.15.6799-6810.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Maurer T, Garrenton LS, Oh A, Pitts K, Anderson DJ, Skelton NJ, Fauber BP, Pan B, Malek S, Stokoe D, Ludlam MJ, Bowman KK, Wu J, Giannetti AM, Starovasnik MA, Mellman I, Jackson PK, Rudolph J, Wang W, Fang G. Small-molecule ligands bind to a distinct pocket in Ras and inhibit SOS-mediated nucleotide exchange activity. Proc Natl Acad Sei. 2012;109:5299–5304. doi: 10.1073/pnas.1116510109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Brunger AT, Milburn MV, Tong L, deVos AM, Jancarik J, Yamaizumi Z, Nishimura S, Ohtsuka E, Kim SH. Crystal structure of an active form of RAS protein, a complex of a GTP analog and the HRAS p21 catalytic domain. Proc Natl Acad Sei. 1990;87:4849–4853. doi: 10.1073/pnas.87.12.4849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Sondek J, Lambright DG, Noel JP, Hamm HE, Sigler PB. GTPase mechanism of Gproteins from the 1.7-A crystal structure of transducin [alpha] - GDP AIF-4. Nature. 1994;372:276–279. doi: 10.1038/372276a0. [DOI] [PubMed] [Google Scholar]
- [82].Tong LA, de Vos AM, Milburn MV, Kim SH. Crystal structures at 2.2 A resolution of the catalytic domains of normal ras protein and an oncogenic mutant complexed with GDP. J Mol Biol. 1991;217:503–516. doi: 10.1016/0022-2836(91)90753-s. [DOI] [PubMed] [Google Scholar]
- [83].Pai EF, Kabsch W, Krengel U, Holmes KC, John J, Wittinghofer A. Structure of the guanine-nucleotide-binding domain of the Ha-ras oncogene product p21 in the triphosphate conformation. Nature. 1989;341:209–214. doi: 10.1038/341209a0. [DOI] [PubMed] [Google Scholar]
- [84].Quinlan MP, Settleman J. Isoform-specific ras functions in development and cancer. Future Oncol. 2009;5:105–116. doi: 10.2217/14796694.5.1.105. [DOI] [PubMed] [Google Scholar]
- [85].Plowman SJ, Hancock JF. Ras signaling from plasma membrane and endomembrane microdomains. Biochim Biophys Acta. 2005;1746:274–283. doi: 10.1016/j.bbamcr.2005.06.004. [DOI] [PubMed] [Google Scholar]
- [86].Umanoff H, Edelmann W, Pellicer A, Kucherlapati R. The murine N-ras gene is not essential for growth and development. Proc Natl Acad Sci. 1995;92:1709–1713. doi: 10.1073/pnas.92.5.1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Koera K, Nakamura K, Nakao K, Miyoshi J, Toyoshima K, Hatta T, Otani H, Aiba A, Katsuki M. K-ras is essential for the development of the mouse embryo. Oncogene. 1997;15:1151–1159. doi: 10.1038/sj.onc.1201284. [DOI] [PubMed] [Google Scholar]
- [88].Prior IA, Hancock JF. Compartmentalization of Ras proteins. J Cell Sci. 2001;114:1603–1608. doi: 10.1242/jcs.114.9.1603. [DOI] [PubMed] [Google Scholar]
- [89].Omerovic J, Prior IA. Compartmentalized signalling: Ras proteins and signalling nanoclusters. FEBS J. 2009;276:1817–1825. doi: 10.1111/j.1742-4658.2009.06928.x. [DOI] [PubMed] [Google Scholar]
- [90].Hancock JF, Prior IA. Electron microscopic imaging of Ras signaling domains. Methods. 2005;37:165–172. doi: 10.1016/j.ymeth.2005.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Aran V, Prior IA. Compartmentalised Ras signaling differentially contributes to phenotypic outputs. Cell Signal. 2013 doi: 10.1016/j.cellsig.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Abankwa D, Hanzal-Bayer M, Ariotti N, Plowman SJ, Gorfe AA, Parton RG, McCammon JA, Hancock JF. A novel switch region regulates H-ras membrane orientation and signal output. EMBO J. 2008;27:727–735. doi: 10.1038/emboj.2008.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Abankwa D, Gorfe AA, Inder K, Hancock JF. Ras membrane orientation and nanodomain localization generate isoform diversity. Proc Natl Acad Sci. 2010;107:1130–1135. doi: 10.1073/pnas.0903907107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Abankwa D, Gorfe AA, Hancock JF. Ras nanoclusters: molecular structure and assembly. Semin Cell Dev Biol. 2007;18:599–607. doi: 10.1016/j.semcdb.2007.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Harding A, Hancock JF. Ras nanoclusters: combining digital and analog signaling. Cell Cycle. 2008;7:127–134. doi: 10.4161/cc.7.2.5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Tian T, Harding A, Inder K, Plowman S, Parton RG, Hancock JF. Plasma membrane nanoswitches generate high-fidelity Ras signal transduction. Nat Cell Biol. 2007;9:905–914. doi: 10.1038/ncb1615. [DOI] [PubMed] [Google Scholar]
- [97].Prior IA, Hancock JF. Ras trafficking, localization and compartmentalized signalling. Semin Cell Dev Biol. 2012;23:145–153. doi: 10.1016/j.semcdb.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Diaz JF, Wroblowski B, Engelborghs Y. Molecular dynamics simulation of the solution structures of Ha-ras-p21 GDP and GTP complexes: flexibility, possible hinges, and levers of the conformational transition. Biochemistry. 1995;34:12038–12047. doi: 10.1021/bi00037a047. [DOI] [PubMed] [Google Scholar]
- [99].Foley CK, Pedersen LG, Charifson PS, Darden TA, Wittinghofer A, Pai EF, Anderson MW. Simulation of the solution structure of the H-ras p21-GTP complex. Biochemistry. 1992;31:4951–4959. doi: 10.1021/bi00136a005. [DOI] [PubMed] [Google Scholar]
- [100].Chen JM, Lee G, Murphy RB, Carty RP, Brandt-Rauf PW, Friedman E, Pincus MR. Comparison of the computed structures for the phosphate-binding loop of the p21 protein containing the oncogenic site Gly 12 with the X-ray crystallographic structures for this region in the p21 protein and EFtu. A model for the structure of the p21 protein in its oncogenic form. J Biomol Struct Dyn. 1989;6:859–875. doi: 10.1080/07391102.1989.10506518. [DOI] [PubMed] [Google Scholar]
- [101].Mello LV, van Aalten DM, Findlay JB. Comparison of ras-p21 bound to GDP and GTP: differences in protein and ligand dynamics. Protein Eng. 1997;10:381–387. doi: 10.1093/protein/10.4.381. [DOI] [PubMed] [Google Scholar]
- [102].Dykes DC, Friedman FK, Dykes SL, Murphy RB, Brandt-Rauf PW, Pincus MR. Molecular dynamics of the H-ras gene-encoded p21 protein; identification of flexible regions and possible effector domains. J Biomol Struct Dyn. 1993;11:443–458. doi: 10.1080/07391102.1993.10508009. [DOI] [PubMed] [Google Scholar]
- [103].Futatsugi N, Tsuda M. Molecular dynamics simulations of Gly-12—>Val mutant of p21(ras): dynamic inhibition mechanism. Biophys J. 2001;81:3483–3488. doi: 10.1016/S0006-3495(01)75979-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Friedman ZY, Devary Y. Dissection of the GTPase mechanism of Ras protein by MD analysis of Ras mutants. Proteins. 2005;59:528–533. doi: 10.1002/prot.20423. [DOI] [PubMed] [Google Scholar]
- [105].Lukman S, Grant BJ, Gorfe AA, Grant GH, McCammon JA. The distinct conformational dynamics of K-Ras and H-Ras A59G. PLoS Comput Biol. 2010;6 doi: 10.1371/journal.pcbi.1000922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Kobayashi C, Saito S. Relation between the conformational heterogeneity and reaction cycle of Ras: molecular simulation of Ras. Biophys J. 2010;99:3726–3734. doi: 10.1016/j.bpj.2010.09.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Matsumoto K, Shima F, Muraoka S, Araki M, Flu L, Ijiri Y, Flirai R, Liao J, Yoshioka T, Kumasaka T, Yamamoto M, Tamura A, Kataoka T. Critical roles of interactions among switch I-preceding residues and between switch II and its neighboring alpha-helix in conformational dynamics of the GTP-bound Ras family small GTPases. J Biol Chem. 2011;286:15403–15412. doi: 10.1074/jbc.M110.204933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Shima F, Ijiri Y, Muraoka S, Liao J, Ye M, Araki M, Matsumoto K, Yamamoto N, Sugimoto T, Yoshikawa Y, Kumasaka T, Yamamoto M, Tamura A, Kataoka T. Structural basis for conformational dynamics of GTP-bound Ras protein. J Biol Chem. 2010;285:22696–22705. doi: 10.1074/jbc.M110.125161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Araki M, Shima F, Yoshikawa Y, Muraoka S, Ijiri Y, Nagahara Y, Shirono T, Kataoka T, Tamura A. Solution structure of the state 1 conformer of GTP-bound H-Ras protein and distinct dynamic properties between the state 1 and state 2 conformers. J Biol Chem. 2011;286:39644–39653. doi: 10.1074/jbc.M111.227074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Ye M, Shima F, Muraoka S, Liao J, Okamoto H, Yamamoto M, Tamura A, Yagi N, Ueki T, Kataoka T. Crystal structure of M-Ras reveals a GTP-bound “off” state conformation of Ras family small GTPases. J Biol Chem. 2005;280:31267–31275. doi: 10.1074/jbc.M505503200. [DOI] [PubMed] [Google Scholar]
- [111].Ensign DL, Webb LJ. Factors determining electrostatic fields in molecular dynamics simulations of the Ras/effector interface. Proteins. 2011;79:3511–3524. doi: 10.1002/prot.23095. [DOI] [PubMed] [Google Scholar]
- [112].Kiel C, Aydin D, Serrano L. Association rate constants of ras-effector interactions are evolutionarily conserved. PLoS Comput Biol. 2008;4:el000245. doi: 10.1371/journal.pcbi.1000245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Baussand J, Kleinjung J. Specific Conformational States of Ras GTPase upon Effector Binding. J Chem Theory Comput. 2013;9:738–749. doi: 10.1021/ct3007265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Resat H, Straatsma TP, Dixon DA, Miller JH. The arginine finger of RasGAP helps Gln-61 align the nucleophilic water in GAP-stimulated hydrolysis of GTP. Proc Natl Acad Sc. 2001;98:6033–6038. doi: 10.1073/pnas.091506998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Soares TA, Miller JH, Straatsma TP. Revisiting the structural flexibility of the complex p21ras-GTP: The catalytic conformation of the molecular switch II. Proteins: Structure, Function, and Bioinformatics. 2001;45:297–312. doi: 10.1002/prot.1150. [DOI] [PubMed] [Google Scholar]
- [116].Farrar CT, Ma J, Singel DJ, Halkides CJ. Structural changes induced in p21Ras upon GAP-334 complexation as probed by ESEEM spectroscopy and molecular-dynamics simulation. Structure. 2000;8:1279–1287. doi: 10.1016/s0969-2126(00)00532-3. [DOI] [PubMed] [Google Scholar]
- [117].Grant BJ, Gorfe AA, McCammon JA. Ras conformational switching: simulating nucleotide-dependent conformational transitions with accelerated molecular dynamics. PLoS Comput Biol. 2009;5:el000325. doi: 10.1371/journal.pcbi.1000325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Diaz JF, Wroblowski B, Schlitter J, Engelborghs Y. Calculation of pathways for the conformational transition between the GTP- and GDP-bound states of the Ha-ras-p21 protein: calculations with explicit solvent simulations and comparison with calculations in vacuum. Proteins. 1997;28:434–451. [PubMed] [Google Scholar]
- [119].Ma J, Karplus M. Molecular switch in signal transduction: reaction paths of the conformational changes in ras p21. Proc Natl Acad Sci. 1997;94:11905–11910. doi: 10.1073/pnas.94.22.11905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Hamelberg D, Mongan J, McCammon JA. Accelerated molecular dynamics: a promising and efficient simulation method for biomolecules. J Chem Phys. 2004;120:11919–11929. doi: 10.1063/1.1755656. [DOI] [PubMed] [Google Scholar]
- [121].Grant BJ, McCammon JA, Gorfe AA. Conformational selection in G-proteins: lessons from Ras and Rho. Biophys J. 2010;99:L87–89. doi: 10.1016/j.bpj.2010.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Stein A, Rueda M, Panjkovich A, Orozco M, Aloy P. A systematic study of the energetics involved in structural changes upon association and connectivity in protein interaction networks. Structure. 2011;19:881–889. doi: 10.1016/j.str.2011.03.009. [DOI] [PubMed] [Google Scholar]
- [123].Nussinov R, Tsai CJ, Ma B. The underappreciated role of allostery in the cellular network. Annu Rev Biophys. 2013;42:169–189. doi: 10.1146/annurev-biophys-083012-130257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Nussinov R, Tsai CJ, Xin F, Radivojac P. Allosteric post-translational modification codes. Trends Biochem Sci. 2012;37:447–455. doi: 10.1016/j.tibs.2012.07.001. [DOI] [PubMed] [Google Scholar]
- [125].Kar G, Keskin O, Gursoy A, Nussinov R. Allostery and population shift in drug discovery. Curr Opin Pharmacol. 2010;10:715–722. doi: 10.1016/j.coph.2010.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Klink BU, Scheidig AJ. New insight into the dynamic properties and the active site architecture of H-Ras p21 revealed by X-ray crystallography at very high resolution. BMC Struct Biol. 2010;10:38. doi: 10.1186/1472-6807-10-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].O'Connor C, Kovrigin EL. Global conformational dynamics in ras. Biochemistry. 2008;47:10244–10246. doi: 10.1021/bi801076c. [DOI] [PubMed] [Google Scholar]
- [128].Ma J, Karplus M. Ligand-induced conformational changes in ras p21: a normal mode and energy minimization analysis. J Mol Biol. 1997;21:114–131. doi: 10.1006/jmbi.1997.1313. [DOI] [PubMed] [Google Scholar]
- [129].Vogel A, Reuther G, Roark MB, Tan KT, Waldmann H, Feller SE, Huster D. Backbone conformational flexibility of the lipid modified membrane anchor of the human N-Ras protein investigated by solid-state NMR and molecular dynamics simulation. Biochim Biophys Acta. 2010;1798:275–285. doi: 10.1016/j.bbamem.2009.09.023. [DOI] [PubMed] [Google Scholar]
- [130].Vogel A, Tan KT, Waldmann H, Feller SE, Brown MF, Huster D. Flexibility of ras lipid modifications studied by 2H solid-state NMR and molecular dynamics simulations. Biophys J. 2007;93:2697–2712. doi: 10.1529/biophysj.107.104562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Huster D, Vogel A, Katzka C, Scheidt HA, Binder H, Dante S, Gutberlet T, Zschomig O, Waldmann H, Arnold K. Membrane insertion of a lipidated ras peptide studied by FTIR, solid-state NMR, and neutron diffraction spectroscopy. J Am Chem Soc. 2003;125:4070–4079. doi: 10.1021/ja0289245. [DOI] [PubMed] [Google Scholar]
- [132].Vogel A, Katzka CP, Waldmann H, Arnold K, Brown MF, Huster D. Lipid modifications of a Ras peptide exhibit altered packing and mobility versus host membrane as detected by 2H solid-state NMR. J Am Chem Soc. 2005;127:12263–12272. doi: 10.1021/ja051856c. [DOI] [PubMed] [Google Scholar]
- [133].Reuther G, Tan KT, Kohler J, Nowak C, Pampel A, Arnold K, Kuhlmann J, Waldmann H, Huster D. Structural model of the membrane-bound C terminus of lipid-modified human N-ras protein. Angew Chem Int Ed Engl. 2006;45:5387–5390. doi: 10.1002/anie.200504266. [DOI] [PubMed] [Google Scholar]
- [134].Reuther G, Tan K-T, Vogel A, Nowak C, Arnold K, Kuhlmann J, Waldmann H, Huster D. The Lipidated Membrane Anchor of Full Length N-Ras Protein Shows an Extensive Dynamics as Revealed by Solid-State NMR Spectroscopy. J Am Chem Soc. 2006;128:13840–13846. doi: 10.1021/ja063635s. [DOI] [PubMed] [Google Scholar]
- [135].Gorfe AA, Baron R, McCammon JA. Water-membrane partition thermodynamics of an amphiphilic lipopeptide: an enthalpy-driven hydrophobic effect. Biophys J. 2008;95:3269–3277. doi: 10.1529/biophysj.108.136481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Roy S, Plowman S, Rotblat B, Prior IA, Muncke C, Grainger S, Parton RG, Henis YI, Kloog Y, Hancock JF. Individual palmitoyl residues serve distinct roles in H-ras trafficking, microlocalization, and signaling. Mol Cell Biol. 2005;25:6722–6733. doi: 10.1128/MCB.25.15.6722-6733.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Abankwa D, Gorfe AA, Hancock JF. Mechanisms of Ras membrane organization and signalling: Ras on a rocker. Cell Cycle. 2008;7:2667–2673. doi: 10.4161/cc.7.17.6596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Abankwa D, Vogel H. A FRET map of membrane anchors suggests distinct microdomains of heterotrimeric G proteins. J Cell Sei. 2007;120:2953–2962. doi: 10.1242/jcs.001404. [DOI] [PubMed] [Google Scholar]
- [139].Kapoor S, Triola G, Vetter IR, Erlkamp M, Waldmann H, Winter R. Revealing conformational substates of lipidated N-Ras protein by pressure modulation. Proc Natl Acad Sei. 2012;109:460–465. doi: 10.1073/pnas.1110553109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].Mazhab-Jafari MT, Marshall CB, Stathopulos PB, Kobashigawa Y, Stambolic V, Kay LE, Inagaki F, Ikura M. Membrane-dependent modulation of the mTOR activator Rheb: NMR observations of a GTPase tethered to a lipid-bilayer nanodisc. J Am Chem Soc. 2013;135:3367–3370. doi: 10.1021/ja312508w. [DOI] [PubMed] [Google Scholar]
- [141].Guldenhaupt J, Rudack T, Bachler P, Mann D, Triola G, Waldmann H, Kotting C, Gerwert K. N-Ras forms dimers at POPC membranes. Biophys J. 2012;103:1585–1593. doi: 10.1016/j.bpj.2012.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142].Nicolini C, Baranski J, Schlummer S, Palomo J, Lumbierres-Burgues M, Kahms M, Kuhlmann J, Sanchez S, Gratton E, Waldmann H, Winter R. Visualizing association of N-ras in lipid microdomains: influence of domain structure and interfacial adsorption. J Am Chem Soc. 2006;128:192–201. doi: 10.1021/ja055779x. [DOI] [PubMed] [Google Scholar]
- [143].Weise K, Kapoor S, Denter C, Nikolaus J, Opitz N, Koch S, Triola G, Herrmann A, Waldmann H, Winter R. Membrane-mediated induction and sorting of K-Ras microdomain signaling platforms. J Am Chem Soc. 2011;133:880–887. doi: 10.1021/ja107532q. [DOI] [PubMed] [Google Scholar]
- [144].Risselada HJ, Marrink SJ. The molecular face of lipid rafts in model membranes. Proc Natl Acad Sei U S A. 2008;105:17367–17372. doi: 10.1073/pnas.0807527105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [145].Domanski J, Marrink SJ, Schafer LV. Transmembrane helices can induce domain formation in crowded model membranes. Biochim Biophys Acta. 2012;1818:984–994. doi: 10.1016/j.bbamem.2011.08.021. [DOI] [PubMed] [Google Scholar]
- [146].Schafer LV, Marrink SJ. Partitioning of lipids at domain boundaries in model membranes. Biophys J. 2010;99:L91–93. doi: 10.1016/j.bpj.2010.08.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [147].de Jong DH, Lopez CA, Marrink SJ. Molecular view on protein sorting into liquid-ordered membrane domains mediated by gangliosides and lipid anchors. Faraday Discuss. 2013;161:347–363. doi: 10.1039/c2fd20086d. [DOI] [PubMed] [Google Scholar]
- [148].Niv H, Gutman O, Kloog Y, Henis YI. Activated K-Ras and H-Ras display different interactions with saturable nonraft sites at the surface of live cells. J Cell Biol. 2002;157:865–872. doi: 10.1083/jcb.200202009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149].Prior IA, Harding A, Yan J, Sluimer J, Parton RG, Hancock JF. GTP-dependent segregation of H-ras from lipid rafts is required for biological activity. Nat Cell Biol. 2001;3:368–375. doi: 10.1038/35070050. [DOI] [PubMed] [Google Scholar]
- [150].Grant BJ, Lukman S, Hocker HJ, Sayyah J, Brown JH, McCammon JA, Gorfe AA. Novel allosteric sites on Ras for lead generation. PLoS One. 2011;6:e25711. doi: 10.1371/journal.pone.0025711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [151].Sun Q, Burke JP, Phan J, Burns MC, Olejniczak ET, Waterson AG, Lee T, Rossanese OW, Fesik SW. Discovery of small molecules that bind to K-Ras and inhibit Sos-mediated activation. Angew Chem Int Ed Engl. 2012;51:6140–6143. doi: 10.1002/anie.201201358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [152].Rosnizeck IC, Spoerner M, Harsch T, Kreitner S, Filchtinski D, Herrmann C, Engel D, Konig B, Kalbitzer HR. Metal-bis(2-picolyl)amine complexes as state 1(T) inhibitors of activated Ras protein. Angew Chem Int Ed Engl. 2012;51:10647–10651. doi: 10.1002/anie.201204148. [DOI] [PubMed] [Google Scholar]
- [153].Shima F, Yoshikawa Y, Ye M, Araki M, Matsumoto S, Liao J, Hu L, Sugimoto T, Ijiri Y, Takeda A, Nishiyama Y, Sato C, Muraoka S, Tamura A, Osoda T, Tsuda K.-i., Miyakawa T, Fukunishi H, Shimada J, Kumasaka T, Yamamoto M, Kataoka T. In silico discovery of small-molecule Ras inhibitors that display antitumor activity by blocking the Ras–effector interaction. Proc Natl Acad Sci. 2013 doi: 10.1073/pnas.1217730110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154].Cho JK, Park JH, Piggott AM, Salim AA, Gorfe AA, Parton RG, Capon RJ, Lacey E, Hancock JF. Staurosporines disrupt phosphatidylserine trafficking and mislocalize Ras proteins. J Biol Chem. 2012;287(52):43573–43584. doi: 10.1074/jbc.M112.424457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [155].Dharini v.d.H., Cho K-J, Ma X, Chigurupati S, Parton RG, Hancock JF. Fendiline inhibits K-Ras plasma membrane localization and blocks K-Ras signal transmission. Mol Cell Biol. 2013;33(2):237–251. doi: 10.1128/MCB.00884-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [156].Zimmermann G, Papke B, Ismail S, Vartak N, Chandra A, Hoffmann M, Hahn SA, Triola G, Wittinghofer A, Bastiaens PIH, Waldmann H. Small molecule inhibition of the KRAS–PDEd interaction impairs oncogenic KRAS signalling. Nature. 2013;497(7451):638–642. doi: 10.1038/nature12205. [DOI] [PubMed] [Google Scholar]