Summary
The relationship between three-dimensional organization of the genome and gene-regulatory networks is poorly understood. Here, we examined long-range chromatin interactions genome-wide in embryonic stem cells (ESCs), iPSCs, and fibroblasts, and uncover a pluripotency-specific genome organization that is gradually re-established during reprogramming. Our data confirm that long-range chromatin interactions are primarily associated with the spatial segregation of open and closed chromatin, defining overall chromosome conformation. Additionally, we identified two further levels of genome organization in ESCs characterized by co-localization of regions with high pluripotency factor occupancy and strong enrichment for Polycomb proteins/H3K27me3, respectively. Underlining the independence of these networks and their functional relevance for genome organization, loss of the Polycomb protein Eed diminishes interactions between Polycomb-regulated regions without altering overarching chromosome conformation. Together, our data highlight a pluripotency-specific genome organization in which Nanog and H3K27me3 occupy distinct nuclear spaces and reveal a role for cell type-specific gene-regulatory networks in genome organization.
Introduction
Chromosome conformation capture (3C)- based technologies (de Wit and de Laat, 2012; Dekker et al., 2002) have led to a new paradigm wherein gene regulation can be studied in the context of the three-dimensional (3D) organization of the genome (Bickmore and van Steensel, 2013). Recent work has demonstrated an organizational hierarchy to metazoan genome structure (Gibcus and Dekker, 2013). At the smallest scale, up to a few hundred kilobases (kb) of linear DNA, enhancers and promoters come into physical contact to establish cell type- specific expression programs (Sanyal et al., 2012; Shen et al., 2012; Smallwood and Ren, 2013). These interactions are maintained by the Cohesin complex, which can be recruited to interphase chromatin via the mediator complex (Kagey et al., 2010) and cell-type specific transcription factors (Denholtz and Plath, 2012; Wei et al., 2013). In mammals, promoter-enhancer interactions are confined to topologically-associating domains (TADs), which typically represent ~1 megabase (Mb) cell type-invariant, self-associating genomic regions whose boundaries are enriched for the insulator protein CTCF (Dixon et al., 2012; Nora et al., 2012).
As a second level of the organizational hierarchy, TADs appear to function as the fundamental modular unit of gene regulation and genome organization, with changes in gene expression and nuclear lamina association durin differentiation often occurring in a TAD-wide manner (Dixon et al., 2012; Nora et al., 2012; Shen et al., 2012). A third level in the organizational hierarchy occurs as a result of the preferential co-localization of specific TADs. These interactions can be identified as long-range chromatin contacts between genomic regions many Mb apart on the same chromosome (cis or intrachromosomal), or on different chromosomes (trans or interchromosomal) (Hakim et al., 2013; Hakim et al., 2011; Noordermeer et al., 2011; Osborne et al., 2004; Schoenfelder et al., 2010; Simonis et al., 2006). Although spatially co-localizing distal genomic regions have been reported to be enriched for the sequence motifs of specific transcription factors (Schoenfelder et al., 2010), co-expressed genes (Osborne et al., 2004), or co-regulated genes (Hakim et al., 2013; Noordermeer et al., 2011), the relationship between long-range chromatin interactions and the regulatory features enriched in the co-localizing regions is poorly understood.
In this study, we explored long-range (distal) chromatin contacts in mouse embryonic stem cells (ESCs) using 4C-seq to understand the relationship between chromatin contacts and gene-regulatory networks that govern cell identity. We define the distal chromatin interactions made by a variety of genomic ‘bait’ regions representing diverse chromatin and pluripotency transcription factor binding profiles, and extend our findings genome-wide. We further examine how genome organization changes in the absence of a critical chromatin regulator, and upon differentiation and transcription factor-induced reprogramming of differentiated cells into induced pluripotent stem cells (iPSCs). Together, our data define a previously unappreciated hierarchy in the organization of long-range chromatin contacts and reveal that distal genomic regions sharing common gene-regulatory features co-localize within the 3D-space of the nucleus.
Results
Experimental approach to studying chromatin contacts
To investigate long-range chromatin interactions between genomic regions Mb away on the same or on different chromosomes in ESCs, we performed 3C coupled to high throughput sequencing (4C-seq) (Splinter et al., 2012) for 16 bait regions (Table S1). 4C-seq allows one to identify any mappable genomic region in close physical proximity to a specific genomic (bait) region within a population of cells at the moment of fixation by means of proximity-based ligation of juxtaposed DNA fragments. The product is a library of chimeric DNA fragments containing the bait region and its interacting DNA partner(s) ligated at a restriction site, in our case HindIII, that can be identified by high-throughput sequencing (Experimental Procedures).
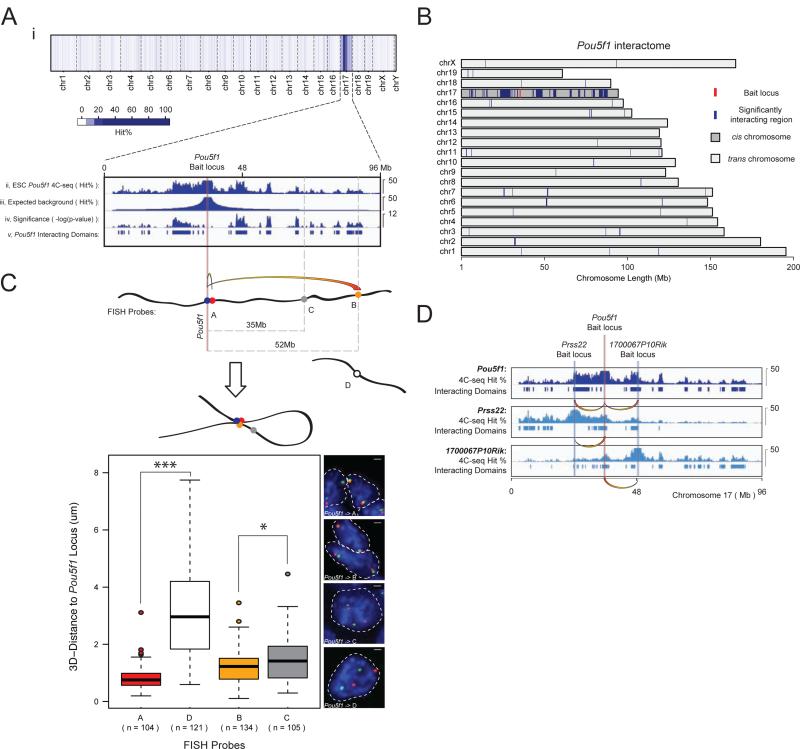
Our 4C-seq data were highly reproducible across biological and technical replicates utilizing distinct primer pairs for amplification of ligated fragments and different HindIII fragments within bait regions as anchor points, and passed stringent quality control requirements (Figures S1A/D/E, S2A, Tables S2 and S3). As a result, replicate data sets for each bait locus were pooled for downstream analysis (Experimental Procedures). To obtain a semi-quantitative measure of interactions, we calculated an average hit probability, referred to as ‘hit percentage’, within 200kb windows along the genome (Experimental Procedures). For all of our baits, the hit percentage was higher in cis than in trans, as exemplified by the 4C-seq interactome of the Pou5f1-encoding region (Figure 1Ai). Furthermore, as expected for the nature of chromosome conformation (Lieberman-Aiden et al., 2009), intrachromosomal interactions decay rapidly as a function of genomic distance to the bait along the linear DNA (Figure 1Aii).
Figure 1. Long-range chromatin contacts of the Pou5f1 bait region in ESCs.
A, Pooled 4C-seq data set, depicting the interaction profile of the Pou5f1 locus located on chromosome (chr) 17 in mouse ESCs. (i) Average hit probability (Hit%) within 200kb windows tiled across the genome. (ii) Hit% in cis (thresholded as displayed) was compared to an empirically modeled background hit% (iii) using the binomial test. (iv) Binomial test results and (v) significantly interacting domains of the Pou5f1 bait region (based on −log(p-values)>=1.8). The vertical red line and associated gray bar denote the Pou5f1 bait locus and the extended 1Mb bait region, respectively, which is always excluded from downstream analysis of the Pou5f1 interactome.
B, Significantly interacting domains (blue) of the Pou5f1 bait region (red), genome-wide.
C, DNA FISH-based 3D distance measurements between the Pou5f1 locus and genomic regions marked by A-C (in cis) or by D (in trans). Based on 4C-seq data, A and B interact with the Pou5f1 locus, but C, located in an intervening genomic region closer to Pou5f1 on linear DNA than B, does not. Boxes demarcate the interquartile range (IQR) and median, and whiskers +/−1.5 times the IQR. *=p-value<0.05, ***=p-value<0.001, Wilcoxon rank-sum test.
D, Two genomic regions containing the gene Prss22 and 1700067P10Rik were identified as interacting partners of the Pou5f1 locus. 4C-seq experiments using Prss22 and 1700067P10Rik as bait regions confirmed their interaction with Pou5f1. Bait regions, 4C-seq hit%, and interacting domains are shown.
See also Figure S1.
Significantly interacting regions were identified as those 200kb windows that showed a markedly higher hit probability than expected based on a binomial test (Figures 1Aii-v/B, S3A, Tables S1/S4, Experimental Procedures). To identify distal interactions in cis, we empirically modeled the expected background hit probability as a function of distance (Figure 1Aiii). Since trans interactions show no such positional biases, the average hit probability across each trans-chromosome was used for the expected background level of these interactions. False discovery rates (FDRs) were estimated using simulated data (Table S4, Experimental Procedures).
To test the reliability of our analysis and 4C-seq data, we partitioned our Pou5f1 4C-seq replicates into two equal subsets, pooled the libraries within each subset, and ran each pooled subset through our analysis pipeline. We obtained significantly overlapping intra- and interchromosomal interactomes from the partitioned data sets (Figure S1B), confirming the quality of our data and the robust nature of our analysis.
Several additional approaches were employed to further validate our 4C-seq-defined chromatin interactions and rule out technical biases affecting data generated by 4C-seq. These include 3C confirmation of ligation products (Figure S1F); 3D fluorescence in-situ hybridization (FISH) demonstrating that an interacting region 52Mb away from the Pou5f1 bait region is significantly closer to the bait in 3D-space of the ESC nucleus than a non-interacting region located only 35Mb away (Figure 1C), reciprocal 4C-seq (Figure 1D); control 4C-seq experiments with unfixed cells and genomic DNA showing no significant interactions (Figures S1A/C); the demonstration of comparable mappability inside and outside our interacting regions (Figure S1G); and the comparison of our 4C-seq data with a previously published genome-wide ESC interactome based on Hi-C (Dixon et al., 2012) showing strong correlation (Figure 3H, Table S5) (Experimental Procedures).
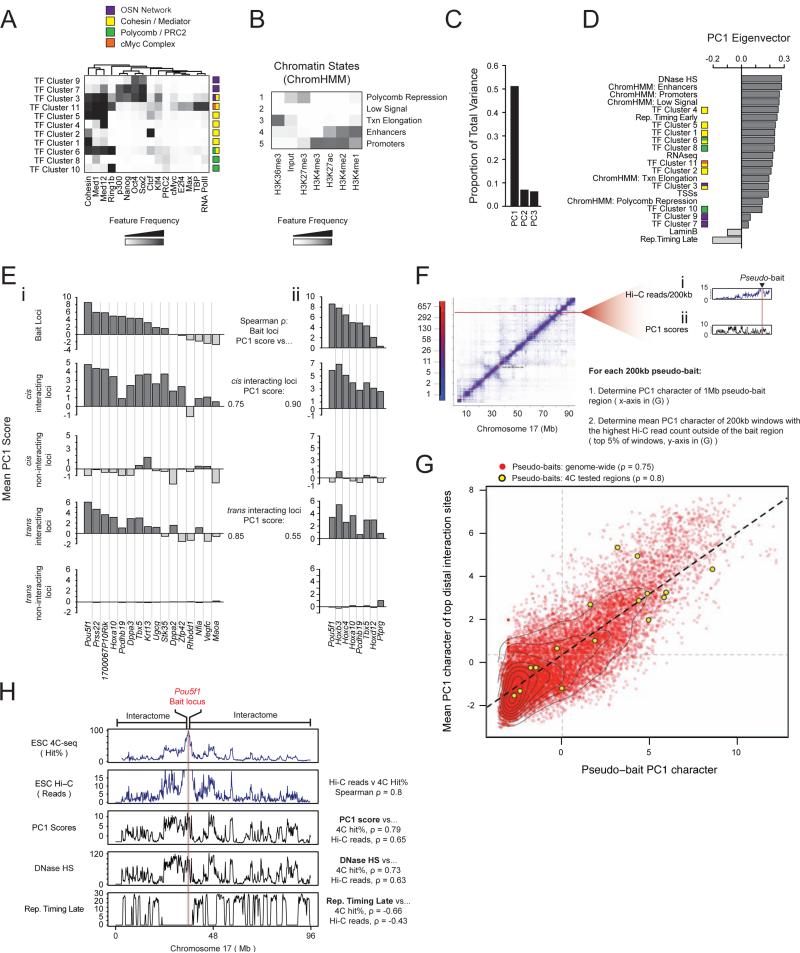
Figure 3. Interactions between regions with similar open/closed chromatin are an intrinsic aspect of chromosome conformation.
A, Transcription factor (TF) clusters defined using k-means clustering at 1kb resolution for noted ESC data sets, annotated based on feature frequency as represented by the heatmap. The color legend identifies specific gene-regulatory networks.
B, Chromatin states were determined based on the six indicated histone modifications in ESCs by a multivariate hidden Markov model, at 200bp resolution (Ernst and Kellis, 2012). Grey scale denotes the frequency with which a given histone mark is found at genomic positions corresponding to the chromatin state.
C, PCA was performed on ESC chromatin states and TF clusters from (A) and (B), as well as RNA-seq expression data, DnaseI hypersensitivity, LaminB binding, early and late DNA replication timing (Rep. timing), and density of transcriptional state sites (TSSs). Proportion of total variance in genomic features described by each principal component is shown.
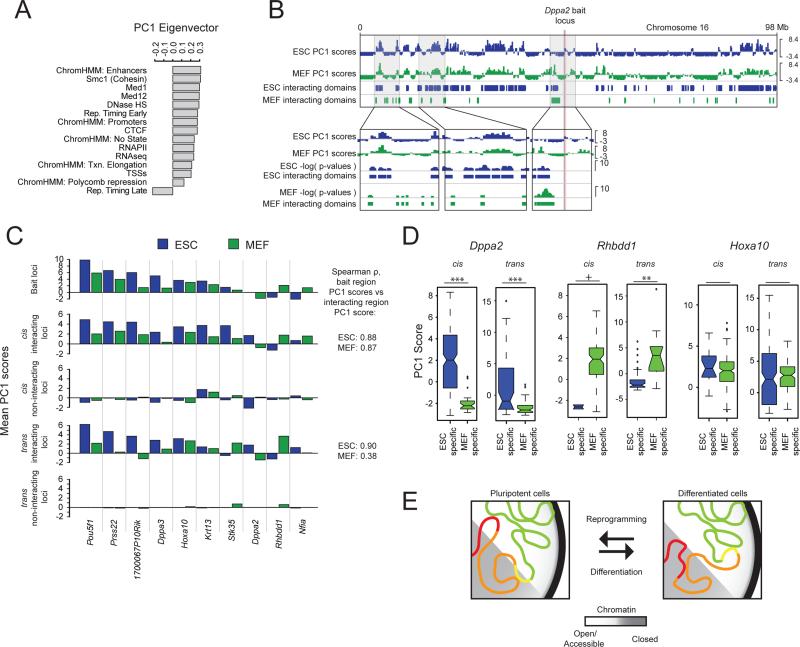
D, PC1 eigenvector ranked by genomic feature contribution.
E, (i) Top to bottom: mean PC1 score within the 1Mb bait region centered on each listed bait's locus; interacting regions in cis; non-interacting regions in cis; interacting regions in trans; non-interacting regions in trans. Spearman rho's give the rank correlation between the mean PC1 score of the 1Mb bait regions and their interactomes in both cis and trans. (ii) Identical analysis to (i) with a partially overlapping set of baits, for an independently derived ESC line, which is discussed in Figure 7 as Eed+/+ ESC line.
F, Schematic of genome-wide, Hi-C-based, pseudo-4C analysis. (i) Each extracted row of the Hi-C contact matrix, adapted from (Dixon et al., 2012), represents the interactome of one pseudo-bait. (ii) Plot of the PC1 character of the same chromosome. The mean PC1 score within the respective 1Mb pseudo-bait region (Pseudo-bait PC1 character) and the mean PC1 score within the pseudo-bait's interactome (top 5% of 200kb windows ranked by reads and excluding 1Mb pseudo bait region), were obtained and plotted as a red point in the scatterplot shown in (G). Pseudo-bait regions corresponding to genomic regions that we used as baits in our 4C-seq analysis in (Ei) are plotted in yellow (4C-bait loci).
G, Result of the analysis described in (F). 4C-bait loci show a similar trend when analyzed based on Hi-C data as in (Ei) based on 4C-seq. Correlations between bait and interactome PC1 scores are noted. The Hi-C data are also summarized by the regression line in black, the mean bait and interactome PC1 scores are demarcated by vertical and horizontal grey lines, respectively. Contour lines represent data density.
H, Top to bottom: comparison of the Pou5f1 4C-seq-based cis-interactome (hit%), the Pou5f1 cis-interactome defined by Hi-C read counts, PC1 scores, DNaseI HS, and late DNA replication timing along chr17. Specific correlation values are specified.
See also Figures S3 and S4.
A pluripotency- specific organization of the mouse genome
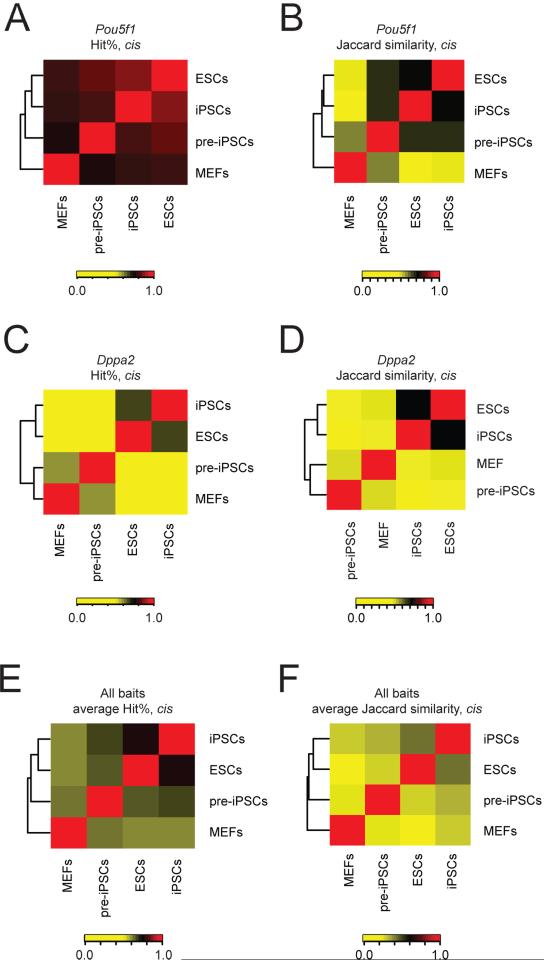
The Pou5f1 gene encodes the transcription factor Oct4 that is essential for the establishment and maintenance of mouse ESCs and specifically expressed in the pluripotent state (Nichols et al., 1998). The initial analysis of our Pou5f1 4C-seq data revealed an extensive intrachromosomal interaction network of this bait region (Figure 1A), raising the question of how the spatial interactions of this important locus are regulated. As a first step towards understanding this question, we determined whether the chromatin interactions of this genomic region change upon differentiation. 4C-seq in mouse embryonic fibroblasts (MEFs) demonstrated that the Pou5f1 interactome in MEFs is distinct from that of ESCs, both in terms of hit probability (Figures 2A, S2A) and interacting domains (Figure 2B) across the cis chromosome, indicating that changes in expression and chromatin state that are associated with the silencing of the Pou5f1 locus during differentiation (Feldman et al., 2006) coincide with changes to that locus’ interactions in 3D space. To expand the purview of this analysis, we determined the interactomes of nine additional baits in MEFs for which we already had generated ESC profiles by 4C-seq, and found that the interactomes of all these baits differed significantly between ESCs and MEFs (Figures 2E/F, S2A, Table S1). We noted that the chromatin contacts of the Dppa2 bait region are much more different between MEFs and ESCs than those of the other examined bait regions (Figures 2C/D, S2A). Consistent with this result, it has been shown that the Dppa2 region repositions towards the nuclear periphery and switches cis-interaction preferences from regions of early DNA replication to regions of late DNA replication during ESC differentiation (Hiratani et al., 2010; Takebayashi et al., 2012). Together, these data highlight an ESC-specific organization of the genome, and suggest that changes in 3D chromatin interactions during differentiation are regulated at a locus-specific level.
Figure 2. Long-range chromatin contacts change during differentiation and reprogramming of somatic cells to iPSCs.
A, Unsupervised hierarchical clustering of Spearman rank correlation values of the hit% within 200kb windows along the cis chromosome (chr17) for Pou5f1 4C-seq-derived interactomes in ESCs, iPSCs, pre-iPSCs, and MEFs. Color key = Spearman rho values.
B, Unsupervised hierarchical clustering of Jaccard similarity coefficients for the overlap of interacting domains of the Pou5f1 bait region in cis, between ESCs, iPSCs, pre-iPSCs, and MEFs. Color key = Jaccard similarity values.
C, As in (A), except for the Dppa2 bait.
D, As in (B), except for the Dppa2 bait.
E, As in (A), except averaged across eight different bait loci (Pou5f1, Stk35, 1700067P10Rik, Nfia, Dppa3, Rhbdd1, Hoxa10, and Dppa2).
F, As in (B), except averaged for the eight different bait loci named in (E).
See also Figure S2.
We next tested whether the 3D organization of the MEF genome can be reset to an ESC-like state by transcription factor-mediated reprogramming to iPSCs (Takahashi and Yamanaka, 2006). To this end, we determined the interactomes of eight of our bait regions, including the Pou5f1 locus, for two additional cell types that represent distinct stages of the reprogramming process (Table S1). First, we performed 4C-seq on faithfully reprogrammed, MEF-derived iPSCs, and found that the long-range chromatin contacts in iPSCs are highly similar to those of ESCs, in terms of hit probability across the cis chromosome (Figures 2A/C/E, S2A), and interacting domains in cis and in trans to the bait region (Figures 2B/D/F; S2B). Second, we examined chromatin interactions in pre-iPSCs that represent a late reprogramming stage at which the pluripotency expression and chromatin program is not yet fully induced (Sridharan et al., 2009). The long-range chromatin interactomes in these cells are typically distinct from those in ESCs and iPSCs, as well as those in MEFs (Figures 2, S2), indicating that the re-organization of chromatin contacts is not complete at this late stage of reprogramming, in line with previous findings detailing the long-range interactions of the Nanog locus (Apostolou et al., 2013). Together, these data show that the large-scale changes in genome organization that occur during differentiation are gradually reset to an ESC-like state during reprogramming to iPSCs, likely in association with progressive changes in chromatin and transcription states (Sridharan et al., 2009). The cell type-specificity of long-range chromatin interactions contrasts with the largely cell-type invariant TAD structure of metazoan genomes (Dixon et al., 2012; Nora et al., 2012; Sexton et al., 2012), suggesting that the organization of long-range chromatin contacts is a key feature of cell identity.
Open/closed chromatin as the foundation of genome organization in ESCs
The finding that changes in distal chromatin contacts occur during differentiation and reprogramming suggested a link between genome organization and genomic features that establish and maintain cell identity. To investigate such an association in the context of ESCs, we compiled a compendium of genomic features available for this cell type (Table S6). Specifically, we considered the binding profiles of the basic transcriptional machinery (TBP, RNA polymerase II), co-activators (Mediator and p300), and architectural proteins (Cohesin and CTCF). We also included transcriptional regulators of three important gene-regulatory networks, such as the pluripotency transcription factor network (Oct4, Sox2, Nanog, and Klf4), transcription factors that cooperate with cMyc (cMyc, Max, E2F4), and the repressive Polycomb protein network (Ring1b and Polycomb Repressive Complex 2 (PRC2)) (Young, 2011). Since regulatory genomic regions exhibit extensive co-occupancy by these factors, we grouped them into 11 groups (clusters) based on co-binding at a 1kb resolution (Figure 3A, Experimental Procedures). We also took into account the combinatorial nature of histone modifications by summarizing the relationship of six histone marks in terms of four functionally distinct chromatin states (ChromHMM states) (Ernst and Kellis, 2012) that are associated with Polycomb repression, transcriptional elongation, enhancers, and promoters. Regions lacking these histone modifications were assigned to a ‘low signal’ state (Figure 3B, Experimental Procedures). In addition, DNA replication timing, DNaseI hypersensitivity, gene density (in terms of transcriptional start sites (TSSs)), RNA-seq- based expression data, as well as LaminB association were considered.
Principal components analysis (PCA) was used to aggregate the resulting 22 linear genomic data sets, reducing their high dimensionality (Experimental Procedures). The first three principal components (PCs) were retained for downstream analyses (Figure 3C). To explore the relationship between genomic features and genome organization in ESCs, we next compared the linear genomic character represented by the PCs to the 4C interactome data for each of the 16 bait regions analyzed in ESCs (Figures S3A, 1A , Table S1).
The first PC (PC1) captures 51% of the variance across all features (Figure 3C) and distinguishes open, accessible chromatin and closed, inaccessible chromatin (Figure 3D). Specifically, regions of the genome with positive PC1 scores are characterized by high gene density, DNaseI hypersensitivity, binding of Cohesin, the basic transcriptional machinery, and transcription factors, as well as active or Polycomb-repressed chromatin states. In contrast, regions of the genome with negative PC1 scores are strongly depleted for these features, and instead are LaminB- associated and replicate their DNA late in S phase (Figure 3D). Notably, the 1Mb regions surrounding each bait locus had widely differing open/closed chromatin character as defined by their PC1 scores (Figure 3Ei, top panel). Strikingly, we found that the mean PC1 scores for the 1Mb bait regions correlated strongly with the mean PC1 scores of the corresponding interactomes, in both cis and trans (Figure 3Ei, Spearman rho=0.75 and 0.85, respectively). That is, baits with highly open and accessible chromatin preferentially interact with regions of the genome with similarly high PC1 scores. By contrast, the interactomes of closed chromatin baits, i.e. with negative PC1 scores, preferentially co-localize with genomic regions of similarly low PC1 scores in cis and trans. These findings were confirmed by a 4C-seq analysis for a partially overlapping set of bait regions in a second, independent ESC line (Figure 3Eii).
To test whether this trend holds genome-wide, we analyzed a previously published Hi-C dataset for genome-wide chromatin interactions in mouse ESCs (Dixon et al., 2012) in a similar manner (Experimental Procedures). For the Hi-C-based analysis, each 200kb region of the genome was treated as a ‘pseudo-bait’ and its intrachromosomal interactome was extracted (Figure 3F). Genome-wide, we found a striking positive correlation between the mean PC1 scores of the extended 1Mb pseudo-bait regions and the mean PC1 scores of their most frequently interacting intrachromosomal sites (Figure 3G), corroborating and extending our 4C-seq-based findings. A comparison of the profiles of the most and least likely interacting regions further demonstrated that genomic regions with very open, high PC1 character, for instance the Pou5f1 region, interact extensively with most other genomic regions with similarly high PC1 scores on the chromosome in cis and do not interact with genomic regions of negative PC1 scores (Figures 3G, S3Ci). In contrast, genomic regions of closed chromatin character, defined by negative PC1 scores, avoid regions of high PC1 character and appear to interact selectively with only a subset of PC1- negative genomic regions (Figures S3Ci). This may reflect a tethering of closed chromatin regions to the nuclear lamina, thus limiting their sampling of distal interactions. The positive correlation between bait and interactome character in terms of their PC1 scores persisted when the 10 Mb surrounding the bait region were excluded (Figure S3Di), demonstrating that interacting regions far away from the bait in cis also show an association with chromatin of similar open/closed character.
Taken together, we conclude that genomic loci with similar PC1 character preferentially interact (or co-localize) within the 3D space of the ESC nucleus, both in cis and in trans. The Hi-C- based results indicate that these associations are a general, genome-wide feature of long-range chromatin interactions. Our findings suggest that the strong interaction preferences between regions of similar PC1 character are an intrinsic aspect of overall chromosome conformation, in line with previous findings (Imakaev et al., 2012; Lieberman-Aiden et al., 2009; Simonis et al., 2006). Our data further demonstrate that interaction preferences in ESCs are not accurately described by a binary model of spatial segregation between open and closed chromatin states into two genome-wide compartments, where genomic regions with open chromatin character may co-localize with any other open chromatin region and vice versa (Lieberman-Aiden et al., 2009). Instead, the ESC interactome follows a more graduated model, where highly open chromatin regions predominantly interact with regions of similarly high PC1 scores, mid PC1 regions with other mid to low PC1 regions, and negative PC1 regions predominantly with other closed chromatin regions, supporting findings in other cell types (Imakaev et al., 2012). Notably, PC1 scores across the ESC genome are continuous rather than discrete (Figure S3Bi), and correlate with the continuum of ESC contact frequencies.
Genomic regions enriched for Oct4/Sox2/Nanog and Polycomb, respectively, frequently co-localize in ESCs
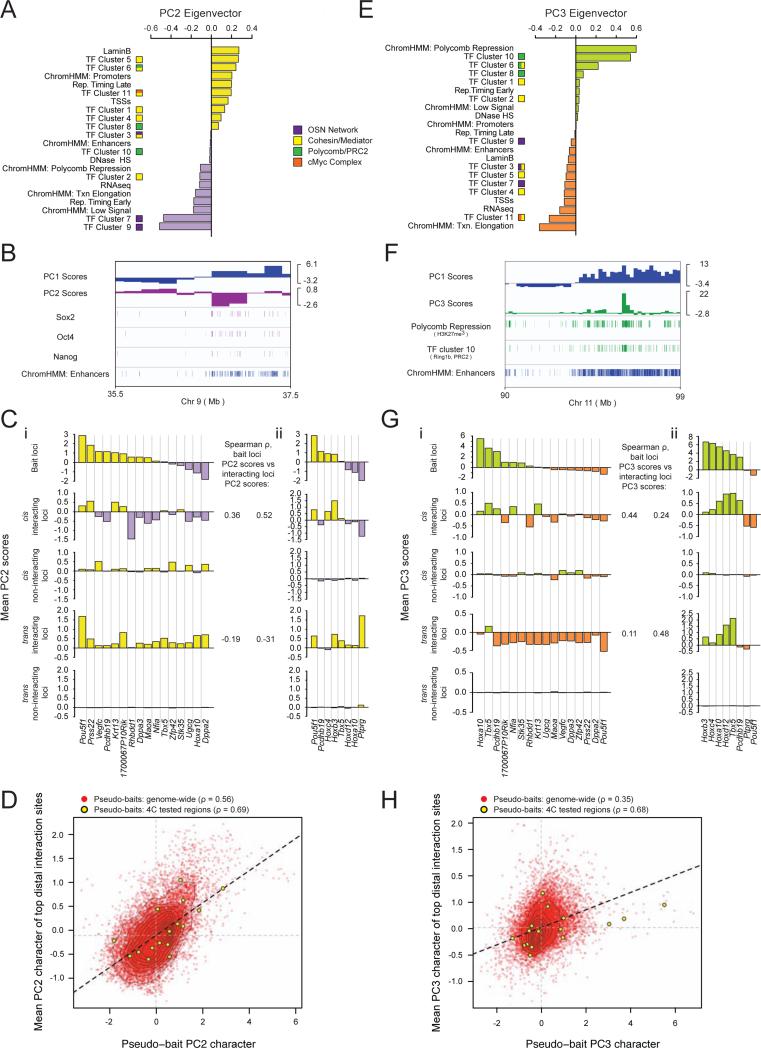
Next, we wanted to explore the extent to which long-range interactions are associated with specific transcriptional networks, beyond their association with the large-scale open/closed chromatin properties demonstrated by the correlation of interactomes with the genomic PC1 character. Since PC1- positive regions reflect enrichment for multiple features with diverse functionalities, we reasoned that an interactome's PC1 character may not reflect any specific mechanistic role of PC1-enriched features, but is most likely a consequence of the overarching chromosome conformation framework (Figures S4A-D). Therefore, we considered the second and third principal components (PC2 and PC3), which account for seven and six percent of the variance in genome features, respectively (Figure 3C). Importantly, these components capture regions of the genome enriched for the previously described gene-regulatory networks in ESCs (Figure 4A/E). Specifically, genomic regions with positive PC2 scores are exceptionally enriched for Mediator and Cohesin binding, as captured by transcription factor (TF) clusters 5, 6, and 11; while regions with negative PC2 scores are enriched for binding of factors belonging to the pluripotency network including Nanog, Oct4, and Sox2, as well as p300, which co-localize as TF clusters 7 and 9 (Figures 4A). Figure 4B displays the relationship between PC1 and negative PC2 scores in terms of these features: whereas genomic regions of open chromatin (defined by positive PC1 values) generally have a higher level of pluripotency factor binding than those with negative PC1 scores, genomic regions with negative PC2 scores have an increased density of pluripotency factor co-occupancy over and beyond what is explained by their PC1 state.
Figure 4. Regions of shared transcriptional network occupancy preferentially interact.
A, PC2 eigenvector with individual feature contributions. TF clusters and chromatin states as in Figure 3.
B, Integrative Genomics Viewer tracks of a representative genomic region with PC1 and PC2 scores; Sox2, Oct4, and Nanog occupancy; and enhancer density.
C, (i) Top to bottom: mean PC2 score within the 1Mb region centered on each bait's locus; interacting regions in cis; non-interacting regions in cis; interacting regions in trans; non-interacting regions in trans. Spearman rho's give the correlation between the baits’ PC2 and the interactomes’ PC2 character across all analyzed baits in cis and trans. (ii) Identical analysis to (i) except for an independently derived ESC line discussed in Figure 7 as Eed+/+ ESCs, with a partially overlapping set of bait loci.
D, Genome-wide pseudo-4C analysis of Hi-C data as described in Figure 3G, except for PC2.
E, PC3 eigenvector with individual feature contributions.
F, Integrative Genomics Viewer tracks showing a representative genomic region with PC1 and PC3 scores; H3K27me3 and TF cluster 10 (Ring1b/PRC2) enrichment, as well as enhancer density.
G, As in (C), except for PC3 scores.
H, As in (D), except for PC3 scores.
See also Figures S3 and S4.
Conversely, regions of the genome with positive PC3 values are highly enriched for occupancy by the Polycomb protein complexes PRC1 (Ring1b) and PRC2 as well as histone H3K27me3. Negative PC3 regions, on the other hand, seemingly capture the recently described super-enhancers associated with highly transcribed genes in pluripotent cells (Whyte et al., 2013), and represent a strong enrichment for active transcriptional elongation along with dense occupation by Mediator, Cohesin, the cMyc complex, and the pluripotency transcription factors Oct4, Sox2, and Nanog (Figures 4E, S4E/F). Genomic regions with positive or negative PC3 scores have an elevated density of their characteristic features above what is explained by their PC1 scores (Figure 4F, data not shown). We conclude that PC2 and PC3 describe a finer level of chromatin structure associated with specific transcription factor- and chromatin-regulatory networks that is not captured by the open and closed chromatin character defined by PC1.
To examine whether the genomic states captured by PC2 and PC3 are associated with long-range chromatin contacts, we examined the PC2 and PC3 scores within our 16 ESC bait regions and their respective interactomes in an analogous manner to our analysis of PC1 in Figure 3. Importantly, bait regions exhibited widely different PC2 character, and the mean PC2 score of each bait's intrachromosomal interactome showed a strong concordance with the mean PC2 score of the 1Mb bait region itself (Figure 4Ci). This observation was confirmed for additional bait regions in a second ESC line (Figure 4Cii) and, remarkably, also for cis-interactions genome-wide based on Hi-C data (Figures 4D, S3Cii/Dii). The PC2-based analysis demonstrates that genomic regions with particularly strong Mediator and Cohesin binding preferentially co-localize in cis, as do regions extremely highly enriched for Oct4, Sox2, and Nanog binding without a concomitant extreme accumulation of Mediator and Cohesin (captured by TF clusters 7 and 9). Notably, however, the co-localization of pluripotency factor-enriched, PC2-negative genomic regions was not evident for trans interactions (Figure 4C).
The comparison of bait and interactome PC3 scores demonstrated a preferential co-localization of the Polycomb/H3K27me3 enriched, PC3-positive genomic regions (captured by TF clusters 6, 8, and 10, and the Polycomb Repression ChromHMM state), both in cis and in trans (Figure 4Gi). For example, Hox loci, which belong to the most strongly Polycomb- occupied and H3K27me3- enriched (PC3-positive) regions in the ESC genome, were found to interact with other regions characterized by high H3K27me3/Polycomb occupancy in cis and trans (Figure 4Gi). The co-localization of distal genomic regions extensively occupied by Polycomb proteins and H3K27me3 may be analogous to the frequent association of Polycomb response elements in Drosophila (Bantignies and Cavalli, 2011), and may therefore represent an evolutionarily conserved feature of genome organization linked to gene regulation. Conversely, bait loci with negative PC3 values that are strongly enriched for ESC super enhancers co-localize with genomic regions of similarly PC3-negative character, both in cis and trans (Figure 4Gi). These trends were confirmed in a second ESC line (Figure 4Gii) and extended to Hi-C data (Figure 4H, S3Ciii/Diii), showing them to be a genome-wide phenomena.
Together, these results argue that regions of the genome enriched for specific gene-regulatory features preferentially co-localize within the 3D space of the nucleus, raising the possibility that specific transcriptional and chromatin-regulatory networks are involved in mediating long-range chromatin contacts in ESCs.
Spatial segregation of Nanog and H3K27me3 in the ESC nucleus
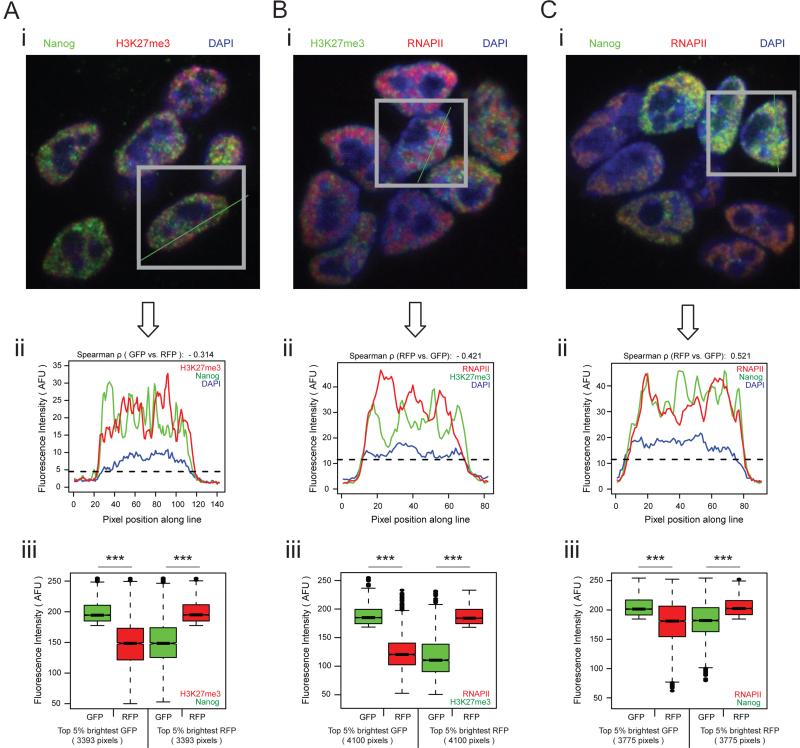
The preferential interactions of genomic regions with positive and negative PC3 scores, respectively (i.e. Polycomb protein versus super enhancer/Oct4/Sox2/Nanog enriched regions), suggested a segregation of genomic regions with opposing PC3 character into distinct compartments in the nucleus. To test this hypothesis, we examined the co-localization of Nanog (PC3-negative), RNA polymerase II (PC3-negative), and H3K27me3 (PC3-positive) in the ESC nucleus by immunostaining. Image analysis showed that Nanog and RNA polymerase II, respectively, have a distinct localization pattern from H3K27me3, with sites strongly enriched for H3K27me3 displaying weak Nanog and RNA polymerase II accumulation and vice versa (Figures 5A/B). By contrast, RNA Polymerase II and Nanog overlap more extensively, albeit not perfectly (Figure 5C), consistent with their differential contribution to PC2 and PC3 scores. For instance, Nanog occupancy is strongly captured by TF cluster 7 and RNA polymerase binding by TF cluster 11, which have similar contributions to PC3, but opposing contributions to PC2 (Figures 4A/E). Overall, the immunofluorescence localization patterns support the spatial segregation of functionally distinct transcription factor- and chromatin-regulatory networks in the ESC nucleus, and are consistent with the co-localization of distinct gene-regulatory modules detected by our chromosome conformation capture-based analysis.
Figure 5. Nanog and H3K27me3 segregate in the ESC nucleus.
A, (i) Image of ESCs immunostained with antibodies against Nanog (green) and H3K27me3 (red). Nuclei are stained with DAPI (blue). (ii) Red and green pixel intensities along the line in (i) for all pixels whose DAPI signals were above the indicated threshold (dotted line). (iii) Quantile normalized fluorescence intensity distribution of the top 5% brightest nuclear (but non-nucleolar) green pixels (GFP) and the normalized red pixel intensity (RFP) at the corresponding position (left); and of the top 5% brightest nuclear (but non-nucleolar) red pixels and the normalized distribution of green pixel intensity at the corresponding position (right), for all cells in the ESC colony in (i). Box and whisker demarcations as in Figure 1C. ***=p-value<2E-16, Wilcoxon rank-sum test.
B, As in (A), except for H3K27me3 (green) and RNAPII (red).
C, As in (A), except for Nanog (green) and RNAPII (red).
Changes in open/closed chromatin character mirror changes in genome organization during differentiation
Our analysis revealed a close relationship between chromatin character and spatial interactions, with the combination of linear genomic features summarized by PC1 (open/closed chromatin) showing the strongest association with the fundamental organization of chromatin interactions. Based on these results, we predicted that dramatic changes in open/closed chromatin character that occur during differentiation should coincide with strong changes in interactome character.
To test this, we examined differences in chromatin interactions between ESCs and MEFs with respect to changes in their open/closed chromatin character. To this end, we performed PCA on concatenated ESC and MEF genomes using linear genomic feature data sets that were available for both cell types and do not include cell-type specific transcription factors (Figure 6A; Table S6, Experimental Procedures). This resulted in a new PC1 eigenvector that allowed the comparison of PC1 scores for ten bait regions and their interactomes across both cell types (Figures S5A-D). We found that, as in ESCs, regions with similar PC1 character in MEFs preferentially co-localize (Figure 6C). Furthermore, changes in bait character between MEFs and ESCs are generally associated with similar changes in their respective interactomes (Figures 6B-D). For instance, the Dppa2 locus participates in extensive interactions with genomic regions of positive PC1 character in ESCs, in accordance with its early replicating and highly transcribed state in pluripotent cells (Takebayashi et al., 2012). In MEFs, the Dppa2 bait region displays a PC1-negative, repressed, late replicating state, and the MEF-specific interactions likewise exhibit negative PC1 scores, both in cis and in trans (Figures 6B-D, S5E). In contrast, the Rhbdd1 bait region transitions from negative to positive PC1 scores from ESCs to MEFs, and its MEF-specific interactions have significantly higher PC1 scores than the ESC-specific interactions (Figures 6C/D). Conversely, bait regions without a change in PC1 character do not change interaction preferences with regards to PC1 scores (Figure 6C/D, Hoxa10).
Figure 6. Changes in open/closed chromatin character between ESCs and MEFs correspond to changes in interaction preferences.
A, PCA was performed for concatenated ESC+MEF data and included the indicated features. The resulting PC1 eigenvector is depicted.
B, Top to bottom: Integrative Genomics Viewer tracks showing the PC1 scores along chr16 in ESCs (blue) and MEFs (green), and the 4C-seq- defined cis-interacting domains of the Dppa2 locus, in ESCs and MEFs. Zoom-ins highlight the switch in PC1 bait character of the Dppa2 locus between MEFs and ESCs (right box) and corresponding changes in interaction preferences (left two boxes).
C, Top to bottom: mean PC1 score within the 1Mb region centered on each listed bait's locus; interacting regions in cis; non-interacting regions in cis; interacting regions in trans; non-interacting regions in trans, for ESC data (blue) and MEF data (green). Spearman rho's give the rank correlation between the PC1 bait character and interactome character per cell type, across all analyzed baits in cis and trans.
D, PC1 score distributions of ESC and MEF-specific, significantly interacting domains of Dppa2, Rhbdd1, and Hoxa10. Box and whisker demarcations as in Figure 1C, notches ≈ 95% confidence interval around medians. +p-value=0.099; **p-value<0.01; ***p-value<0.001; Wilcoxon rank-sum test.
E, Chromatin interaction model, wherein large-scale changes in chromatin interactions mirror changes in open/closed chromatin (PC1) character upon ESC differentiation. Note, for instance, the different interactions of the genomic regions marked in red and yellow, switching between open and closed chromatin.
See also Figures S5.
Together, these data support a model where the open/closed chromatin character is the strongest predictor of interaction preferences between distal genomic loci, and long-range chromatin interaction preferences are subject to change during differentiation in concordance with changes to the PC1 nature of the regions in question (Figure 6E). Of note, although the Pou5f1 gene itself becomes repressed during differentiation (Feldman et al., 2006), the extended 1Mb Pou5f1 bait region is strongly positive in PC1 character in MEFs (Figure 6C), likely explaining the more limited difference in spatial interactions between ESCs and MEFs for this bait region compared to the the Dppa2 bait (Figure 2), which shows a more dramatic change in PC1 character upon differentiation.
The preferential co-localization of Polycomb-enriched genomic regions is Eed-dependent
Our data demonstrated that genomic regions are more likely to contact each other when they share strong enrichment of similar regulatory proteins (Figure 4). Therefore, we considered testing the functional importance of a specific gene-regulatory network for long-range chromatin interactions. To this end, we determined the long-range chromatin contacts in ESCs lacking the protein Eed, a subunit of Polycomb complex PRC2 that is required for all genomic H3K27me3 (Montgomery et al., 2005). We examined particularly the interactions of bait regions with positive PC3 scores, which capture high occupancy by PRC2 and H3K27me3 (Table S1). Importantly, despite the complete loss of H3K27me3 (Figure S6A), Eed−/− ESCs continue to express pluripotency-specific transcription factors including Oct4, Sox2, and Nanog at normal levels, maintain their ability to self-renew, and do not spontaneously differentiate when cultured appropriately (Chamberlain et al., 2008), allowing us to test the role of PRC2 in genome organization without a change in cell identity.
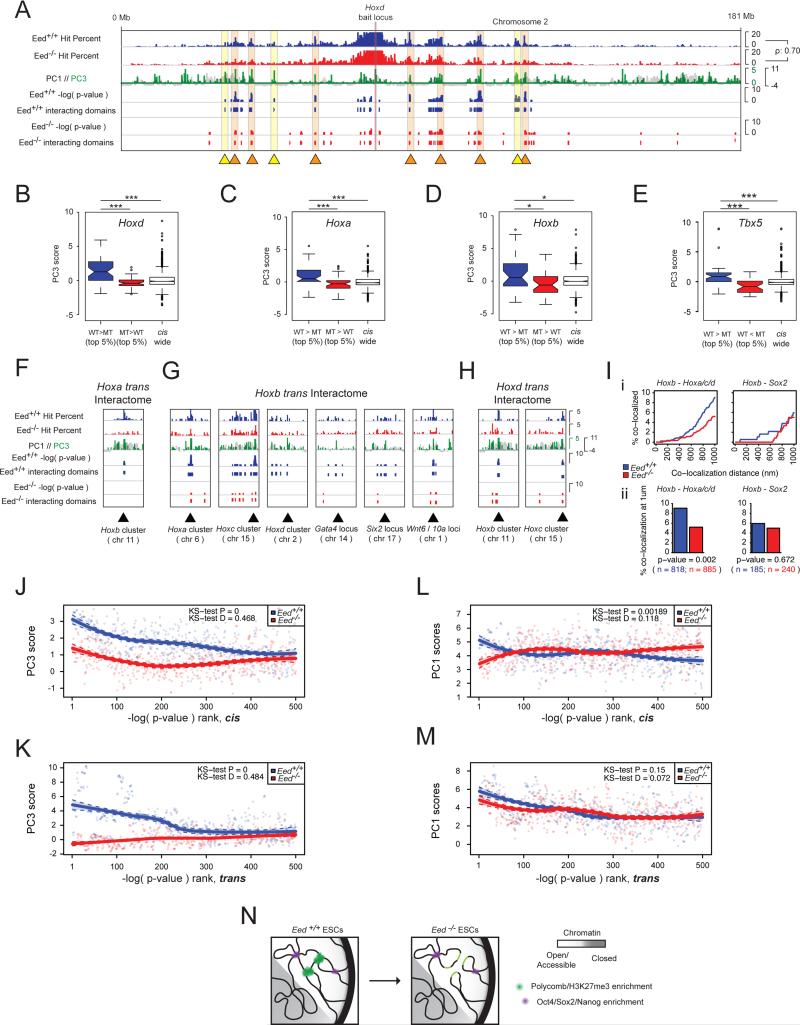
Notably, 4C-seq analysis showed that the intrachromosomal long-range interactions of the Hoxd cluster, a Polycomb-targeted genomic region with highly positive PC3 scores, showed a strong correlation between the Eed+/+ and the Eed−/− ESC lines (Figure 7A). However, despite this overall similarity, numerous intrachromosomal interactions present in Eed+/+ ESCs are lost or have a less significant interactions by p-values, indicative of a reduced interaction frequency, in Eed−/− ESCs (Figure 7A, yellow and orange highlights, respectively). A visual inspection of chromatin contacts indicated that these losses and reductions appear to occur at regions of high Polycomb enrichment in wildtype ESCs, as defined by high PC3 scores (Figure 7A). In agreement with this, long-range chromatin interactions of the Hoxd cluster that specifically occur in Eed+/+ ESCs have a significantly more positive PC3 character than those that are specific for Eed−/− ESCs (Figures 7B). This result extends to other Polycomb-regulated regions such as the Hoxa and Hoxb clusters and the Tbx5 locus (Figures 7C-E, S6D).
Figure 7. Eed is required for the co-localization of Polycomb-occupied genomic regions.
A, Integrative Genomics Viewer tracks showing the Hoxd12 interactome in cis in terms of hit% for Eed+/+ (blue) and Eed−/− (red) ESCs, chromosome-wide PC1 scores (black) overlaid with PC3 scores (green, positive values shown only), binomial test −log(p-values), and interacting domains. Regions that lose significant interactions with the Hoxd12 bait upon Eed ablation are marked with yellow triangles and shading; those that do not lose interactions with the Hoxd12 locus upon Eed ablation, but show a decrease in interaction strength with orange triangles and shading. The Spearman rho shows the rank correlation of the hit% between Eed+/+ and Eed−/− ESCs.
B, Eed+/+ and Eed−/− ESCs Hoxd12 4C-seq hit% tracks from (A) were subtracted and the 200kb windows with the top and bottom 5% of resulting values were used to define regions of the cis-chromosome that showed stronger interactions in Eed+/+ (WT>MT) and Eed−/− (MT>WT) ESCs, respectively. The PC3 score distribution of these genomic regions and of the entire chromosome are shown. Box and whisker demarcation as in Figure 1C, notches ≈ 95% confidence interval around medians. ***=p-value<0.001, Wilcoxon rank-sum test.
C-E, As in (B), but for the Hoxa10, Hoxb3, and Tbx5 4C-seq cis interactomes. *=p-value<0.05.
F-H, Trans interactions between the indicated (PC3-positive) Hox loci in Eed+/+ and Eed−/− ESCs, displayed as in (A). Hoxa refers to results of the Hoxa10 bait locus, Hoxb to Hoxb3, Hoxc to Hoxc4, Hoxd to Hoxd12.
I, DNA FISH analysis of the trans interactions between Hox clusters. (i) Cumulative frequency distribution plots of co-localization frequencies between Hoxb3 (chr11) and the other three Hox loci (Hoxa10-chr6, Hoxc4-chr15, Hoxd12-chr2) (left), as well as between Hoxb3 and the Sox2 (chr3) locus (right), with co-localization distances noted on the x-axis, measured in Eed+/+ (blue) and Eed−/− (red) ESCs. (ii) Co-localization frequencies at 1um for Hoxb3 and the other Hox loci (left), as well as for Hoxb3 and Sox2 (right), derived from (i). n=FISH signal pairs analyzed in both (i) and (ii); p-value from two-tailed Fisher exact test.
J, The cis interactomes of the six PC3-positive (Polycomb/H3K27me3 enriched) bait loci (Hoxa10, Hoxb3, Hoxc4, Hoxd12, Pcdhb19, Tbx5, see Figure 4Gii) were ranked by −log(p-value) for both Eed+/+ (blue) and Eed−/− (red) ESCs, and the 500 top genomic sites plotted against their average PC3 scores in wildtype ESCs. Loess regression was used for curve generation. KS=Kolmogorov–Smirnov test to determine the probability that the two underlying probability distributions differ (D = KS-test D statistic).
K, As in (J), but for the trans interactomes.
L, As in (J), except for PC1 scores.
M, As in (K), except for PC1 scores.
N, Chromatin interaction model, wherein in the absence of Eed, the frequency of interactions between regions with high PC3 scores is reduced, but large-scale chromosome conformation is largely conserved.
See also Figures S6.
A comparison of chromatin interactions in trans between Eed+/+ and the Eed−/− ESCs revealed that the Hox clusters interact with each other as well as with other regions of high Polycomb/H3K27me3 enrichment that encode developmental regulators in wildtype ESCs (Figures 7F-H). Importantly, many of these interchromosomal contacts are also lost or reduced in the absence of Eed (Figures 7F-H). For instance, interactions of the Hoxb cluster with the Hoxa, Hoxc, and Hoxd clusters are observed in wildtype ESCs, and are diminished in knockout cells (Figures 7F-H). Consistent with the 4C-seq data, we found that the co-localization frequency between the Hoxb cluster and the Hoxa, Hoxc, and Hoxd clusters in wildtype ESCs was significantly higher than in knockout ESCs, as measured by FISH (Figures 7I, S6B/C).
To explore whether the absence of Eed specifically affects chromatin contacts that occur between genomic regions characterized by positive PC3 scores, we examined the chromatin character of the most significantly interacting distal regions across six PC3-positive baits (Table S1). We found that in Eed+/+ ESCs, the highest-ranking (and likely most frequent) interactions fall within genomic regions with highly positive PC3 and PC1 scores and with less strongly positive PC2 scores, both in cis and trans (Figures 7J-M, S6E). In the absence of Eed, the strongest chromatin contacts no longer occur with regions that are highly positive for PC3 in wildtype ESCs (Figure 7J/K), although they still take place between distal genomic regions of similarly positive PC1 and PC2 scores, both in cis and trans (Figures 7L/M, S6E). The corollary to this finding is demonstrated by two bait regions that are not enriched for Polycomb binding (Pou5f1 and Ptprg): they do not show a similarly dramatic difference in interaction preferences between Eed+/+ and Eed−/− ESCs with regards to any of the three principal components (Figure S6F-I). These data indicate that loss of PRC2 and H3K27me3 specifically alters the co-association of PC3-positive genomic regions, but does not dramatically affect spatial interactions associated with PC1 and PC2 character.
Based these data we conclude that Eed is required for the establishment and/or stable maintenance of inter- and intrachromosomal chromatin interactions between Polycomb-occupied, PC3-positive regions in ESCs. Our data also suggest that the overall chromosome topology does not dramatically change upon loss of Eed (Figures 7A/I). Additionally, the results indicate that regions that are spatially interacting and Polycomb protein-bound in wildtype ESCs remain confined by a similar chromosome topology in the absence of Eed, but their interaction frequency, i.e. their proximity, is dramatically reduced (Figures 7F-I, model in 7N).
Discussion
Our work describes a pluripotency-specific organization of the mouse genome and suggests a model where distal regions of the genome bound by similar regulatory proteins co-localize within the 3D space of the ESC nucleus. Based on our data, we propose a model with two layers of regulation for long-range chromatin contacts in ESCs: (i) We posit that, at the largest scale, the open/closed chromatin character (described by the PC1 character of the genome) defines the regions of the genome that have the potential to come into close spatial proximity with one another, both intra- and interchromosomally, which is intricately linked to the overall folding of the chromosome and in agreement with other recent findings (Imakaev et al., 2012; Lieberman-Aiden et al., 2009). (ii) Our data also suggest that on a finer scale, and within the constraints established by the open/closed chromatin architecture, genomic regions are more likely to contact each other when they share strong enrichment of similar regulatory proteins, such as binding by the pluripotency transcription factors Oct4, Sox2, and Nanog with Mediator and Cohesin (represented by negative PC3 scores), or of the repressive Polycomb complex (captured by positive PC3 scores).
Our results demonstrate that the depletion of a single gene-regulatory network in ESCs specifically affects long-range interactions of genomic regions particularly strongly enriched for occupancy by this network (in our case PRC2 and H3K27me3), without altering the global interaction network associated with open/closed chromatin character. A potential explanation for the limited effect on overall chromatin interactions may be that the chromosomal conformation chassis is likely maintained by many combinatorially acting regulatory factors that probably involve numerous interactions mediated by Cohesin (Apostolou et al., 2013; Phillips-Cremins et al., 2013). Notably, in addition to our description of PRC2 as critical regulator of specific long-range chromatin interactions (Figure 7), a functional requirement for the transcription factor Klf4 in the maintenance of long-range chromatin contacts in ESCs has recently been reported (Wei et al., 2013), extending previous findings that demonstrated a requirement for Oct4 in the organization of short-range chromatin interactions within the Nanog locus (Levasseur et al., 2008).
The role of cell-type specific gene-regulatory networks in defining specific long-range chromatin interactions potentially allows TADs enriched for specific gene regulatory features to co-localize in the 3D space of the nucleus. Interestingly, while our results demonstrate that Polycomb complexes are important for long-range chromatin contacts associated with the regulation of Polycomb targets in mammals, it has recently been shown that the TAD structure within a specific locus, the X-chromosome inactivation center, is not affected by the Eed knockout (Nora et al., 2012), indicating different regulatory mechanisms at the different hierarchies of genome organization. It is also interesting to speculate that the interaction between the Hoxd and Hoxc clusters may provide a mechanism for how the non-coding RNA HOTAIR, encoded within the Hoxc cluster, finds its target genes within the Hoxd cluster located on a different chromosome (Rinn et al., 2007), by exploiting 3D conformation of the genome, in a manner similar to that employed by long-noncoding RNA Xist (Engreitz et al., 2013).
Based on the data presented here and other reports (Hakim et al., 2011; Noordermeer et al., 2011), we suggest that cell type-specific gene-regulatory networks generally guide specific spatial interactions within the context of a cell type-specific chromosome folding pattern that relates to the open/closed chromatin character. A preferential co-localization of distal genomic regions with similar regulatory networks tens of mega-bases apart on the same chromosome, and in trans, suggests a previously unappreciated role for transcriptional networks in influencing the 3D positional preferences of chromatin in mammalian cells, which may represent an evolutionarily conserved interaction between eukaryotic genome organization and gene regulation (Sexton et al., 2012; Tanizawa et al., 2010). We speculate that this organizational hierarchy facilitates the recruitment of regulatory proteins, and potentially non-coding RNAs, to their genomic target sites and the establishment of chromatin environments, which are both critical for the efficient regulation of gene expression.
Experimental Procedures
4C-seq and 3C
4C-seq libraries listed in Table S1 were prepared as described (Splinter et al., 2012) using the primers given in Table S2, and sequenced on Illumina machines. Reads that aligned to unique HindIII sites in the genome (build mm9) with at most two mismatches were retained. Read distribution statistics for all libraries are given in Table S3. 3C libraries were prepared in an analogous manner to 4C libraries, ending with the first ligation step and amplified with primers listed in Table S2.
Data analysis
For each 4C-seq library, read counts at each unique HindIII site were collapsed to a ‘hit’ to reduce effects of clonal amplification. Replicate libraries that passed stringent quality control were pooled for downstream analysis by calculating the probability of a hit at each HindIII site across all replicates. Next, we determined the average hit probability within 200 kilobase (kb) windows tiled along each chromosome for each pooled data set, referring to this as the ‘hit percentage’. A binomial test with different background models for cis and trans interactions was used to identify significantly interacting regions of each bait locus.
For PCA, 31 linear genomic features (summarized in Table S6) were transformed into five chromatin states and 11 TF clusters (Figures 3A/B), and together with six additional features aggregated within 200kb windows across the genome to obtain a feature density matrix. To describe the linear genomic feature state of ESCs and MEFs, vectors containing feature density within 200kb windows for each cell type were concatenated, allowing PCA to be conducted on the combined feature matrix (Table S6). To obtain a PC score enrichment value for each 4C bait, the mean PC score within the 200kb bait window and the four flanking windows was calculated (i.e. five 200kb bait windows = bait region). The PC score enrichment within the bait's interactome was calculated as the mean PC score within 200kb windows that overlapped 4C positive domains (as determined by the binomial test) by at least 25%, excluding the five bait windows. The rankings of the bait and interactome PC score enrichment values were correlated using Spearman rho statistics.
For the Hi-C data comparison, normalized, mouse ESC Hi-C interaction matrices based on 40kb bins were downloaded from the Ren Lab website (Dixon et al., 2012) and re-binned into 200kb bins to match the resolution of our 4C and feature data. Each 200kb window of re-binned Hi-C data was treated as pseudo-bait and its intrachromosomal interactome was extracted from the chromosome-wide contact matrix. Bait and interactome PC scores were calculated as described above for 4C-seq interactomes, except that the interactome was defined as those 5% of 200kb windows that had the highest read count, excluding the 1Mb bait region.
FISH and immunostaining analysis
3D FISH and immunostaining were done following standard methods.
Supplementary Material
Highlights.
*Long-range chromatin contacts in pluripotent cells differ from those in somatic cells
*Distal genomic regions with extensive Oct4/Sox2/Nanog binding co-localize in ESCs
*Distal Polycomb protein/H3K27me3-enriched genomic regions frequently interact in ESCs
*Spatial clustering of Polycomb/H3K27me3-enriched genomic regions requires Eed
Acknowledgements
We thank Shawn Cokus for advice, Sanjeet Patel for RNA-seq data analysis, and Terry Magnuson for Eed−/− ESCs. KP is supported by the UCLA Broad Center of Regenerative Medicine and Stem Cell Research, NIH (DP2OD001686; P01GM099134), and CIRM (RN1-00564); MP by NIH (P01GM099134), NSF (HYS-1066293), and the hospitality of the Aspen Center for Physics; GB by the Whitcome Pre-doctoral Training Program; MD by fellowships from the UCLA Broad Center of Regenerative Medicine and Stem Cell Research, the UCLA Graduate Division, and CIRM; CC by a Leukemia and Lymphoma Research Fellowship 10040; JE by funds from the UCLA David Geffen School of Medicine; and WdL by a ERC Starting Grant (209700).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author contribution
MD, GB: designed research, performed experiments, analyzed data, wrote the paper; CC: performed experiments; ES and WdL: trained MD in 4C-seq library preparation in 2008; JE: analytic tools; MP: supervision, data analysis, KP: designed research, analyzed data, wrote the paper, and supervision of overall project.
Accession Number
Our 4C-seq data are available at GEO under GSE50029.
Please see the Supplemental Experimental Procedures for more detail on all experimental procedures and analysis.
References
- Apostolou E, Ferrari F, Walsh RM, Bar-Nur O, Stadtfeld M, Cheloufi S, Stuart HT, Polo JM, Ohsumi TK, Borowsky ML, et al. Genome-wide chromatin interactions of the Nanog locus in pluripotency, differentiation, and reprogramming. Cell stem cell. 2013;12:699–712. doi: 10.1016/j.stem.2013.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bantignies F, Cavalli G. Polycomb group proteins: repression in 3D. Trends in genetics : TIG. 2011;27:454–464. doi: 10.1016/j.tig.2011.06.008. [DOI] [PubMed] [Google Scholar]
- Bickmore WA, van Steensel B. Genome Architecture: Domain Organization of Interphase Chromosomes. Cell. 2013;152:1270–1284. doi: 10.1016/j.cell.2013.02.001. [DOI] [PubMed] [Google Scholar]
- Chamberlain SJ, Yee D, Magnuson T. Polycomb repressive complex 2 is dispensable for maintenance of embryonic stem cell pluripotency. Stem Cells. 2008;26:1496–1505. doi: 10.1634/stemcells.2008-0102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit E, de Laat W. A decade of 3C technologies: insights into nuclear organization. Genes & development. 2012;26:11–24. doi: 10.1101/gad.179804.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker J, Rippe K, Dekker M, Kleckner N. Capturing chromosome conformation. Science. 2002;295:1306–1311. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- Denholtz M, Plath K. Pluripotency in 3D: genome organization in pluripotent cells. Current opinion in cell biology. 2012;24:793–801. doi: 10.1016/j.ceb.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, Ren B. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engreitz JM, Pandya-Jones A, McDonel P, Shishkin A, Sirokman K, Surka C, Kadri S, Xing J, Goren A, Lander ES, et al. The Xist lncRNA Exploits Three-Dimensional Genome Architecture to Spread Across the X Chromosome. Science. 2013 doi: 10.1126/science.1237973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst J, Kellis M. ChromHMM: automating chromatin-state discovery and characterization. Nature methods. 2012;9:215–216. doi: 10.1038/nmeth.1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman N, Gerson A, Fang J, Li E, Zhang Y, Shinkai Y, Cedar H, Bergman Y. G9a-mediated irreversible epigenetic inactivation of Oct-3/4 during early embryogenesis. Nat Cell Biol. 2006;8:188–194. doi: 10.1038/ncb1353. [DOI] [PubMed] [Google Scholar]
- Gibcus JH, Dekker J. The Hierarchy of the 3D Genome. Molecular cell. 2013;49:773–782. doi: 10.1016/j.molcel.2013.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakim O, Sung MH, Nakayamada S, Voss TC, Baek S, Hager GL. Spatial congregation of STAT binding directs selective nuclear architecture during T-cell functional differentiation. Genome research. 2013;23:462–472. doi: 10.1101/gr.147652.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakim O, Sung MH, Voss TC, Splinter E, John S, Sabo PJ, Thurman RE, Stamatoyannopoulos JA, de Laat W, Hager GL. Diverse gene reprogramming events occur in the same spatial clusters of distal regulatory elements. Genome research. 2011;21:697–706. doi: 10.1101/gr.111153.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiratani I, Ryba T, Itoh M, Rathjen J, Kulik M, Papp B, Fussner E, Bazett-Jones DP, Plath K, Dalton S, et al. Genome-wide dynamics of replication timing revealed by in vitro models of mouse embryogenesis. Genome research. 2010;20:155–169. doi: 10.1101/gr.099796.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imakaev M, Fudenberg G, McCord RP, Naumova N, Goloborodko A, Lajoie BR, Dekker J, Mirny LA. Iterative correction of Hi-C data reveals hallmarks of chromosome organization. Nature methods. 2012;9:999–1003. doi: 10.1038/nmeth.2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL, Ebmeier CC, Goossens J, Rahl PB, Levine SS, et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010;467:430–435. doi: 10.1038/nature09380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levasseur DN, Wang J, Dorschner MO, Stamatoyannopoulos JA, Orkin SH. Oct4 dependence of chromatin structure within the extended Nanog locus in ES cells. Genes & development. 2008;22:575–580. doi: 10.1101/gad.1606308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery ND, Yee D, Chen A, Kalantry S, Chamberlain SJ, Otte AP, Magnuson T. The murine polycomb group protein Eed is required for global histone H3 lysine-27 methylation. Current biology : CB. 2005;15:942–947. doi: 10.1016/j.cub.2005.04.051. [DOI] [PubMed] [Google Scholar]
- Nichols J, Zevnik B, Anastassiadis K, Niwa H, Klewe-Nebenius D, Chambers I, Scholer H, Smith A. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell. 1998;95:379–391. doi: 10.1016/s0092-8674(00)81769-9. [DOI] [PubMed] [Google Scholar]
- Noordermeer D, de Wit E, Klous P, van de Werken H, Simonis M, Lopez-Jones M, Eussen B, de Klein A, Singer RH, de Laat W. Variegated gene expression caused by cell-specific long-range DNA interactions. Nat Cell Biol. 2011;13:944–951. doi: 10.1038/ncb2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nora EP, Lajoie BR, Schulz EG, Giorgetti L, Okamoto I, Servant N, Piolot T, van Berkum NL, Meisig J, Sedat J, et al. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature. 2012;485:381–385. doi: 10.1038/nature11049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne CS, Chakalova L, Brown KE, Carter D, Horton A, Debrand E, Goyenechea B, Mitchell JA, Lopes S, Reik W, et al. Active genes dynamically colocalize to shared sites of ongoing transcription. Nature genetics. 2004;36:1065–1071. doi: 10.1038/ng1423. [DOI] [PubMed] [Google Scholar]
- Phillips-Cremins JE, Sauria ME, Sanyal A, Gerasimova TI, Lajoie BR, Bell JS, Ong CT, Hookway TA, Guo C, Sun Y, et al. Architectural protein subclasses shape 3D organization of genomes during lineage commitment. Cell. 2013;153:1281–1295. doi: 10.1016/j.cell.2013.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanyal A, Lajoie BR, Jain G, Dekker J. The long-range interaction landscape of gene promoters. Nature. 2012;489:109–113. doi: 10.1038/nature11279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenfelder S, Sexton T, Chakalova L, Cope NF, Horton A, Andrews S, Kurukuti S, Mitchell JA, Umlauf D, Dimitrova DS, et al. Preferential associations between co-regulated genes reveal a transcriptional interactome in erythroid cells. Nature genetics. 2010;42:53–61. doi: 10.1038/ng.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sexton T, Yaffe E, Kenigsberg E, Bantignies F, Leblanc B, Hoichman M, Parrinello H, Tanay A, Cavalli G. Three-dimensional folding and functional organization principles of the Drosophila genome. Cell. 2012;148:458–472. doi: 10.1016/j.cell.2012.01.010. [DOI] [PubMed] [Google Scholar]
- Shen Y, Yue F, McCleary DF, Ye Z, Edsall L, Kuan S, Wagner U, Dixon J, Lee L, Lobanenkov VV, et al. A map of the cis-regulatory sequences in the mouse genome. Nature. 2012;488:116–120. doi: 10.1038/nature11243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonis M, Klous P, Splinter E, Moshkin Y, Willemsen R, de Wit E, van Steensel B, de Laat W. Nuclear organization of active and inactive chromatin domains uncovered by chromosome conformation capture-on-chip (4C). Nature genetics. 2006;38:1348–1354. doi: 10.1038/ng1896. [DOI] [PubMed] [Google Scholar]
- Smallwood A, Ren B. Genome organization and long-range regulation of gene expression by enhancers. Current opinion in cell biology. 2013;25:387–394. doi: 10.1016/j.ceb.2013.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Splinter E, de Wit E, van de Werken HJ, Klous P, de Laat W. Determining long-range chromatin interactions for selected genomic sites using 4C-seq technology: From fixation to computation. Methods. 2012 doi: 10.1016/j.ymeth.2012.04.009. [DOI] [PubMed] [Google Scholar]
- Sridharan R, Tchieu J, Mason MJ, Yachechko R, Kuoy E, Horvath S, Zhou Q, Plath K. Role of the murine reprogramming factors in the induction of pluripotency. Cell. 2009;136:364–377. doi: 10.1016/j.cell.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Takebayashi S, Dileep V, Ryba T, Dennis JH, Gilbert DM. Chromatin-interaction compartment switch at developmentally regulated chromosomal domains reveals an unusual principle of chromatin folding. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:12574–12579. doi: 10.1073/pnas.1207185109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanizawa H, Iwasaki O, Tanaka A, Capizzi JR, Wickramasinghe P, Lee M, Fu Z, Noma K. Mapping of long-range associations throughout the fission yeast genome reveals global genome organization linked to transcriptional regulation. Nucleic acids research. 2010;38:8164–8177. doi: 10.1093/nar/gkq955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Z, Gao F, Kim S, Yang H, Lyu J, An W, Wang K, Lu W. Klf4 organizes long-range chromosomal interactions with the oct4 locus in reprogramming and pluripotency. Cell stem cell. 2013;13:36–47. doi: 10.1016/j.stem.2013.05.010. [DOI] [PubMed] [Google Scholar]
- Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH, Rahl PB, Lee TI, Young RA. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153:307–319. doi: 10.1016/j.cell.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young RA. Control of the embryonic stem cell state. Cell. 2011;144:940–954. doi: 10.1016/j.cell.2011.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.