Abstract
There is increasing interest in the use of edited proton magnetic resonance spectroscopy for the detection of GABA in the human brain. At a recent meeting held at Cardiff University, a number of spectroscopy groups met to discuss the acquisition, analysis and interpretation of GABA-edited MR spectra. This paper aims to set out the issues discussed at this meeting, reporting areas of consensus around parameters and procedures in the field and highlighting those areas where differences remain. It is hoped that this paper can fulfill two needs, providing a summary of the current ‘state-of-the-art’ in the field of GABA-edited MRS at 3 T using MEGA-PRESS and a basic guide to help researchers new to the field to avoid some of the pitfalls inherent in the acquisition and processing of edited MRS for GABA.
Keywords: GABA, Edited MRS, MEGA-PRESS, MRS analysis
Introduction
Magnetic resonance spectroscopy (MRS) provides a non invasive technique to measure neurometabolites in vivo. In particular a large number of studies into methods to accurately and reliably measure the neurotransmitters glutamate and GABA (γ-aminobutyric acid) have taken place, a few of which are referenced here (Edden and Barker, 2007; Evans et al., 2010; Hancu, 2009; Henry et al., 2010; Hurd et al., 2004; Jang et al., 2005; Jensen et al., 2005a; Mullins et al., 2008; Rothman et al., 1993; Waddell et al., 2007). GABA in particular has proven to be difficult to reliably measure in-vivo with standard single voxel techniques, in large part due to the spectral overlap of the main GABA peaks with peaks of other neurotransmitters which are present in much greater concentrations, in particular the creatine (Cr) peak at 3.0 ppm. While high field (3 T–7 T) short echo sequences have shown some promise in allowing detection of GABA (Hu et al., 2007; Mekle et al., 2009; Napolitano et al., 2012; Stagg et al., 2011), recent technical advances and increased availability of spectral editing sequences have resulted in a rapid growth in the use of edited proton MRS to detect the inhibitory neurotransmitter GABA in both the healthy and diseased brain (Puts and Edden, 2012). In response to this development, researchers from several spectroscopy groups met in August 2011 to discuss current practice for the use of the MEGA-PRESS sequence for GABA-edited MRS (Edden and Barker, 2007; Mescher et al., 1998; Rothman et al., 1993; Terpstra et al., 2002). The purpose of the meeting was to present recent findings (Boy et al., 2010, 2011; Foerster et al., 2012a, 2012b; Michels et al., 2012; O'Gorman et al., 2011b; Petrou et al., 2012; Puts et al., 2011) and to discuss issues of acquisition, processing and quantification encountered in performing these studies.
This paper focuses on MEGA-PRESS (Mescher et al., 1996, 1998) editing for GABA at 3 T, as this is currently the most widely used MRS technique for quantifying GABA and therefore the most promising ground on which to build consensus. This paper is not intended to be a prescriptive rule book for MEGA-PRESS or GABA acquisition and analysis; rather, it is intended as a useful guide to current “minimal-best” practice as reached from consensus amongst researchers in the MRS field present at the meeting. Although it is likely that some of the discussion presented here may generalize to different editing strategies and field strengths, this paper does not cover the entire range of alternate acquisition schemes for GABA measurement which includes 2D J-resolved MRS (Jensen et al., 2005b; Ryner et al., 1995), alterations in TE or sequence timing parameters (Hu et al., 2007; Mullins et al., 2008; Thompson and Allen, 2001), unedited spectra at 7 T (Mekle et al., 2009), CT-PRESS (Mayer et al., 2006) or other editing sequences (Choi et al., 2005) (for a broader review see Puts and Edden (2012)). However we believe that applied research into GABAergic function in both the healthy and diseased brain will benefit greatly from standardization of acquisition and analysis practices, allowing the quantitative comparison of data from different brain regions, studies, and scanner platforms.
MEGA-PRESS
MEGA-PRESS (MEshcher-GArwood Point RESolved Spectroscopy), named after the authors who first proposed the MEGA suppression scheme, is quickly becoming the standard technique used in MRS measurements of GABA. It allows GABA signals to be separated from the stronger overlying signals of other metabolites by taking advantage of known couplings within the GABA molecule. Scalar coupling is an inter-action between different hydrogen nuclei within a molecule, transmitted through the bonding electron network, which alters the appearance of the spectrum and the time-evolution of spins during an experiment. In the context of MEGA-PRESS, applying an RF pulse to one coupled spin can modify the time-evolution of a coupling partner and therefore the appearance of the corresponding peak in the spectrum. We refer the interested reader to de Graaf (2007) and Keeler (2011) for further explanation of scalar coupling that is beyond the scope of this article.
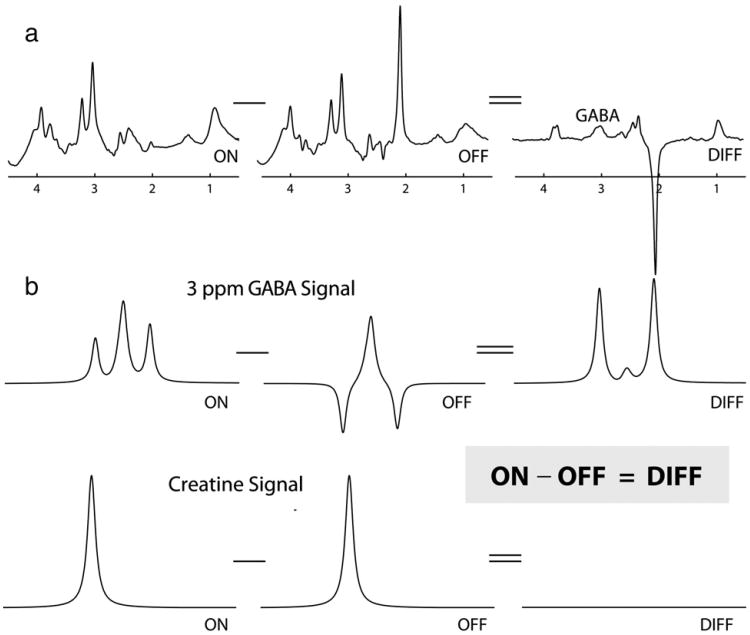
A difference-edited technique, MEGA-PRESS involves the collection of two interleaved datasets which differ in their treatment of the GABA spin system. In one dataset, an editing pulse is applied to GABA spins at 1.9 ppm in order to selectively refocus the evolution of J-coupling to the GABA spins at 3 ppm (often referred to as ‘ON’). In the other, the inversion pulse is applied elsewhere so that the J-coupling evolves freely throughout the echo time (often referred to as ‘OFF’). The majority of peaks in the spectrum are unaffected by the editing pulses, so subtraction of the refocused ON spectrum from the non-refocused OFF spectrum removes all these peaks from the spectrum and retains only those peaks that are affected by the editing pulses. This process is demonstrated in Fig. 1. Thus, in-vivo, the edited spectrum contains signals close to 1.9 ppm (those directly affected by the pulses), the GABA signal at 3 ppm (coupled to GABA spins at 1.9 ppm), the combined glutamate/ glutamine/glutathione (Glx) peaks at 3.75 ppm (coupled to the Glx resonances at approximately 2.1 ppm), and J-coupled macromolecular (MM) peaks. The ON and OFF spectra are generally collected in an interleaved fashion to limit the impact of subject and hardware instabilities, and subtraction is performed in post-processing. For more specific information regarding the MEGA-PRESS technique and its acquisition and post-processing, readers are directed towards the following articles (Edden and Barker, 2007; Evans et al., 2010; Henry et al., 2010; Mescher et al., 1998; Near et al., 2011; O'Gorman et al., 2011b; Terpstra et al., 2002).
Fig. 1.
Schematic diagram of MEGA-PRESS editing for GABA. (a) Editing pulses applied at 1.9 ppm modulate the shape of the GABA signals at 3 ppm (b). Subtracting scans acquired without these pulses (labeled OFF) from scans acquired with the editing pulses (ON) removes overlying creatine signals from the edited spectrum, revealing the GABA signal in the difference spectrum (labeled DIFF). (b) shows the effect of editing pulses on signals at 3 ppm only.
MEGA-PRESS has become the most widely used technique for MRS measurements of GABA, largely due to ease of implementation within pre-existing PRESS sequences and the distribution of a research sequence by at least one major vendor. Implementing MEGA-PRESS within a stock PRESS sequence at the simplest level involves adding two RF pulses and altering the timing of the gradient pulses. MEGA-PRESS was published contemporaneously with the BASING method (Star-Lack et al., 1998) which similarly adds two editing pulses to the PRESS sequence. The distinction between the two lies mainly in the choice of gradient scheme used for coherence transfer pathway selection; both allow for simultaneous water suppression, a feature that is often not used due to the development of excellent pre-saturation methods. Many GABA experiments being applied currently could be loosely described either as MEGA- or BASING-edited. They are possibly almost uniformly referred to as MEGA-PRESS because that method was originally applied to editing GABA, whereas BASING was originally applied to lactate.
Acquisition
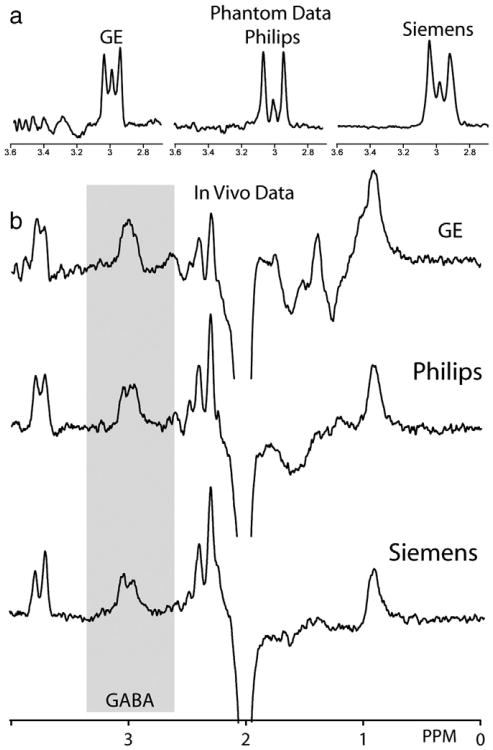
Almost inevitably, the implementation and application of the MEGA-PRESS method differs between makes and models of scanners, field strength, operating systems and research groups. However, the acquired spectra share a number of stereotypical characteristics, as shown in Fig. 2. The differences in implementation originate largely from differences in the timing, slice profile and bandwidth of slice-selective pulses used in the base PRESS experiment and differences in the timing and bandwidth of the editing pulses. Despite these differences in sequence implementation, spectra from all three major MR system vendors are sufficiently similar to be amenable to a common series of processing and analysis steps (Fig. 2).
Fig. 2.
Comparison of GABA-edited spectra across three vendors. The Siemens data is acquired with a vendor-distributed sequence, whereas the GE and Philips sequences are customer implementations by and available from RAEE. (a) Phantom data (acquired in a 10 mM GABA solution in phosphate-buffered saline) show similar edited signal in each case. Note that the commonly anticipated ‘pseudo-doublet’ is not observed in any implementation, and that implementations differ significantly in the extent to which the ‘center peak’ is edited. (b) In-vivo edited spectra. Typical parameters are: TE 68 ms; TR 2 s; 3×3×3 cm3 voxel; acquisition time 10 min; editing pulses applied at 1.9 ppm (ON) and 7.46 ppm (OFF).
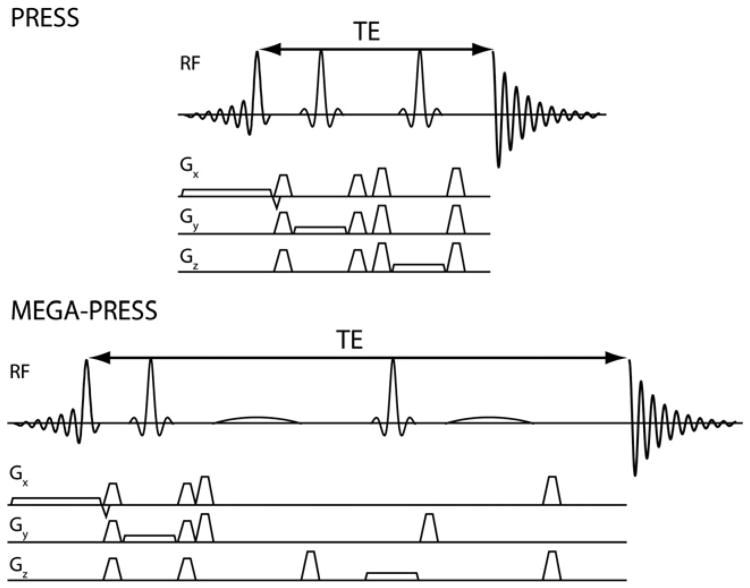
The following discussion of the intricacies of MEGA-PRESS implementation assumes that the MEGA-PRESS sequence (an example is shown in Fig. 3b) consists of two editing pulses added into a base PRESS sequence (an example is shown in Fig. 3a), usually the vendor-default PRESS implementation.
Fig. 3.
Representative PRESS (a) and MEGA-PRESS (b) pulse sequences, showing the addition of the editing pulses symmetrically around the second 180° pulse.
Basic parameters
Echo times (TE) are largely standardized to the 68 ms duration suggested in the original GABA editing paper (Rothman et al., 1993). Repetition times vary between groups, resulting in variable degrees of T1-weighting of the signals; this should be considered in the light of the significant MM contribution to the edited signal, which is discussed further below.
PRESS timing
At medium and long echo times, the first spin echo duration (TE1) of the PRESS sequence is typically kept as short as possible to minimize the degree of excitation of multiple-quantum coherence pathways. The value of TE1 varies between vendors due to differences in maximum B1 strength, maximum gradient strength and slew rate, and gradient areas chosen to achieve slice-selection. Changing the relative values of TE1 and TE2 (while maintaining TE) will modulate the appearance of the edited GABA signal detected (see Fig. 2 for in vitro examples) as it does for other J-coupled metabolite peaks (Thompson and Allen, 2001).
PRESS pulses
It has been widely reported that losses in editing efficiency occur due to finite bandwidth slice-selective refocusing in edited PRESS (Edden and Barker, 2007; Near et al., 2011). The bandwidth of slice-selective refocusing pulses (which is generally inherited from the base PRESS sequence) is determined by the maximum B1 available and the RF waveform used. The slice-profile of these pulses (i.e. how rectangular the slice selection is) will therefore also impact the appearance of the acquired doublet. This is one of the reasons for slightly different phantom spectral results between different vendor systems (Fig. 2a). This figure highlights the fact that even under optimum conditions and when using a phantom, the edited spectra acquired deviate from the theoretical ideal, and small changes in sequence parameters can have unintended consequences in the final acquired spectrum. This highlights the need to be exact when stipulating the RF pulses used in numerical simulations.
Editing pulse timing
In order to fully refocus the evolution of coupling during TE, and therefore maximize editing efficiency, the two editing pulses should be separated by TE/2 (as shown in Fig. 3b). However this optimal timing constrains the maximum duration of the editing pulses and may not always be adopted. The absolute timing of the first editing pulse is often set to be halfway between the slice-selective excitation pulse (specifically the zero-phase time point during the pulse) and the second slice-selective refocusing pulse. The second editing pulse is often set halfway between that second excitation pulse and the end of the echo time. Combined with the differences in resulting edited spectra seen in Fig. 3, it would therefore seem prudent for the researcher to be fully aware of the timing schemes used in their implementation of MEGA-PRESS, and how they may differ when comparing results to other vendors or versions of the sequence.
Editing pulse bandwidth
The bandwidth of editing pulses, a key parameter as it determines the degree of co-editing of macromolecular signal (see Section 7 “Contamination of spectra by co-edited macromolecular signal”), is determined by both the RF waveform used for editing and the duration of the editing pulses. The editing pulse duration is in turn determined by the remaining period of the echo time not already occupied by PRESS localization pulses and gradients. Where greater maximum B1 is available, slice selective pulses can be shorter and editing pulses longer, resulting in narrower bandwidth editing pulses with less co-editing of macromolecular signals.
Phase cycling
Although pulsed field gradients are used in the PRESS sequence for localization and coherence transfer pathway selection, localization is improved with additional phase cycling. It is an unresolved question whether phase cycling for localization (which can greatly improve water suppression, for example) is a higher priority than rapid (e.g. contiguous) interleaving of ON and OFF spectra for MEGA-PRESS subtraction. For ease of implementation, phase cycling of between 2 and 16 steps is often prioritized over interleaving, but the phase cycling and edit ON/OFF interleaving scheme should be selected carefully to minimize subtraction artefacts resulting from potential drift or motion between collection of the ON and OFF lines. The degree of phase cycling also impacts the time resolution of data available for pre-processing frequency- and phase-correction (see “Pre-processing of data before fitting”) in some data export formats.
Voxel size
Typically, the voxel size used in MEGA-PRESS is large when compared to that used for other neuroimaging modalities (for example, fMRI). Primarily, the large voxel size is necessary to offset the inherent low signal to noise ratio (SNR) for GABA (reported to be between 0.7 and 1.4 mM/cm3 in concentration (Govindaraju et al., 2000; Petroff, 2002; Rothman et al., 1993)), and so voxels on the order of 3×3×3 cm3 are commonly used as a compromise between localization and signal quality. As with all spectroscopic techniques when SNR may be limited, reductions in voxel size may require increases in acquisition times and vice versa to ensure data reliability.
Unsuppressed water
To allow concentration reference to tissue water, unsuppressed water spectra are acquired after the metabolite spectra as a separate scan (or as part of the standard PRESS acquisition). Typically 16 spectral averages are collected for the water reference scan, allowing for a full phase cycle, but as long as spectral quality is good, any number should suffice.
Pre-processing of data before fitting
Processing of edited single voxel MRS data follows the same workflow as that for unedited single voxel measurements: if a phased array receive coil is used, data from the individual coils are phase-corrected and combined either using the default vendor approach or off-line; exponential line broadening (3–4 Hz is typical) is usually applied (unless LCModel is to be used for analysis); Fourier transformation is applied; and frequency- and phase-correction are applied to time-resolved frequency domain data prior to temporal averaging and subsequent fitting (Zhu et al., 1992). As the data are usually collected as a series of ON and OFF editing pairs they are already in a format that facilitates within-scan correction of each pair for any potential frequency drift. Indeed, frequency correction of this type is particularly beneficial as it improves line width in the difference spectrum (Waddell et al., 2007) and can reduce or remove subtraction artefacts associated with frequency and phase instability.
The details of frequency correction vary extensively between groups. Typically a high-signal-to-noise peak is fitted to determine the frequency and phase corrections to be applied to each spectral pair across the entire acquisition. While the residual water peak is often chosen for frequency and phase corrections due to its higher SNR and spectral separation from GABA signals, the frequency and phase of the residual water signal may respond in an unpredictable manner to any frequency instability, and the efficacy of this approach varies according to the degree of water suppression applied (which differs between vendors). One method for dealing with this frequency instability involves the addition of an interleaved water navigator to the MEGA-PRESS sequence (Bhattacharyya et al., 2007). Creatine (Cr) can also be used as a frequency and phase reference, and may be preferable to the water peak due to the common location of origin of the GABA and Cr signals. However, Cr has lower SNR than water, and the form of the Cr signal is expected to differ between OFF and ON scans due to changes in the underlying GABA signal at the same chemical shift. For this reason, pair-wise corrections (applying the same correction parameters to both spectra in each ON–OFF pair) should be applied if Cr is to be used as a frequency and phase reference (Evans et al., 2012), except in cases where large frequency drifts are present, since frequency correction in the presence of large frequency drifts may result in incomplete subtraction of choline and creatine signals. The N-acetyl-d-aspartate (NAA) signal does not appear in ON spectra, so could only be used with a pair-wise correction based on the OFF spectra (at the cost of temporal resolution). An additional benefit of frequency correction is the possibility of using the magnitude of frequency shifts as an exclusion criterion for data confounded by excessive subject motion (Bhattacharyya et al., 2007).
Once the frequency- and phase-corrections have been applied, time averaging is performed and the edited difference spectrum calculated. In addition, the sum of the editing OFF spectra alone can be calculated, essentially a standard press sequence, so that other metabolites such as NAA, Cr and Cho can be quantified either as the internal quantification reference (see below) or for additional metabolic information. Alternatively, reference Cr levels can be calculated from the sum of edit ON and OFF spectra, with the advantage of improved SNR (relative to the edit OFF spectra alone).
Signal quantification and fitting
As with unedited spectroscopy, there are a number of tools available for fitting and quantification of GABA concentration from edited spectra. Fitting methods that use predefined models of spectral peaks corresponding to the spectrum of expected neurometabolites, called basis sets, such as LCModel (Provencher, 1993, 2001), TARQUIN (Wilson et al., 2010), and QUEST in jMRUI (Naressi et al., 2001; Stefan et al., 2009), originally designed for unedited spectra, can be applied to edited spectra with appropriate modifications to basis sets and control parameters. Some tools, which are custom-written for GABA-MRS analysis, such as Gannet (available through gabamrs.blogspot.com), can perform both preprocessing and fitting. Table 1 summarizes the pre-processing and fitting steps commonly used in the various MRS quantification tools.
Table 1.
Summary description of analysis methods.
| Fitting method | AMARES (jMRUI) | Gannet | LC-MODEL | Tarquin |
|---|---|---|---|---|
| DATA format | GE, Philips, Siemens | GE, Philips, Siemens | GE, Philips, Siemens | GE, Philips, Siemens |
| Frequency correction | Optional automatic processing | Automatic | Optional pre-processing step. Not part of the LC Model package | Optional automatic processing |
| Water subtraction | Optional semi-automatic processing | Optional automatic processing | Not performed | Optional automatic |
| Calculation of edited spectrum | Manual | Automatic | Pre-processing required | Automatic |
| GABA model | User choice – singlet or doublet, prior knowledge for Gaussian or Lorentzian | Singlet, Gaussian | Basis set uses simulated or phantom spectra. | Two Gaussian singlets |
| Concentration estimates | User choice of reference to water, Cr NAA | Automatic reference to water and Cr | Automatic reference to water or user choice of NAA, Cr | Automatic reference to water or user choice of Cr, NAA |
| Quality measures | SD of residual | SD of residual, rejection of data with poor quality, or excessive motion | CRLB | SD of residuals |
| Analysis of non-edited spectrum | Single, or double peak model | Single peak model | Metabolite basis set analysis | Metabolite basis set analysis |
Where used, basis sets should be acquired experimentally, or simulated using full 3-dimensional voxel simulation (rather than hard-pulse or on-resonance approximations), with the exact pulse widths and timings of the individual experiment used as approximations may fail to capture the significant spatial variation in intensity and multiplet structure of the edited signal that occur throughout the voxel (see e.g. (Edden and Barker, 2007)). Software such as VESPA (scion.duhs.duke.edu, n.d.) is freely available and has been used by several groups to simulate basis spectra. However, validation of simulations against phantom data is advised as there is on-going debate as to the values of the couplings in GABA and the extent to which two-bond proton–proton couplings need to be included (Govindaraju et al., 2000; Kreis and Bolliger, 2012). A database of simulated spectra for different sequence implementations would be useful for future researchers and would promote common analysis pathways. There has been mention of such a database on both the TARQUIN and VESPA users mailing lists, and the VESPA project has a repository for user contributions http://scion.duhs.duke.edu/vespa/contrib, although as yet no metabolite results for any sequence have been contributed.
Using the AMARES fitting routine in jMRUI is another option where prior knowledge and a degree of manual user intervention can be used to fit both the GABA and other metabolite peaks as a mixture of single Gaussian or Lorentzian peaks.
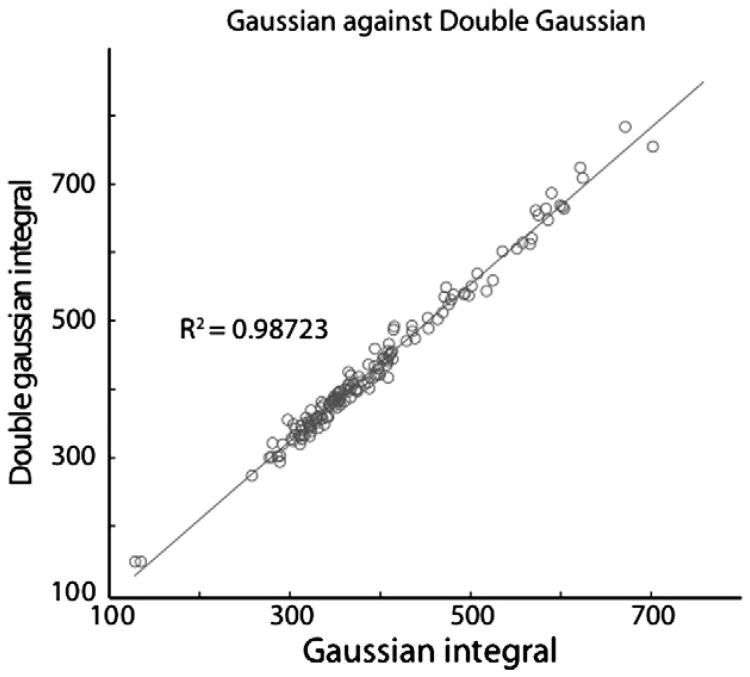
While the simple triplet model of the 3-ppm GABA signal supports the use of a pseudo-doublet model for GABA fitting, both simulations and phantom data show significant contribution from the center peak (as seen in Figs. 1 and 2), and the appearance of in-vivo spectra from different implementations vary in the extent to which a splitting is seen in-vivo, with estimates varying from 20% to 60% of spectra showing doublet character. Note that small-to-moderate Cr subtraction artefacts can easily be misinterpreted as ‘true’ doublet splittings, and the use of doublet character as a benchmark for spectral quality cannot be universally applied. Trials of model-type within one pipeline (GANNET) suggest that the integral that results from fitting by either a Gaussian singlet or doublet model agree with an R2 of 0.9872 for a dataset of 144 spectra from occipital, sensorimotor and DLPFC (as shown in Fig. 4). This is perhaps not surprising given the general line broadening seen in in vivo data, and the presence of underlying macromolecule peaks (discussed in greater detail later), and highlights that even with edited spectra, discrimination of individual spectral peaks is difficult in vivo.
Fig. 4.
The effect of fit model on the estimation of GABA: Analysis was performed on 144 in-vivo datasets using both single Gaussian and double Gaussian functions to fit the GABA peak. The high correlation between the resulting integral values demonstrates that, for in-vivo data, fitting a single Gaussian is equivalent to using a more complex model.
If LCModel is to be used to fit edited spectra, the default fit parameters (including an unconstrained baseline and the default macromo-lecular fitting) are not recommended as these may lead to variable results, often with minimal detection of GABA due to LCModel apportioning the majority of the peak at 3.0 ppm to the MM component. The use of LCModel for fitting edited spectra is limited by the fact that its default settings tend to assume a spectrum of sharp peaks superimposed on a broad positive baseline. In in vivo edited spectra, the GABA signal is not particularly narrow (compared to the MM signal) and the majority of the MM baseline is removed upon editing. These problems can be addressed either by constraining the baseline (by setting VITRO=T or NOBASE=T in LCModel) or by explicitly modeling the co-edited macromolecules in the basis set (see below) and are not limited to LCModel. However, LCModel is viewed in some circles as the default tool for the analysis of 1H-MRS spectra and many literature examples of variable or inappropriate fitting arise from misuse of LCModel (e.g. Fig. 5 in Taki et al., 2009).
TARQUIN is an MRS analysis package that uses a linear combination of basis functions to fit spectra, similar to LCModel. The fitting is performed in the time-domain and simulated basis sets based on quantum calculations can be automatically generated to match common acquisition protocols. However, for edited MEGA-PRESS data TARQUIN uses a simple predefined basis set which models the GABA peak as two single Gaussian peaks. Full simulation of the MEGA-PRESS sequence is under development by the developers of TARQUIN.
The reliability of these various fitting/analysis methods has been assessed in a recent study investigating the (within-session) test–retest reproducibility of GABA concentrations in a group of healthy adult volunteers (O'Gorman et al., 2011b). Results from this study indicate that the reproducibility of GABA quantification is similar across methods (see Table 2). The reliability of GABA quantification was assessed from MEGA-PRESS data acquired from a 25×40×30 mm3 voxel in the dorsolateral prefrontal cortex in sixteen healthy adults (age 25–38 years; with signed informed consent and local ethics board approval) (O'Gorman et al., 2011b). Four consecutive resting MEGA-PRESS spectra were acquired from each participant and water-scaled GABA concentrations were derived with LCModel, jMRUI, TARQUIN and Gannet. For each fitting method, the reliability was quantified as the coefficient of variation of the GABA concentrations derived for each participant, averaged across the subject group. Coefficients of variation ranged from 7% to 9% for all fitting methods (Table 2). Further details with regard to the processing methods are given in Table 1.
Table 2.
Within-session reliability of different analysis methods.
| Analysis method | GABA test–retest reproducibility (% CV from consecutive scans) |
|---|---|
| AMARES | 9% |
| Gannet | 8% |
| LCModel | 7% |
| TARQUIN | 8% |
While MEGA-PRESS has been shown to be reliable in vivo, researchers are recommended to validate the technique on their systems against phantoms of known concentration, and to perform this step on a regular basis as part of a normal quality assurance protocol. Validation of in vivo measures is of course difficult as apart from other MRS techniques, other non-invasive methods to measure neuronal GABA are non-existent. This may be considered one potential downfall of any MRS methods, in that ground truth for concentration is not known, and all measures by their nature are relative to some proposed reference, whose concentration may itself be based on assumptions and estimations. However, the use of appropriate assumptions, and literature values for key constants (e.g. T1, T2, and water concentrations) as discussed in the next section is widely accepted, and should provide measures that are comparable between research groups.
Concentration estimation and the use of Internal Standards
The calculation of concentration values from fitted edited spectra follows the same principles (and suffers from the same limitations) as for standard single-voxel MRS. In general, concentrations are calculated relative to an experimentally acquired internal reference of (an assumed) known concentration. In published studies to date, three references have been used – Cr, Water and NAA. Each has its merits: Cr and NAA have the advantage that they are acquired during the MEGA-PRESS scan, rather than as a separate scan at the beginning or end of the GABA acquisition, so potential effects of subject motion are minimized. Using Cr as a reference is familiar amongst the clinical community and has been shown to perform well (Bogner et al., 2010). In addition, the main Cr resonance is very close to the edited GABA signal; therefore chemical shift displacement issues will be negligible. The water signal has higher SNR and is more easily modeled, although chemical shift effects (Howe et al., 1993; Weinreb et al., 1985) mean that the acquired water signal (in some implementations) may be from a different location than the acquired voxel. Within the Gannet pipeline, referencing to both Cr and water is used (when water data are provided), which is useful in establishing that it is the GABA numerator, rather than the denominator, that drives observed effects as changes in NAA, Cr and water signals are all well documented in disease.
Water-scaled GABA concentrations can be estimated in institutional units from metabolite peak amplitudes according to the following equation:
| (1) |
where SGABA and SH2O are the averaged raw GABA and water signals, respectively, [H2O] is the brain water concentration (e.g. 55,550 mmol/l), VISH2O is the water visibility (e.g. 0.65 in white matter), eff is the editing efficiency (0.5), TR is the repetition time, T1H2O is the T1 of water (1.1 s) (Wansapura et al., 1999), T2H2O is the T2 of water (0.095 s) (Wansapura et al., 1999), T1GABA is the T1 of GABA (0.8 s), T2GABA is the T2 of GABA (0.13 s) (Träber et al., 2004), and MMcor is a macromolecular correction factor given by the fraction of GABA thought to occupy the GABA+peak (0.45).
The use of water as a concentration reference for spectroscopy has been discussed extensively elsewhere, and while it has many advantages, users are recommended to acquaint themselves fully with current practices and cautions (Gasparovic et al., 2006). The most important issue with regard to quantification using water as an internal reference is the accurate correction for partial volume effects within the voxel – this is especially important when performing group comparisons or correlations with other parametric measures. It is most important to correct the measured GABA signal for the CSF-fraction of the voxel, such that the quoted GABA pseudo-concentration is per unit brain tissue, excluding CSF (Kreis et al., 1993). Correction of the water reference signal for apparent water density and relaxivity in each tissue type is recommended (Gasparovic et al., 2006). Most current image analysis packages for structural/anatomical neuroimaging data have the ability to perform tissue segmentation, the reliability of which will depend on several factors, not the least of which is the quality of the anatomical images collected. While ensuring correct registration of the voxel of interest with the anatomical brain structures is seen as the biggest hurdle, the importance of performing tissue segmentation and correction was universally acknowledged in all discussions, especially if water was used as a concentration reference. Readers are directed to Alger (2010) for a detailed discussion of the issues inherent in quantitative measures in MRS.
Correction of GABA measurements for voxel gray matter (GM) and white matter (WM) fraction is more controversial. While some groups have found that the concentration of GABA is two times higher in gray matter than white matter (Jensen et al., 2005a; Petroff et al., 1988, 1989), it is probably more appropriate to utilize GM:WM ratios explicitly as covariates in any statistical analysis rather than to attempt to correct measures based on these reported differences in concentration between tissue types – indeed it is not obvious what such corrected quantities would correspond to, as they no longer represent concentrations. Corrections for voxel GM:WM fraction invariably also ignore the heterogeneous excitation of GABA signal within the voxel (Edden and Barker, 2007) and are as likely to inject tissue-fraction-driven effects into the data as they are to correct for these effects.
Contamination of spectra by co-edited macromolecular signal
As with any spectra acquired with a short-to-medium TE, macro-molecular (MM) contamination represents a major area of concern. Unfortunately, spectral editing does not separate GABA signal from at least one MM component, arising from spins at 3 ppm coupled to spins at 1.7 ppm which are affected by the editing pulses. As such reduction of the MM signal component is perhaps one of the most important areas for development in the measurement of GABA.
Three main approaches have been proposed to separate GABA from co-edited MM signals: direct measurement of the MM baseline through metabolite nulling (Behar et al., 1994; Hofmann et al., 2001; McLean et al., 2004); symmetric editing-based suppression of MM (Henry et al., 2001; Mescher et al., 1998; Rothman et al., 1993) and fitting of data with GABA and MM basis functions (Murdoch and Dydak, 2011). Currently each method has some detrimental effect on data quality or time of acquisition, thus the most common approach to date has been to accept MM contamination as a limitation of the MEGA-PRESS method at 3 T. A discussion of the three main techniques applied currently, and their caveats follows.
Direct measurement of the MM baseline through the use of an inversion pulse prior to the PRESS sequence to cause metabolite nulling has been proposed for normal short echo MRS as well as MEGA-PRESS studies (Behar et al., 1994; Hofmann et al., 2001; McLean et al., 2004). However using this technique to account for the MM contributions has several disadvantages; (i) it increases experiment time, (ii) in the MEGA-PRESS implementation it reduces the amplitude of the peak at 3.0 ppm, reducing fit reliability, (iii) it increases the noise level in the resulting MM-corrected GABA spectrum by √2 (assuming the residual noise of the metabolite-nulled acquisition is the same as that of the GABA acquisition), and, (iv) it requires the subtraction of two non-interleaved measurements, resulting in a greater sensitivity to participant motion. Pilot data (O'Gorman et al., 2011a) show that metabolite-nulling significantly reduces the reliability of GABA measures (with CRLBs increasing from 20 to 44%) and reduces the sensitivity of MEGA-PRESS GABA levels. These factors would suggest that metabolite nulling is not a useful technique for control of MM contributions to the MEGA-PRESS experiment.
Symmetric suppression of MM signals is a promising approach, and has been shown to be somewhat effective. Caution is advised when applied at 3 T however as the editing pulses commonly used are insufficiently selective to suppress MM without also significantly impacting on the GABA signals via co-editing. Recent work has suggested that the use of a slightly increased echo time over the typical 68 ms in MEGA-PRESS allows for editing pulses that are sufficiently selective to avoid this co-editing (Edden et al., 2012). Validation of this technique, or others with more selective editing pulses is one potentially rewarding area of future development.
Finally, explicitly modeling the MM contributions in the basis set is advantageous in that this method does not require additional scans (Murdoch and Dydak, 2011). However, fitting of edited spectra for both GABA and MM is heavily dependent on data quality and fitting constraints, and reproducibility data from one group suggests that modeling the co-edited macromolecules may introduce additional variability into the estimated GABA levels. While simple to implement, other fitting methods like AMARES, and Gannet do not yet allow for simulation of MM or accurate modeling of the baseline beyond a linear fit. In this case fitting of the edited peak at 3.0 ppm as either a singlet or a doublet will yield an estimate for GABA that should be reported as having a significant contribution from MM, often referred to as GABA+ in the literature.
While unfortunate, the contribution of MM to the GABA signal must be acknowledged, and the likelihood of MM driving observed results must be assessed on a case-by-case basis. For example, studies that hypothesize strong relationships between GABA-MRS measures and behavioral function (e.g. Edden et al., 2009; Puts et al., 2011; Yoon et al., 2010) from the outset, or examine experimentally-induced changes within session due to a task (Floyer-Lea et al., 2006; Michels et al., 2012) are perhaps less likely to be confounded by MM. However, group studies comparing patients with neurotypical controls may be affected by MM changes between groups, particularly in light of recent data suggesting that aging may influence the macromolecular contribution (Aufhaus et al., 2012). As such work on methods to account for MM contributions is an important area of development in GABA measures.
While MM contributions are an acknowledged problem, there is as yet no real consensus on the best way to deal with them, and those methods that are available are not yet widely applied. This widespread failure to account for MM, while perhaps understandable given the limitations of the methods available, stands as the greatest single limitation of the area to date and should be acknowledged as such. While problematic, accounting for MM contribution also represents an area of considerable on-going work, both in the MEGA-PRESS community and within MRS in general. All those interested in using MRS to measure GABA should follow these future advances closely.
Future developments
Methodological developments continue, both in terms of acquisition and processing, and this review is only intended to present a snapshot of a rapidly developing field. Quantification of GABA at fields higher than 3 T, although not widely available, will benefit from increased SNR, increased spectral dispersion and increased selectivity of editing pulses, advancing our understanding of GABAs role in the brain even further. There is also considerable work being done on non-edited MRS techniques to detect GABA reliably (Hu et al., 2007; Napolitano et al., 2012) at 3 T, however, MEGA-PRESS is still the most commonly applied MRS technique for GABA measurement, warranting this review and discussion of appropriate implementation and current problems with this technique.
Absolute quantification in meaningful concentration units is one useful goal, requiring the measurement of GABA relaxation times (Edden et al., 2011), editing efficiency, improved handling of MM contamination, and extensive validation of both acquisition and processing steps. However, much like other MRS measures at present, until a standard practice is settled upon by the wider community most GABA measures should be thought of as provisional or institutional estimates, and not true absolute concentrations when making comparisons between studies at different sites. Provided the same techniques are applied however, studies from the same sites, should be directly comparable.
Determining a standardized approach can be problematic in any academic field where several differing view points vie for prominence however a summary of the consensus reached at the MEGA-PRESS meeting regarding minimal best practice follows. Data should be collected at the standard TE of 68 ms although recent data suggests that the use of 80 ms allows for better MM removal through the use of symmetric-editing (Edden et al., 2012). Phase cycling of at least 2 steps should be performed, and allows for repeated interleaving of ON and OFF edited spectra. The interleaving of spectra allows for frequency correction and monitoring across a spectral acquisition and is recommended to both improve line width of the final spectra, and to catch subject motion which may lead to subtraction artefacts. Researchers should also be aware of the differences in timing and pulse shapes between vendors and sequence versions and the effects this may have on the data acquired. Voxel size, or acquisition time can be tailored to the question at hand, but should also be tested to ensure they provide adequate signal to noise for reliable fitting. There are several processing and fitting implementations available, if basis sets are used, models from phantoms or from full simulation with appropriately modeled RF pulses are recommended over simpler approximations, however use of either a single or double Gaussian peak for GABA has been shown to produce equally reliable results. Concentration referencing to an unsuppressed water scan is recommended, although the Cr and NAA peaks from the unedited spectrum can also be used as long as whichever is used should be prominently mentioned when reporting the results. If water is used, appropriate correction for tissue content and relaxation differences is required. Finally, MM contributions need to be addressed, either through acknowledgement of their contribution to the GABA signal reported, or through the use of some technique to control for these. While several techniques exist, researchers are advised to be aware of the caveats and potential pitfalls of each as discussed previously.
The measurement of GABA concentration using edited MRS at 3 T is an extremely powerful approach for investigating the role of GABAergic inhibition in healthy brain function and the pathophysiology of neurological and psychiatric disease. Development of common acquisition and analysis strategies across sites and vendors will enhance the interpretability of the literature and widen uptake of the methodology to sites without pre-existing MRS expertise. It is our hope that this report can form the foundation of such a common acquisition, processing and analysis framework.
Acknowledgments
This work is funded in part by: The University Research Priority Program “Integrative Human Physiology” at the University of Zurich The Welsh Institute of Cognitive Neuroscience NIH grants P41 EB015909 and R21 NS077300.
Cardiff Symposium on MRS of GABA, 22-23 August 2011: Dr. Matthew J. Brookes. Sir Peter Mansfield Magnetic Resonance Centre, Nottingham University, UK
Adrian Garcia – University of Birmingham, Birmingham, UK
Bradley R. Foerster, MD – Department of Radiology, University of Michigan, Ann Arbor, MI & Ann Arbor VA Healthcare System, Ann Arbor, MI
Myria Petrou, MA MBChB MS, Department of Radiology, University of Michigan, Ann Arbor, MI
Darren Price – Sir Peter Mansfield Magnetic Resonance Centre, Nottingham University, UK
Bhavana S. Solanky – NMR Research Unit, Department of Neuroinflammation, University College London, Institute of Neurology, UK
Inês R. Violante – Institute of Biomedical Research in Light and Image, Faculty of Medicine, University of Coimbra, Coimbra, Portugal
Steve Williams – Biomedical Imaging Institute, School of Cancer and Enabling Sciences, Manchester University, Manchester, UK
Martin Wilson – Cancer Sciences, University of Birmingham, Birmingham, UK
Appendix 1.
Processing pipelines
A number of MRS research groups were represented at the Cardiff meeting, all at differing stages of development of a processing pipeline. Illustrative examples of the processing steps for two of the three main clinical MRI system manufacturers are presented below to outline current procedures. Areas of similarity in each will then be discussed and considered in terms of appropriateness and utility. It should be pointed out that the MEGA-PRESS sequences are not currently standard “product” sequences for these manufacturers and are distributed as research patches or sequences specific to platform.1
The example pipeline includes steps which can be thought of as reflecting best-practice beliefs amongst the participants and formed the basis for round table discussion on day two of the meeting. We have set out below discussion of those aspects of these processing steps that are most important to consider, highlighting both areas of consensus and those where further refinement and consideration of the technical issues is recommended. Note that for the AMARES, LCModel, and TARQUIN pipelines, automated drift correction is not applied but can be implemented as an additional pre-processing step using custom software.
A1.1. AMARES/jMRUI
GE pre-processing
In SAGE, data are coil combined and phase corrected, using the first point of the FID of the unsuppressed water signal for each coil. The coil-combined unsuppressed water lines are then averaged and saved to a sage data file for subsequent processing, and the coil-combined metabolite lines are subtracted and saved to a separate sage data file. (If Cr or other metabolite information is desired then the coil-combined unedited lines are saved to a different data file.) Automated drift correction is not applied. The binary sage data files are converted to text files for subsequent processing in jMRUI.
Philips pre-processing
Data are exported from the scanner as *.SDAT and *.SPAR files and opened in jMRUI as a time series of editing pairs (ON and OFF spectra). The ON and OFF spectra are 180° out of phase, so to produce the edited spectrum a simple addition across all the spectra is required. Phase correction may be required. If individual editing pairs are summed first, then the inverted NAA peak can be used to check for frequency drifts before summation to produce the final edited spectrum. HSVD removal of any residual water can then be performed and 4 Hz Gausian apodization applied.
Processing
The residual water signal is removed by Hankel SVD. GABA levels are evaluated with the AMARES algorithm, after manually defining the center frequency and width of the GABA peak. GABA is modeled as a Gaussian singlet (phase 0°) and NAA is modeled as a single inverted Lorentzian peak (phase 180°). GABA levels are quantified either relative to NAA, unsuppressed water or creatine (modeled as a single Lorentzian in the unedited lines). Alternatively, GABA/Cr can be calculated by multiplying the GABA/NAA ratio from the edited spectrum by the NAA/Cr concentration calculated by LCModel from the unedited lines (Donahue et al., 2010).
Quantification
Water-scaled GABA concentrations must be calculated manually, but can be estimated in institutional units using Eq. (1).
A1.2. Gannet
Pre-processing
None required.
Processing
Coil-combination, phasing, apodization, and frequency correction are performed automatically (see Table 1 for details). GABA is modeled as a single Gaussian superimposed on a linear baseline, and both the water-scaled GABA concentration and the GABA/Cr ratio are calculated.
Quantification
Water-scaled GABA concentrations are calculated according to Eq. (1), taking into account the editing efficiency and approximate macromolecular contributions to the GABA+ peak.
A1.3. LCModel
GE pre-processing
Data are coil combined and phase corrected in SAGE, as described above for AMARES/jMRUI. The coil-combined unsuppressed water lines are then averaged and saved to a sage data file for subsequent processing, and the coil-combined metabolite lines are subtracted and saved to a separate sage data file. No apodization is performed. Automated drift correction is not applied. The binary sage data files are converted to LCModel .RAW format.
Philips pre-processing
SDAT files are converted to LCModel .RAW format and averaged. (Since ON and OFF spectra are acquired 180° out of phase, the MEGA-PRESS difference spectrum can be derived from a sum of all spectra, so prior subtraction is not necessary). If frequency correction is to be applied it should be done before the averaging step. No apodization is performed.
Processing
The (subtracted) metabolite and water files are processed in LCModel using either an experimental or simulated basis set including basis spectra for GABA, glutamate, glutamine, Glx, NAA and glutathione, either with the control parameters VITRO=T, NOBASE=T, or with the coedited macromolecular signal included in the basis set (Murdoch and Dydak, 2011).
Quantification
Water-scaled GABA concentrations are calculated according to a modified version of Eq. (1). The editing efficiency and macromolecular correction factors are not applied automatically.
A1.4. Tarquin
Pre-processing
None required.
Processing
Coil-combination and phasing are performed automatically (see Table 1 for details). Frequency correction can be applied, with the option to use both the Cr and Cho peaks, or the Cr peak alone. GABA is modeled as two Gaussian singlets, and both the water-scaled GABA concentration and the GABA/Cr ratio are calculated. A simple single Gausian model is also included for the MM peak.
Quantification
Water-scaled GABA concentrations are calculated according to a modified version of Eq. (1), excluding the relaxation correction since Tarquin partially accounts for relaxation effects by using a scaling factor in its estimate. Readers are advised that this scaling factor is set by default, and may not take into account the true relaxation effects for MEGA-PRESS sequences and so should either explicitly set this scaling factor to one, or correct for its effect post hoc. The editing efficiency and macromolecular correction factors are not applied automatically.
Footnotes
These sequences are freely available from Dr Edden (raee2@jhu.edu).
References
- Alger JR. Quantitative proton magnetic resonance spectroscopy and spectroscopic imaging of the brain. Top Magn Reson Imaging. 2010;21(2):115–128. doi: 10.1097/RMR.0b013e31821e568f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aufhaus E, Weber-Fahr W, Sack M, Tunc-Skarka N, Oberthuer G, Hoerst M, Meyer-Lindenberg A, Boettcher U, Ende G. Absence of changes in GABA concentrations with age and gender in the human anterior cingulate cortex: a MEGA-PRESS study with symmetric editing pulse frequencies for macromolecule suppression. Magn Reson Med. 2012 doi: 10.1002/mrm.24257. http://dx.doi.org/10.1002/mrm.24257. [DOI] [PubMed]
- Behar KL, Rothman DL, Spencer DD, Petroff OA. Analysis of macromolecule resonances in 1H NMR spectra of human brain. Magn Reson Med. 1994;32(3):294–302. doi: 10.1002/mrm.1910320304. [DOI] [PubMed] [Google Scholar]
- Bhattacharyya PK, Lowe MJ, Phillips MD. Spectral quality control in motioncorrupted single-voxel J-difference editing scans: an interleaved navigator approach. Magn Reson Med. 2007;58(4):808–812. doi: 10.1002/mrm.21337. [DOI] [PubMed] [Google Scholar]
- Bogner W, Gruber S, Doelken M, Stadlbauer A, Ganslandt O, Boettcher U, Trattnig S, et al. In vivo quantification of intracerebral GABA by single-voxel (1)H-MRS – how reproducible are the results? Eur J Radiol. 2010;73(3):526–531. doi: 10.1016/j.ejrad.2009.01.014. http://dx.doi.org/10.1016/j.ejrad.2009.01.014. [DOI] [PubMed] [Google Scholar]
- Boy F, Evans CJ, Edden RAE, Singh KD, Husain M, Sumner P. Individual differences in subconscious motor control predicted by GABA concentration in SMA. Curr Biol. 2010;20(19):1779–1785. doi: 10.1016/j.cub.2010.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boy F, Evans CJ, Edden RAE, Lawrence AD, Singh KD, Husain M, Sumner P. Dorsolateral prefrontal gamma-aminobutyric acid in men predicts individual differences in rash impulsivity. BPS. 2011;70(9):866–872. doi: 10.1016/j.biopsych.2011.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi IY, Lee SP, Shen J. In vivo single-shot three-dimensionally localized multiple quantum spectroscopy of GABA in the human brain with improved spectral selectivity. J Magn Reson. 2005;172(1):9–16. doi: 10.1016/j.jmr.2004.09.021. [DOI] [PubMed] [Google Scholar]
- de Graaf RA. In Vivo NMR Spectroscopy: Principles and Techniques. 2nd. Wiley-Interscience; 2007. [Google Scholar]
- Donahue MJ, Near J, Blicher JU, Jezzard P. Baseline GABA concentration and fMRI response. NeuroImage. 2010;53(2):392–398. doi: 10.1016/j.neuroimage.2010.07.017. http://dx.doi.org/10.1016/j.neuroimage.2010.07.017. [DOI] [PubMed] [Google Scholar]
- Edden RAE, Barker PB. Spatial effects in the detection of gamma-aminobutyric acid: improved sensitivity at high fields using inner volume saturation. Magn Reson Med. 2007;58(6):1276–1282. doi: 10.1002/mrm.21383. [DOI] [PubMed] [Google Scholar]
- Edden RAE, Muthukumaraswamy SD, Freeman TCA, Singh KD. Orientation discrimination performance is predicted by GABA concentration and gamma oscillation frequency in human primary visual cortex. J Neurosci. 2009;29(50):15721–15726. doi: 10.1523/JNEUROSCI.4426-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edden RA, Intrapiromkul J, Zhu H, Cheng Y, Barker PB. Measuring T(2) in vivo with J-difference editing: application to GABA at 3 Tesla. J Magn Reson Imaging. 2011;35(1):229–234. doi: 10.1002/jmri.22865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edden R, Puts N, Barker PB. Macromolecule-suppressed GABA-edited magnetic resonance spectroscopy at 3T. Magn Reson Med. 2012;68(3):657–661. doi: 10.1002/mrm.24391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans CJ, Puts NAJ, Robson SE, Boy F, McGonigle DJ, Sumner P, Singh KD, et al. Diurnal stability of gamma-aminobutyric acid concentration in visual and sensorimotor cortex. J Magn Reson Imaging. 2010;31(1):204–209. doi: 10.1002/jmri.21996. [DOI] [PubMed] [Google Scholar]
- Evans CJ, Puts NA, Robson SE, Boy F, McGonigle DJ, Sumner P, Singh KD, Edden RA. Subtraction artifacts and frequency (Mis-)alignment in J-difference GABA editing. J Magn Reson Imaging. 2012 doi: 10.1002/jmri.23923. http://dx.doi.org/10.1002/jmri.23923 (Epub ahead of print) [DOI] [PubMed]
- Floyer-Lea A, Wylezinska M, Kincses T, Matthews PM. Rapid modulation of GABA concentration in human sensorimotor cortex during motor learning. J Neurophysiol. 2006;95(3):1639–1644. doi: 10.1152/jn.00346.2005. [DOI] [PubMed] [Google Scholar]
- Foerster BR, Callaghan BC, Petrou M, Edden RA, Chenevert TL, Feldman EL. Decreased motor cortex gamma-aminobutyric acid in amyotrophic lateral sclerosis. Neurology. 2012a;78(20):1596–1600. doi: 10.1212/WNL.0b013e3182563b57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foerster BR, Petrou M, Edden RAE, Sundgren PC, Schmidt-Wilcke T, Lowe SE, Harte SE, et al. Reduced insular γ-aminobutyric acid in fibromyalgia. Arthritis Rheum. 2012b;64(2):579–583. doi: 10.1002/art.33339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasparovic C, Song T, Devier D, Bockholt HJ, Caprihan A, Mullins PG, Posse S, et al. Use of tissue water as a concentration reference for proton spectroscopic imaging. Magn Reson Med. 2006;55(6):1219–1226. doi: 10.1002/mrm.20901. http://dx.doi.org/10.1002/mrm.20901. [DOI] [PubMed] [Google Scholar]
- Govindaraju V, Young K, Maudsley AA. Proton NMR chemical shifts and coupling constants for brain metabolites. NMR Biomed. 2000;13(3):129–153. doi: 10.1002/1099-1492(200005)13:3<129::aid-nbm619>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- Hancu I. Optimized glutamate detection at 3T. J Magn Reson Imaging. 2009;30(5):1155–1162. doi: 10.1002/jmri.21936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry PG, Dautry C, Hantraye P, Bloch G. Brain GABA editing without macromolecule contamination. Magn Reson Med. 2001;45(3):517–520. doi: 10.1002/1522-2594(200103)45:3<517::aid-mrm1068>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- Henry ME, Lauriat TL, Shanahan M, Renshaw PF, Jensen JE. Accuracy and stability of measuring GABA, glutamate, and glutamine by proton magnetic resonance spectroscopy: a phantom study at 4 Tesla. J Magn Reson. 2010:1–9. doi: 10.1016/j.jmr.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann L, Slotboom J, Boesch C, Kreis R. Characterization of the macromolecule baseline in localized (1)H-MR spectra of human brain. Magn Reson Med. 2001;46(5):855–863. doi: 10.1002/mrm.1269. [DOI] [PubMed] [Google Scholar]
- Howe FA, Stubbs M, Rodrigues LM, Griffiths JR. An assessment of artefacts in localized and non-localized 31P MRS studies of phosphate metabolites and pH in rat tumours. NMR Biomed. 1993;6(1):43–52. doi: 10.1002/nbm.1940060108. [DOI] [PubMed] [Google Scholar]
- Hu J, Yang S, Xuan Y, Jiang Q, Yang Y, Haacke EM. Simultaneous detection of resolved glutamate, glutamine, and γ-aminobutyric acid at 4T. J Magn Reson. 2007;185(2):204–213. doi: 10.1016/j.jmr.2006.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurd R, Sailasuta N, Srinivasan R, Vigneron DB, Pelletier D, Nelson SJ. Measurement of brain glutamate using TE-averaged PRESS at 3T. Magn Reson Med. 2004;51(3):435–440. doi: 10.1002/mrm.20007. [DOI] [PubMed] [Google Scholar]
- Jang DP, Lee JM, Lee E, Park S, Kim JJ, Namkoong K, Yoon KJ, et al. Interindividual reproducibility of glutamate quantification using 1.5-T proton magnetic resonance spectroscopy. Magn Reson Med. 2005;53(3):708–712. doi: 10.1002/mrm.20387. [DOI] [PubMed] [Google Scholar]
- Jensen JE, deB Frederick B, Renshaw PF. Grey and white matter GABA level differences in the human brain using two-dimensional, J-resolved spectroscopic imaging. NMR Biomed. 2005a;18(8):570–576. doi: 10.1002/nbm.994. [DOI] [PubMed] [Google Scholar]
- Jensen JE, Frederick BD, Wang L, Brown J, Renshaw PF. Two-dimensional, J-resolved spectroscopic imaging of GABA at 4 Tesla in the human brain. Magn Reson Med. 2005b;54(4):783–788. doi: 10.1002/mrm.20644. [DOI] [PubMed] [Google Scholar]
- Keeler J. Understanding NMR Spectroscopy. 2nd. Wiley; 2011. [Google Scholar]
- Kreis R, Bolliger CS. The need for updates of spin system parameters, illustrated for the case of γ-aminobutyric acid. NMR Biomed. 2012;25:1401–1403. doi: 10.1002/nbm.2810. http://dx.doi.org/10.1002/nbm.2810. [DOI] [PubMed] [Google Scholar]
- Kreis R, Ernst T, Ross BD. Development of the human brain: in vivo quantification of metabolite and water content with proton magnetic resonance spectroscopy. Magn Reson Med. 1993;30(4):424–437. doi: 10.1002/mrm.1910300405. [DOI] [PubMed] [Google Scholar]
- Mayer D, Kim DH, Adalsteinsson E, Spielman DM. Fast CT-PRESS-based spiral chemical shift imaging at 3 Tesla. Magn Reson Med. 2006;55(5):974–978. doi: 10.1002/mrm.20853. [DOI] [PubMed] [Google Scholar]
- McLean MA, Simister RJ, Barker GJ, Duncan JS. Discrimination between neurochemical and macromolecular signals in human frontal lobes using short echo time proton magnetic resonance spectroscopy. Faraday Discuss. 2004;126:93–102. doi: 10.1039/b304938h. discussion 169–83. [DOI] [PubMed] [Google Scholar]
- Mekle R, Mlynárik V, Gambarota G, Hergt M, Krueger G, Gruetter R. MR spectroscopy of the human brain with enhanced signal intensity at ultrashort echo times on a clinical platform at 3T and 7T. Magn Reson Med. 2009;61(6):1279–1285. doi: 10.1002/mrm.21961. [DOI] [PubMed] [Google Scholar]
- Mescher M, Tannus A, Johnson MO, Garwood M. Solvent suppression using selective echo dephasing. J Magn Reson Ser A. 1996;123(2):226–229. [Google Scholar]
- Mescher M, Merkle H, Kirsch J, Garwood M, Gruetter R. Simultaneous in vivo spectral editing and water suppression. NMR Biomed. 1998;11(6):266–272. doi: 10.1002/(sici)1099-1492(199810)11:6<266::aid-nbm530>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- Michels L, Martin E, Klaver P, Edden R, Zelaya F, Lythgoe DJ, Lüchinger R, et al. Frontal GABA levels change during working memory. PLoS One. 2012;7(4):e31933. doi: 10.1371/journal.pone.0031933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullins PG, Chen H, Xu J, Caprihan A, Gasparovic C. Comparative reliability of proton spectroscopy techniques designed to improve detection of J-coupled metabolites. Magn Reson Med. 2008;60(4):964–969. doi: 10.1002/mrm.21696. [DOI] [PubMed] [Google Scholar]
- Murdoch JB, Dydak U. Modeling MEGA-PRESS macromolecules for a better grasp of GABA. Presented at the Intl Soc Mag Reson Med, Montreal. 2011;19:1394. [Google Scholar]
- Napolitano A, Kockenberger W, Auer DP. Reliable gamma aminobutyric acid measurement using optimized PRESS at 3 T. T Magn Reson Med. 2012 doi: 10.1002/mrm.24397. http://dx.doi.org/10.1002/mrm.24397. [DOI] [PubMed]
- Naressi A, Couturier C, Devos JM, Janssen M, Mangeat C, de Beer R, Graveron-Demilly D. Java-based graphical user interface for the MRUI quantitation package. Magn Reson Mater Phys. 2001;12(2–3):141–152. doi: 10.1007/BF02668096. [DOI] [PubMed] [Google Scholar]
- Near J, Simpson R, Cowen P, Jezzard P. Efficient gamma-aminobutyric acid editing at 3T without macromolecule contamination: MEGA-SPECIAL. NMR Biomed. 2011;24(10):1277–1285. doi: 10.1002/nbm.1688. [DOI] [PubMed] [Google Scholar]
- O'Gorman RL, Edden RA, Michels L, Murdoch JB, Martin E. Presented at the Intl Soc Mag Reson Med. Vol. 19. Montreal, Canada: 2011a. Precision and repeatability of in vivo GABA and glutamate quantification; p. 3434. [Google Scholar]
- O'Gorman RL, Michels L, Edden RA, Murdoch JB, Martin E. In vivo detection of GABA and glutamate with MEGA-PRESS: reproducibility and gender effects. J Magn Reson Imaging. 2011b;33(5):1262–1267. doi: 10.1002/jmri.22520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petroff OAC. Book review: GABA and glutamate in the human brain. Neuroscientist. 2002;8(6):562–573. doi: 10.1177/1073858402238515. [DOI] [PubMed] [Google Scholar]
- Petroff OA, Ogino T, Alger JR. High-resolution proton magnetic resonance spectroscopy of rabbit brain: regional metabolite levels and postmortem changes. J Neurochem. 1988;51(1):163–171. doi: 10.1111/j.1471-4159.1988.tb04850.x. [DOI] [PubMed] [Google Scholar]
- Petroff OA, Spencer DD, Alger JR, Prichard JW. High-field proton magnetic resonance spectroscopy of human cerebrum obtained during surgery for epilepsy. Neurology. 1989;39(9):1197–1202. doi: 10.1212/wnl.39.9.1197. [DOI] [PubMed] [Google Scholar]
- Petrou M, Pop-Busui R, Foerster BR, Edden RA, Callaghan BC, Harte SE, Harris RE, et al. Altered excitation–inhibition balance in the brain of patients with diabetic neuropathy. Acad Radiol. 2012;19(5):607–612. doi: 10.1016/j.acra.2012.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provencher SW. Estimation of metabolite concentrations from localized in vivo proton NMR spectra. Magn Reson Med. 1993;30(6):672–679. doi: 10.1002/mrm.1910300604. [DOI] [PubMed] [Google Scholar]
- Provencher SW. Automatic quantitation of localized in vivo 1H spectra with LCModel. NMR Biomed. 2001;14(4):260–264. doi: 10.1002/nbm.698. [DOI] [PubMed] [Google Scholar]
- Puts NAJ, Edden RAE. In vivo magnetic resonance spectroscopy of GABA: a methodological review. Prog Nucl Magn Reson Spectrosc. 2012;60(C):29–41. doi: 10.1016/j.pnmrs.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puts NA, Edden RA, Evans CJ, McGlone F, McGonigle DJ. Regionally specific human GABA concentration correlates with tactile discrimination thresholds. J Neurosci. 2011;31(46):16556–16560. doi: 10.1523/JNEUROSCI.4489-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothman DL, Petroff OA, Behar KL, Mattson RH. Localized 1H NMR measurements of gamma-aminobutyric acid in human brain in vivo. Proc Natl Acad Sci U S A. 1993;90(12):5662–5666. doi: 10.1073/pnas.90.12.5662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryner LN, Sorenson JA, Thomas MA. Localized 2D J-resolved 1H MR spectroscopy: strong coupling effects in vitro and in vivo. Magn Reson Imaging. 1995;13(6):853–869. doi: 10.1016/0730-725x(95)00031-b. [DOI] [PubMed] [Google Scholar]
- scion.duhs.duke.edu,d.scion.duhs.duke.edu. Retrieved June 9, 2012, from http://scion.duhs.duke.edu.
- Stagg CJ, Bestmann S, Constantinescu AO, Moreno LM, Allman C, Mekle R, Woolrich M, et al. Relationship between physiological measures of excitability and levels of glutamate and GABA in the human motor cortex. J Physiol. 2011;589(Pt 23):5845–5855. doi: 10.1113/jphysiol.2011.216978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Star-Lack J, Spielman D, Adalsteinsson E, Kurhanewicz J, Terris DJ, Vigneron DB. In vivo lactate editing with simultaneous detection of choline, creatine, NAA, and lipid singlets at 1.5 T using PRESS excitation with applications to the study of brain and head and neck tumors. J Magn Reson. 1998;133(2):243–254. doi: 10.1006/jmre.1998.1458. [DOI] [PubMed] [Google Scholar]
- Stefan D, Cesare FD, Andrasescu A, Popa E, Lazariev A, Vescovo E, Strbak O, et al. Quantitation of magnetic resonance spectroscopy signals: the jMRUI software package. Meas Sci Technol. 2009;20(10):104035. [Google Scholar]
- Taki MM, Harada M, Mori K, Kubo H, Nose A, Matsuda T, Nishitani H. High gamma-aminobutyric acid level in cortical tubers in epileptic infants with tuberous sclerosis complex measured with the MEGA-editing J-difference method and a three-Tesla clinical MRI Instrument. Neuroimage. 2009;47(4):1207–1214. doi: 10.1016/j.neuroimage.2009.05.060. [DOI] [PubMed] [Google Scholar]
- Terpstra M, Ugurbil K, Gruetter R. Direct in vivo measurement of human cerebral GABA concentration using MEGA-editing at 7 Tesla. Magn Reson Med. 2002;47(5):1009–1012. doi: 10.1002/mrm.10146. [DOI] [PubMed] [Google Scholar]
- Thompson RB, Allen PS. Response of metabolites with coupled spins to the STEAM sequence. Magn Reson Med. 2001;45(6):955–965. doi: 10.1002/mrm.1128. [DOI] [PubMed] [Google Scholar]
- Träber F, Block W, Lamerichs R, Gieseke J, Schild HH. 1H metabolite relaxation times at 3.0 Tesla: measurements of T1 and T2 values in normal brain and determination of regional differences in transverse relaxation. J Magn Reson Imaging. 2004;19(5):537–545. doi: 10.1002/jmri.20053. [DOI] [PubMed] [Google Scholar]
- Waddell KW, Avison MJ, Joers JM, Gore JC. A practical guide to robust detection of GABA in human brain by J-difference spectroscopy at 3 T using a standard volume coil. Magn Reson Imaging. 2007;25(7):1032–1038. doi: 10.1016/j.mri.2006.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wansapura JP, Holland SK, Dunn RS, Ball WS. NMR relaxation times in the human brain at 3.0 tesla. J Magn Reson Imaging. 1999;9(4):531–538. doi: 10.1002/(sici)1522-2586(199904)9:4<531::aid-jmri4>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- Weinreb JC, Brateman L, Babcock EE, Maravilla KR, Cohen JM, Horner SD. Chemical shift artifact in clinical magnetic resonance images at 0.35 T. AJR Am. J Roentgenol. 1985;145(1):183–185. doi: 10.2214/ajr.145.1.183. [DOI] [PubMed] [Google Scholar]
- Wilson M, Reynolds G, Kauppinen RA, Arvanitis TN, Peet AC. A constrained least-squares approach to the automated quantitation of in vivo 1H magnetic resonance spectroscopy data. Magn Reson Med. 2010;65(1):1–12. doi: 10.1002/mrm.22579. [DOI] [PubMed] [Google Scholar]
- Yoon JH, Maddock RJ, Rokem A, Silver MA, Minzenberg MJ, Ragland JD, Carter CS. GABA concentration is reduced in visual cortex in schizophrenia and correlates with orientation-specific surround suppression. J Neurosci. 2010;30(10):3777–3781. doi: 10.1523/JNEUROSCI.6158-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu G, Gheorghiu D, Allen PS. Motional degradation of metabolite signal strengths when using STEAM: a correction method. NMR Biomed. 1992;5(4):209–211. doi: 10.1002/nbm.1940050408. [DOI] [PubMed] [Google Scholar]