Abstract
Key message
In this study, we identified several genes, which potentially contribute to phenological variation in the grapevine. This may help to maintain consistent yield and suitability of particular varieties in future climatic conditions.
Abstract
The timing of major developmental events in fruit crops differs with cultivar, weather conditions and ecological site. This plasticity results also in diverse levels of fruitfulness. Identifying the genetic factors responsible for phenology and fertility variation may help to improve these traits to better match future climates. Two Vitis vinifera populations, an F1 progeny of Syrah × Pinot Noir and a phenological core collection composed of 163 cultivars, were evaluated for phenology and fertility subtraits during three to six growing seasons in the same geographical location. The phenotypic variability in the core collection mostly overlapped with that observed in the F1 progeny and several accessions had exceeding values of phenological response. The progeny population was used together with SSR and SNP markers to map quantitative trait loci (QTLs). This allowed us to detect nine QTLs related to budburst, flowering beginning, the onset of ripening (véraison) and total fertility, explaining from 8 to 44 % of phenotypic variation. A genomic region on chromosome 15 was associated with budburst and véraison and two QTLs for fruitfulness were located on chromosomes 3 and 18. Several genes potentially affecting fertility and the timing of fruit development were proposed, based on their position and putative function. Allelic variation at these candidate loci may be explored by sampling accessions from the core collection.
Electronic supplementary material
The online version of this article (doi:10.1007/s00122-013-2170-1) contains supplementary material, which is available to authorized users.
Introduction
Phenological differences among genotypes may affect the majority of biological phenomena, such as plant germination, flowering and pollination, fruit ripening, colour changing and leaf fall, as well as animal migration and breeding. From individual physiology to global metabolic changes, with regard to interspecific relationships, all these processes have periodic cycles and are influenced by the timing of environmental events (Schnelle 1955; Lieth 1974; Sparks and Menzel 2002; Wilczek et al. 2010). Recently, there has been an increasing interest in how alterations of phenology may affect plant adaptation to environment, as a number of studies have documented phenological responses to global climate change, which has also effects on human activities such as agriculture, forestry and viticulture (e.g. Chuine et al. 2004; Jones et al. 2005; Webb et al. 2007; van Leeuwen et al. 2008; Caffarra and Eccel 2011; Chew et al. 2012).
With regard to viticulture, there is a varying degree of phenotypic plasticity in grapevine phenology (Sadras et al. 2009; Dal Santo et al. 2013a). The key developmental stages, budburst, flowering and timing of harvest, are driven mainly by temperature and differ greatly with variety, climate and geographical location. For instance, the cultivar Pinot Noir can ripen together with Syrah in cold regions, but earlier than Syrah in warm regions (Dry 1983; Coombe 1988; van Leeuwen et al. 2008; Jackson 2008). This plasticity results also in different levels of fertility (fruitfulness) and yield (Sadras et al. 2009; Nicotra et al. 2010; Anderson et al. 2012). Several studies have shown that the growth rate and composition of the grape can be affected by climate change, representing a risk to present and future fruit production (e.g. Coombe 1988; Schultz 2000; Duchêne and Schneider 2005; Jones et al. 2005; Brunet et al. 2007; Jackson 2008; Webb et al. 2008; Duchêne et al. 2010; Keller 2010). Therefore, to maintain suitability of particular varieties and consistency in yield and wine styles, grape growers need to consider altering the balance of cultivars from specific regions or developing new cultivars with improved traits to better match future climate conditions (Schultz 2000; Webb et al. 2010; Hannah et al. 2013).
Exploiting the phenotypic and genetic differences between grapevines may allow for successful grape cultivation over a range of climate types and provides possibilities for traditional breeding, as well as identification of target genes for marker-assisted selection (Martinez-Zapater et al. 2010). One way to identify the location of key genes with reference to specific markers and the sequenced genome is by discovering quantitative trait loci (QTLs), which indicate regions of a genome that contribute to trait variation. For instance, QTLs related to fertility, growth and phenology have been mapped from F1 segregating progenies obtained by crossing two grapevine cultivars, and this has been used to start identification of candidate genes (Costantini et al. 2008; Doligez et al. 2010; Duchêne et al. 2010). Although only a handful of these QTLs have been characterized, and their structure and interactions are complex (Doligez et al. 2010; Martinez-Zapater et al. 2010), the potential adaptive benefits of exploiting the variation in phenological response were recently demonstrated using the progeny of cultivars Riesling and Gewürztraminer (Duchêne et al. 2010, 2012).
In this study, we evaluated the phenological and fruitfulness variability within two populations of wine grapes planted in the same location: a germplasm core collection composed of different cultivated varieties and a segregating population derived from a cross between cultivars Syrah and Pinot Noir. Next, we performed QTL mapping in the biparental population and detected several regions in the grapevine genome correlated with the phenotypic variation in budburst, flowering, the onset of ripening (véraison) and total fertility. Finally, we proposed and discussed several candidate genes based on a Gene Ontology (GO) term enrichment and functional annotation analysis.
Materials and methods
Plant material and climatic description of the study site
The grapevine populations analysed in this study belong to the FEM grape germplasm collection located in San Michele all’Adige (Trentino, Italy). The first population is a Vitis vinifera “phenological core collection” (core P) composed of 163 various cultivar accessions (listed in supplementary Table S1), replicated five times and selected as the most representative samples of genetic and agro-morphological diversity (Emanuelli et al. 2013). The second population is a progeny derived from a cross between grapevine varieties Syrah and Pinot Noir. It comprises 170 F1 individuals which were used for linkage map construction (Costantini et al. in prep). All plants of these two populations were grafted on the rootstock Kober 5BB at the FEM experimental field “Giaroni”, and then uniformly pruned and trained according to the Guyot system.
San Michele all’Adige has a humid continental climate classified as Dfb (snow, fully humid, warm summer) under the Köppen-Geiger climate classification system (Kottek et al. 2006). Weather records were obtained from a meteorological station located close to the FEM experimental field (elevation 205 m above sea level, 46.189° N, 11.134° E). The station recorded daily observations of maximum, minimum, and average temperatures and precipitation in 2005–2011 (supplementary Table S2). Climate characteristics for this period showed that precipitation averaged 90 mm during the growing season with a minimum of 68 mm in 2006 and a maximum of 131 mm in 2008. The average growing season temperature in the region was 18.1 °C (17–18 °C within all years). The average maximum temperature during the growing season was 23.2 °C (22.2 °C in 2008 and 24.8 °C in 2006). The average minimum temperature during the growing season was 11.8 °C with a low of 10.3 °C in 2010 to a high of 12.9 °C in 2006.
Phenotypic assessment
Six developmental stages were defined based on the modified E-L system for grapevine phenological classification (Coombe 1995). These stages comprised: budburst (BB, stage E–L 4), when 50 % of the shoots had the leaf tips visible; beginning of flowering (FB, stage E–L 20) corresponding to 10 % of flower caps off; end of flowering (FE, stage E–L 26), when flower cap fall was complete; beginning of véraison (VB, stage E–L 34), when berries started to soften; end of véraison (VE stage E–L 37) in which all berries were soft; and ripening (R, stage E–L 38), when juice extracted from the berries had 18 degrees Brix (a measurement of the sugar content in a solution). Phenotypic assessment was recorded from 3 to 6 years during 2005–2011, depending on trait and population analysed.
Plant observations and the daily climate data were summarized for the growing season from April to October, since growing season averages are adequate to explain the phenology of grapevine (Jones et al. 2005). The dates of each phenological stage were converted into day of the year (DOY), i.e. the number of days after January 1 on which the plant attained each developmental stage. For the progeny population, we calculated also the temperature accumulated over time, referred to as heatsum, which determines the rate of spring development of plants. Heatsum is the accumulation of growing degree days (GDD) up to the date of a phenological event. One GDD is equal to one degree above the base temperature during 24 h. A commonly used heat accumulation index is the Winkler index, where GDD is the sum of the differences between the mean daily air temperature and 10 °C threshold temperature over the active period from April to October (Amerine and Winkler 1944; Hunter and Lechowicz 1992; Ghelardini et al. 2006).
Berry clusters were counted during stage E–L 29 to calculate bud fruitfulness, i.e. the first measure of yield potential, expressed as the total fertility index and estimated by dividing the total number of clusters by the total number of growing shoots per plant. The fertility, phenology and climate data were analysed using the program SPSS 17.0 (SPSS Statistics for Windows, Version 17.0, SPSS Inc., Chicago, IL, USA).
QTL analysis
For QTL identification, we used a previously constructed genetic linkage map based on the genotyping of 652 SSR and SNP markers in 170 F1 individuals from the progeny of Syrah and Pinot Noir (Costantini et al. in prep., updated from Troggio et al. 2007). In the case of the phenological traits, two types of phenotypic datasets were used for each year: one based on DOY and the other based on GDD. Exemplary correlations between DOY and GDD for 3 years (2008–2010) are shown in supplementary Fig. S1. The QTL analysis was performed in MapQTL 6.0 (van Ooijen 2009) with the simple interval mapping and multiple QTL mapping (MQM) functions (with step size 1 cM). Using these methods, background markers were selected to take over the role of the putative QTL as cofactors and reduce the residual variance. The LOD profiles from interval mapping were inspected and the marker closest to each LOD peak was selected as the cofactor to perform the MQM mapping. Several cycles were performed to obtain the potentially maximum number of cofactors for the MQM analysis. These cofactors were then subjected to backward elimination procedure, which leaves out one cofactor at a time to create a subset of markers. The retained set of cofactors was used for further rounds of MQM. In the final LOD profile, QTLs were declared significant, if the maximum LOD exceeded the linkage group and/or genome-wide LOD threshold (calculated using 1,000 permutations) and mean error rate was lower than 0.05. Each QTL was characterized by its LOD score and percentage of phenotypic variation explained in the mapping population. Further, a non-parametric Kruskal–Wallis test was performed to provide support to the marker–trait associations separately for each season. This test is regarded as the non-parametric equivalent to the one-way analysis of variance and indicates that the results of the QTL mapping are not influenced by segregation distortion or non-normal distribution of particular traits (Lehmann 1975; van Ooijen 2009). Confidence intervals were estimated in cM and corresponded to an LOD score drop of one on either side of the likelihood peak. The physical positions of these intervals are provided relative to the genome sequence of Pinot Noir clone ENTAV115 (Velasco et al. 2007).
Candidate gene selection
We selected candidate genes from among functionally annotated genes located within three QTL intervals on chromosomes 3, 15 and 18. The reference genome PN40024 (12× assembly) was used to extract version 1 of the gene predictions (12×v1; Jaillon et al. 2007; Forcato 2010; Grimplet et al. 2012). The physical positions of the QTL intervals based on this reference genome were as follows (in bp): for chromosome 3: 6,686,683–8,843,323, for chromosome 15: 13,060,296–16,414,837 and for chromosome 18: 10,665,387–13,879,246. To choose the candidate genes the intervals were tested for GO annotation enrichment using agriGO (Du et al. 2010). The statistical significance of functional enrichment within the intervals was evaluated using the hypergeometric distribution. A GO term was significantly enriched in the QTL interval, if the p value was less than 0.05 in comparison with the dataset of all gene transcripts annotated in the reference genome (available in agriGO). The genes were grouped into eight functional categories: cellular process, development, metabolism, regulation, response to stimulus, signalling, transport and diverse functions (supplementary Tables S3, S4 and S5).
Results
Distribution of the phenological and fertility traits in the core collection and the QTL mapping population
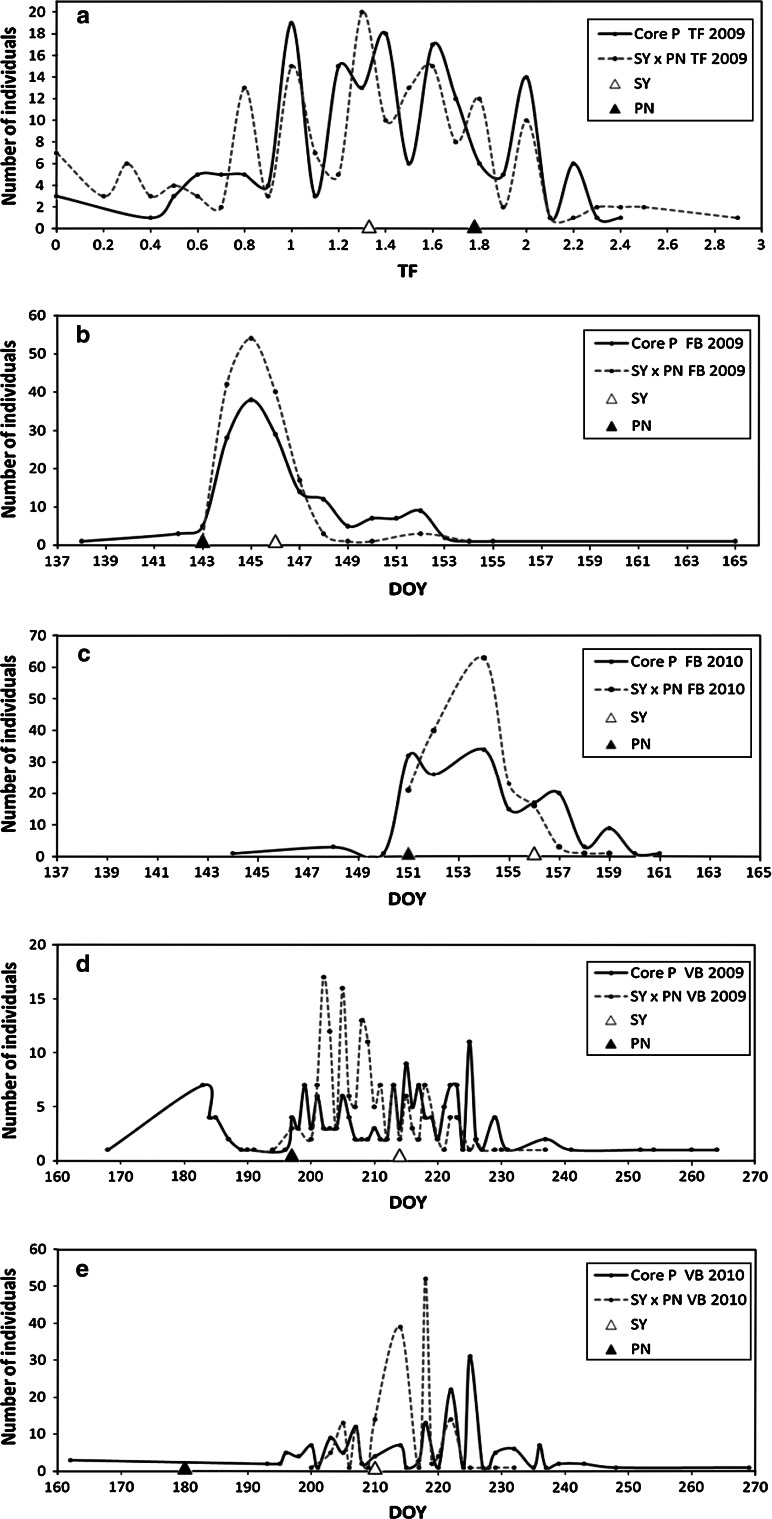
We observed non-normal distribution of the data for all traits and seasons, except for total fertility in 2009 (in both populations; Fig. 1a). The six phenological events defined in this study varied between cultivars of the “phenological collection” (core P), as well as in the progeny of the Syrah × Pinot Noir mapping population (SY × PN). The window of time in which each event typically occurred in the phenological collection among the analysed years was greater for véraison beginning and véraison end (both 75 days), as well as ripening (64 days), than for budburst (22 days), flowering beginning (19 days) and flowering end (16 days; Table 1). These values in the mapping population were as follows: 17 days for budburst, 12 days for flowering beginning, 14 days for flowering end, 39 days for véraison beginning, 47 days for véraison end and 31 days for ripening. Syrah was the earlier parent for budburst, while Pinot Noir was the earlier parent for flowering, véraison and ripening. Most of the progeny showed on average the timing of phenological events situated between both parents, but transgressive segregation was observed as well (Fig. 1b–e).
Fig. 1.
Comparison of phenotypic variability in two grapevine populations: the “phenological core collection” (Core P) and the progeny of a cross between Syrah and Pinot Noir (SY × PN). Exemplary frequency plots for: a total fertility index (TF) in 2009 and b–e the timing of flowering beginning (FB) and véraison beginning (VB) in 2009 and 2010, respectively; DOY day of year in Julian days
Table 1.
Descriptive statistics and comparison of the phenotypic data from the grapevine “phenological core collection” (Core P) and the F1 progeny of Syrah and Pinot Noir (SY × PN) gathered during the same years (the timing of main phenological stages in DOY-days of year)
| Trait | Years | Core P | SY × PN | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N | Min | Max | Range | Mean | SD | N | Min | Max | Range | Mean | SD | ||||||||
| BB | 2008 | 162 | 96 | Apr 07 | 123 | May 02 | 27 | 109 | Apr 18 | 5 | 168 | 100 | Apr 09 | 117 | Apr 26 | 17 | 110 | Apr 19 | 4 |
| 2009 | 162 | 93 | Apr 03 | 114 | Apr 24 | 21 | 102 | Apr 11 | 4 | 168 | 96 | Apr 06 | 114 | Apr 24 | 18 | 100 | Apr 10 | 3 | |
| 2010 | 161 | 99 | Apr 09 | 116 | Apr 26 | 17 | 106 | Apr 16 | 4 | 169 | 99 | Apr 09 | 116 | Apr 26 | 17 | 106 | Apr 16 | 4 | |
| 22 | 106 | Apr 16 | 17 | 106 | Apr 16 | ||||||||||||||
| FB | 2006 | 163 | 147 | May 27 | 167 | Jun 16 | 20 | 157 | Jun 06 | 4 | 148 | 148 | May 28 | 165 | Jun 14 | 17 | 156 | Jun 05 | 2 |
| 2007 | 162 | 134 | May 14 | 155 | Jun 04 | 21 | 141 | May 21 | 3 | 167 | 131 | May 11 | 142 | May 22 | 11 | 139 | May 19 | 4 | |
| 2008 | 162 | 145 | May 24 | 165 | Jun 13 | 20 | 156 | Jun 04 | 4 | 166 | 151 | May 30 | 162 | Jun 10 | 11 | 155 | Jun 03 | 2 | |
| 2009 | 162 | 138 | May 18 | 155 | Jun 04 | 17 | 146 | May 26 | 3 | 164 | 143 | May 23 | 154 | Jun 03 | 11 | 145 | May 25 | 2 | |
| 2010 | 161 | 144 | May 24 | 161 | Jun 10 | 17 | 154 | Jun 03 | 3 | 168 | 151 | May 31 | 159 | Jun 08 | 8 | 154 | Jun 03 | 2 | |
| 19 | 151 | May 31 | 12 | 150 | May 30 | ||||||||||||||
| FE | 2006 | 163 | 163 | Jun 12 | 174 | Jun 23 | 11 | 168 | Jun 17 | 2 | 148 | 155 | Jun 04 | 174 | Jun 23 | 19 | 167 | Jun 16 | 2 |
| 2007 | 162 | 145 | May 25 | 162 | Jun 11 | 17 | 152 | Jun 01 | 5 | 167 | 134 | May 14 | 156 | Jun 05 | 22 | 150 | May 30 | 3 | |
| 2008 | 162 | 153 | Jun 01 | 168 | Jun 16 | 15 | 162 | Jun 10 | 2 | 166 | 158 | Jun 06 | 165 | Jun 13 | 7 | 160 | Jun 08 | 1 | |
| 2009 | 162 | 141 | May 21 | 160 | Jun 09 | 19 | 153 | Jun 02 | 3 | 164 | 147 | May 27 | 160 | Jun 09 | 13 | 150 | May 30 | 2 | |
| 2010 | 161 | 151 | May 31 | 167 | Jun 16 | 16 | 159 | Jun 08 | 3 | 168 | 154 | Jun 03 | 163 | Jun 12 | 9 | 157 | Jun 06 | 2 | |
| 16 | 158 | Jun 07 | 14 | 156 | Jun 05 | ||||||||||||||
| VB | 2006 | 163 | 190 | Jul 09 | 253 | Sep 10 | 63 | 220 | Aug 08 | 14 | 148 | 204 | Jul 23 | 239 | Aug 27 | 35 | 223 | Aug 11 | 7 |
| 2007 | 162 | 183 | Jul 02 | 262 | Sep 19 | 79 | 207 | Jul 26 | 16 | 166 | 194 | Jul 13 | 221 | Aug 09 | 27 | 208 | Jul 27 | 6 | |
| 2008 | 161 | 186 | Jul 07 | 263 | Sep 19 | 77 | 219 | Aug 06 | 14 | 166 | 188 | Jul 06 | 241 | Aug 28 | 53 | 220 | Aug 07 | 7 | |
| 2009 | 162 | 183 | Jul 02 | 264 | Sep 21 | 81 | 211 | Jul 30 | 15 | 160 | 190 | Jul 09 | 237 | Aug 25 | 47 | 209 | Jul 28 | 8 | |
| 2010 | 160 | 193 | Jul 12 | 269 | Sep 26 | 76 | 218 | Aug 06 | 13 | 166 | 200 | Jul 19 | 232 | Aug 20 | 32 | 214 | Aug 02 | 6 | |
| 75 | 216 | Aug 04 | 39 | 215 | Aug 03 | ||||||||||||||
| VE | 2006 | 163 | 204 | Jul 23 | 276 | Oct 03 | 72 | 243 | Aug 31 | 16 | 148 | 228 | Aug 16 | 247 | Sep 04 | 19 | 241 | Aug 29 | 5 |
| 2007 | 162 | 199 | Jul 18 | 261 | Sep 18 | 62 | 233 | Aug 21 | 17 | 167 | 201 | Jul 20 | 236 | Aug 24 | 35 | 222 | Aug 10 | 8 | |
| 2008 | 161 | 203 | Jul 21 | 281 | Oct 07 | 78 | 241 | Aug 28 | 19 | 164 | 212 | Jul 30 | 274 | Sep 30 | 62 | 238 | Aug 25 | 12 | |
| 2009 | 161 | 197 | Jul 16 | 276 | Oct 03 | 79 | 233 | Aug 21 | 23 | 159 | 198 | Jul 17 | 267 | Sep 24 | 69 | 227 | Aug 15 | 15 | |
| 2010 | 160 | 198 | Jul 17 | 284 | Oct 11 | 86 | 232 | Aug 20 | 18 | 165 | 207 | Jul 26 | 258 | Sep 15 | 51 | 226 | Aug 14 | 9 | |
| 75 | 235 | Aug 23 | 47 | 231 | Aug 19 | ||||||||||||||
| R | 2006 | 163 | 215 | Aug 03 | 276 | Oct 03 | 61 | 253 | Sep 10 | 14 | 88 | 249 | Sep 06 | 262 | Sep 19 | 13 | 257 | Sep 14 | 5 |
| 2007 | 162 | 212 | Jul 31 | 275 | Oct 02 | 63 | 246 | Sep 03 | 19 | 143 | 248 | Sep 05 | 267 | Sep 24 | 19 | 250 | Sep 07 | 5 | |
| 2008 | 162 | 225 | Aug 12 | 294 | Oct 20 | 69 | 263 | Sep 19 | 18 | 166 | 233 | Aug 20 | 263 | Sep 19 | 30 | 247 | Sep 03 | 8 | |
| 2010 | 160 | 237 | Aug 25 | 299 | Oct 26 | 62 | 258 | Sep 15 | 18 | 88 | 237 | Aug 25 | 299 | Oct 26 | 62 | 247 | Sep 04 | 14 | |
| 64 | 256 | Sep 13 | 31 | 250 | Sep 07 | ||||||||||||||
| TF | 2007 | 163 | 0.00 | 2.89 | 2.89 | 1.64 | 0.42 | 170 | 0.00 | 3.00 | 3.00 | 1.52 | 0.60 | ||||||
| 2008 | 163 | 0.00 | 2.60 | 2.60 | 1.39 | 0.48 | 170 | 0.00 | 3.00 | 3.00 | 1.67 | 0.54 | |||||||
| 2009 | 163 | 0.00 | 2.40 | 2.40 | 1.37 | 0.48 | 170 | 0.00 | 2.86 | 2.86 | 1.26 | 0.58 | |||||||
| 2.63 | 1.47 | 2.95 | 1.48 | ||||||||||||||||
BB budburst, FB flowering beginning, FE flowering end, VB véraison beginning, VE véraison end, R ripening, TF total fertility index)
Further evaluation of these two populations in the analysed period of 3 to 6 years indicated that in core P budburst occurred on average on April 16, with a range between the earliest and the latest variety of 23 days (Magaratch and Garganega, respectively; supplementary Table S1). The highest year-to-year budburst variability was observed for varieties Datal, Goyura and Terret Noir (SD ± 8), and the lowest for Fertilia, Humagne, Morrastel, Neretta cuneese and Ortega (SD ± 1). In the SY × PN mapping population, the average budburst occurred also on April 16.
Flowering started in the phenological core collection on May 31 on average, with a range of 15 days (the earliest was Léon Millot and the latest Bombino bianco). The among-years variability in flowering beginning was the highest for Cegled szepe (SD ± 11) and the lowest for Beogradska Rana, Bombino bianco, Dunkelfelder, Kanzler Feld O, Maiolica, Petit meslier and Zweigelt blau (SD ± 4). In the SY × PN progeny, on average, flowering began on May 30.
Flowering ended in the core P on average on June 7, with a range of 13 days (the earliest was Léon Millot and the latest Airen, Alarije, Albana, Bombino bianco, Coda di volpe bianca, Parellada and Trebbiano Toscano). The highest year-to-year flowering end variability was observed for varieties Arnsburger, Ehrenfelser, Muscat delecta, Ortega, Sicilien and Turan (SD ± 9), and the lowest for Bombino bianco and Parellada (SD ± 4). In the SY × PN mapping population, flowering ended on average on June 5.
The average date of véraison beginning occurred in the observed 163 varieties on August 4, ranging for 68 days (the earliest accession was Turan and the latest Ohanés). The highest among-years variation of véraison beginning was observed for Malvasia di candia aromatica (SD ± 19) and the lowest for Coda di volpe bianca, Fertilia, Jacquère, Maiolica and Monja (SD ± 3). In the SY × PN mapping population, véraison started on average on August 3.
Véraison ended in the core collection on August 23 on average, with a range of 65 days (the earliest was Madelaine angevine × Calabrese and the latest Raboso Piave). The year-to-year variability in véraison end was highest for Ohanés (SD ± 28) and lowest for Léon Millot (SD ± 3). In the SY × PN progeny, véraison ended on August 19 on average.
Ripening (harvest) dates for the observed varieties of the phenological collection averaged on September 13, with a range of 54 days between the earliest variety Nektar and the latest variety Ohanés. The highest variation among years in this stage was observed for Cegled szepe and Léon Millot (SD ± 31), and the lowest for Petit Meslier (SD ± 4). Dates of harvest for the SY × PN progeny averaged on September 7.
Intervals between the main phenological events are also an important measure of developmental timing. The 163 grapevine varieties in core P revealed an average interval from budburst to flowering end of 54 days. The shortest interval between these two events was 44 days (in accessions Blauer Gelbhoelzer and Garganega) and the longest 60 days (in varieties Early Muscat and Perlon). The interval between budburst and véraison beginning was 110 days on average, with a range of 62 days from the shortest average interval of 88 days in varieties Beogradska Rana and Turan to 150 days in the variety Ohanés. The period from flowering beginning to véraison beginning averaged 64 days, with a range of 64 days for the analysed years (the variety Turan had the shortest average interval of 40 days, while Ohanés had the longest average interval of 104 days). The time from flowering beginning to ripening for the 163 cultivars averaged 102 days. This interval varied from the shortest for Beogradska Rana, Contessa and Nektar (77 days) to the longest for Aspiran noir and Ohanés (126 days). The total ripening stage from véraison beginning to harvest had an average of 38 days, with a range of 53 days. Coda di volpe bianca and Pinot meunier had the shortest average interval of 20 days, while Boglarka had an average 73 day interval. The length of the interval from budburst to ripening for the region studied covered the period from early April to late October and averaged 147 days across cultivars in the collection. This interval characterized the time needed for each plant to ripen and varied by 51 days on average (from 123 days for varieties Nektar, Pinot meunier and Sicilien to 174 days for Dattier noir).
The total fertility coefficient was estimated as the number of flower clusters per number of shoots. The average fertility index value in the phenological core collection was 1.47. The highest average fertility index value 2.2 was obtained for varieties Charmont, Schiras Samling and Segalin, and the lowest average fertility index value 0.39 was obtained for the variety Braghina. The highest year-to-year variability of this parameter was observed for cultivars Malvar and Montonico bianco (SD ± 1.0), and the lowest for Coarna neagra and Contessa (SD ± 0.0). In the mapping population, the average fertility index in these same growing seasons (2007–2009) was 1.48 and ranged from 0.33 to 2.53.
Spearman rank-order correlations between the analysed traits within each year were significant at p < 0.01 in the core P collection and at p < 0.05 in the SY × PN population. In general, correlations in the core collection were around twice higher than in the mapping population. The strongest correlations in both populations were between flowering beginning and flowering end, as well as between véraison beginning and véraison end (for both correlations r = 0.8 and 0.7, in the core collection and the progeny, respectively). We observed strong positive correlations for budburst and flowering beginning in both populations (r = 0.7 and 0.6 in the core collection and the progeny, respectively) and for the pairs budburst–véraison beginning and véraison beginning–ripening in the core collection (r = 0.6 and 0.7, respectively). Furthermore, the timing of véraison end and ripening was highly correlated in the core collection (r = 0.75). Associations of total fertility index with timing of phenological events were slightly negative and averaged around r = −0.2; however, these were significant only in the core collection (Table 2). We did not consider correlations observed in only 1 year, as well as discordant correlations over different years.
Table 2.
Average significant Spearman correlation between the phenological subtraits and fertility in the grapevine “phenological core collection” (below diagonal, p < 0.01) and the SY × PN progeny (above diagonal, p < 0.05)
| Trait | BB | FB | FE | VB | VE | R | TF |
|---|---|---|---|---|---|---|---|
| BB | 0.564 | 0.456 | 0.246 | 0.198 | NS | −0.166a | |
| FB | 0.660 | 0.682 | 0.305 | 0.293 | NS | NS | |
| FE | 0.663 | 0.823 | 0.292 | 0.331 | NS | NS | |
| VB | 0.580 | 0.601 | 0.574 | 0.660 | 0.448 | NS | |
| VE | 0.505 | 0.537 | 0.504 | 0.820 | 0.420 | NS | |
| R | 0.356 | 0.494 | 0.465 | 0.682 | 0.751 | NS | |
| TF | NS | −0.256 | −0.228 | −0.286 | −0.228 | −0.230 |
TF total fertility index, BB budburst, FB flowering beginning, FE flowering end, VB véraison beginning, VE véraison end, R ripening
aSignificant in 2 years only
QTL detection
Integrating the phenotype and genotype data from the Syrah and Pinot Noir progeny allowed us to detect nine QTLs related to phenological and fertility traits within six grapevine chromosomes: one for budburst (BB) on chromosome 15, one for flowering beginning (FB) on chromosome 7, five for véraison (VB, VE) on chromosomes 2, 15 and 17, and two for fertility (TF) on chromosomes 3 and 18 (Table 3). In the case of QTLs for phenology, both types of phenotype datasets, i.e. the one based on DOY and the other based on GDD were compared and gave similar results. We discovered three strong QTLs for véraison beginning with LOD scores exceeding genome-wide significance levels in two to five growing seasons. The locus on chromosome 2 was detected using datasets from five consecutive years and explained 11.3–21.0 % of phenotypic variation. Two interesting QTLs, located on chromosomes 15 and 17, explained 13.5–18.3 % and 7.9–14.2 % of trait variation, respectively. For véraison end two regions were discovered on chromosomes 2 and 15, which overlapped with the QTLs detected for véraison beginning. These two loci explained 14.0–43.7 % and 10.6–18.2 % of phenotypic variation, respectively. The other suggestive QTLs were identified for flowering beginning on chromosome 7, explaining 8.3–11.0 % of trait variation, and for budburst on chromosome 15, explaining 7.9–10.7 % of phenotypic variation and overlapping with the region discovered for véraison beginning. In the case of fruitfulness, the phenotypic data were surveyed over six consecutive years (2006–2011). This allowed us to identify two suggestive regions on chromosomes 3 and 18, consistent in two to three seasons and explaining 17.0–20.1 % and 8.3–9.1 % of phenotypic variation, respectively. We did not detect any stable QTL using datasets of flowering end and ripening.
Table 3.
Main QTLs for phenology and fertility subtraits identified using phenotypic data collected during at least 2 years
| Trait | Chr. | Associated marker | cM | Dataset | LOD Max | 5 % Chr. | 5 % GW | KW test (p) | % variance explained | Marker 1 | Marker 2 | CI (cM) | CI (bp) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BB | 15 | SNP15017 | 25.99 | 2008 | 2.93 | 2.7 | 4.3 | NS | 7.9 | SNP7251 | SNP6095 | 23.22–26.43 | 10,306,970–11,049,526 |
| 2009 | 3.69 | 2.7 | 4.8 | 0.1 | 10.7 | ||||||||
| FB | 7 | SNP5022 | 42.94 | 2008 | 4.15 | 2.9 | 4.4 | 0.0005 | 11.0 | VMC8D11 | SNP4094 | 40.91–46.62 | 16,401,285–17,046,939 |
| 2009 | 3.24 | 2.9 | 4.5 | 0.005 | 8.3 | ||||||||
| VMC8D11 | 40.91 | 2010 | 3.29 | 2.9 | 4.4 | 0.05 | 9.2 | SNP6082 | SNP5022 | 39.98–42.94 | 16,871,868–17,131,388 | ||
| VB | 2 | SNP4067 | 15.66 | 2007 | 9.08 | 2.8 | 4.5 | 0.0001 | 21.0 | UDV27 | VMC6F1 | 8.66–18.76 | 3,117,421–4,925,368 |
| 17.01 | 2009 | 10.32 | 2.5 | 4.3 | 0.0001 | 20.5 | |||||||
| VMC6F1 | 18.76 | 2006 | 4.69 | 2.8 | 4.5 | 0.005 | 12.9 | SNP4067 | 2010D07R | 17.01–27.20 | 4,703,600–5,737,052 | ||
| 21.76 | 2010 | 5.87 | 2.9 | 4.3 | 0.0001 | 13.2 | |||||||
| SNP4123 | 31.31 | 2008 | 5.82 | 2.7 | 4.9 | 0.001 | 11.3 | 2010D07R | VMC2C10.1 | 27.20–33.62 | 5,737,052–8,284,867 | ||
| 15 | SNP6063 | 18.10 | 2007 | 6.92 | 2.8 | 4.5 | 0.0001 | 17.1 | SNP6116 | SNP7251 | 15.10–23.22 | 10,306,970–12,346,018 | |
| SNP7251 | 21.51 | 2010 | 4.92 | 2.7 | 4.3 | 0.0001 | 13.5 | SNP6063 | SNP4061 | 18.51–27.82 | 10,126,670–12,036,121 | ||
| SNP4061 | 27.82 | 2008 | 8.12 | 2.6 | 4.9 | 0.0001 | 18.3 | SNP7251 | VVIP33 | 23.22–33.31 | 9,926,097–10,306,970 | ||
| 2009 | 5.63 | 2.6 | 4.3 | 0.0001 | 13.8 | ||||||||
| 17 | SNP8084 | 39.47 | 2009 | 3.29 | 2.8 | 4.3 | NS | 7.9 | SNP8071 | SNP7177 | 37.48–45.43 | 5,006,342–6,265,765 | |
| 2010 | 6.20 | 2.8 | 4.3 | 0.1 | 14.0 | ||||||||
| SNP8021 | 51.92 | 2007 | 3.68 | 2.9 | 4.5 | 0.0005 | 9.8 | SNP7177 | SNP7270 | 45.43–57.47 | 3,954,375–5,047,079 | ||
| 52.92 | 2008 | 5.63 | 2.7 | 4.9 | 0.0005 | 14.2 | |||||||
| VE | 2 | 1074J14R | 27.61 | 2010 | 5.68 | 2.6 | 4.3 | 0.05 | 14.0 | 2010D07R | VMC2C10.1 | 27.20–33.62 | 5,662,834–8,284,867 |
| SNP4045 | 45.33 | 2007 | 16.33 | 2.8 | 4.5 | 0.0005 | 30.8 | SNP8148 | VMC7G3 | 41.00–49.48 | 9,207,951–18,083,459 | ||
| SNP7234 | 45.95 | 2009 | 9.13 | 2.8 | 4.4 | 0.0001 | 22.8 | ||||||
| SNP7054 | 46.58 | 2008 | 14.90 | 2.7 | 4.4 | 0.0005 | 43.7 | ||||||
| 15 | SNP6116 | 15.10 | 2010 | 5.21 | 2.7 | 4.3 | 0.0001 | 14.0 | SNP0056 | SNP6063 | 12.50–18.51 | 12,020,146–12,644,676 | |
| 17.10 | 2007 | 6.74 | 2.8 | 4.5 | 0.0001 | 18.2 | |||||||
| SNP6063 | 18.10 | 2008 | 3.58 | 2.6 | 4.4 | 0.01 | 10.6 | SNP6116 | SNP7251 | 15.10–23.22 | 10,306,970–12,346,018 | ||
| TF | 3 | SNP5050 | 21.12 | 2006 | 5.41 | 2.8 | 4.9 | 0.005 | 20.1 | SNP3031 | SNP7132 | 16.57–22.67 | 7,184,057–7,777,664 |
| 20.12 | 2008 | 3.89 | 2.7 | 4.4 | 0.05 | 17.0 | |||||||
| 18 | SNP7048 | 59.39 | 2009 | 3.37 | 3.0 | 4.3 | 0.05 | 9.1 | VVIN83 | SNP3032 | 51.39–63.23 | 9,543,849–12,480,055 | |
| 59.54 | 2010 | 3.20 | 3.0 | 4.4 | 0.1 | 8.5 | |||||||
| SNP7055 | 60.54 | 2011 | 3.04 | 2.9 | 4.4 | 0.05 | 8.3 | SNP7048 | 59.54–63.23 | 10,864,719–12,480,055 |
Position of the SSR and SNP markers flanking the QTL confidence intervals (CI) is based on the reference genome Pinot Noir clone ENTAV115 (Velasco et al. 2007)
Candidate genes for development and fertility
We further focused on the QTLs related to budburst and véraison on chromosome 15, as well as to fertility on chromosomes 3 and 18. The number of functionally annotated genes in the QTL confidence intervals ranged from 89 (chromosome 3) to 210 (chromosome 18; supplementary Tables S3, S4 and S5). Among these, we selected and discussed several candidate genes that may contribute to grapevine development and fruitfulness, including genes essential for cell growth and coding for transcription factors and signalling molecules (Table 4).
Table 4.
Candidate genes for grapevine phenology and fertility annotated in the PN40024 genome sequence
| Chr. | Functional category | Gene annotation | Gene unique ID | Position | References |
|---|---|---|---|---|---|
| 15 | Metabolism | Chalcone and stilbene synthase VvCHS2 | VIT_15s0021g02170 | 13,099,190–13,100,695 | Parage et al. (2012) |
| Glutathione S-transferase Z2 (GSTZ2) | VIT_15s0048g00950 | 15,085,283–15,090,953 | Edwards et al. (2000); Braidot et al. (2008) | ||
| Regulation | Scarecrow transcription factor 6 (SCL6) | VIT_15s0048g00270 | 14,396,828–14,399,794 | Llave et al. (2002); Unver et al. (2010) | |
| Homeodomain GLABROUS1 (HDG1) | VIT_15s0048g02000 | 16,133,315–16,138,857 | Nakamura et al. (2006) | ||
| Signalling | Beta expansin VvEXPB3 | VIT_15s0021g02670 | 13,673,378–13,674,771 | Cosgrove (2000); Dal Santo et al. (2013b) | |
| Beta expansin VvEXPB4 | VIT_15s0021g02700 | 13,735,914–13,737,731 | |||
| 3 | Cellular process | Mitotic checkpoint protein BUB3 | VIT_03s0091g00710 | 7,149,908–7,160,638 | Caillaud et al. (2009) |
| Xyloglucan:xyloglucosyl transferase (XTH10) | VIT_03s0088g00650 | 8,842,942–8,844,322 | Nunan et al. (2001); Bourquin et al. (2002); | ||
| Metabolism | Sinapoylglucose:malate sinapoyltransferase (SMT) | VIT_03s0091g01200 | 7,855,565–7,861,870 | Lehfeldt et al. (2000); Bienert et al. (2012) | |
| Serine carboxypeptidase S10 | VIT_03s0091g01240 | 7,903,958–7,907,570 | |||
| Sinapoylglucose-choline O-sinapoyltransferase (SCT) | VIT_03s0091g01270 | 7,935,398–7,938,767 | |||
| Serine carboxypeptidase S10 | VIT_03s0091g01290 | 7,951,175–7,955,690 | |||
| Serine carboxypeptidase SCPL17 | VIT_03s0088g00160 | 8,195,668–8,197,771 | |||
| Serine carboxypeptidase S10 | VIT_03s0088g00260 | 8,252,549–8,257,129 | |||
| Signalling | Phytosulfokine PSK2 | VIT_03s0088g00290 | 8,315,170–8,315,924 | Motose et al. (2009) | |
| 18 | Cellular process | Endo-1,4-beta-glucanase | VIT_18s0001g14040 | 12,086,327–12,090,970 | Nunan et al. (2001); Buchanan et al. (2012) |
| Regulation | Zinc finger (CCCH-type) family protein | VIT_18s0001g15570 | 13,682,439–13,700,040 | Schmitz et al. (2005); Wang et al. (2008) | |
| Signalling | Cytokinin dehydrogenase 5 precursor | VIT_18s0001g13200 | 11,256,653–11,261,569 | Fortes et al. (2011) | |
| Calmodulin binding protein, IQD32 | VIT_18s0001g13870 | 11,862,607–11,871,541 | Kline et al. (2010) | ||
| Clavata 1 receptor kinase (CLV1) | VIT_18s0001g14610 | 12,668,387–12,671,744 | Clark et al. (1997); Durbak and Tax (2011) | ||
| ABA-responsive element-binding protein 3 (AREB3) | VIT_18s0001g14890 | 12,936,974–12,937,903 | Kline et al. (2010) |
Discussion
Phenotypic evaluation
Phenology and fertility were evaluated in two populations of cultivated grapevine with a potentially different level of variation: the germplasm collection and the offspring derived from a cross between two cultivars Syrah and Pinot Noir (the QTL mapping population). While we observed that the distributions of total fertility, budburst and timing of ripening had a similar shape in both populations, the distribution of flowering and véraison time in the mapping population generally displayed narrow peaks with many individuals finishing the particular developmental stage close to the average timing (Fig. 1b–e). Differences in the range of timing between the core collection and the QTL mapping population were more pronounced with the dates of véraison and ripening than with budburst or flowering (i.e. the DOY ranges of véraison beginning, véraison end and ripening were around twice larger for the core collection than for the mapping population, while the dates of budburst and flowering were similar in both populations). This could be explained by high phenotypic diversity in the core collection, resulting from the presence of specific accessions, such as the early ripening Nektar and the late ripening Ohanés (Supplementary Table S1).
The pairwise Spearman correlations between traits in this study confirmed that as the plants continue the growth cycle, each next event is more strongly correlated to the previous event. In former studies, it was observed that correlations between véraison and ripening dates were very high (Jones et al. 2005; Bock et al. 2011; Tomasi et al. 2011). However, in those studies budbreak was not significantly correlated with successive stages of development and thus it was regarded as an event influenced by the variable weather conditions early in the season. We, in turn, observed that the timing of budburst in both analysed populations was significantly correlated with the timing of flowering and the start of véraison (Table 2). This may suggest that genes underlying these traits are located within the same QTL regions.
The total fertility index in the present research was obtained by dividing the total number of fruit clusters by the total number of shoots per plant. It seems that climatic conditions during the observed growing seasons had an effect on the values of this parameter for all individuals in the core collection and in the segregating progeny of Syrah × Pinot Noir. Former comparative studies of fertility among grapevine varieties indicated that the differences in fruitfulness could be due to variation in cultivars, environmental factors (especially air temperature), as well as grafting and training methods (Sommer et al. 2000). Furthermore, this trait may be subjected to the growing conditions of the plant during the previous season. For instance, compared to well-exposed shoots, shoots which develop in dense shade are more likely to have nodes with less fruitful shoots during the following season (Sánchez and Dokoozlian 2005). Fertility, however, may also be affected by other factors, such as the number of flower clusters on the plant and the number of buds which were left after dormant pruning (Sansavini and Fanigliulo 1998; Morris and Main 2010). Based on our observations in the present survey during three and six growing seasons (in the core collection and the progeny population, respectively), it can be concluded that the studied trait is not genetically stable and depends on external conditions. All the plants in our experimental fields were pruned uniformly in each season; nevertheless, it may be noticed that there were quite high differences in rainfall and temperature means as well as flowering time during the analysed years, which might have affected fruitfulness (Supplementary Tables S1 and S2). The significant differences among seasons were revealed for both populations by the ANOVA test (data not shown).
QTL detection and selection of candidate genes
The QTL mapping methods typically rely on the assumption that the phenotype follows a normal distribution for each QTL genotype. In our case, almost all phenotype datasets displayed a non-normal distribution. In general, the interval mapping procedure (including the multiple QTL model and cofactor selection) is quite robust against deviations from normality (van Ooijen 2009). Therefore, we performed this method together with a maximum likelihood mixture model and the permutation test based on the actual data, rather than assuming normality. We further tested if the results of interval mapping were not influenced by non-normal distributions of the data using the non-parametric Kruskal–Wallis analysis. With these approaches, we were able to detect nine QTLs on chromosomes 2, 3, 7, 15, 17 and 18.
The QTL for flowering time on chromosome 7 has already been found using the progeny from a cross between Gewürztraminer and Riesling (Duchêne et al. 2012). This locus contains several genes implicated in the flowering process, such as VvFT (FLOWERING LOCUS T) and VvSVP1 (SHORT VEGETATIVE PHASE 1) (Carmona et al. 2007; Diaz-Riquelme et al. 2009). Other QTLs for this trait were found in progenies from different biparental crosses on chromosomes 1, 2, 6, 14, 15 and 18 (Costantini et al. 2008; Carreño Ruiz 2012; Duchêne et al. 2012). Likewise, the QTLs for véraison beginning and véraison end on chromosome 2 have been discovered previously in different studies, along with other QTLs on chromosomes 1, 3, 5, 6, 16 and 18 (Costantini et al. 2008; Carreño Ruiz 2012; Duchêne et al. 2012). The region discovered on chromosome 2 co-localized with the locus responsible for berry colour, which carries genes VvMybA1 and VvMybA2 involved in the regulation of anthocyanin biosynthesis (Kobayashi et al. 2004; Fournier-Level et al. 2010). The QTL on chromosome 17 has also been detected in the progeny of Ruby Seedless × Moscatuel as related to véraison and berry colour (Carreño Ruiz 2012). Here, we focused on the QTLs which have not been investigated yet. Below, we discuss several candidate genes underlying the QTLs for budburst and véraison located on chromosome 15, and for fertility on chromosomes 3 and 18 (Table 4).
On chromosome 15 we identified two overlapping QTLs related to véraison beginning and véraison end. These two intervals overlapped as well with the QTL for the start of budburst. Some other QTLs for budburst have recently been detected on chromosomes 4, 12 and 19 in different biparental populations (Carreño Ruiz 2012; Duchêne et al. 2012).
Among the genes on chromosome 15, we found several transcription factors implicated in bud and fruit development. For example, a plant-specific scarecrow-like transcription factor 6 (SCL6) is a member of the GRAS gene family, which controls a wide range of developmental processes, including hormone signalling and bud formation (Llave et al. 2002; Unver et al. 2010; Schulze et al. 2010). Another gene HDG1 belongs to the class IV HD-ZIP gene family. Some genes of this family are involved in epidermal development and accumulation of anthocyanin in the shoot (Nakamura et al. 2006).
Cell elongation is an important process in plant development and it is driven by cell wall loosening and turgor pressure. Cell wall remodelling depends on a complex association of physical, chemical and enzymatic processes that are controlled by hormones and environmental factors. One group of genes regulating cell wall architecture is the expansin family. Two expansin genes are located in this QTL region: VvEXPB3 and VvEXPB4 (Cosgrove 2000; Dal Santo et al. 2013b).
Other candidate genes for véraison time in this interval include VvCHS2, which catalyzes the first step of flavonoid biosynthesis (Parage et al. 2012), and GSTZ2 coding for glutathione S-transferase, which may be involved in the transport of flavonoids (Edwards et al. 2000; Braidot et al. 2008).
Several QTLs for fertility (fruitfulness) in grapevine were previously detected on chromosomes 5, 8 and 14 (Fanizza et al. 2005; Doligez et al. 2010). Here, we identified two QTLs for fertility, which were stable in at least two growing seasons: on chromosomes 3 and 18. Among the genes on chromosome 3, we found several with highly enriched GO terms, such as a cluster of genes coding for serine carboxypeptidase-like peptides (SCP, SCPL) (Table 4). These genes have been isolated from several plant species and some of them function as acyltransferases and lyases. They may be involved in a broad range of biochemical pathways, including those of secondary metabolite biosynthesis, herbicide conjugation and germination-associated degradation of seed protein reserves. Thus, they may be vital for normal plant growth and development, synthesis of compounds that protect plants against UV light and pathogens and resistance to natural and artificial xenobiotics (Lehfeldt et al. 2000; Fraser et al. 2005; Bienert et al. 2012). In grapevine, genes from this cluster on chromosome 3 have been identified as candidate glucose-acyltransferases (VvGAT-like) acting in the proanthocyanidin biosynthetic pathway. Proanthocyanidins are secondary metabolites belonging to flavonoids, which play a major role in plant protection against biotic and abiotic stresses (Carrier et al. 2013). As such, variation in these genes might influence grapevine fertility.
This QTL region harbours another key gene required in many aspects of cell wall biosynthesis: XTH, a member of xyloglucan endotransglycosylases/hydrolases. These enzymes are active in xylem and phloem fibres at the stage of secondary wall formation. They reconstruct primary walls, probably by creating and reinforcing the connections between the primary and secondary wall layers, and these cross-links may play an important role in preventing further cell expansion (Bourquin et al. 2002).
Other candidate molecules in this region include the cell cycle arrest protein BUB3, which functions in a molecular complex controlling cell division (Caillaud et al. 2009) and a precursor of phytosulfokine (PSK2), which is a sulfated peptide hormone required for the proliferation and differentiation of plant cells (Motose et al. 2009).
When searching amongst the 210 genes on chromosome 18, we found several candidate molecules potentially contributing to grapevine fruitfulness, such as transcriptional factors. For example, one candidate gene in this region belongs to a CCCH-type zinc finger family and members of this family have been shown to play diverse roles in plant developmental processes and environmental responses, e.g. in the physiological control of female fertility at the level of early embryonic development (Schmitz et al. 2005; Wang et al. 2008).
There are several genes in this interval which are involved in the response to abscisic acid (ABA). ABA is a hormone that controls the overall plant response to environmental stresses. This molecule influences also the onset of grape berry ripening (Gambetta et al. 2010). Transcription factor AREB3 is an ABA response element DNA-binding protein, which is required in initiating the long-term changes in gene expression induced by ABA (Kline et al. 2010). Calmodulin binding protein IQD32 is a phosphopeptide significantly altered in response to ABA treatment. Calcium signalling plays an important role in plants for coordinating a wide range of developmental processes and responses to environmental change. The frequently predicted nuclear localization of IQD proteins suggests that they link calcium signalling pathways to the regulation of gene expression (Abel et al. 2005; Kline et al. 2010). For example SUN, a member of the IQD gene family in tomato, influences floral and fruit morphology (Wu et al. 2011).
Also cytokinins are essential plant hormones that control various aspects of plant growth and development, such as cell division and flower and fruit formation. These molecules are involved in berry set and in growth promotion, and tend to inhibit ripening. One of the candidates located on chromosome 18 codes for a cytokinin dehydrogenase, active in the maintenance of optimal cytokinin concentration via their degradation (Fortes et al. 2011).
Another key regulator of cellular events, which is located in this QTL, is the LRR receptor kinase CLAVATA1 (CLV1). This gene and its homologues are involved in plant development and environmental responses. CLV1 may function as a signal transduction component that acts in the communication of cell division. In Arabidopsis, this kinase plays a critical role in the maintenance of the stem cells in shoot apical meristems and regulates fruit development (Clark et al. 1997; Durbak and Tax 2011).
The last proposed candidate in this interval, Endo-(1,4)-β-glucanase, has been implicated in the breakdown of cell walls during processes observed in normal growth and development, including floral abscission and fruit ripening (Nunan et al. 2001; Buchanan et al. 2012).
Conclusions
In the present research, we compared phenotypic variation in phenology and fertility in two grapevine populations: (1) the collection of different major and minor grapevine cultivars, selected over years and grown locally or worldwide for fruit production and (2) the progeny derived from a new experimental cross between two varieties, Syrah and Pinot Noir, maintained in the field for several years. Although these two populations have different degrees of relationship among individuals, we found that the range of phenotypic variation for our traits of interest in the progeny population covered in the most part the variation recorded for the core collection, which was selected specifically to maximize the variability in the timing of key developmental stages. In addition, as expected, we detected several accessions in the core collection with the earlier or later dates for budburst, flowering and harvest, compared to the progeny population. Nevertheless, the SY × PN progeny could be used as a QTL mapping population to identify loci explaining this phenotypic variation. For the phenological traits, we tried to isolate the genetic and climatic influence (namely, air temperature) by using the DOY, as well as the GDD phenotypic dataset. Ultimately, we identified nine minor and major QTLs related to budburst, flowering beginning, véraison beginning, véraison end and total fertility. The genomic region on chromosome 15 contains overlapping QTLs for budburst and véraison, and harbours genes underlying fruit development, including expansins (VvEXPB3, VvEXPB4) and enzymes involved in biosynthesis and transport of flavonoids (VvCHS2, GSTZ2). For fertility, two QTLs were located on chromosomes 3 and 18. Among the genes potentially affecting grape fruitfulness are the complex of serine carboxypeptidase-like genes (SCPL), xyloglucan:xyloglucosyl transferase XTH10, protein kinase CLV1 and ABA-responsive molecules (AREB3, IQD32). The next step will be to study polymorphism within these candidate loci and to further investigate their relationship with trait variation by analysing multiple accessions from the core collection. The long-term objective of this research is to provide information on the genetic basis of these traits and to facilitate selection of varieties adapted to atmospheric conditions of a specific geographic region.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
We thank the staff of the Grapevine Breeding Platform for assistance in the field and Stefano Corradini (CTT-Unità Sistema Informativo Geografico) for providing the meteorological data. This work was carried out in the framework of the ERA-NET Plant Genomics (ERA-PG 074B GRASP GRAPE WINE) with the financial support provided by the Autonomous Province of Trento (Accordo di Programma).
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical standards
The authors declare that the experiments of this study comply with current local laws.
References
- Abel S, Savchenko T, Levy M. Genome-wide comparative analysis of the IQD gene families in Arabidopsis thaliana and Oryza sativa. BMC Evol Biol. 2005;5:72. doi: 10.1186/1471-2148-5-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amerine MA, Winkler AJ. Composition and quality of musts and wines of California grapes. Hilgardia. 1944;15:493–675. [Google Scholar]
- Anderson JT, Inouye DW, McKinney AM, Colautti RI, Mitchell-Olds T. Phenotypic plasticity and adaptive evolution contribute to advancing flowering phenology in response to climate change. Proc R Soc B. 2012;279:3843–3852. doi: 10.1098/rspb.2012.1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bienert MD, Delannoy M, Navarre C, Boutry M. NtSCP1 from tobacco is an extracellular serine carboxypeptidase III that has an impact on cell elongation. Plant Physiol. 2012;158:1220–1229. doi: 10.1104/pp.111.192088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock A, Sparks T, Estrella N, Menzel A. Changes in the phenology and composition of wine from Franconia, Germany. Clim Res. 2011;50(1):69–81. doi: 10.3354/cr01048. [DOI] [Google Scholar]
- Bourquin V, Nishikubo N, Abe H, Brumer H, Denman S, Eklund M, Christiernin M, Teeri TT, Sundberg B, Mellerowicz EJ. Xyloglucan endotransglycosylases have a function during the formation of secondary cell walls of vascular tissues. Plant Cell. 2002;14:3073–8308. doi: 10.1105/tpc.007773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braidot E, Zancani M, Petrussa E, Peresson C, Bertolini A, Patui S, Macrì F, Vianello A. Transport and accumulation of flavonoids in grapevine (Vitis vinifera L.) Plant Signal Behav. 2008;3(9):626–632. doi: 10.4161/psb.3.9.6686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunet M, Jones PD, Sigró J, Saladié O, Aguilar E, Moberg A, Della-Marta PM, Lister D, Walther A, López D. Temporal and spatial temperature variability and change over Spain during 1850–2005. J Geophys Res. 2007;112:D12117. doi: 10.1029/2006JD008249. [DOI] [Google Scholar]
- Buchanan M, Burton RA, Dhugga KS, Rafalski AJ, Tingey SV, Shirley NJ, Fincher GB. Endo-(1,4)-β-Glucanase gene families in the grasses: temporal and spatial co-transcription of orthologous genes. BMC Plant Biol. 2012;12(1):235. doi: 10.1186/1471-2229-12-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caffarra A, Eccel E. Projecting the impacts of climate change on the phenology of grapevine in a mountain area. Austr J Grape Wine R. 2011;17:52–61. doi: 10.1111/j.1755-0238.2010.00118.x. [DOI] [Google Scholar]
- Caillaud MC, Paganelli L, Lecomte P, Deslandes L, Quentin M, Pecrix Y, Le Bris M, Marfaing N, Abad P, Favery B. Spindle assembly checkpoint protein dynamics reveal conserved and unsuspected roles in plant cell division. PLoS ONE. 2009;4:e6757. doi: 10.1371/journal.pone.0006757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmona MJ, Calonje M, Martinez-Zapater JM. The FT/TFL1 gene family in grapevine. Plant Mol Biol. 2007;63:637–650. doi: 10.1007/s11103-006-9113-z. [DOI] [PubMed] [Google Scholar]
- Carreño Ruiz I (2012) Identificación de regiones cromosómicas implicadas en el control genético de caracteres de interés para la mejora genética de la uva de mesa. Proyecto de investigación, Universidad de Murcia
- Carrier G, Huang YF, Le Cunff L, Fournier-Level A, Vialet S, Souquet JM, Cheynier V, Terrier N, This P (2013) Selection of candidate genes for grape proanthocyanidin pathway by an integrative approach. Plant Physiol Biochem. doi:10.1016/j.plaphy.2013.04.014 [DOI] [PubMed]
- Chew YH, Wilczek AM, Williams M, Welch SM, Schmitt J, Halliday KJ. An augmented Arabidopsis phenology model reveals seasonal temperature control of flowering time. New Phytol. 2012;194(3):654–665. doi: 10.1111/j.1469-8137.2012.04069.x. [DOI] [PubMed] [Google Scholar]
- Chuine I, Yiou P, Viovy N, Seguin B, Daux V, Le Roy Ladurie E. Grape ripening as a past climate indicator. Nature. 2004;432:289–290. doi: 10.1038/432289a. [DOI] [PubMed] [Google Scholar]
- Clark SE, Williams RW, Meyerowitz EM. The CLAVATA1 gene encodes a putative receptor kinase that controls shoot and floral meristem size in Arabidopsis. Cell. 1997;89:575–585. doi: 10.1016/S0092-8674(00)80239-1. [DOI] [PubMed] [Google Scholar]
- Coombe BG (1988) Grape phenology. In: Coombe BG, Dry PR (eds) Viticulture, vol 1, Resources. Winetitles, Adelaide, pp 139–153
- Coombe BG. Adoption of a system for identifying grapevine growth stages. Aust J Grape Wine Res. 1995;1:104–110. doi: 10.1111/j.1755-0238.1995.tb00086.x. [DOI] [Google Scholar]
- Cosgrove DJ. Loosening of plant cell walls by expansins. Nature. 2000;407:321–326. doi: 10.1038/35030000. [DOI] [PubMed] [Google Scholar]
- Costantini L, Battilana J, Lamaj F, Fanizza G, Grando MS. Berry and phenology-related traits in grapevine (Vitis vinifera L.): from quantitative trait loci to underlying genes. BMC Plant Biol. 2008;8:38. doi: 10.1186/1471-2229-8-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dal Santo S, Tornielli GB, Zenoni S, Fasoli M, Farina L, Anesi A, Guzzo F, Delledonne M, Pezzotti M. The plasticity of the grapevine berry transcriptome. Genome Biol. 2013;14:R54. doi: 10.1186/gb-2013-14-6-r54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dal Santo S, Vannozzi A, Tornielli GB, Fasoli M, Venturini L, Pezzotti M, Zenoni S. Genome-wide analysis of the expansin gene superfamily reveals grapevine-specific structural and functional characteristics. PLoS ONE. 2013;8(4):e62206. doi: 10.1371/journal.pone.0062206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Riquelme J, Lijavetzky D, Martinez-Zapater JM, Carmona MJ. Genome-wide analysis of MIKCC-type MADS Box genes in grapevine. Plant Physiol. 2009;149:354–369. doi: 10.1104/pp.108.131052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doligez A, Bertrand Y, Dias S, Grolier M, Ballester JF, Bouquet A, This P. QTLs for fertility in table grape. Tree Genet Genomes. 2010;6(3):413–422. doi: 10.1007/s11295-009-0259-0. [DOI] [Google Scholar]
- Dry PR (1983) Recent advances in Australian viticulture. Advances in viticulture and oenology for economic gain. In: Lee TH, Somers TC (eds) Proceedings of Fifth Aust Wine Industry Tech Conference, 29 Nov–1 Dec 1983, Perth, WA, Aust Wine Res Institute, vol 1984, pp 9–23
- Du Z, Zhou X, Ling Y, Zhang Z, Su Z. agriGO: a GO analysis toolkit for the agricultural community. Nucleic Acids Res. 2010;38:W64–W70. doi: 10.1093/nar/gkq310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duchêne E, Schneider C. Grapevine and climatic changes: a glance at the situation in Alsace. Agron Sustain Dev. 2005;25:93–99. doi: 10.1051/agro:2004057. [DOI] [Google Scholar]
- Duchêne E, Huard F, Dumas V, Schneider C, Merdinoglu D. The challenge of adapting grapevine varieties to climate change. Clim Res. 2010;41:193–204. doi: 10.3354/cr00850. [DOI] [Google Scholar]
- Duchêne E, Butterlin G, Dumas V, Merdinoglu D. Towards the adaptation of grapevine varieties to climate change: QTLs and candidate genes for developmental stages. Theor Appl Genet. 2012;124(4):623–635. doi: 10.1007/s00122-011-1734-1. [DOI] [PubMed] [Google Scholar]
- Durbak AR, Tax FE. CLAVATA signaling pathway receptors of Arabidopsis regulate cell proliferation in fruit organ formation as well as in meristems. Genetics. 2011;189:177–194. doi: 10.1534/genetics.111.130930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards R, Dixon DP, Walbot V. Plant glutathione S-transferases: enzymes with multiple functions in sickness and in health. Trends Plant Sci. 2000;5:193–198. doi: 10.1016/S1360-1385(00)01601-0. [DOI] [PubMed] [Google Scholar]
- Emanuelli F, Lorenzi S, Grzeskowiak L, Catalano V, Stefanini M, Troggio M, Myles S, Martinez-Zapater JM, Zyprian E, Moreira FM, Grando MS. Genetic diversity and population structure assessed by SSR and SNP markers in a large germplasm collection of grape. BMC Plant Biol. 2013;13:39. doi: 10.1186/1471-2229-13-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanizza G, Lamaj F, Costantini L, Chaabane R. QTL analysis for fruit yield components in table grapes (Vitis vinifera) Theor Appl Genet. 2005;111:658–664. doi: 10.1007/s00122-005-2016-6. [DOI] [PubMed] [Google Scholar]
- Forcato C (2010) Gene prediction and functional annotation in the Vitis vinifera genome. In Università Degli Studi Di Padova. PhD Thesis
- Fortes AM, Agudelo-Romero P, Silva MS, Ali K, Sousa L, Maltese F, Choi YH, Grimplet J, Martinez-Zapater JM, Verpoorte R, Pais MS. Transcript and metabolite analysis in Trincadeira cultivar reveals novel information regarding the dynamics of grape ripening. BMC Plant Biol. 2011;11:149. doi: 10.1186/1471-2229-11-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fournier-Level A, Lacombe T, Le Cunff L, Boursiquot JM, This P. Evolution of the VvMybA gene family, the major determinant of berry colour in cultivated grapevine (Vitis vinifera L.) Heredity. 2010;104(4):351–362. doi: 10.1038/hdy.2009.148. [DOI] [PubMed] [Google Scholar]
- Fraser CM, Rider LW, Chapple C. An expression and bioinformatics analysis of the Arabidopsis serine carboxypeptidase-like gene family. Plant Physiol. 2005;138:1136–1148. doi: 10.1104/pp.104.057950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambetta GA, Matthews MA, Shaghasi TH, McElrone AJ, Castellarin SD. Sugar and abscisic acid signaling orthologs are activated at the onset of ripening in grape. Planta. 2010;232:219–234. doi: 10.1007/s00425-010-1165-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghelardini L, Falusi M, Santini A. Variation in timing of bud-burst of Ulmus minor clones from different geographical origins. Can J Forest Res. 2006;36:1982–1991. doi: 10.1139/x06-092. [DOI] [Google Scholar]
- Grimplet J, Van Hemert J, Carbonell-Bejerano P, Diaz-Riquelme J, Fennell A, Pezzotti M, Martinez-Zapater JM. Comparative analysis of grapevine whole-genome predictions, functional annotation and categorization of the predicted gene sequences. BMC Res notes. 2012;5:213. doi: 10.1186/1756-0500-5-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannah L, Roehrdanz PR, Ikegami M, Shepard AV, Shaw MR, Tabor G, Zhi L, Marquet PA, Hijmans RJ. Climate change, wine, and conservation. PNAS. 2013 doi: 10.1073/pnas.1210127110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter AF, Lechowicz MJ. Predicting the timing of budburst in temperate trees. J Appl Ecol. 1992;25:597–604. doi: 10.2307/2404467. [DOI] [Google Scholar]
- Jackson RS. Wine science: principles and applications. San Diego: Elsevier; 2008. [Google Scholar]
- Jaillon O, Aury JM, Noel B, Policriti A, Clepet C, Casagrande A, Choisne N, Aubourg S, Vitulo N, Jubin C, Vezzi A, Legeai F, Hugueney P, Dasilva C, Horner D, Mica E, Jublot D, Poulain J, Bruyère C, Billault A, Segurens B, Gouyvenoux M, Ugarte E, Cattonaro F, Anthouard V, Vico V, Del Fabbro C, Alaux M, Di Gaspero G, Dumas V, Felice N, Paillard S, Juman I, Moroldo M, Scalabrin S, Canaguier A, Le Clainche I, Malacrida G, Durand E, Pesole G, Laucou V, Chatelet P, Merdinoglu D, Delledonne M, Pezzotti M, Lecharny A, Scarpelli C, Artiguenave F, Pè ME, Valle G, Morgante M, Caboche M, Adam-Blondon A-F, Weissenbach J, Quétier F, Wincker P. The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature. 2007;449:463–467. doi: 10.1038/nature06148. [DOI] [PubMed] [Google Scholar]
- Jones GV, White MA, Cooper OR, Storchmann K. Climate change and global wine quality. Clim Change. 2005;73:319–343. doi: 10.1007/s10584-005-4704-2. [DOI] [Google Scholar]
- Keller M. Managing grapevines to optimise fruit development in a challenging environment: a climate change primer for viticulturists. Aust J Grape Wine Res. 2010;16:56–69. doi: 10.1111/j.1755-0238.2009.00077.x. [DOI] [Google Scholar]
- Kline KG, Sussman MR, Jones AM. Abscisic acid receptors. Plant Physiol. 2010;154:479–482. doi: 10.1104/pp.110.160846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi S, Goto-Yamamoto N, Hirochika H. Retrotransposon-induced mutations in grape skin color. Science. 2004;304:982. doi: 10.1126/science.1095011. [DOI] [PubMed] [Google Scholar]
- Kottek M, Grieser J, Beck C, Rudolf B, Rubel F. World map of Koeppen-Geiger climate classification updated. Meteorol Z. 2006;15:259–263. doi: 10.1127/0941-2948/2006/0130. [DOI] [Google Scholar]
- Lehfeldt C, Shirley AM, Meyer K, Ruegger MO, Cusumano JC, Viitanen PV, Strack D, Chapple C. Cloning of the SNG1 gene of Arabidopsis reveals a role for a serine carboxypeptidase-like protein as an acyltransferase in secondary metabolism. Plant Cell. 2000;12:1295–1306. doi: 10.1105/tpc.12.8.1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann E. Nonparametrics. New York: McGraw-Hill; 1975. [Google Scholar]
- Lieth H, editor. Phenology and seasonality modelling. New York: Springer; 1974. [Google Scholar]
- Llave C, Xie Z, Kasschau KD, Carrington JC. Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science. 2002;297:2053–2056. doi: 10.1126/science.1076311. [DOI] [PubMed] [Google Scholar]
- Martinez-Zapater JM, Carmona MJ, Diaz-Riquelme J, Fernandez L, Lijavetzky D. Grapevine genetics after the genome sequence: challenges and limitations. Aust J Grape Wine Res. 2010;16:33–46. doi: 10.1111/j.1755-0238.2009.00073.x. [DOI] [Google Scholar]
- Morris JR, Main GL. Response of Concord grapevines to varied shoot positioning and pruning methods in a warm, long-season growing region. Am J Enol Vitic. 2010;61:201–213. doi: 10.5344/ajev.2010.10005. [DOI] [Google Scholar]
- Motose H, Iwamoto K, Endo S, Demura T, Sakagami Y, Matsubayashi Y, Moore KL, Fukuda H. Involvement of phytosulfokine in the attenuation of stress response during the transdifferentiation of Zinnia mesophyll cells into tracheary elements. Plant Phys. 2009;150:437–447. doi: 10.1104/pp.109.135954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura M, Katsumata H, Abe M, Yabe N, Komeda Y, Yamamoto K, Takahashi T. Characterisation of the class IV homeodomain-leucine zipper gene family in Arabidopsis. Plant Physiol. 2006;141:1363–1375. doi: 10.1104/pp.106.077388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicotra AB, Atkin OK, Bonser SP, Davidson A, Finnegan EJ, Mathesius U, Poot P, Purugganan MD, Richards CL, Valladares F, van Kleunen M. Plant phenotypic plasticity in a changing climate. Trends Plant Sci. 2010;15:684–692. doi: 10.1016/j.tplants.2010.09.008. [DOI] [PubMed] [Google Scholar]
- Nunan KJ, Davies C, Robinson SP, Fincher GB. Expression patterns of cell wall-modifying enzymes during grape berry development. Planta. 2001;214:257–264. doi: 10.1007/s004250100609. [DOI] [PubMed] [Google Scholar]
- Parage C, Tavares R, Rety S, Baltenweck-Guyot R, Poutaraud A, Renault L, Heintz D, Lugan R, Marais G, Aubourg S, Hugueney P. Structural, functional, and evolutionary analysis of the unusually large stilbene synthase gene family in grapevine. Plant Physiol. 2012;160:1407–1419. doi: 10.1104/pp.112.202705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadras VO, Reynolds M, De la Vega AJ, Petrie PR, Robinson R. Phenotypic plasticity of yield and phenology in wheat, sunflower and grapevine. Field Crops Res. 2009;110:242–250. doi: 10.1016/j.fcr.2008.09.004. [DOI] [Google Scholar]
- Sánchez LA, Dokoozlian NK. Bud microclimate and fruitfulness in Vitis vinifera L. Am J Enol Vitic. 2005;56:319–329. [Google Scholar]
- Sansavini S, Fanigliulo G. Bud fertility and effects of pruning of seedless grapevine varieties Centennial Seedless and Sugraone [Basilicata - Vitis vinifera L.] Rivista di Frutticoltura e di Ortofloricoltura. 1998;60(2):55–60. [Google Scholar]
- Schmitz RJ, Hong L, Michaels S, Amasino RM. FRIGIDA-ESSENTIAL 1 interacts genetically with FRIGIDA and FRIGIDA-LIKE 1 to promote the winter-annual habit of Arabidopsis thaliana. Development. 2005;132:5471–5478. doi: 10.1242/dev.02170. [DOI] [PubMed] [Google Scholar]
- Schnelle F. Pflanzen-Phänologie. Leipzig: Akademische Verlagsgesellschaft; 1955. [Google Scholar]
- Schultz HR. Climate change and viticulture: a European perspective on climatology, carbon dioxide and UV-B effects. Austr J Grape Wine R. 2000;6:2–12. doi: 10.1111/j.1755-0238.2000.tb00156.x. [DOI] [Google Scholar]
- Schulze S, Schäfer BN, Parizotto EA, Voinnet O, Theres K. LOST MERISTEMS genes regulate cell differentiation of central zone descendants in Arabidopsis shoot meristems. Plant J. 2010;64:668–678. doi: 10.1111/j.1365-313X.2010.04359.x. [DOI] [PubMed] [Google Scholar]
- Sommer KJ, Islam MT, Clingeleffer PR. Light and temperature effects on shoot fruitfulness in Vitis vinifera L. cv. Sultana: influence of trellis type and grafting. Austr J Grape Wine R. 2000;6:99–108. doi: 10.1111/j.1755-0238.2000.tb00167.x. [DOI] [Google Scholar]
- Sparks TH, Menzel A. Observed changes in seasons: an overview. Int J Climatol. 2002;22:1715–1725. doi: 10.1002/joc.821. [DOI] [Google Scholar]
- Tomasi D, Jones GV, Giust M, Lovat L, Gaiotti F. Grapevine phenology and climate change: relationships and trends in the Veneto Region of Italy for 1964–2009. Am J Enol Vitic. 2011;62:329–339. doi: 10.5344/ajev.2011.10108. [DOI] [Google Scholar]
- Troggio M, Malacarne G, Coppola G, Segala C, Cartwright DA, Pindo M, Stefanini M, Mank R, Moroldo M, Morgante M, Grando MS, Velasco R. A dense single-nucleotide-polymorphism-based genetic linkage map of grapevine (Vitis vinifera L.) anchoring Pinot Noir bacterial artificial chromosome contigs. Genetics. 2007;176:2637–2650. doi: 10.1534/genetics.106.067462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unver T, Parmaksız İ, Dündar E. Identification of conserved micro-RNAs and their target transcripts in opium poppy (Papaver somniferum L.) Plant Cell Rep. 2010;29:757–769. doi: 10.1007/s00299-010-0862-4. [DOI] [PubMed] [Google Scholar]
- Van Leeuwen C, Garnier C, Agut C, Baculat B, Barbeau G, Besnard E, Bois B, Boursiquot JM, Chuine I, Dessup T, Dufourcq T, Garcia-Cortazar I, Marguerit E, Monamy C, Koundouras S, Payan JC, Parker A, Renouf V, Rodriguez-Lovelle B, Roby JP, Tonietto J, Trambouze W (2008) Heat requirements for grapevine varieties is essential information to adapt plant material in a changing climate. In: Proceedings of VIIth International Terroir Congress, 20–23 May, Nyon, Switzerland, vol 1, pp 222–227
- van Ooijen JW (2009) MapQTL® 6: Software for the mapping of quantitative trait loci in experimental populations of diploid species. Wageningen, Netherlands, Kyazma BV
- Velasco R, Zharkikh A, Troggio M, Cartwright DA, Cestaro A, Pruss D, Pindo M, FitzGerald LM, Vezzulli S, Reid J, Malacarne G, Iliev D, Coppola G, Wardell B, Micheletti D, Macalma TM, Facci M, Mitchell JT, Perazzolli M, Eldredge G, Gatto P, Oyzerski R, Moretto M, Gutin N, Stefanini M, Chen Y, Segala C, Davenport C, Demattè L, Mraz A, Battilana J, Stormo K, Costa F, Tao Q, Si-Amour A, Harkins T, Lackey A, Perbost C, Taillon B, Stella A, Solovyev V, Fawcett JA, Sterck L, Vandepoele K, Grando MS, Toppo S, Moser C, Lanchbury J, Bogden R, Skolnick M, Sgaramella V, Bhatnagar SK, Fontana P, Gutin A, Van de Peer Y, Salamini F, Viola R. High quality draft consensus sequence of the genome of a heterozygous grapevine variety. PLoS ONE. 2007;2:e1326. doi: 10.1371/journal.pone.0001326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Guo YH, Wu CG, Yang GD, Li YY, Zheng CH. Genome-wide analysis of CCCH zinc finger family in Arabidopsis and rice. BMC Genomics. 2008;9:44. doi: 10.1186/1471-2164-9-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb LB, Whetton PH, Barlow EWR. Modelled impact of future climate change on the phenology of winegrapes in Australia. Austr J Grape Wine Res. 2007;13:165–175. doi: 10.1111/j.1755-0238.2007.tb00247.x. [DOI] [Google Scholar]
- Webb LB, Whetton PH, Barlow EWR. Climate change and winegrape quality in Australia. Clim Res. 2008;36:99–111. doi: 10.3354/cr00740. [DOI] [Google Scholar]
- Webb LB, Dunn G, Barlow EWR. Adapting agriculture to climate change: preparing Australian agriculture, forestry and fisheries for the future. In: Stokes CJ, Howden SM, editors. Viticulture. Melbourne: CSIRO Publishing; 2010. [Google Scholar]
- Wilczek AM, Burghardt LT, Cobb AR, Cooper MD, Welch SM, Schmitt J. Genetic and physiological bases for phenological responses to current and predicted climates. Philos Trans R Soc B Biol Sci. 2010;365:3129–3147. doi: 10.1098/rstb.2010.0128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Xiao H, Cabrera A, Meulia T, van der Knaap E. SUN regulates vegetative and reproductive organ shape by changing cell division patterns. Plant Physiol. 2011;157:1175–1186. doi: 10.1104/pp.111.181065. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.