Abstract
Tight junctions, or zonula occludens, are the most apical component of the junctional complex and provide one form of cell–cell adhesion in epithelial and endothelial cells. Nearly 90% of malignant tumors are derived from the epithelium. Loss of cell–cell adhesion is one of the steps in the progression of cancer to metastasis. At least three main tight junction family proteins have been discovered: occludin, claudin, and junctional adhesion molecule (JAM). Claudins are the most important structural and functional components of tight junction integral membrane proteins, with at least 24 members in mammals. They are crucial for the paracellular flux of ions and small molecules. Overexpression or downregulation of claudins is frequently observed in epithelial-derived cancers. However, molecular mechanisms by which claudins affect tumorigenesis remain largely unknown. As the pivotal proteins in epithelial cells, altered expression and distribution of different claudins have been reported in a wide variety of human malignancies, including pancreatic, colonic, lung, ovarian, thyroid, prostate, esophageal, and breast cancers. In this review, we will give the readers an overall picture of the changes in claudin expression observed in various cancers and their mechanisms of regulation. Downregulation of claudins contributes to epithelial transformation by increasing the paracellular permeability of nutrients and growth factors to cancerous cells. In the cases of upregulation of claudin expression, the barrier function of the cancerous epithelia changes, as they often display a disorganized arrangement of tight junction strands with increased permeability to paracellular markers. Finally, we will summarize the literature suggesting that claudins may become useful biomarkers for cancer detection and diagnosis as well as possible therapeutic targets for cancer treatment.
Keywords: tight junctions, claudins, human cancers
Introduction to tight junctions and cell–cell adhesion
Cell-to-cell adhesion in epithelial cell sheets is maintained mainly through two types of junctions: adherens junction and tight junction. A number of studies have focused their attention on the transmembrane protein of the adherens junction.1,2 Only in recent years has the importance of tight junction proteins in epithelial cell proliferation, survival, apoptosis, and differentiation been recognized.
Tight junctions are involved in cell-to-cell adhesion and serve two major functions in epithelial cell layers: the barrier (or gate) function and the fence function. The barrier function of tight junctions regulates the passage of ions, water, and various macromolecules, even cancer cells, through paracellular spaces. Thus, the barrier function is relevant to edema, jaundice, diarrhea, and blood-borne metastasis. On the other hand, the fence function maintains the cell polarity. In other words, tight junctions work as a fence to prevent the intermixing of molecules in the apical membrane with those in the lateral membrane. This fence function is highly involved in cancer cell biology because loss of cell polarity occurs in many types of cancer cells. The fence and barrier functions of the tight junction have a common feature of compartmentalization: the fence function is performed at the subcellular level and the barrier function is performed at the organ level. Finally, because of the ability of tight junction proteins to recruit signaling proteins, tight junctions have also been hypothesized to be involved in the regulation of cell proliferation, differentiation, and many other cellular functions.3,4
Introduction to claudins
Claudins were first cloned and named in 1998 by Mikio et al.5 The name “claudin” comes from the Latin word “claudere” (“to close”), suggesting the barrier role of these proteins. Claudins are a multigene family and encode tetraspan membrane proteins that are crucial structural and functional components of tight junctions. Claudins have important roles in regulating paracellular permeability and maintaining cell polarity in epithelial and endothelial cell sheets.6,7 There are currently 24 claudin members in mammals, but claudin-13 is missing in humans. Claudin members are divided into classic and non-classic claudins based on their sequence similarity. Sequence similarity is determined by the alignment and phylogenetic tree analysis of whole-length sequences of claudins. Classic claudins include claudins 1–10, 14, 15, 17, and 19, and non-classic claudins contain claudins 11–13, 16, 18, and 20–24.8 Classic claudins exhibit a much stronger sequence homology than non-classic claudins. The subdivision of claudin members into classic and non classic groups was initially suggested from the sequence analysis of mouse claudin proteins. This analysis was subsequently extended to human claudins.7,8
In recent years, the roles of claudin proteins in human physiology and pathophysiology, including carcinoma development, are just beginning to be unraveled. Several claudin knockout mouse models have been generated over the past 10 years, and their diverse phenotypes clearly demonstrate the importance of claudin proteins in maintaining the tissue integrity and homeostasis in various organs. Although the underlying mechanisms of claudin regulation in various tissues and their exact roles in normal physiology as well as in disease states are being elucidated, more research work remains to be done.9–14 In this review, we discuss the conceptual framework concerning claudins and their potential implications in cancer. We anticipate that, in the next several years, our understanding of the potential role of claudins in the regulation of tumorigenesis will be significantly increased, which may, in turn, provide new approaches for the targeted therapy.
Role of claudins in tight junctions
Tight junctions seal the paracellular cleft of epithelia and endothelia and form crucial barriers between tissue compartments. Tight junctions consist of integral membrane proteins including occludin, claudins, and tight junction-associated proteins, such as ZO-1. Recent studies demonstrate the multilateral interactions between tight junction proteins.3,4,6 Of these molecules, claudins are responsible for the formation of tight junction strands and are connected to the actin cytoskeleton mediated by ZO-1.3,4,6
Claudins are the most important components of tight junctions. They form the paracellular barrier that controls the flux of ions and small molecules in the intercellular space between epithelial cells. Claudins have four transmembrane domains with N- and C-termini in the cytoplasm. The first extracellular loop consists of about 53 amino acids and the second one is much smaller, with about 24 amino acids. The N-terminal end is usually very short (four to ten amino acids), while the C-terminal end ranges from 21 to 63 amino acids and is necessary for the localization of these proteins in the tight junctions (see Figure 1). It is anticipated that the cysteines of an individual claudin can form intramolecular or intermolecular disulfide bonds. All human claudins (with the exception of claudin-12) contain a motif at the C-terminus that let them bind to PDZ (PSD-95/DLG/ZO-1) domains of scaffold proteins.15–17
Figure 1.
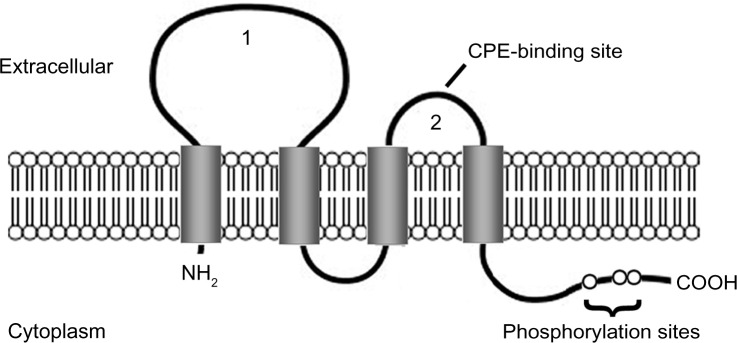
Claudin protein structure.
Notes: The claudin protein consists of four transmembrane domains and two extracellular loops (1 and 2). The N- and C-termini are located in the cytoplasm. The second extracellular loop has a binding site for Clostridium perfringens enterotoxin (CPE) in claudin-3 and -4. The C-terminal region contains phosphorylation sites that may be involved in protein–protein interactions and signal transduction.
Claudins are critical for sealing the epithelial sheets and controlling paracellular flux of ions and small molecules. Claudins are present in normal tissues, hyperplastic conditions, benign neoplasms, and cancers that exhibit epithelial differentiation. Loss of claudin expression has been reported in several malignancies. Differential expression of various claudin family members in cancer can potentially be used to confirm the histologic identity of certain types of cancer and exclude others.7,9–13
The expression pattern of claudins is highly tissue-specific, and most tissues express multiple claudins. Claudins can interact with claudins from adjacent cells in a homotypic or heterotypic fashion to form tight junctions. Many studies suggest that the combination of claudins determines the selectivity and permeability of tight junctions in a given tissue.6–8 Claudins can be polymerized together between epithelial cells to form an adhesive structure.
Role of claudins and tight junctions in cancer
The function of claudins in the maintenance of normal epithelial cell homeostasis has been well studied; however, their role in the process of tumorigenesis is less clear. The exact biological significance of altered claudin expression in cancer remains largely unknown.
Cancer cells can spread from the primary site to other parts of the body, a process known as metastasis. During cancer metastasis, several important steps need to be undertaken. An important step in cancer progression is the epithelial–mesenchymal transition. During this process, epithelial cells downregulate cell–cell adhesion structures, alter their polarity, reorganize their cytoskeleton, and become isolated and motile.18 Tight junctions are involved in this cancer metastatic process. Claudin-6 decreases in breast invasive ductal carcinomas and is inversely correlated with lymph node metastasis.19 Reduced expression of claudin-7 is correlated with higher tumor grade and locoregional and distant metastases, including locoregional recurrences.20 It has been reported that claudin-2 level is elevated in liver metastases. The first claudin-2 extracellular loop is essential for mediating tumor cell–hepatocyte interaction and the ability of breast cancer cells to form liver metastases in vivo.21 Claudin-3 and -4 control tumor growth and metastases by sustaining expression of E-cadherin and limiting β-catenin signaling.22
The association between altered claudin expression and cancer has been widely reported since the discovery of claudins.23,24 As shown in Table 1,25–90 the level of claudin-1 has been found to be reduced in breast cancer as well as in colon cancer.28,30 Claudin-7 has also been found to be downregulated in invasive breast cancer and in head and neck cancers.66,68 These reports of decreased tight junction protein expression in cancer are consistent with the generally accepted idea that tumorigenesis is accompanied by the disruption of tight junctions, a process that may play an important role in the loss of cohesion, invasiveness, and lack of differentiation observed in cancer cells. Paradoxically, other studies have shown that certain claudin proteins are upregulated in cancer (see Table 1).29,67 In fact, many published studies reported an overexpression of claudins in various cancers (see Table 1).35,36,56,58 For example, gene expression study of ovarian cancer showed that claudin-3 and -4 were among the most highly upregulated genes in this cancer. Several additional reports have since confirmed the high expression of these two claudins in ovarian cancer. In addition, claudin-3 and -4 have also been reported to have an increased expression in other cancers, such as breast, prostate, and pancreatic cancers.
Table 1.
Changes in claudin protein or mRNA expression in human tumor samples
| Protein | Tumor type | Expression | Reference |
|---|---|---|---|
| Claudin-1 | Colonic | ↑ | 29 |
| Squamous cell carcinoma | ↑ | 31 | |
| Cervical | ↑ | 32 | |
| Gastric | ↑ | 35 | |
| Esophageal | ↑ | 36 | |
| Biliary tract | ↑ | 37 | |
| Keratinized (pearl) | ↑ | 38 | |
| Thyroid | ↑ | 39 | |
| Squamous cell carcinoma of the tongue | ↑ | 40 | |
| Urothelial carcinoma | ↑ | 41 | |
| Hepatoblastoma | ↑ | 42 | |
| Squamous cell carcinoma of the lung | ↑ | 45 | |
| Pancreatic | ↑ | 46 | |
| Breast | ↓ | 28 | |
| Colonic | ↓ | 30 | |
| Glioblastoma multiforme | ↓ | 33 | |
| Melanoma | ↓ | 34 | |
| Hepatocellular carcinoma | ↓ | 43 | |
| Adenocarcinoma of the lung | ↓ | 45 | |
| Claudin-2 | Esophageal | ↑ | 36 |
| Colonic | ↑ | 48,49 | |
| Hepatoblastoma | ↑ | 51 | |
| Lung | ↑ | 26 | |
| Breast | ↓ | 47 | |
| Cervical | ↓ | 50 | |
| Prostate | ↓ | 52 | |
| Claudin-3 | Gastric | ↑ | 35 |
| Esophageal | ↑ | 36 | |
| Urothelial carcinoma | ↑ | 41 | |
| Renal cell carcinoma | ↑ | 44 | |
| Breast | ↑ | 53 | |
| Colonic | ↑ | 54 | |
| Prostate | ↑ | 55 | |
| Ovarian | ↑ | 56 | |
| Pancreatic | ↑ | 57 | |
| Breast | ↓ | 47 | |
| Hepatoblastoma | ↓ | 51 | |
| Claudin-4 | Squamous cell carcinoma | ↑ | 31 |
| Esophageal | ↑ | 36 | |
| Biliary tract | ↑ | 37 | |
| Squamous cell carcinoma of the tongue | ↑ | 40 | |
| Urothelial carcinoma | ↑ | 41 | |
| Renal cell carcinoma | ↑ | 44 | |
| Breast | ↑ | 53 | |
| Colonic | ↑ | 54 | |
| Prostate | ↑ | 55 | |
| Ovarian | ↑ | 56 | |
| Gastric | ↑ | 58 | |
| Pancreatic | ↑ | 60 | |
| Thyroid | ↑ | 63 | |
| Lung | ↑ | 27 | |
| Hepatoblastoma | ↓ | 42 | |
| Breast | ↓ | 47 | |
| Gastric | ↓ | 59 | |
| Colonic | ↓ | 61 | |
| Cervical | ↓ | 62 | |
| Claudin-5 | Adenocarcinoma of the lung | ↑ | 45 |
| Pancreatic | ↑ | 57 | |
| Lung | ↑ | 27 | |
| Glioblastoma multiforme | ↓ | 33 | |
| Squamous cell carcinoma of the lung | ↓ | 45 | |
| Breast | ↓ | 47 | |
| Claudin-6 | Gastric adenocarcinoma | ↑ | 35 |
| Breast (cell) | ↓ | 64 | |
| Claudin-7 | Cervical | ↑ | 32 |
| Squamous cell carcinoma of the tongue | ↑ | 40 | |
| Urothelial carcinoma | ↑ | 41 | |
| Thyroid | ↑ | 63 | |
| Breast | ↑ | 67 | |
| Colonic | ↑ | 69,70 | |
| Gastric | ↑ | 73 | |
| Esophageal | ↑ | 74 | |
| Chromophobe renal cell carcinoma | ↑ | 76 | |
| Ovarian | ↑ | 78 | |
| Hepatocellular carcinoma (mRNA) | ↑ | 79 | |
| Hepatoblastoma | ↓ | 42 | |
| Breast | ↓ | 66 | |
| Head and neck squamous cell carcinoma | ↓ | 68 | |
| Colonic | ↓ | 71,72 | |
| Esophageal | ↓ | 75 | |
| Uterus | ↓ | 77 | |
| Lung | ↓ | 25 | |
| Claudin-8 | Biliary tract | ↓ | 37 |
| Colonic (mRNA) | ↓ | 80 | |
| Claudin-9 | Gastric adenocarcinoma (cell) | ↑ | 65 |
| Claudin-10 | Hepatocellular carcinoma (mRNA) | ↑ | 81 |
| Lung adenocarcinomas | ↑ | 82 | |
| Thyroid papillary (mRNA) | ↑ | 83 | |
| Biliary tract | ↓ | 37 | |
| Claudin-11 | Bladder (mRNA) | ↓ | 84 |
| Claudin-12 | Colonic (mRNA) | ↑ | 80 |
| Claudin-16 | Breast (mRNA) | ↑ | 90 |
| Ovarian (mRNA) | ↑ | 85 | |
| Breast | ↓ | 86 | |
| Claudin-18 | Lung adenocarcinomas | ↑ | 82 |
| Pancreatic | ↑ | 88 | |
| Cholangiocarcinoma | ↑ | 88 | |
| Gastric (mRNA) | ↓ | 87 | |
| Claudin-23 | Gastric (mRNA) | ↓ | 89 |
Abbreviation: mRNA, messenger RNA.
The loss of claudins and other tight junction proteins in cancer has been interpreted as a mechanism for the loss of cell adhesion and an important step in the progression of cancer to metastasis. On the other hand, as discussed previously, several claudins, including claudin-3 and -4 are often upregulated in many types of cancer (Table 1), suggesting that these proteins may have a positive effect on tumorigenesis. Recent research has shown that, at least in the case of ovarian cells, the expression of claudin-3 and -4 may lead to an increase in cell invasion, motility, and survival, all of which are characteristics important for metastasis.91 Consistent with these in vitro findings is a report that claudin-4 expression in pancreatic intraductal papillary mucinous neoplasms was associated with a more invasive phenotype.92 Similarly, expression of claudin-3 and -4 was observed in advanced ovarian cancer but not in ovarian cystadenomas.93,94 It has been reported, however, that re-expression of claudin-1 in breast cancer cells can lead to the increased apoptosis in three-dimensional cultures.95 Therefore, the functions of claudins in cancer may be highly tissue-specific and may depend on the type and stage of cancer.
Several other claudin members have been reported to play an important role in various types of tissues; however, their roles in tumorigenesis are unknown. For example, CLDN13 is a mouse-specific gene mainly expressed in tissues associated with hematopoietic function.96 Claudin-13 messenger RNA (mRNA) and protein are observed in colon, but they are undetectable in the rest of the intestines.97 Mutations in CLDN14 gene are known to be involved in autosomal recessive deafness in humans.98 Baker et al recently reported that CLDN14 heterozygous mice, but not CLDN14 null mice, displayed several blood vessel-related phenotypes, including abnormal distribution of basement membrane laminin around tumor blood vessels, increased intratumoral leakage, and enhanced endothelial cell proliferation.99 Patients with CLDN19 mutations have a high risk of progression to chronic renal disease.100 In human polycystic kidneys, decreased expression and dyslocalization of claudin-19 are noticed, suggesting a possible correlation between claudin-19 and renal disorders.101 Presently, not much is known about tissue distributions or the physiological functions of claudins 20–27. Claudin-21 and claudins 24–27 are expressed in the intestines, stomach, liver, and kidney.102
The mechanisms responsible for the differential expression of claudins in cancer are largely unknown; however, recent studies have identified several growth factors, cytokines, and transcription factors that affect claudin expression.25,103 The tumor-promoting factors, hepatocyte growth factor and epidermal growth factor, have been shown to decrease claudin-7 expression and increase claudin-1, -3, and -4 expressions, respectively.25,103
Critical analysis of the potential for targeting claudin proteins in cancer
In recent years, claudin’s potential value as a target for therapeutic intervention has been increasingly recognized regardless of its roles in tumorigenesis. This is because claudin proteins are expressed at the cell surface and contain two extracellular domains that can serve as potential targeting sites (Figure 1). In addition, many studies (see Table 1) report that some claudins are overexpressed in certain types of cancer while others are downregulated in different types of cancer, thus creating differential expression patterns between tumor and normal cells. Tight junction permeability is often higher in tumor tissues than in normal tissues, which makes claudins more accessible. Therefore, a therapeutic intervention could be achieved.
Due to the high specificity of claudin expression patterns in cancer, it has been suggested that claudins may serve as useful molecular biomarkers for certain cancers. For example, claudin expression may be used as a prognostic indicator because low claudin-1 expression has been shown to be associated with a poor prognosis in stage II colon cancer.30 Claudin-10 expression has also been shown to be an independent prognostic factor for hepatocellular carcinoma recurrence after curative hepatectomy.81,104
It is known that claudin-3 and -4 are receptors for the bacterial toxin Clostridium perfringens enterotoxin (CPE). CPE is a single polypeptide of 35 kDa. Upon binding to its receptors, CPE induces cytolysis through its effects on membrane permeability. The high expression levels of claudin-3 and -4 in multiple types of cancer may provide a unique opportunity for anticancer therapy using CPE. Prostate adenocarcinoma cells expressing claudin-3 and -4 have been shown to be sensitive to CPE-mediated cytolysis.55,105,106 This CPE-mediated cytolysis was specific because prostate cancer cells lacking claudin-3 and -4 were unaffected by CPE treatment. Similar experiments have also shown that breast, ovarian, and pancreatic cancer cells were sensitive to CPE treatment as long as these cancer cells were expressing claudin-3 and/or -4.53,105,107 The challenge, however, is that claudin-3 and/or -4 are also expressed in several normal human tissues, including gut, lungs, and kidneys. This could hinder the use of CPE for potential anti-cancer therapy. Therefore, whether this approach will be useful in the clinical setting remains to be seen.
To overcome this hurdle, it has been suggested that a nontoxic C-terminal CPE fragment could be delivered locally to normal tissues and prevent CPE toxicity during cancer treatment. Other potential problems with the use of CPE in cancer treatment include the possible immune response against CPE in patients receiving the treatment, the level of surface claudin expression, and the penetration of CPE into the tumor mass.53,55,105–107 Therefore, additional studies are required to solve these issues before CPE can be used as a therapeutic approach.
A study has shown that claudin peptides can be internalized by specific and nonspecific pathways.108 The cellular uptake of the claudin-1 peptide follows the clathrin-mediated endocytosis as indicated by inhibitors and respective tracers for colocalization. In addition, macropinocytosis and caveolae-mediated endocytosis of the peptide have also been observed. In contrast, the claudin-5 peptide is mainly internalized via the caveolae-mediated endocytosis as evidenced by the colocalization with respective tracers and vesicle markers, whereas the nonselective macropinocytosis seems to be involved in a less effective manner.108
Since claudins are transmembrane proteins and contain two extracellular loops, they may also offer promising targets for antibody-based therapy. Antibodies that recognize different claudins’ extracellular loops have been produced and shown to specifically bind to claudins on the surface of the cell, providing a proof of principle for this approach. Besides antibody-based treatment, many small molecules or compounds can affect tight junction functions and, therefore, could potentially be used for anticancer therapy.109 For example, it has been reported that hydroxycamptothecin-loaded Fe3O4 nanoparticles induce human HCC827 lung cancer cell apoptosis and disrupt tight junctions by internalizing claudin-1, -3, -4, and -7 proteins and decreasing their protein expressions.110 HangAmDan-B, an anti-invasive agent extracted from eight Korean medical animals and plants, can inhibit the cell migration and invasion of NCI-H460 lung cancer cells and tighten the tight junctions through down-regulating claudin-1, -2, -3, and -4 at both transcriptional and translational levels.111 Butyrate improves the barrier function of intestinal epithelia and has the potential to treat inflammatory bowel disease. It restores the tight junctions in inflammatory bowel disease patients by downregulating the expression of claudin-1 and -2 and upregulating occludin, cingulin, ZO-1, and ZO-2.112
It has been suggested from the gene expression studies of breast and ovarian cancers56,67 that the identification of specific expression patterns of claudin family members could be used as potential biomarkers for these cancers. A cancer biomarker refers to a substance or process that is indicative of the presence of cancer in the body. Not all of the claudins in Table 1 are considered as biomarkers. Up until now, only certain claudins could be an indicative marker for diagnosis and disease progression. For example, claudin-7 may serve as an immunohistochemical biomarker in the differential diagnosis of chromophobe renal cell carcinoma and oncocytoma; claudin-4 could serve as an immunohistochemical biomarker in the differential diagnosis of undifferentiated pancreatic cancers and highly invasive pancreatic cancers; and the expression of claudin-10 in hepatocellular carcinomas can potentially serve to predict disease recurrence after curative hepatectomy.60,76,81 These findings are important because changes in expression patterns of claudins allow for detection, diagnosis, and potential treatment of drug-resistant cancers. Clinical trials are certainly required to establish this potential, but research studies on claudin functions and regulations as well as their roles in cancer are critically needed for providing important insight in relation to normal and neoplastic cellular physiology.
The fact that the expression levels of many claudin family members are altered in various types of cancers and that claudins are localized at the cell surface make claudins potentially attractive molecular targets for cancer therapy. The clinical application of using claudin-targeted therapy, however, will present several challenges. The most important challenge is the systemic toxicity. Because claudins are expressed in the epithelia of most organs in the body, the toxicity issue must be addressed. The small molecules or synthetic compounds designed to disrupt tight junctions may also be a double-edged sword, because an increase in tight junction permeability can enhance the uptake of anticancer agents, but, at the same time, it may also increase the nutritional uptake to promote tumor progression. Therefore, further studies, especially at the animal level, are needed to determine the toxicity issue of claudin-targeted therapy and to evaluate whether this approach can be applied safely and practically in a clinical setting.
Claudins play an indispensable role in tissue homeostasis and barrier formation; our knowledge about the role of claudin proteins in cancer biology is, however, still limited. Nevertheless, with our increasing understanding of specific functions of claudins in various cancers, claudins may serve as promising tumor biomarkers as well as therapeutic targets in the near future.
Conclusion
Claudins are the main structural and functional proteins of tight junctions in epithelial cells and maintain the tissue homeostasis through regulating epithelial barriers, paracellular transport, and signal transduction. Numerous studies have demonstrated the altered expression patterns of different claudin members in a variety of diseases, particularly in cancers. Identifying the specific functions of claudins in tumorigenesis has become one of the main research focuses in the field of cell biology. It is still unclear what the association between altered claudin expression and tumorigenesis is, despite great research efforts in the last decade. With rapid research advancement and newly developed technology, however, the role of claudins in cancer and its clinical applications in cancer detection, diagnosis, and potential therapeutic intervention may become a valid approach in the near future.
Acknowledgments
This work was supported by National Natural Science Foundation of China (81372585) (LD); the National Natural Science Foundation of China (31200581) and the Foundation from Hangzhou Science and Technology Bureau (20120633B26) to ZL, and the National Institute of Health grants (ES016888) to YHC.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Lampugnani MG. Endothelial cell-to-cell junctions: adhesion and signaling in physiology and pathology. Cold Spring Harb Perspect Med. 2012;2((10). pii) doi: 10.1101/cshperspect.a006528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liebner S, Cavallaro U, Dejana E. The multiple languages of endothelial cell-to-cell communication. Arterioscler Thromb Vasc Biol. 2006;26(7):1431–1438. doi: 10.1161/01.ATV.0000218510.04541.5e. [DOI] [PubMed] [Google Scholar]
- 3.Tsukita S, Furuse M, Itoh M. Multifunctional strands in tight junctions. Nat Rev Mol Cell Biol. 2001;2(4):285–293. doi: 10.1038/35067088. [DOI] [PubMed] [Google Scholar]
- 4.Schneeberger EE, Lynch RD. The tight junction: a multifunctional complex. Am J Physiol Cell Physiol. 2004;286(6):C1213–C1228. doi: 10.1152/ajpcell.00558.2003. [DOI] [PubMed] [Google Scholar]
- 5.Furuse M, Fujita K, Hiiragi T, Fujimoto K, Tsukita S. Claudin-1 and -2: novel integral membrane proteins localizing at tight junctions with no sequence similarity to occludin. J Cell Biol. 1998;141(7):1539–1550. doi: 10.1083/jcb.141.7.1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Markov AG. [Claudins as tight junction proteins: the molecular element of paracellular transport] Ross Fiziol Zh Im I M Sechenova. 2013;99(2):175–195. Russian. [PubMed] [Google Scholar]
- 7.Lal-Nag M, Morin PJ. The claudins. Genome Biol. 2009;10(8):235. doi: 10.1186/gb-2009-10-8-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Krause G, Winkler L, Mueller SL, Haseloff RF, Piontek J, Blasig IE. Structure and function of claudins. Biochim Biophys Acta. 2008;1778(3):631–645. doi: 10.1016/j.bbamem.2007.10.018. [DOI] [PubMed] [Google Scholar]
- 9.Escudero-Esparza A, Jiang WG, Martin TA. The Claudin family and its role in cancer and metastasis. Front Biosci (Landmark Ed) 2011;16:1069–1083. doi: 10.2741/3736. [DOI] [PubMed] [Google Scholar]
- 10.Ouban A, Ahmed AA. Claudins in human cancer: a review. Histol Histopathol. 2010;25(1):83–90. doi: 10.14670/HH-25.83. [DOI] [PubMed] [Google Scholar]
- 11.Hewitt KJ, Agarwal R, Morin PJ. The claudin gene family: expression in normal and neoplastic tissues. BMC Cancer. 2006;6:186. doi: 10.1186/1471-2407-6-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morin PJ. Claudin proteins in human cancer: promising new targets for diagnosis and therapy. Cancer Res. 2005;65(21):9603–9606. doi: 10.1158/0008-5472.CAN-05-2782. [DOI] [PubMed] [Google Scholar]
- 13.Singh AB, Sharma A, Dhawan P. Claudin family of proteins and cancer: an overview. J Oncol. 2010;2010:541957. doi: 10.1155/2010/541957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ding L, Lu Z, Foreman O, et al. Inflammation and disruption of the mucosal architecture in claudin-7-deficient mice. Gastroenterology. 2012;142(2):305–315. doi: 10.1053/j.gastro.2011.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Findley MK, Koval M. Regulation and roles for claudin-family tight junction proteins. IUBMB Life. 2009;61(4):431–437. doi: 10.1002/iub.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Will C, Fromm M, Muller D. Claudin tight junction proteins: novel aspects in paracellular transport. Perit Dial Int. 2008;28(6):577–584. [PubMed] [Google Scholar]
- 17.Elkouby-Naor L, Ben-Yosef T. Functions of claudin tight junction proteins and their complex interactions in various physiological systems. Int Rev Cell Mol Biol. 2010;279:1–32. doi: 10.1016/S1937-6448(10)79001-8. [DOI] [PubMed] [Google Scholar]
- 18.Klymkowsky MW, Savagner P. Epithelial-mesenchymal transition: a cancer researcher’s conceptual friend and foe. Am J Pathol. 2009;174(5):1588–1593. doi: 10.2353/ajpath.2009.080545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu X, Jin H, Liu Y, et al. The expression patterns and correlations of claudin-6, methy-CpG binding protein 2, DNA methyltransferase 1, histone deacetylase 1, acetyl-histone H3 and acetyl-histone H4 and their clinicopathological significance in breast invasive ductal carcinomas. Diagn Pathol. 2012;7(1):33. doi: 10.1186/1746-1596-7-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sauer T, Pedersen MK, Ebeltoft K, Naess O. Reduced expression of Claudin-7 in fine needle aspirates from breast carcinomas correlate with grading and metastatic disease. Cytopathology. 2005;16(4):193–198. doi: 10.1111/j.1365-2303.2005.00257.x. [DOI] [PubMed] [Google Scholar]
- 21.Tabariès S, Dupuy F, Dong Z, et al. Claudin-2 promotes breast cancer liver metastasis by facilitating tumor cell interactions with hepatocytes. Mol Cell Biol. 2012;32(15):2979–2991. doi: 10.1128/MCB.00299-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shang X, Lin X, Alvarez E, Manorek G, Howell SB. Tight junction proteins claudin-3 and claudin-4 control tumor growth and metastases. Neoplasia. 2012;14(10):974–985. doi: 10.1593/neo.12942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Swisshelm K, Macek R, Kubbies M. Role of claudins in tumorigenesis. Adv Drug Deliv Rev. 2005;57(6):919–928. doi: 10.1016/j.addr.2005.01.006. [DOI] [PubMed] [Google Scholar]
- 24.Oliveira SS, Morgado-Diaz JA. Claudins: multifunctional players in epithelial tight junctions and their role in cancer. Cell Mol Life Sci. 2007;64(1):17–28. doi: 10.1007/s00018-006-6314-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lu Z, Ding L, Hong H, Hoggard J, Lu Q, Chen YH. Claudin-7 inhibits human lung cancer cell migration and invasion through ERK/MAPK signaling pathway. Exp Cell Res. 2011;317(13):1935–1946. doi: 10.1016/j.yexcr.2011.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ikari A, Sato T, Watanabe R, Yamazaki Y, Sugatani J. Increase in claudin-2 expression by an EGFR/MEK/ERK/c-Fos pathway in lung adenocarcinoma A549 cells. Biochim Biophys Acta. 2012;1823(6):1110–1118. doi: 10.1016/j.bbamcr.2012.04.005. [DOI] [PubMed] [Google Scholar]
- 27.Jung JH, Jung CK, Choi HJ, et al. Diagnostic utility of expression of claudins in non-small cell lung cancer: different expression profiles in squamous cell carcinomas and adenocarcinomas. Pathol Res Pract. 2009;205(6):409–416. doi: 10.1016/j.prp.2008.12.015. [DOI] [PubMed] [Google Scholar]
- 28.Kramer F, White K, Kubbies M, Swisshelm K, Weber BH. Genomic organization of claudin-1 and its assessment in hereditary and sporadic breast cancer. Human Genet. 2000;107(3):249–256. doi: 10.1007/s004390000375. [DOI] [PubMed] [Google Scholar]
- 29.Miwa N, Furuse M, Tsukita S, Niikawa N, Nakamura Y, Furukawa Y. Involvement of claudin-1 in the beta-catenin/Tcf signaling pathway and its frequent upregulation in human colorectal cancers. Oncol Res. 2001;12(11–12):469–476. doi: 10.3727/096504001108747477. [DOI] [PubMed] [Google Scholar]
- 30.Resnick MB, Konkin T, Routhier J, Sabo E, Pricolo VE. Claudin-1 is a strong prognostic indicator in stage II colonic cancer: a tissue microarray study. Mod Pathol. 2005;18(4):511–518. doi: 10.1038/modpathol.3800301. [DOI] [PubMed] [Google Scholar]
- 31.Ouban A, Hamdan H, Hakam A, Ahmed AA. Claudin-1 expression in squamous cell carcinomas of different organs: comparative study of cancerous tissues and normal controls. Int J Surg Pathol. 2012;20(2):132–138. doi: 10.1177/1066896911424488. [DOI] [PubMed] [Google Scholar]
- 32.Lee JW, Lee SJ, Seo J, et al. Increased expressions of claudin-1 and claudin-7 during the progression of cervical neoplasia. Gynecol Oncol. 2005;97(1):53–59. doi: 10.1016/j.ygyno.2004.11.058. [DOI] [PubMed] [Google Scholar]
- 33.Liebner S, Fischmann A, Rascher G, et al. Claudin-1 and claudin-5 expression and tight junction morphology are altered in blood vessels of human glioblastoma multiforme. Acta Neuropathol. 2000;100(3):323–331. doi: 10.1007/s004010000180. [DOI] [PubMed] [Google Scholar]
- 34.Cohn ML, Goncharuk VN, Diwan AH, Zhang PS, Shen SS, Prieto VG. Loss of claudin-1 expression in tumor-associated vessels correlates with acquisition of metastatic phenotype in melanocytic neoplasms. J Cutan Pathol. 2005;32(8):533–536. doi: 10.1111/j.0303-6987.2005.00324.x. [DOI] [PubMed] [Google Scholar]
- 35.Resnick MB, Gavilanez M, Newton E, et al. Claudin expression in gastric adenocarcinomas: a tissue microarray study with prognostic correlation. Hum Pathol. 2005;36(8):886–892. doi: 10.1016/j.humpath.2005.05.019. [DOI] [PubMed] [Google Scholar]
- 36.Gyõrffy H, Holczbauer A, Nagy P, et al. Claudin expression in Barrett’s esophagus and adenocarcinoma. Virchows Arch. 2005;447(6):961–968. doi: 10.1007/s00428-005-0045-9. [DOI] [PubMed] [Google Scholar]
- 37.Németh Z, Szász AM, Tátrai P, et al. Claudin-1, -2, -3, -4, -7, -8, and -10 protein expression in biliary tract cancers. J Histochem Cytochem. 2009;57(2):113–121. doi: 10.1369/jhc.2008.952291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morita K, Tsukita S, Miyachi Y. Tight junction-associated proteins (occludin, ZO-1, claudin-1, claudin-4) in squamous cell carcinoma and Bowen’s disease. Br J Dermatol. 2004;151(2):328–334. doi: 10.1111/j.1365-2133.2004.06029.x. [DOI] [PubMed] [Google Scholar]
- 39.Fluge O, Bruland O, Akslen LA, Lillehaug JR, Varhaug JE. Gene expression in poorly differentiated papillary thyroid carcinomas. Thyroid. 2006;16(2):161–175. doi: 10.1089/thy.2006.16.161. [DOI] [PubMed] [Google Scholar]
- 40.Bello IO, Vilen ST, Niinimaa A, Kantola S, Soini Y, Salo T. Expression of claudins 1, 4, 5, and 7 and occludin, and relationship with prognosis in squamous cell carcinoma of the tongue. Human Pathol. 2008;39(8):1212–1220. doi: 10.1016/j.humpath.2007.12.015. [DOI] [PubMed] [Google Scholar]
- 41.Nakanishi K, Ogata S, Hiroi S, Tominaga S, Aida S, Kawai T. Expression of occludin and claudins 1, 3, 4, and 7 in urothelial carcinoma of the upper urinary tract. Am J Clin Pathol. 2008;130(1):43–49. doi: 10.1309/U77A6BTEXVCA5D0E. [DOI] [PubMed] [Google Scholar]
- 42.Haid S, Windisch MP, Bartenschlager R, Pietschmann T. Mouse-specific residues of claudin-1 limit hepatitis C virus genotype 2a infection in a human hepatocyte cell line. J Virol. 2010;84(2):964–975. doi: 10.1128/JVI.01504-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Higashi Y, Suzuki S, Sakaguchi T, et al. Loss of claudin-1 expression correlates with malignancy of hepatocellular carcinoma. J Surg Res. 2007;139(1):68–76. doi: 10.1016/j.jss.2006.08.038. [DOI] [PubMed] [Google Scholar]
- 44.Lechpammer M, Resnick MB, Sabo E, et al. The diagnostic and prognostic utility of claudin expression in renal cell neoplasms. Mod Pathol. 2008;21(11):1320–1329. doi: 10.1038/modpathol.2008.116. [DOI] [PubMed] [Google Scholar]
- 45.Paschoud S, Bongiovanni M, Pache JC, Citi S. Claudin-1 and claudin-5 expression patterns differentiate lung squamous cell carcinomas from adenocarcinomas. Mod Pathol. 2007;20(9):947–954. doi: 10.1038/modpathol.3800835. [DOI] [PubMed] [Google Scholar]
- 46.Iacobuzio-Donahue CA, Maitra A, Shen-Ong GL, et al. Discovery of novel tumor markers of pancreatic cancer using global gene expression technology. Discovery of novel tumor markers of pancreatic cancer using global gene expression technology. Am J Pathol. 2002;160(4):1239–1249. doi: 10.1016/S0002-9440(10)62551-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim TH, Huh JH, Lee S, Kang H, Kim GI, An HJ. Down-regulation of claudin-2 in breast carcinomas is associated with advanced disease. Histopathology. 2008;53(1):48–55. doi: 10.1111/j.1365-2559.2008.03052.x. [DOI] [PubMed] [Google Scholar]
- 48.Weber CR, Nalle SC, Tretiakova M, Rubin DT, Turner JR. Claudin-1 and claudin-2 expression is elevated in inflammatory bowel disease and may contribute to early neoplastic transformation. Lab Invest. 2008;88(10):1110–1120. doi: 10.1038/labinvest.2008.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Aung PP, Mitani Y, Sanada Y, Nakayama H, Matsusaki K, Yasui W. Differential expression of claudin-2 in normal human tissues and gastrointestinal carcinomas. Virchows Arch. 2006;448(4):428–434. doi: 10.1007/s00428-005-0120-2. [DOI] [PubMed] [Google Scholar]
- 50.Timmons BC, Mitchell SM, Gilpin C, Mahendroo MS. Dynamic changes in the cervical epithelial tight junction complex and differentiation occur during cervical ripening and parturition. Endocrinology. 2007;148(3):1278–1287. doi: 10.1210/en.2006-0851. [DOI] [PubMed] [Google Scholar]
- 51.Halasz J, Holczbauer A, Paska C, et al. Claudin-1 and claudin-2 differentiate fetal and embryonal components in human hepatoblastoma. Hum Pathol. 2006;37(5):555–561. doi: 10.1016/j.humpath.2005.12.015. [DOI] [PubMed] [Google Scholar]
- 52.Soini Y. Expression of claudins 1, 2, 3, 4, 5 and 7 in various types of tumours. Histopathology. 2005;46(5):551–560. doi: 10.1111/j.1365-2559.2005.02127.x. [DOI] [PubMed] [Google Scholar]
- 53.Kominsky SL, Vali M, Korz D, et al. Clostridium perfringens enterotoxin elicits rapid and specific cytolysis of breast carcinoma cells mediated through tight junction proteins claudin 3 and 4. Am J Pathol. 2004;164(5):1627–1633. doi: 10.1016/S0002-9440(10)63721-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.de Oliveira SS, de Oliveira IM, De Souza W, Morgado-Díaz JA. Claudins upregulation in human colorectal cancer. FEBS Lett. 2005;579(27):6179–6185. doi: 10.1016/j.febslet.2005.09.091. [DOI] [PubMed] [Google Scholar]
- 55.Maeda T, Murata M, Chiba H, et al. Claudin-4-targeted therapy using Clostridium perfringens enterotoxin for prostate cancer. Prostate. 2012;72(4):351–360. doi: 10.1002/pros.21436. [DOI] [PubMed] [Google Scholar]
- 56.Hough CD, Sherman-Baust CA, Pizer ES, et al. Large-scale serial analysis of gene expression reveals genes differentially expressed in ovarian cancer. Cancer Res. 2000;60(22):6281–6287. [PubMed] [Google Scholar]
- 57.Missiaglia E, Blaveri E, Terris B, et al. Analysis of gene expression in cancer cell lines identifies candidate markers for pancreatic tumorigenesis and metastasis. Int J Cancer. 2004;112(1):100–112. doi: 10.1002/ijc.20376. [DOI] [PubMed] [Google Scholar]
- 58.Kwon MJ, Kim SH, Jeong HM, et al. Claudin-4 overexpression is associated with epigenetic derepression in gastric carcinoma. Lab Invest. 2011;91(11):1652–1667. doi: 10.1038/labinvest.2011.117. [DOI] [PubMed] [Google Scholar]
- 59.Lee SK, Moon J, Park SW, Song SY, Chung JB, Kang JK. Loss of the tight junction protein claudin 4 correlates with histological growth-pattern and differentiation in advanced gastric adenocarcinoma. Oncology Rep. 2005;13(2):193–199. [PubMed] [Google Scholar]
- 60.Michl P, Buchholz M, Rolke M, et al. Claudin-4: a new target for pancreatic cancer treatment using Clostridium perfringens enterotoxin. Gastroenterology. 2001;121(3):678–684. doi: 10.1053/gast.2001.27124. [DOI] [PubMed] [Google Scholar]
- 61.Ueda J, Semba S, Chiba H, et al. Heterogeneous expression of claudin-4 in human colorectal cancer: decreased claudin-4 expression at the invasive front correlates cancer invasion and metastasis. Pathobiology. 2007;74(1):32–41. doi: 10.1159/000101049. [DOI] [PubMed] [Google Scholar]
- 62.Sobel G, Szabó I, Páska C, et al. Changes of cell adhesion and extracellular matrix (ECM) components in cervical intraepithelial neoplasia. Pathology Oncol Res. 2005;11(1):26–31. doi: 10.1007/BF03032402. [DOI] [PubMed] [Google Scholar]
- 63.Tzelepi VN, Tsamandas AC, Vlotinou HD, Vagianos CE, Scopa CD. Tight junctions in thyroid carcinogenesis: diverse expression of claudin-1, claudin-4, claudin-7 and occludin in thyroid neoplasms. Mod Pathol. 2008;21(1):22–30. doi: 10.1038/modpathol.3800959. [DOI] [PubMed] [Google Scholar]
- 64.Wu Q, Liu Y, Ren Y, et al. Tight junction protein, claudin-6, down-regulates the malignant phenotype of breast carcinoma. Eur J Cancer Prev. 2010;19(3):186–194. doi: 10.1097/CEJ.0b013e328337210e. [DOI] [PubMed] [Google Scholar]
- 65.Zavala-Zendejas VE, Torres-Martinez AC, Salas-Morales B, Fortoul TI, Montaño LF, Rendon-Huerta EP. Claudin-6, 7, or 9 overexpression in the human gastric adenocarcinoma cell line AGS increases its invasiveness, migration, and proliferation rate. Cancer Invest. 2011;29:1–11. doi: 10.3109/07357907.2010.512594. [DOI] [PubMed] [Google Scholar]
- 66.Kominsky SL, Argani P, Korz D, et al. Loss of the tight junction protein claudin-7 correlates with histological grade in both ductal carcinoma in situ and invasive ductal carcinoma of the breast. Oncogene. 2003;22(13):2021–2033. doi: 10.1038/sj.onc.1206199. [DOI] [PubMed] [Google Scholar]
- 67.Nacht M, Ferguson AT, Zhang W, et al. Combining serial analysis of gene expression and array technologies to identify genes differentially expressed in breast cancer. Cancer Res. 1999;59(21):5464–5470. [PubMed] [Google Scholar]
- 68.Al Moustafa AE, Alaoui-Jamali MA, Batist G, et al. Identification of genes associated with head and neck carcinogenesis by cDNA microarray comparison between matched primary normal epithelial and squamous carcinoma cells. Oncogene. 2002;21(17):2634–2640. doi: 10.1038/sj.onc.1205351. [DOI] [PubMed] [Google Scholar]
- 69.Darido C, Buchert M, Pannequin J, et al. Defective claudin-7 regulation by Tcf-4 and Sox-9 disrupts the polarity and increases the tumorigenicity of colorectal cancer cells. Cancer Res. 2008;68(11):4258–4268. doi: 10.1158/0008-5472.CAN-07-5805. [DOI] [PubMed] [Google Scholar]
- 70.Kuhn S, Koch M, Nübel T, et al. A complex of EpCAM, claudin-7, CD44 variant isoforms, and tetraspanins promotes colorectal cancer progression. Mol Cancer Res. 2007;5(6):553–567. doi: 10.1158/1541-7786.MCR-06-0384. [DOI] [PubMed] [Google Scholar]
- 71.Oshima T, Kunisaki C, Yoshihara K, et al. Reduced expression of the claudin-7 gene correlates with venous invasion and liver metastasis in colorectal cancer. Oncology Rep. 2008;19(4):953–959. [PubMed] [Google Scholar]
- 72.Nakayama F, Semba S, Usami Y, Chiba H, Sawada N, Yokozaki H. Hypermethylation-modulated downregulation of claudin-7 expression promotes the progression of colorectal carcinoma. Pathobiology. 2008;75(3):177–185. doi: 10.1159/000124978. [DOI] [PubMed] [Google Scholar]
- 73.Johnson AH, Frierson HF, Zaika A, et al. Expression of tight-junction protein claudin-7 is an early event in gastric tumorigenesis. Am J Pathol. 2005;167(2):577–584. doi: 10.1016/S0002-9440(10)62999-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Montgomery E, Mamelak AJ, Gibson M, et al. Overexpression of claudin proteins in esophageal adenocarcinoma and its precursor lesions. Appl Immunohistochem Mol Morphol. 2006;14(1):24–30. doi: 10.1097/01.pai.0000151933.04800.1c. [DOI] [PubMed] [Google Scholar]
- 75.Usami Y, Chiba H, Nakayama F, et al. Reduced expression of claudin-7 correlates with invasion and metastasis in squamous cell carcinoma of the esophagus. Hum Pathol. 2006;37(5):569–577. doi: 10.1016/j.humpath.2005.12.018. [DOI] [PubMed] [Google Scholar]
- 76.Hornsby CD, Cohen C, Amin MB, et al. Claudin-7 immunohistochemistry in renal tumors: a candidate marker for chromophobe renal cell carcinoma identified by gene expression profiling. Arch Pathol Lab Med. 2007;131(10):1541–1546. doi: 10.5858/2007-131-1541-CIIRTA. [DOI] [PubMed] [Google Scholar]
- 77.Li X, Li Y, Qiu H, Wang Y. Downregulation of claudin-7 potentiates cellular proliferation and invasion in endometrial cancer. Oncol Lett. 2013;6(1):101–105. doi: 10.3892/ol.2013.1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Dahiya N, Becker KG, Wood WH, 3rd, Zhang Y, Morin PJ. Claudin-7 is frequently overexpressed in ovarian cancer and promotes invasion. PloS One. 2011;6(7):e22119. doi: 10.1371/journal.pone.0022119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Brokalaki EI, Weber F, Sotiropoulos GC, Daoudaki M, Cicinnati VR, Beckebaum S. Claudin-7 expression in hepatocellular carcinoma. Transplant Proc. 2012;44(9):2737–2740. doi: 10.1016/j.transproceed.2012.09.009. [DOI] [PubMed] [Google Scholar]
- 80.Grone J, Weber B, Staub E, et al. Differential expression of genes encoding tight junction proteins in colorectal cancer: frequent dysregulation of claudin-1, -8 and -12. Int J Colorectal Dis. 2007;22(6):651–659. doi: 10.1007/s00384-006-0197-3. [DOI] [PubMed] [Google Scholar]
- 81.Cheung ST, Leung KL, Ip YC, et al. Claudin-10 expression level is associated with recurrence of primary hepatocellular carcinoma. Clin Cancer Res. 2005;11(2 Pt 1):551–556. [PubMed] [Google Scholar]
- 82.Merikallio H, Pääkkö P, Harju T, Soini Y. Claudins 10 and 18 are predominantly expressed in lung adenocarcinomas and in tumors of nonsmokers. Int J Clin Exp Pathol. 2011;4(7):667–673. [PMC free article] [PubMed] [Google Scholar]
- 83.Aldred MA, Huang Y, Liyanarachchi S, et al. Papillary and follicular thyroid carcinomas show distinctly different microarray expression profiles and can be distinguished by a minimum of five genes. J Clin Oncol. 2004;22(17):3531–3539. doi: 10.1200/JCO.2004.08.127. [DOI] [PubMed] [Google Scholar]
- 84.Awsare NS, Martin TA, Haynes MD, Matthews PN, Jiang WG. Claudin-11 decreases the invasiveness of bladder cancer cells. Oncology Rep. 2011;25(6):1503–1509. doi: 10.3892/or.2011.1244. [DOI] [PubMed] [Google Scholar]
- 85.Rangel LB, Sherman-Baust CA, Wernyj RP, Schwartz DR, Cho KR, Morin PJ. Characterization of novel human ovarian cancer-specific transcripts (HOSTs) identified by serial analysis of gene expression. Oncogene. 2003;22(46):7225–7232. doi: 10.1038/sj.onc.1207008. [DOI] [PubMed] [Google Scholar]
- 86.Martin TA, Harrison GM, Watkins G, Jiang WG. Claudin-16 reduces the aggressive behavior of human breast cancer cells. J Cell Biochem. 2008;105(1):41–52. doi: 10.1002/jcb.21797. [DOI] [PubMed] [Google Scholar]
- 87.Sanada Y, Oue N, Mitani Y, Yoshida K, Nakayama H, Yasui W. Down-regulation of the claudin-18 gene, identified through serial analysis of gene expression data analysis, in gastric cancer with an intestinal phenotype. J Pathol. 2006;208(5):633–642. doi: 10.1002/path.1922. [DOI] [PubMed] [Google Scholar]
- 88.Shinozaki A, Shibahara J, Noda N, et al. Claudin-18 in biliary neoplasms. Its significance in the classification of intrahepatic cholangiocarcinoma. Virchows Arch. 2011;459(1):73–80. doi: 10.1007/s00428-011-1092-z. [DOI] [PubMed] [Google Scholar]
- 89.Katoh M, Katoh M. CLDN23 gene, frequently down-regulated in intestinal-type gastric cancer, is a novel member of CLAUDIN gene family. Int J Mol Med. 2003;11(6):683–689. [PubMed] [Google Scholar]
- 90.Kuo SJ, Chien SY, Lin C, Chan SE, Tsai HT, Chen DR. Significant elevation of CLDN16 and HAPLN3 gene expression in human breast cancer. Oncology Rep. 2010;24(3):759–766. doi: 10.3892/or_00000918. [DOI] [PubMed] [Google Scholar]
- 91.Agarwal R, D’Souza T, Morin PJ. Claudin-3 and claudin-4 expression in ovarian epithelial cells enhances invasion and is associated with increased matrix metalloproteinase-2 activity. Cancer Res. 2005;65(16):7378–7385. doi: 10.1158/0008-5472.CAN-05-1036. [DOI] [PubMed] [Google Scholar]
- 92.Tsutsumi K, Sato N, Cui L, et al. Expression of claudin-4 (CLDN4) mRNA in intraductal papillary mucinous neoplasms of the pancreas. Mod Pathol. 2011;24(4):533–541. doi: 10.1038/modpathol.2010.218. [DOI] [PubMed] [Google Scholar]
- 93.Rangel LB, Agarwal R, D’Souza T, et al. Tight junction proteins claudin-3 and claudin-4 are frequently overexpressed in ovarian cancer but not in ovarian cystadenomas. Clin Cancer Res. 2003;9(7):2567–2575. [PubMed] [Google Scholar]
- 94.English DP, Santin AD. Claudins overexpression in ovarian cancer: potential targets for Clostridium perfringens enterotoxin (CPE) based diagnosis and therapy. Int J Mol Sci. 2013;14(5):10412–10437. doi: 10.3390/ijms140510412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hoevel T, Macek R, Swisshelm K, Kubbies M. Reexpression of the TJ protein CLDN1 induces apoptosis in breast tumor spheroids. Int J Cancer. 2004;108(3):374–383. doi: 10.1002/ijc.11571. [DOI] [PubMed] [Google Scholar]
- 96.Thompson PD, Tipney H, Brass A, et al. Claudin 13, a member of the claudin family regulated in mouse stress induced erythropoiesis. PloS One. 2010;5(9):e12667. doi: 10.1371/journal.pone.0012667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Fujita H, Chiba H, Yokozaki H, et al. Differential expression and subcellular localization of claudin-7, -8, -12, -13, and -15 along the mouse intestine. J Histochem Cytochem. 2006;54(8):933–944. doi: 10.1369/jhc.6A6944.2006. [DOI] [PubMed] [Google Scholar]
- 98.Wilcox ER, Burton QL, Naz S, et al. Mutations in the gene encoding tight junction claudin-14 cause autosomal recessive deafness DFNB29. Cell. 2001;104(1):165–172. doi: 10.1016/s0092-8674(01)00200-8. [DOI] [PubMed] [Google Scholar]
- 99.Baker M, Reynolds LE, Robinson SD, et al. Stromal Claudin14-heterozygosity, but not deletion, increases tumour blood leakage without affecting tumour growth. PloS One. 2013;8(5):e62516. doi: 10.1371/journal.pone.0062516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Claverie-Martin F, García-Nieto V, Loris C, et al. RenalTube Group Claudin-19 mutations and clinical phenotype in Spanish patients with familial hypomagnesemia with hypercalciuria and nephrocalcinosis. PloS One. 2013;8(1):e53151. doi: 10.1371/journal.pone.0053151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lee NP, Tong MK, Leung PP, et al. Kidney claudin-19: localization in distal tubules and collecting ducts and dysregulation in polycystic renal disease. FEBS Lett. 2006;580(3):923–931. doi: 10.1016/j.febslet.2006.01.019. [DOI] [PubMed] [Google Scholar]
- 102.Günzel D, Yu AS. Claudins and the modulation of tight junction permeability. Physiol Rev. 2013;93(2):525–569. doi: 10.1152/physrev.00019.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Walsh SV, Hopkins AM, Nusrat A. Modulation of tight junction structure and function by cytokines. Adv Drug Deliv Rev. 2000;41(3):303–313. doi: 10.1016/s0169-409x(00)00048-x. [DOI] [PubMed] [Google Scholar]
- 104.Huang GW, Ding X, Chen SL, Zeng L. Expression of claudin 10 protein in hepatocellular carcinoma: impact on survival. J Cancer Res Clin Oncol. 2011;137(8):1213–1218. doi: 10.1007/s00432-011-0987-z. [DOI] [PubMed] [Google Scholar]
- 105.Walther W, Petkov S, Kuvardina ON, et al. Novel Clostridium per-fringens enterotoxin suicide gene therapy for selective treatment of claudin-3- and -4-overexpressing tumors. Gene Ther. 2012;19(5):494–503. doi: 10.1038/gt.2011.136. [DOI] [PubMed] [Google Scholar]
- 106.Long H, Crean CD, Lee WH, Cummings OW, Gabig TG. Expression of Clostridium perfringens enterotoxin receptors claudin-3 and claudin-4 in prostate cancer epithelium. Cancer Res. 2001;61(21):7878–7881. [PubMed] [Google Scholar]
- 107.Cocco E, Casagrande F, Bellone S, et al. Clostridium perfringens enterotoxin carboxy-terminal fragment is a novel tumor-homing peptide for human ovarian cancer. BMC Cancer. 2010;10:349. doi: 10.1186/1471-2407-10-349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Zwanziger D, Staat C, Andjelkovic AV, Blasig IE. Claudin-derived peptides are internalized via specific endocytosis pathways. Ann N Y Acad Sci. 2012;1257:29–37. doi: 10.1111/j.1749-6632.2012.06567.x. [DOI] [PubMed] [Google Scholar]
- 109.Kominsky SL. Claudins: emerging targets for cancer therapy. Expert Rev Mol Med. 2006;8(18):1–11. doi: 10.1017/S1462399406000056. [DOI] [PubMed] [Google Scholar]
- 110.Zhang G, Ding L, Renegar R, et al. Hydroxycamptothecin-loaded Fe3O4 nanoparticles induce human lung cancer cell apoptosis through caspase-8 pathway activation and disrupt tight junctions. Cancer Sci. 2011;102(6):1216–1222. doi: 10.1111/j.1349-7006.2011.01930.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Choi YJ, Shin DY, Lee YW, et al. Inhibition of cell motility and invasion by HangAmDan-B in NCI-H460 human non-small cell lung cancer cells. Oncology Rep. 2011;26(6):1601–1608. doi: 10.3892/or.2011.1440. [DOI] [PubMed] [Google Scholar]
- 112.Plöger S, Stumpff F, Penner GB, et al. Microbial butyrate and its role for barrier function in the gastrointestinal tract. Ann N Y Acad Sci. 2012;1258:52–59. doi: 10.1111/j.1749-6632.2012.06553.x. [DOI] [PubMed] [Google Scholar]


