Abstract
Bioremediation is a technique that uses microbial metabolism to remove pollutants. Various techniques and strategies of bioremediation (e.g., phytoremediation enhanced by endophytic microorganisms, rhizoremediation) can mainly be used to remove hazardous waste from the biosphere. During the last decade, this specific technique has emerged as a potential cleanup tool only for metal pollutants. This situation has changed recently as a possibility has appeared for bioremediation of other pollutants, for instance, volatile organic compounds, crude oils, and radionuclides. The mechanisms of bioremediation depend on the mobility, solubility, degradability, and bioavailability of contaminants. Biodegradation of pollutions is associated with microbial growth and metabolism, i.e., factors that have an impact on the process. Moreover, these factors have a great influence on degradation. As a result, recognition of natural microbial processes is indispensable for understanding the mechanisms of effective bioremediation. In this review, we have emphasized the occurrence of endophytic microorganisms and colonization of plants by endophytes. In addition, the role of enhanced bioremediation by endophytic bacteria and especially of phytoremediation is presented.
Keywords: Bioremediation, Greenhouse gases, Plant, Endophytes
Introduction
Many chemical compounds found in the Earth's atmosphere act as “greenhouse gases.” Greenhouse gases absorb infrared radiation and trap heat in the atmosphere, thereby enhancing the natural greenhouse effect defined as global warming. They may be caused by emissions associated with some human activities or occur naturally, and include a number of gases such as carbon dioxide, methane, nitrous oxide, and halogenated compounds. From year to year, the emission of these gases increases due to the changes in the economic output, extended energy consumption, increasing emission from landfills, livestock, rice farming, septic processes, and fertilizers as well as other factors. Nowadays, we look for modern, cheap, and promising solutions to decrease emission of greenhouse gases into the Earth's atmosphere. Therefore, techniques used for bioremediation of environmental contaminants are gaining considerable momentum.
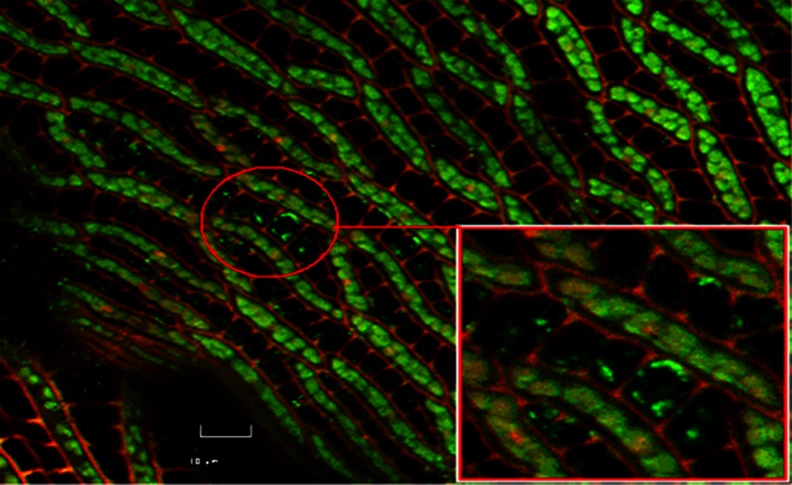
One of the approaches is phytoremediation, in which living green plants in situ are used. They have the ability of decreasing and/or removing contaminants from soil, water, sediments, and air. In phytoremediation processes, selected or engineered microorganisms have been recently used in order to enhance phytoremediation. Numerous studies have demonstrated that endophytic microorganisms can accelerate these processes efficiently by interacting closely with their host plants (Khan and Dotty 2011; Li et al. 2012). These microorganisms reside inside both specific plant tissues and the root cortex or the xylem (Fig. 1). They also systematically colonize the plant by the vascular or apoplast system. Endophytes can also colonize dead and hollow hyaline cells of the plant genus Sphagnum (Fig. 1).
Fig. 1.
Methanotrophic bacteria colonizing the hyaline cells of gametophytes Sphagnum sp., the Live/Dead®BacLight™ kit, Invitrogen (Stępniewska et al. 2013)
The huge variety of the metabolic pathways employed by endophytes makes them valuable tools for bioremediation, which can be used for assimilation of methane, fixation of nitrogen, bioremediation of pollutants (e.g., pesticides, herbicides, insecticides, petrochemicals, polychlorobiphenyls, phenols/chlorophenols), and biotransformation of organic substances, for example propylene to epoxypropane and production of chiral alcohols (Gai et al. 2009; Kim et al. 2012). On the other hand, endophytic microorganisms can produce secondary metabolites that may have an influence on antifungal and antibacterial properties, plant hormones, or their precursors such as plant growth factors, vitamins B12 (Ivanova et al. 2006) and B1 (Mercado–Blanco and Bakker 2007; Simons et al. 1997), and bioprotectants (Trotsenko and Khmelenina 2002).
The aim of this review was to present the potential use of the plant–endophyte system in bioremediation of greenhouse gas pollutions (particularly methane, carbon dioxide) as a method of mitigation of environmental problems without any need to excavate contaminated soil and dispose of it elsewhere. Furthermore, another aim of this paper was to emphasize the scope, magnitude, and complexity of endophytic activity in these studies.
Endophytic microorganisms
Endophytes are defined as microorganisms (fungi, bacteria) that colonize living, internal tissues of plants without causing any immediate, negative effects. The term endophyte was first introduced in 1886 by De Bary for microorganisms (fungi, yeast, and bacteria) colonizing internal plant tissues (De Bary 1884). In 1887, Victor Gallipe postulated that soil microorganisms can penetrate healthy plant tissues; therefore, recognition of colonization mechanisms is so valuable (Galippe 1887). However, those early results were dismissed due to an overall belief that microorganisms discovered inside tissues constitute pollution resulting from the isolation process (Smith 1911). One hundred twenty years later, in 1986, Carrol changed his view of endophytic organisms. He postulated that fungi which cause asymptomatic infections entirely within the tissues of the host plants are endophytes (Carroll 1986). Petrini (1991) viewed them as all organisms living in plant organisms that can colonize tissues without any macroscopically visible symptoms. Hirsch and Braun (1992) described endobionts as a group of microorganisms colonizing tissues without any visible consequences of infection (latent pathogens). One of the latest definitions of endophytes was proposed by Posada and Vega (2005) who used this term to describe all organisms inhabiting different internal parts of plants, including seeds.
The first studies of the biology of Agrostemma githago L. endophytes started by Darnell in 1904 were focused on species richness and abundance, but not on their interactions (Tan and Zou 2001).
Between 1933 and 1989, intensive development of research on endophytes took place, focused particularly on identification of different grass species endophytes (Clay and Schardl 2002; Latch et al. 1985; Saha et al. 1987; Sampson 1938; White 1987). At the end of 1977–1983, great progress in the knowledge of coniferous trees was made (Carroll et al. 1977; Carroll and Carroll 1978; Carroll and Petrini 1983). A significant contribution to the research on endophytic microorganisms was made by Petrini, who examined deciduous trees such as willow and oak (Petrini and Petrini 1985; Petrini 1991, 1996).
In 1998, Schulz and coworkers introduced leaf imprint as a new method for checking the isolation protocols, aiming to eliminate epiphytic organisms (Schulz et al. 1998). Sánchez and Márquez (2008) used this approach as an excellent sterilization method for isolation of endophytes from one kind of grass (Dactylis glomerata L.). The method has been further developed (Arnold et al. 2001, 2007; Suryanarayanan and Kumaresan 2000).
Currently, a substantial body of research on endophytes is focused on the methods of isolation, biodiversity, secondary metabolites, and especially mechanisms of the interaction between the endophyte and the host.
Occurrence of endophytes
The isolation of endophytic organisms from almost all known plants is shown in a large number of literature reports. There are approximately 300,000 plant species living on the Earth, and each individual plant can be the host to one or even more kinds of endophytes (Petrini 1991; Strobel and Daisy 2003; Huang et al. 2007).
They may be isolated from roots, stems, leaves, and inflorescences of weeds, fruit plants, and important vegetables (Bulgari et al. 2012; Bhore et al. 2010; Munif et al. 2012). Endophytic bacteria have been isolated from monocotyledonous plants, e.g., Liliaceae, grass, zea, rice, and orchids (Gangwar and Kaur 2009; Kelemu et al. 2011; Lin et al. 2012; Miyamoto et al. 2004; Peng et al. 2006; Rogers et al. 2012), as well as dicotyledonous plants, for instance oak (Basha et al. 2012; Ma et al. 2013). Some endophytes have been characterized from different tree species, for example oak, pear tree, Sorbus aucuparia, and Betula verrucosa (Krid et al. 2010; Scortichini and Loreti 2007). The existence of endophytes has also been confirmed in beets, corn, bananas, tomatoes, and rice roots (Brown et al. 1999; Cao et al. 2005, 2005; Altalhi 2009; Pereira et al. 1999).
These organisms, classified as Bacillus sp., Enterobacter sp., and Sporosarcina aquimarina (Rylo sona Janarthine et al. 2011), have been found in roots of some coastal mangrove pioneer plants (Avicennia marina).
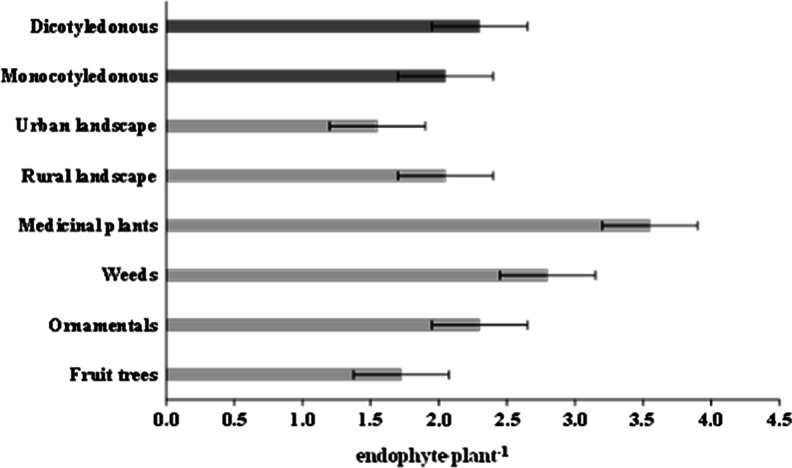
Ting and his coworkers (2009) have performed an analysis of prevalence of Fusarium oxysporum fungi in terms of the plant type, group, and their environmental setting. The aim of the determination of diversity was to present endophytes from various beneficial plant species (fruit, ornamentals, weeds, medicinal plants). Antagonistic endophytes were shown to be mainly fungal endophytes, and they were found primarily in weed and medicinal plant samples. The highest rate of occurrence of endophytes was observed in the medicinal plants (three endophytic organisms per plant), while the weeds were characterized by a lower rate of prevalence (2.4 endophytes per plant) (Ting et al. 2009; Fig. 2).
Fig. 2.
Endophyte recovery rate (endophyte/plant) for plants sampled from various plant types, landscapes, and plant groups. Bars indicate standard error of the means; modified from Ting et al. (2009)
Colonization of plants by endophytes
Endophytic bacteria show a tremendous diversity not only in plant hosts, but also in bacterial taxa (Bacon and Hinton 2006; Hardoim et al. 2008; Vendan et al. 2010). Some hosts are reported to have several endophytes, and the latter may have a wide host range. Therefore, several different species of endophytes can be isolated from a single plant. It is said that the diversity of endophytic communities in the endosphere is regulated by stochastic events, which are influenced by deterministic processes of colonization in turn (Battin et al. 2007). It should be added that the microenvironment of soil has an influence on the colonization of plant endophytes by diverse bacteria and their community composition (Hardoim et al. 2008).
It has been postulated that the early step in the colonization of a plant may depend on absorption of soil aggregates, biodiversity of plants and their physiology, as well as microbial prevalence (Hardoim et al. 2008). The main factors that may regulate microbial colonization include the plant genotype, the growth stage, the physiological status, the type of plant tissues, some soil environmental conditions, as well as some agricultural practices (Conrath et al. 2006; Singh et al. 2009). Moreover, the microbial metabolic pathways of colonization may play an important role as determinants of endophyte diversity. For example, the rate of motile bacteria isolated from the interior part of roots was approximately fivefold higher than that of bacteria in the soil tightly adhering to the roots (Czaban et al. 2007). It has been proved that the ability of soil bacteria to approach plant roots is induced by chemotaxis and the efficiency in microcolony formation. These are the key factors that determine the success of bacteria to become endophytic (Bacilio-Jiménez et al. 2003). The process of plant colonization by endophytic microorganisms is a complex phenomenon. It includes recognition of the host, spore germination, penetration, colonization, and maintenance of endophytes in the host cells (Van Antwerpen et al. 2002). Diverse sources of endophytic microorganisms have been shown. They can be contained in seeds and vegetative planting material, since they originate from the surrounding natural environment such as the rhizosphere and phyllosphere. The processes of colonization depend on several biotic and abiotic factors. It has been shown that they include physical and biological characteristics of the host plant, temperature, humidity conditions, and seasonal fluctuations of other cohabiting microorganisms (Quadt-Hallman et al. 1997).
Overall population densities of endophytic microorganisms may be variable. It is said that the microbial population densities are positively correlated with both the growth stage and changes from the young phase to maturity in plants. For example, the highest rate of endophytic colonization by Solanum tuberosum L. cv. Bartina has been found in the senescent stage, i.e., 6.93 colony forming units (CFU) g−1 of dry weight (DW) in comparison to young plants 4.67 CFU g−1 of dry weight. There is substantial evidence showing that the population density of endophytes depends on the host genotypes (Singh et al. 2009). The highest population density of Pseudomonas striata (133,334 CFU g−1 DW) has been noted in Zea mays L. cv. PRO 311, while the lowest density of the P. striata community (50 CFU g−1 DW) has been found in another species of corn—zea Kiran. Likewise, the total population density of Piriformospora indica in the root of Z. mays Mahikanchan, 247.334 CFU g−1 DW, was higher than that in Z. mays Seedtech which was 48.666 CFU g−1 DW (Singh et al. 2009). Seghers and coworkers (2004) have shown that agricultural practices can affect the composition of the endophyte community. Application of organic fertilizers resulted in increased total population density of endophytes, primarily of type I methanotrophs (Seghers et al. 2004).
The role of endophytic microorganisms in bioremediation
The collaboration between the plant and endophytes can play a key role in the degradation of hazardous contaminants in the rhizosphere. Recently, a promising area of exploitation of endophytic bacteria for phytoremediation of contaminated environments has been described. The advantages and disadvantages of removal of toxic metals, different organic and voltaic substances, greenhouse gases as well as mixed contaminants are listed in Table 1 (Doty 2008).
Table 1.
Advantages and disadvantages of phytoremediation (Doty 2008)
| Advantages | Disadvantages |
|---|---|
| Low cost | Inhibition of plant growth by poor soil quality |
| In situ | Contaminant, phytotoxicity |
| Solar-powered technology | Unknown effects of biodegradation products |
| Maintains in top soil | A slower method |
| Large social acceptance | Lack of the metabolic capacity of the plant to deal with |
| Nondestructive to the soil structure | High levels of these contaminants |
Bacterial endophytes might function more effectively than bacteria added to the soil because they participate in a process known as bioaugmentation (Newman and Reynol 2005). Large numbers of bacterial strains isolated from grapevine (Vitis vinifera L.) plants were resistant to lead, mercury, nickel, zinc, and manganese (Altalhi 2009). In their study, the authors Guo et al. (2010) showed that the endophytic bacterium Bacillus sp. reduced cadmium to approximately 94 % in the presence of industrially used metabolic inhibitors N,N′-dicyclohexylcarbodiimide (specific ATPase inhibitor, DCC) or 2,4-dinitrophenol (DNP). Similarly, inoculation with endophytic bacteria, Serratia nematodiphila LRE07, alleviated growth inhibition in Solanum nigrum L. in the presence of cadmium (Wan et al. 2012).
Ma et al. (2011) isolated Ni-resistant endophytic bacteria from tissues of Alyssum serpyllifolium growing in serpentine soils in Braganca in the northeast part of Portugal. Inoculation of Brassica juncea seeds with this strain significantly increased the plant biomass. Bioremediation of heavy metals involving endophytic bacteria L14 (EB L14) isolated from a cadmium hyperaccumulator Solanum nigrum L. has been described by Chen et al. (2012).
The endophytic microbial community may also assist in phytoremediation of petroleum. Preference for petroleum-degrading bacteria in the root interior has been illustrated with an example of plants growing in petroleum-contaminated soil (Siciliano et al. 2001). van Aken and coworkers (2004) have indicated that Methylobacterium populum sp. nov. strain BJ001 isolated from poplar trees is able to degrade energetic compounds such as 2,4,6-trinitrotoluene (TNT), hexahydro-1,3,5-trinitro- 1,3,5-triazine (HMX), and hexahydro-1,3,5- trinitro-1,3,5-triazine (RDX). Mineralization of about 60 % of RDX to carbon dioxide was observed within 2 months' time. The bioremediation potential during degradation of xenobiotic compounds by three strains of Pseudomonas sp. isolated from xylem sap of poplar trees was tested by Germaine et al. (2004). Recently, Oliveira et al. (2012) have isolated three strains from Cerrado plants exhibiting the capacity for degradation of different fractions of petroleum, diesel oil, and gasoline.
Over the recent years, much more attention has been focused on the application of endophytic bacteria for phytoremediation. Burkholderia cepacia L.S.2.4 bacteria genetically modified by introduction of a pTOM toluene-degradation plasmid of B. cepacia G4, a natural endophyte of yellow lupine, were used for phytoremediation of toluene (Barac et al. 2004). The recombinant strain induced strong (up to 50–70 %) degradation of toluene. Germaine and colleagues (2009) described inoculation of the pea (Pisum sativum) with a genetically modified bacterial endophyte that naturally possessed the ability to degrade 2,4-dichlorophenoxyacetic acid.
The results showed that the plants inoculated with Pseudomonas putida VM1441(pNAH7) had a higher degradation capacity of up to 40 % for 2,4-dichlorophenoxyacetic acid from the soil (Germaine et al. 2009). The first in situ inoculation of poplar trees growing on a trichloroethylene (TCE)-contaminated site with TCE-degrading strain P. putida W619-TCE was done by (Weyens et al. 2009). This kind of inoculation resulted in a 90 % reduction of TCE evapotranspiration under the field conditions. This promising result was obtained after introduction of P. putida W619-TCE to poplar trees, as a root endophyte. Probably, the TCE metabolic activity in the members of the poplar's endogenous endophytic population was obtained by further horizontal gene transfer (Weyens et al. 2009). In subsequent studies, Weyens et al. (2010) used engineered endophytes for improving phytoremediation of environments contaminated by organic pollutants and toxic metals. The yellow lupine was inoculated with B. cepacia VM1468 possessing (a) the pTOM-Bu61 plasmid coding for constitutive trichloroethylene degradation and (b) the ncc-nre Ni resistance/sequestration. Inoculation with B. cepacia M1468 into plants resulted in a decrease in Ni and TCE phytotoxicity, which was reflected by a 30 % increase in root biomass and up to a 50 % decrease in the activities of enzymes involved in antioxidative defense in the roots. In addition, the decreasing trend in TCE evapotranspiration showed about a fivefold higher Ni uptake observed after inoculation of plants (Weyens et al. 2010). Bioaugmentation of two grass species (Festuca arundinacea Schreb. and Festuca pratensis Huds) with endophytic fungi Neotyphodium coenophialum and Neotyphodium uncinatum resulted in PAH and TPH removal from the plant rhizosphere of 80–84 and 64–72 %, respectively, compared with 56 and 31 % in control plants (Soleimani et al. 2010). At the same time, Chen and coworkers (2012) demonstrated that culturable endophytes in aquatic plants have the potential to enhance in situ phytoremediation. This was one of the first studies aimed at isolation and comparison of culturable endophytic bacteria among different aquatic plants showing great diversity of microorganisms dominated by Gammaproteobacteria. Ho et al. (2012) isolated endophytic bacteria tolerating aromatic compounds from plants predominantly occurring in constructed wetlands, including reed (Phragmites australis) and water spinach (Ipomoea aquatica). Achromobacter xylosoxidans strain F3B was chosen for in planta studies using Arabidopsis thaliana as a model plant. It promoted removal of catechol or phenol pollutants (Ho et al. 2012). Kang and colleagues (2012) reported a novel endophyte from the hybrid poplar (Populus deltoides × P. nigra). This unique endophyte, identified as Enterobacter sp. PDN3, showed high tolerance to TCE up to 55.3 μM (Kang et al. 2012). This strategy is promising for improvement of the efficiency of phytoremediation of volatile organic contaminants. Furthermore, recombinant endophytic bacteria are easier in application than genetic plants because their strains can successfully colonize multiple plants. In addition, other benefits to plants such as nitrogen fixation, phosphate solubilization, and stress tolerance have been observed (Dimkpa et al. 2009; Doty et al. 2009; Gai et al. 2009; Jing et al. 2007; Li et al. 2012).
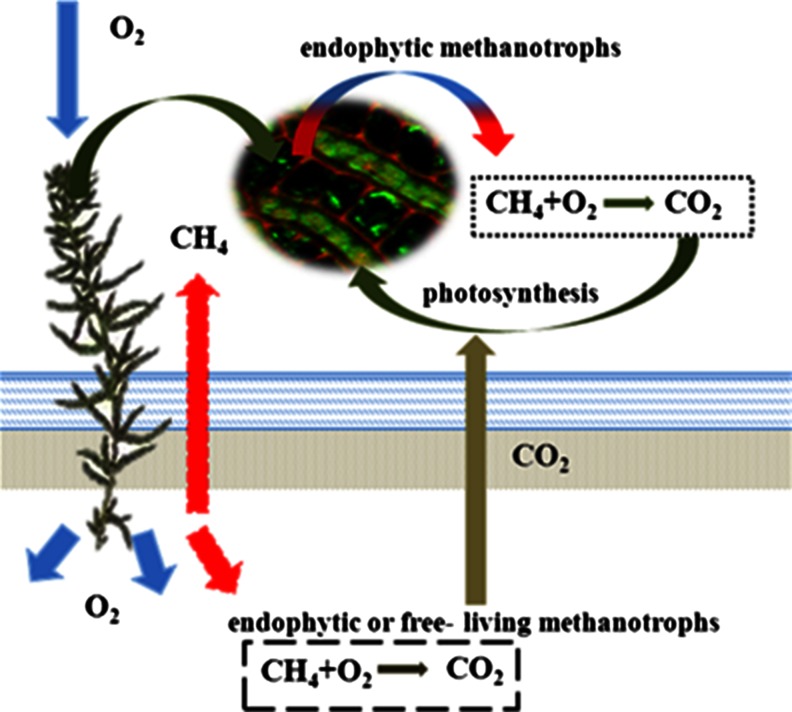
Besides bioremediation of volatile organic compounds, a lot of research has been focused on greenhouse gas emissions (particularly methane and carbon dioxide) depending on the composition of vegetation (Chen and Murrell 2010; Parmentier et al. 2011; Goraj et al. 2013; López et al. 2013). Methane and carbon dioxide are the main greenhouse gases (IPCC 2007). Thus, at the time of the global warming effect, reduction of the methane concentration in the atmosphere, both from natural and anthropogenic sources, is very important. One of the most recent discoveries has shown that the endophytic methanotrophic bacteria found in moss tissues belong to the genus Sphagnum (Raghoebarsing et al. 2005). It has been demonstrated that methanotrophs inhabiting Sphagnum spp., e.g., Methylocella palustris and Methylocapsa acidiphila, oxidize methane to carbon dioxide, which is later used by Sphagnum plants in the process of photosynthesis (Raghoebarsing et al. 2005; Stępniewska et al. 2013; Fig. 1). This discovery substantially changed the description of the carbon cycle in peat ecosystems and at the same time the global carbon cycle. In this way, methanotrophic endophytes inhabiting Sphagnum spp. can act as a natural methane filter that can reduce CH4 and CO2 emission from peatlands by up to 50 % (Kip et al. 2012; Fig. 3). Other field studies have shown the potential ability of the plant–methanotrophic bacteria systems to reduce methane emission up to 77 %, depending on the season and the host plant (Goraj et al. 2013). Furthermore, isolated endophytes from Sphagnum spp. could colonize crops and promote their growth. Molecular genetic analysis has shown that the dominant endophytic groups belong to the genera Burkholderia, Pseudomonas, Flavobacterium, Serratia, and Collimonas. Shcherbakov and colleagues (2013) have suggested that the isolated strain can be a promising object for the development of effective growth-promoting and protective microbiological preparations to be used in agriculture. Furthermore, the endophytes inhabiting Sphagnum spp. can be used for the inoculation of plants inhabiting an artificial wetland system used to treat mixed contaminations (for example heavy metals, different organic contaminations, and greenhouse gases). A majority of artificial wetland systems use the common reed Phragmites sp., cattail Typha sp., and willow Salix sp. that can be components of indigenous peatland flora (Moshiri 1993).
Fig. 3.
The role of endophytic methanotrophs in peatlands (modified from Kip et al. 2012)
Recent studies have indicated the big potential of plants in the remediation of polluted sites. The excellence of adaptation abilities and promising remediation efficiencies strongly imply the superiority of endophytes in the bioremediation of mixed contamination at their low concentrations. It could be useful for developing an efficient metal removal system (Li et al. 2012). On the other hand, the adaptation abilities and the remediation efficiencies of endophytic microorganisms still need further understanding and recognition for practical applications.
Concluding remarks and future perspective
The enormous importance of studies on the endophytic system is related to the connection between the specific metabolic abilities and the use of innovative microbial sources which are valuable in biotechnology nowadays. For instance, endophytic microorganisms can synthesize bioactive metabolites in different diseases, ensuring biological control of induced systemic resistance (ISR) and systemic-acquired resistance (SAR) factors, which may reduce plant pathogens. Endophytic microorganisms may accelerate phytoremediation or bioremediation processes.
The best way to extend the knowledge is to conduct research in the following areas:
The practical application of bioremediation, particularly phytoremediation techniques
A better understanding of plant–endophyte interactions and the dynamics of endophytic microorganisms (growth population and biodiversity)
The possibility of exploitation of woody plants for phytoremediation
Determination of the biodegradation rate of contaminants
Stress tolerance in plants
Focus on endophytic microorganisms degrading multiple metal or organic contaminants by phytoremediation
Construction of wetlands for remediation and using microbes to enhance native plants for restoration
Acknowledgments
This work was supported by the National Science Centre grant in Poland (no. 2011/01/N/NZ9/06811 and N 305 29 94 40).
References
- Altalhi AD. Plasmids profiles, antibiotic and heavy metal resistance incidence of endophytic bacteria isolated from grapevine (Vitis vinifera L.) Afr J Biotechnol. 2009;8:5873–5882. [Google Scholar]
- Arnold AE, Maynard Z, Gilbert GS. Fungal endophytes in dicotyledonous neotropical trees: patterns of abundance and diversity. Mycol Res. 2001;105:1502–1507. doi: 10.1017/S0953756201004956. [DOI] [Google Scholar]
- Arnold AE, Henk DA, Eells RA, Lutzoni F, Vilgalys R. Diversity and phylogenetic affinities of foliar fungal endophytes in loblolly pine inferred by culturing and environmental PCR. Mycologia. 2007;99:185–206. doi: 10.3852/mycologia.99.2.185. [DOI] [PubMed] [Google Scholar]
- Bacilio-Jiménez M, Aguilar-Flores S, Ventura-Zapata E, Perez-Campos E, Bouquelet S, Zenteno E. Chemical characterization of root exudates from rice (Oryza sativa) and their effects on the chemotactic response of endophytic bacteria. Plant Soil. 2003;249:271–277. doi: 10.1023/A:1022888900465. [DOI] [Google Scholar]
- Bacon CW, Hinton DM. Bacterial endophytes: the endophytic niche, its occupants, and its utility. In: Gnanamanickam SS, editor. Plant-associated bacteria. Dordrecht: Springer; 2006. pp. 155–194. [Google Scholar]
- Barac T, Taghavi S, Borremans B, Provoost A, Oeyen L, Colpaert JV, Vangronsveld J, van der Lelie D. Engineered endophytic bacteria improve phytoremediation of water-soluble, volatile, organic pollutants. Nat Biotechnol. 2004;22:583–588. doi: 10.1038/nbt960. [DOI] [PubMed] [Google Scholar]
- Basha NS, Ogbaghebriel A, Yemane K, Zenebe M. Isolation and screening of endophytic fungi from Eritrean traditional medicinal plant Terminalia brownii leaves for antimicrobial activity. IJGP. 2012;6:40–44. [Google Scholar]
- Battin TJ, Sloan WT, Kjelleberg S, Daims H, Head IM, Curtis TP, Eberl L. Microbial landscapes: new paths to biofilm research. Nat Rev Microbiol. 2007;5:76–81. doi: 10.1038/nrmicro1556. [DOI] [PubMed] [Google Scholar]
- Bhore SJ, Nithya R, Loh CY. Screening of endophytic bacteria isolated from leaves of Sambung Nyawa [Gynura procumbens (Lour.) Merr.] for cytokinin-like compounds. Bioinformation. 2010;5:191–197. doi: 10.6026/97320630005191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown KB, Hyde KD, Guest DI. Preliminary studies on endophytic fungal communities of Musa acuminata species complex in Hong Kong and Australia. Fungal Divers. 1999;1:27–51. [Google Scholar]
- Bulgari D, Bozkurt AI, Casati P. Endophytic bacterial community living in roots of healthy and ‘Candidatus Phytoplasma mali’-infected apple (Malus domestica, Borkh.) trees. A Van Leeuw. 2012;102:677–687. doi: 10.1007/s10482-012-9766-3. [DOI] [PubMed] [Google Scholar]
- Cao L, Qui Z, You J, Tan H, Zhou S. Isolation and characterization of endophytic Streptomycete antagonists of Fusarium wilt pathogen from surface-sterilized banana roots. FEMS Microbiol. 2005;247:147–152. doi: 10.1016/j.femsle.2005.05.006. [DOI] [PubMed] [Google Scholar]
- Carroll GE. The biology of the endophytism in plants with particular reference to woody perennials. In: Fokkema NJ, van den Heuvel I, editors. The microbiology of the phyllosphere. Cambridge: Cambridge University Press; 1986. pp. 205–222. [Google Scholar]
- Carroll GC, Carroll FE. Studies on the incidence of coniferous needle endophytes in the Pacific Northwest. Can J Bot. 1978;56:3032–3043. doi: 10.1139/b78-367. [DOI] [Google Scholar]
- Carroll GC, Petrini O. Patterns of substrate utilization by some endophytes from coniferous foliage. Mycologia. 1983;75:53–63. doi: 10.2307/3792923. [DOI] [Google Scholar]
- Carroll FE, Müller E, Sutton BC. Preliminary studies on the incidence of needle endophytes in some European conifers. Sydowia. 1977;29:87–103. [Google Scholar]
- Chen Y, Murrell JC. Methanotrophs in moss. Nat Geosci. 2010;3:595–596. doi: 10.1038/ngeo952. [DOI] [Google Scholar]
- Chen WM, Tang YQ, Mori K, Wu XL. Distribution of culturable endophytic bacteria in aquatic plants and their potential for bioremediation in polluted waters. Aquat Biol. 2012;15:99–110. doi: 10.3354/ab00422. [DOI] [Google Scholar]
- Clay K, Schardl C. Evolutionary origins and ecological consequences of endophyte symbiosis with grasses. Am Nat. 2002;160:99–127. doi: 10.1086/342161. [DOI] [PubMed] [Google Scholar]
- IPCC Climate Change (2007). The physical science basis. Contribution of Working Group I to the Fourth Assessment Report of the Intergovernmental Panel on Climate Change, Solomon S, Qin D, Manning M, eds, Cambridge University Press, Cambridge, UK and New York
- Conrath U, Beckers GJM, Flors V, Garcia-Agustin P, Jakab G, Mauch F, Newman MA, Pieterse CMJ, Poinssot B, Pozo MJ, Pugin A, Schaffrath U, Ton J, Wendehenne D, Zimmerli L, Mauch-Mani B. Priming: getting ready for battle. Mol Plant Microbe Interact. 2006;19:1062–1071. doi: 10.1094/MPMI-19-1062. [DOI] [PubMed] [Google Scholar]
- Czaban J, Gajda A, Wróblewska B. The motility of bacteria from rhizosphere and different zones of winter wheat roots. Pol J Environ. 2007;16:301–308. [Google Scholar]
- De Bary HA. Vergleichende morphologie und biologie der pilze mycetozoen und bacterien. Leipzig: Verlag von Wilhelm Engelmann; 1884. [Google Scholar]
- De Oliveira NC, Rodrigues AA, Alves MIR, Filho NRA, Sadoyama G, Vieira JDG. Endophytic bacteria with potential for bioremediation of petroleum hydrocarbons and derivatives. Afr J Biotechnol. 2012;11:2977–2984. [Google Scholar]
- Dimkpa C, Weinand T, Asch F. Plant–rhizobacteria interactions alleviate abiotic stress conditions. Plant Cell Environ. 2009;32:1682–1694. doi: 10.1111/j.1365-3040.2009.02028.x. [DOI] [PubMed] [Google Scholar]
- Doty SL. Enhancing phytoremediation through the use of transgenics and endophytes. New Phytol. 2008;179:318–333. doi: 10.1111/j.1469-8137.2008.02446.x. [DOI] [PubMed] [Google Scholar]
- Doty SL, Oakley B, Xin G, Kang JW, Singleton G, Khan Z, Vajzovic A, Staley JT. Diazotrophic endophytes of native black cottonwood and willow. Symbiosis. 2009;47:23–33. doi: 10.1007/BF03179967. [DOI] [Google Scholar]
- Gai CS, Lacava PT, Quecine MC, Auriac MC, Lopes JRS, Araújo WL, Miller TA, Azevedo JL. Transmission of Methylobacterium mesophilicum by Bucephalogonia xanthophis for paratransgenic control strategy of citrus variegated chlorosis. J Microbiol. 2009;47:448–454. doi: 10.1007/s12275-008-0303-z. [DOI] [PubMed] [Google Scholar]
- Galippe V. Note sur la présence de micro-organismes dans les tissus végétaux. Comptes Rendus Hebdomadaires des Séances et Mémoires de la Société de Biologie et des ses Filiales et Associées. 1887;39:410–416. [Google Scholar]
- Gangwar M, Kaur G. Isolation and characterization of endophytic bacteria from endorhizosphere of sugarcane and ryegrass. Internet J Microbiol. 2009;7:139–144. [Google Scholar]
- Germaine K, Keogh E, Garcĭa-Cabellos G, Borreans B, van der Lelie D, Barac T, Oeyen L, Vangronsveld J, Moore FP, Moore ERB, Campbell CD, Ryan D, Dowling DN. Colonisation of poplar trees by GFP expressing bacterial endophytes. FEMS Microbiol Ecol. 2004;8:109–118. doi: 10.1016/j.femsec.2003.12.009. [DOI] [PubMed] [Google Scholar]
- Germaine KJ, Keogh E, Ryan D, Dowling DN. Bacterial endophyte-mediated naphthalene phytoprotection and phytoremediation. FEMS Microbiol Lett. 2009;296:226–234. doi: 10.1111/j.1574-6968.2009.01637.x. [DOI] [PubMed] [Google Scholar]
- Goraj W, Kuźniar A, Urban D, Pietrzykowska K, Stępniewska Z. Influence of plant composition on methane emission from Moszne peatland. J Ecol Eng. 2013;14:53–57. doi: 10.5604/2081139X.1031537. [DOI] [Google Scholar]
- Guo H, Luo S, Chen L, Xiao X, Xi Q, Wei W, Zeng G, Liu C, Wan Y, Chen J, He Y. Bioremediation of heavy metals by growing hyperaccumulaor endophytic bacterium Bacillus sp. L14. Bioresour Technol. 2010;101:8599–8605. doi: 10.1016/j.biortech.2010.06.085. [DOI] [PubMed] [Google Scholar]
- Hardoim PR, van Overbeek LS, van Elsas JD. Properties of bacterial endophytes and their proposed role in plant growth. Trends Microbiol. 2008;16:463–471. doi: 10.1016/j.tim.2008.07.008. [DOI] [PubMed] [Google Scholar]
- Hirsch GU, Braun U. Communities of parasitic microfungi. In: Winterhoff W, editor. Handbook of vegetation science: fungi in vegetation science. Dordrecht: Kluwer Academic; 1992. pp. 225–250. [Google Scholar]
- Ho YN, Mathew DC, Hsiao SC, Shih CH, Chien MF, Chiang HM, Huang CC. Selection and application of endophytic bacterium Achromobacter xylosoxidans strain F3B for improving phytoremediation of phenolic pollutants. J Hazard Mater. 2012;15:43–49. doi: 10.1016/j.jhazmat.2012.03.035. [DOI] [PubMed] [Google Scholar]
- Huang WY, Cai YZ, Xing J, Corke H, Sun M. A potential antioxidant resource: endophytic fungi isolated from traditional Chinese medicinal plants. Econ Bot. 2007;61:14–30. doi: 10.1663/0013-0001(2007)61[14:APAREF]2.0.CO;2. [DOI] [Google Scholar]
- Ivanova EG, Fedorov DN, Doronina NV, Trotsenko YA. Production of vitamin B12 in aerobic methylotrophic bacteria. Microbiology. 2006;75:494–496. doi: 10.1134/S0026261706040217. [DOI] [PubMed] [Google Scholar]
- Jing Y-d, He Z-l, Yang X-e. Role of soil rhizobacteria in phytoremediation of heavy metal contaminated soils. J Zhejiang Univ Sci B. 2007;8(192):207. doi: 10.1631/jzus.2007.B0192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang JW, Khan Z, Doty SL. Biodegradation of trichloroethylene by an endophyte of hybrid poplar. Appl Environ Microbiol. 2012;12:3504–3507. doi: 10.1128/AEM.06852-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelemu S, Fory P, Zuleta C, Ricaurte J, Rao I, Lascano C. Detecting bacterial endophytes in tropical grasses of the Brachiaria genus and determining their role in improving plant growth. Afr J Biotechnol. 2011;10:965–976. [Google Scholar]
- Khan Z, Dotty S. Endophyte-assisted phytoremediation. Curr. Topics in Plant Biology. 2011;12:97–105. [Google Scholar]
- Kim T-U, Cho S-H, Han J-H, Shin Y-M, Lee HB, Kim SB. Diversity and physiological properties of root endophytic Actinobacteria in native herbaceous plants of Korea. J Microbiol. 2012;50:50–57. doi: 10.1007/s12275-012-1417-x. [DOI] [PubMed] [Google Scholar]
- Kip N, Fritz C, Langelaan ES, Pan Y, Bodrossy L, Pancotto V, Jetten MSM, Smolders AJP, Op den Camp HJM. Methanotrophic activity and diversity in different Sphagnum magellanicum dominated habitats in the southernmost peat bogs of Patagonia. Biogeosciences. 2012;9:47–55. doi: 10.5194/bg-9-47-2012. [DOI] [Google Scholar]
- Krid S, Rhouma A, Mogou I, Quesada JM, Nesme X, Gargouri A. Pseudomonas savastanoi endophytic bacteria in olive tree knots and antagonistic potential of strains of Pseudomonas fluorescens and Bacillus subtilis. J Plant Pathol. 2010;92:335–341. [Google Scholar]
- Latch GCM, Christensen MJ, Gaynor DL. Aphid detection of endophytic infection in tall fescue. N Z J Agric Res. 1985;28:129–132. doi: 10.1080/00288233.1985.10427006. [DOI] [Google Scholar]
- Li H-Y, Wei D-Q, Shen M, Zhou Z-P. Endophytes and their role in phytoremediation. Fungal divers. 2012;54:11–18. doi: 10.1007/s13225-012-0165-x. [DOI] [Google Scholar]
- Lin L, Ge HM, Yan T, Qin YH, Tan RX. Thaxtomin A-deficient endophytic Streptomyces sp. enhances plant disease resistance to pathogenic Streptomyces scabies. Planta. 2012;236:1849–1861. doi: 10.1007/s00425-012-1741-8. [DOI] [PubMed] [Google Scholar]
- López JC, Quijano G, Souza TS, Estrada JM, Lebrero R, Muñoz R. Biotechnologies for greenhouse gases (CH4, N2O and CO2) abatement: state of the art and challenges. Appl Microbiol Biotechnol. 2013;97:2277–2303. doi: 10.1007/s00253-013-4734-z. [DOI] [PubMed] [Google Scholar]
- Ma Y, Prasad MNV, Rajkumar M, Freitas H. Plant growth promoting rhizobacteria and endophytes accelerate phytoremediation of metalliferous soils. Biotechnol Adv. 2011;29:248–258. doi: 10.1016/j.biotechadv.2010.12.001. [DOI] [PubMed] [Google Scholar]
- Ma L, Cao YH, Cheng MH, Huang Y, Mo MH, Wang Y, Yang JZ, Yang FX. Phylogenetic diversity of bacterial endophytes of Panax notoginseng with antagonistic characteristics towards pathogens of root-rot disease complex. Antonie Leeuwenhoek. 2013 doi: 10.1007/s10482-012-9810-3. [DOI] [PubMed] [Google Scholar]
- Mercado–Blanco J, Bakker PAHM. Interaction between plants and beneficial Pseudomonas spp.: exploiting bacterial traits for crop protection. Antonie Leeuwenhoek. 2007;92:367–389. doi: 10.1007/s10482-007-9167-1. [DOI] [PubMed] [Google Scholar]
- Miyamoto T, Kawahara M, Minamisawa K. Novel endophytic nitrogen-fixing clostridia from the grass Miscanthus sinensis as revealed by terminal restriction fragment length polymorphism analysis. Appl Environ Microb. 2004;70:6580–6586. doi: 10.1128/AEM.70.11.6580-6586.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moshiri GA. Constructed wetlands for water quality improvement. Boca Raton: CRC Press; 1993. [Google Scholar]
- Munif A, Hallmann J, Sikora RA. Isolation of endophytic bacteria from tomato and their biocontrol activities against fungal disease. Microbiol Indones. 2012;6:148–156. doi: 10.5454/mi.6.4.2. [DOI] [Google Scholar]
- Newman LA, Reynol CM. Bacteria and phytoremediation: new uses for endophytic bacteria in plants. Trends Biotechnol. 2005;23:6–8. doi: 10.1016/j.tibtech.2004.11.010. [DOI] [PubMed] [Google Scholar]
- Parmentier FJW, van Huissteden J, Kip N, Op den Camp HJM, Jetten MSM, Maximov TC, Dolman AJ. The role of endophytic methane-oxidizing bacteria in submerged Sphagnum in determining methane emissions of Northeastern Siberian tundra. Biogeosciences. 2011;8:1267–1278. doi: 10.5194/bg-8-1267-2011. [DOI] [Google Scholar]
- Peng G, Wang H, Zhang G, Hou W, Liu Y, Wang ET, Tan Z. Azospirillum melinis sp. nov., a group of diazotrophs isolated from tropical molasses grass. Int J Syst Evol Microbiol. 2006;56:1263–1271. doi: 10.1099/ijs.0.64025-0. [DOI] [PubMed] [Google Scholar]
- Pereira JO, Carneiro-Vieira ML, Azevedo JL. Endophytic fungi from Musa acuminata and their reintroduction into axenic plants. World J Microbiol Biotechnol. 1999;15:37–40. doi: 10.1023/A:1008859823806. [DOI] [Google Scholar]
- Petrini O. Fungal endophytes of tree leaves. In: Fokkema NJ, van den Heuvel I, editors. Microbial ecology of the leaves. Cambridge: Cambridge University Press; 1991. pp. 185–187. [Google Scholar]
- Petrini O. Ecological and physiological aspects of host specificity in endophytic fungi. In: Redlin SC, Carris LM, editors. Endophytic fungi in grasses and woody plants. St. Paul: APS Press; 1996. pp. 87–100. [Google Scholar]
- Petrini L, Petrini O. Xylariaceous fungi as endophytes. Sydowia. Ann Mycol Ser II. 1985;38:216–234. [Google Scholar]
- Posada F, Vega FE. Establishment of the fungal entomopathogen Beauveria bassiana (Ascomycota: Hypocreales) as an endophyte in cocoa seedlings (Theobroma cacao) Mycologia. 2005;97:1195–1200. doi: 10.3852/mycologia.97.6.1195. [DOI] [PubMed] [Google Scholar]
- Quadt-Hallman A, Hallman J, Kloepper JW. Bacterial endophytes in cotton: location and interaction with other plant associated bacteria. Can J Microbiol. 1997;43:254–259. doi: 10.1139/m97-035. [DOI] [Google Scholar]
- Raghoebarsing AA, Alfons JP, Smolders AJP, Schmid MC, Rijpstra WIC, Wolters-Arts M, Derksen J, Jetten MSM, Schouten S, Damste JSS, Lamers LPM, Roelofs JGM, Op den Camp HJM, Strous M. Methanotrophic symbionts provide carbon for photosynthesis in peat bogs. Nature. 2005;436:1153–1156. doi: 10.1038/nature03802. [DOI] [PubMed] [Google Scholar]
- Rogers A, McDonald K, Muehlbauer MF, Hoffman A, Koenig K, Newman L, Taghavi S, van der Lelie D. Inoculation of hybrid poplar with the endophytic bacterium Enterobacter sp. 638 increases biomass but does not impact leaf level physiology. GCB Bioenergy. 2012;4:364–370. doi: 10.1111/j.1757-1707.2011.01119.x. [DOI] [Google Scholar]
- Rylo sona Janarthine S, Eganathan P, Balasubramanian T, Vijayalakshmi S. Endophytic bacteria isolated from the pneumatophores of Avicennia marina. Afr J Microbiol Res. 2011;5:4455–4466. doi: 10.5897/AJMR10.188. [DOI] [Google Scholar]
- Saha DC, Johnson-Cicalese JM, Halisky PM, Heemstra MI, Funk CR. Occurrence and significance of endophytic fungi in the fine fescues. Plant Dis. 1987;71:1021–1024. doi: 10.1094/PD-71-1021. [DOI] [Google Scholar]
- Sampson K. Further observations on the systemic infection of Lolium. T Brit Mycol Soc. 1938;21:84–97. doi: 10.1016/S0007-1536(37)80007-8. [DOI] [Google Scholar]
- Sánchez-Márquez S, Bills GF, Zabalgogeazcoa I. Diversity and structure of the fungal endophytic assemblages from two sympatric coastal grasses. Fungal Divers. 2008;33:87–100. [Google Scholar]
- Schulz B, Guske S, Dammann U, Boyle C. Endophyte-host interactions II. Defining symbiosis of the endophyte-host interaction. Symbiosis. 1998;25:213–227. [Google Scholar]
- Scortichini M, Loreti S. Occurrence of an endophytic, potentially pathogenic strain of Pseudomonas syringae in symptomless wild trees of Corylus avellana l. J Plant Pathol. 2007;89:431–434. [Google Scholar]
- Seghers D, Wittebolle L, Top EM, Verstraete W, Siciliano SD. Impact of agricultural practices on the Zea mays L. endophytic community. Appl Environ Microbiol. 2004;70:1475–1482. doi: 10.1128/AEM.70.3.1475-1482.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shcherbakov AV, Bragina AV, Kuzmina EY, Berg C, Muntyan AN, Makarova NM, Malfanova NV, Cardinale M, Berg G, Chebotar VK, Tikhonovich IA. Endophytic bacteria of Sphagnum mosses as promising objects of agricultural microbiology. Microbiology. 2013;82:306–315. doi: 10.1134/S0026261713030107. [DOI] [PubMed] [Google Scholar]
- Siciliano SD, Fortin N, Mihoc A, Wisse G, Labelle S, Beaumier D, Ouellette D, Roy R, Whyte LG, Banks MK, Schwab P, Lee K, Greer CW. Selection of specific endophytic bacterial genotypes by plants in response to soil contamination. Appl Environ Microbiol. 2001;67:2469–2475. doi: 10.1128/AEM.67.6.2469-2475.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons M, Permentier HP, de Weger LA, Wijffelman CA, Lugtenberg BJJ. Amino acid synthesis is necessary for tomato root colonization by Pseudomonas fluorescens strain WCS365. Mol Plant Microb Interact. 1997;10:102–106. doi: 10.1094/MPMI.1997.10.1.102. [DOI] [Google Scholar]
- Singh G, Singh N, Marwaha TS. Crop genotype and a novel symbiotic fungus influences the root endophytic colonization potential of plant growth promoting rhizobacteria. Physiol Mol Biol Plants. 2009;15:87–92. doi: 10.1007/s12298-009-0009-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith EF (1911) Bacteria in relation to plant diseases. Carnegie Institution of Washington. p. 2
- Soleimani M, Afyuni M, Hajabbasi MA, Nourbakhsh F, Sabzalian MR, Christensen JH. Phytoremediation of an aged petroleum contaminated soil using endophyte infected and non-infected grasses. Chemosphere. 2010;81:1084–1090. doi: 10.1016/j.chemosphere.2010.09.034. [DOI] [PubMed] [Google Scholar]
- Stępniewska Z, Kuźniar A, Pytlak A, Szymczycha J. Detection of methanotrophic endosymbionts in Sphagnum sp. originating from Moszne peat bog (East Poland) Afr J Microbiol Res. 2013;7:1319–1325. [Google Scholar]
- Strobel G, Daisy B. Bioprospecting for microbial endophytes and their natural products. Microbiol Mol Biol R. 2003;67:491–502. doi: 10.1128/MMBR.67.4.491-502.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suryanarayanan TS, Kumaresan V. Endophytic fungi of some halophytes from an estuarine mangrove forest. Mycol Res. 2000;104:1465–1467. doi: 10.1017/S0953756200002859. [DOI] [Google Scholar]
- Tan RX, Zou WX. Endophytes: a rich source of functional metabolites. Nat Prod Rep. 2001;18:448–459. doi: 10.1039/b100918o. [DOI] [PubMed] [Google Scholar]
- Ting ASY, Mah SW, Tee CS. Prevalence of endophytes antagonistic towards Fusarium oxysporum F. sp. Cubense Race 4 in various plants. Am.-Eurasian J Sustain Agric. 2009;3:399–406. [Google Scholar]
- Trotsenko YA, Khmelenina VN. Biology of extremophilic and extremotolerant methanotrophs. Arch Microbiol. 2002;177:123–131. doi: 10.1007/s00203-001-0368-0. [DOI] [PubMed] [Google Scholar]
- van Aken B, Tehrani R, Schnoor JL. Biodegradation of nitro-substituted explosives 2,4,6-trinitrotoluene, hexahydro-1,3,5-trinitro-1,3,5-triazine, and octahydro-1,3,5,7-tetranitro-1,3,5-tetrazocine by a photosymbiotic Methylobacterium sp. associated with poplar tissues (Populus deltoids × nigra DN34) Appl Environ Microbiol. 2004;70:508. doi: 10.1128/AEM.70.1.508-517.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Antwerpen T, Rutherford RS, Vogel JL. Assessment of sugarcane endophytic bacteria and rhizospheric Burkholderia species as antifungal agents. Proc Annu Congr S Afr Sugar Technol Assoc. 2002;76:301–304. [Google Scholar]
- Vendan RT, Yu YJ, Lee SH, Rhee YH. Diversity of endophytic bacteria in ginseng and their potential for plant growth promotion. J Microbiol. 2010;48:559–565. doi: 10.1007/s12275-010-0082-1. [DOI] [PubMed] [Google Scholar]
- Wan Y, Luo S, Chen J, Xiao X, Chen L, Zeng G, Liu C, He Y. Effect of endophyte-infection on growth parameters and Cd-induced phytotoxicity of Cd-hyperaccumulator Solanum nigrum L. Chemosphere. 2012;89:743–750. doi: 10.1016/j.chemosphere.2012.07.005. [DOI] [PubMed] [Google Scholar]
- Weyens N, van der Lelie D, Artois T, Smeets K, Taghavi K, Newman L, Carleer R, Vangronsveld J. Bioaugmentation with engineered endophytic bacteria improves contaminant fate in phytoremediation. Environ Sci Technol. 2009;43:9413–9418. doi: 10.1021/es901997z. [DOI] [PubMed] [Google Scholar]
- Weyens N, Croes S, Dupae J, Newman L, van der Lelie D, Carleer R, Vangronsveld R. Endophytic bacteria improve phytoremediation of Ni and TCE co-contamination. Environ Pollut. 2010;158:2422–2427. doi: 10.1016/j.envpol.2010.04.004. [DOI] [PubMed] [Google Scholar]
- White JF. The widespread distribution of endophytes in the Poaceae. Plant Dis. 1987;71:340–342. doi: 10.1094/PD-71-0340. [DOI] [Google Scholar]