Abstract
The world of non-coding RNAs has only recently started being discovered. For the past 40 years, coding genes, mRNA and proteins have been the center of cellular and molecular biology, and pathologic alterations were attributed to either the aberration of gene sequence or altered promoter activity. It was only after the completion of the human genome sequence that the scientific community started seriously wondering why only a very small portion of the genome corresponded to protein coding genes.
New technologies such as the whole-genome and -transcriptome sequencing demonstrated that at least 90% of the genome is actively transcribed. The identification and cataloguing of multiple kinds of non-coding RNA (ncRNA) has exponentially increased and it is now widely accepted that ncRNAs play major biological roles in cellular physiology, development, metabolism, and are also implicated in a variety of diseases.
The aim of this review is to describe the two major classes (long and short forms) of non-coding RNAs and describe their subclasses in terms of function and their relevance and potential in oral diseases.
Keywords: non-coding RNAs, oral diseases, long non-coding RNA, short non-coding RNA
INTRODUCTION
Since the description of the central dogma of the molecular biology by Crick in 1970 (Crick, 1970), researchers focused on the understanding of the cellular behavior based purely on the description and analysis of mRNA and protein levels. Consequently, observed pathologic alterations were presumed to be based either on the aberration of gene sequence or promoter activity. Recently, especially after the completion of the human genome sequence, the scientific community was surprised by the fact that just a tiny portion of the genome corresponded to protein coding sequences (< 2%) (International Human Genome Sequencing Consortium, 2004, Hattori, 2005) and most of the genome was described as filled with “junk” sequences. Therefore, focal efforts to understand the role of these previously uninvestigated sequences are critical to comprehensive diagnosis, treatment, and prevention of disease.
Technical advances on whole-genome and -transcriptome sequencing demonstrated that at least 90% of the genome is actively transcribed and therefore significantly more complex than previously thought (Kapranov et al., 2005). The description of multiple kinds of non-coding RNA (ncRNA) has exponentially increased and it is now widely accepted that ncRNAs play major biological roles in cellular physiology, development, metabolism, and are also implicated in a variety of diseases. NcRNAs are loosely classified as either small ncRNAs, including small interfering RNAs (siRNAs) and microRNAs (miRNAs) or long ncRNAs (lncRNAs). These ncRNAs have the capacity to modify protein levels by mechanisms independent of transcription, supported by recent evidence that the expression levels of mRNA account for only 37% of protein expression levels (Schwanhausser et al., 2011). Perhaps equally compelling, the epigenetic modifications incurred by these ncRNAs were also shown to be transmittable despite lack of modification to the original DNA sequence. Recent work in mice, worms, pigs and even some pioneer analyses in humans present evidence that trans-generational epigenetic inheritance may also take place, the control of which appears to be based on the specific function of each class of ncRNAs. Together, these recent discoveries indicate that organismal physiology is guided by a number of complex and overlapping mechanisms, and a better understanding of these processes necessitates more dimensional analyses than isolated measurements of DNA, RNA, or protein levels.
In particular, the diagnosis, treatment, and prevention of some pathologic conditions have been limited by our lack of understanding regarding the underlying molecular causes that drive these diseases. Therefore, characterizing the ncRNA profiles of some oral illnesses may be the key to identification of risk factors, diagnosis, and treatment of dental caries, periodontal disease, oral cancer, Sjögren's Syndrome, HIV/AIDS-associated oral diseases and chronic orofacial pain. In this review we will describe the characteristics of the bulk of new ncRNA described (divided in two broader categories: long and short ncRNAs), their function and their relevance for oral pathology.
Long noncoding RNA (lncRNA)
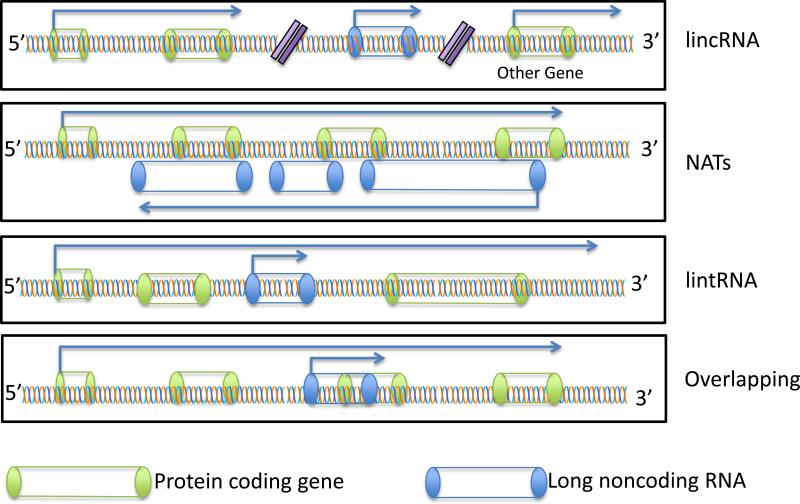
According with the HUGO Gene Nomenclature Committee (HGNC) [HUGO Gene Nomenclature Committee at the European Bioinformatics Institute” http://www.genenames.org], lncRNA correspond to non-protein coding genes that encode transcripts at least 200nt in length. Subtypes of lncRNA are defined by genomic context in relation to the nearest protein-coding gene and include include: antisense (NATs or aRNA), intronic (lintRNA), intergenic (lincRNA) and overlapping (Figure 1 and Table 1). In this review we will describe the first 3 classes of lncRNA.
Figure 1.
Schematic of the genomic context of lncRNAs and a summary of their known functions.
LincRNA: epigenetic changes, maintenance of pluripotency and lineage commitment control.
NATs: alternative splicing, translation control, RNA interference, X-chromosome inactivation.
lintRNA: chromatin modification and organization, guidance of ribonucleoproteins and creation of scaffolds for their assembly. Overlapping: Chromatin modification.
Table 1.
Non-coding RNA types
| Type | Sub Type | Symbol | Size (nt) | Localization | Function | References |
|---|---|---|---|---|---|---|
| Long non-coding RNA | Natural Antisence | NATS or aRNA | ~110-130 | Cytoplasm | Regulates protein localization, mRNA translation and stability | (Mercer & Mattck, 2013, Carrieri et al., 2012, Gong & Maquat, 2011, Willingham et al., 2005) |
| Long Intronic RNA | lintRNA* | 200 to ~100.000 | Nucleus/Cytoplasm | Epigenetic modulator, histone modification, DNA modification | (Mercer & Mattick, 2013, Rinn et al., 2007) | |
| Long intergenic non-coding RNAs | lincRNA | 200 to ~100.000 | Nucleus | Epigenetic modulator, histone modification, DNA modification | (Khalil et al., 2009, Mercer & Mattick, 2013, Tsai et al., 2010) | |
| Short non-coding RNA | Small cytoplasmic | scRNA | ~60-120 | Cytoplasm | Structure Ribonucleoproteins | (Verhagen & Pruijn, 2011, Selvakumar et al., 2012, Perreault et al., 2007) |
| Small nuclear RNA | snRNA | ~150 | Nucleus | RNA biogenesis and processing | (West, 2012) | |
| Small nucleolar RNA | snoRNA | ~130 | Nucleous | Chemically modify, mature, and stabilize rRNAs, Regulate alternative splicing | (Terns & Terns, 2002, Cavaille & Bachellere, 1998, Kishore et al., 2010) | |
| MicroRNA | miRNA | 17-25 | Cytoplasm | Post-transcriptional control protein expression | (Kim et al., 2009) | |
| PIWI-interacting RNAs | piRNA | 25–33 | Cytoplasm | Transposon silencing, heterochromatin modification in germinal cells | (Siomi et al., 2011, Pillai & Chuma, 2012) | |
| MicroRNA offset | moRNA | ~20 | Cytoplasm | Functonal regulators of protein expression (hypothetical) | (Shi et al., 2009, Umbach et al., 2010) | |
| 5'-capped promoter-associated small RNAs | PASRS | 30-35 | Nucleus | Functional regulators of gene transcription (hypothetical) | (Affymetrix ENCODE Transcriptome Project, 2009) | |
| Small Interfering RNAs | siRNA | 21-23 | Cytoplasm | Regulate post-transcriptional expression | (Xia et al., 2013, Lejeune & Allshire, 2011) | |
lintRNA is not the official acronym, use here to simplify text. Stand for Long intronic RNA
Natural antisense transcripts (NATs) or lncRNA antisence (aRNAs)
One of the major groups of lcnRNAs is the natural antisense transcripts (NATs). NATs are transcripts that are complementary to other endogenous RNA transcripts. These NATs can be transcribed either in cis in the same loci but from opposite DNA strands, or in trans at different loci than their target's location. NATs can function at transcriptional and post-transcriptional mechanisms to regulate gene expression. Increasing evidence shows diverse regulatory roles of NATs such as RNA interference (RNAi), alternative splicing, trafficking, genomic imprinting and X-chromosome inactivation (Zhang et al., 2006).
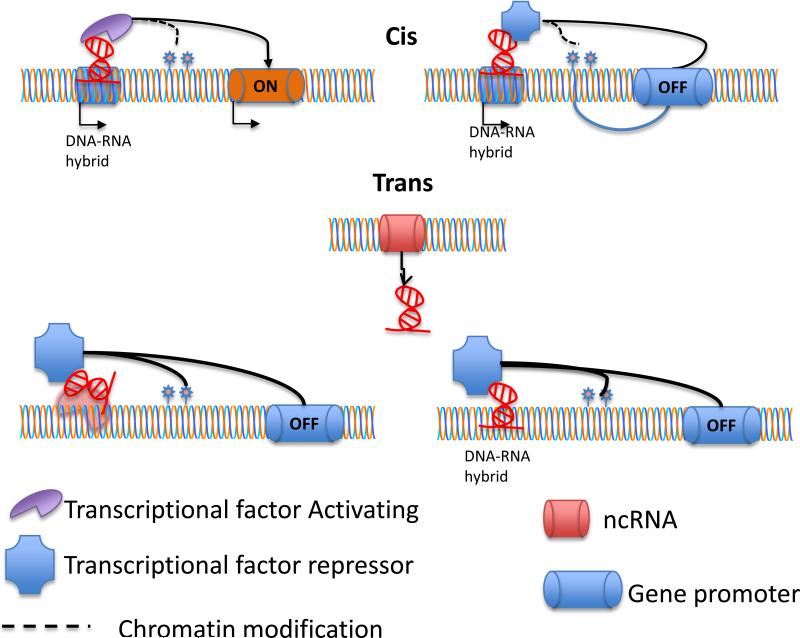
While trans-NATs can usually target multiple transcripts with few mismatches, cis-NATs regulate its target through the formation of double-stranded RNA, resulting in either competitive transcriptional interference (Zhang et al., 2006), RNA masking or gene silencing of RNAi (Tam et al., 2008, Watanabe et al., 2008), or through collisions of RNA polymerase complexes moving in opposite directions within the same locus, leading to premature stopping of the RNA polymerase and degradation of incomplete transcripts (Lapidot & Pilpel, 2006, Osato et al., 2007) (Figure 2). Although about 40 cis-NATs have been experimentally identified, based on the annotated cDNA clones to genome sequences and using the Basic Local Alignment Search Tool from NCBI (BLAST) between complete mRNA and Expressed Sequence Tags (EST) databases, it has been estimated that more than 2000 human, 2400 mouse, and 1000 Drosophila cis-NATs exist in each respective genome (Osato et al., 2007). In general, cis-NATs function as negative regulators with various mechanisms in repression of gene expression. It has been shown that during the development both the Xist and Tisx pair of NATs cause hypermethylation of chromosome and lead to X-chromosome inactivation in females (Li et al., 2006).
Figure 2.
Mechanisms of action of the lncRNAs. Two possible mechanisms are depicted. First, the lncRNAs act on the cis, affecting the same gene that encodes them. Second, lncRNAs act on the trans, controlling the transciption of genes encoded in other genomic regions. In this case, the lncRNAs can also act as a scaffold to link several transcriptionally involved proteins or the lncRNAs can directly bind to the DNA to recruit transcriptional factors.
The expression of human cis-NATs transcribed by RNA Pol II is epigenetically regulated (Conley & Jordan, 2012), and aberrant production of these transcripts has been linked with human disease. Prader-Willi and Angelman syndromes, due to UBE3A expression, are affected due to the presence of a 20 kb NAT which shares complementary sequences of UBE3A (Runte et al., 2001). Mihalich et al, reported patients with endometriosis show low expression of 1B FGF-antisense transcripts, which correlates with endometrial cell proliferation (Mihalich et al., 2003). β-thalassemia is an inherited autosomal recessive anemia, in which the normal a-globin gene (HBA2) is affected due the nearby LUC7L gene. LUC7L is expressed in the opposite direction from the aberrant expression of an antisense HBA2 transcript, which methylates the CpG island of HBA2 and results in silencing gene (Tufarelli et al., 2003) (Figure 2, action in cis). NATs can also positively gene (Tufarelli et al., 2003) (Figure 2, action in cis). NATs can also positively regulate mRNA expression in disease. Modarresi et al., demonstrated that inhibition of brain-derived neurotrophic factor antisense transcript (BDNF-AS) leads to upregulation of BDNF expression and induces neuronal outgrowth and differentiation both in vitro, and in vivo, which is essential for their function and survival (Modarresi et al., 2012). While no direct link between NATs and oral disease has been reported, a number of proposed studies investigating the potential role of NATs in the aberrant regulation of driver mRNAs are being investigated.
Long Intronic RNA (lintRNA[note, this is not official acronym])
Initially described by the group of Verjovski-Almeida (Nakaya et al., 2007, Louro et al., 2008), lintRNAs were identified using a dual approach pairing in silico analysis of the EST sequence in Genbank and a custom made oligoarray platform containing sense and antisense probes for randomly selected totally intronic noncoding transcripts plus the corresponding protein-coding genes. LintRNAs have the characteristic of being totally contained within an intron (Figure 1). In general, lintRNA sequences are poorly conserved and consequently their functional relevance was debated (Pang et al., 2006). However, studies of cross-species microarray hybridization showed tissue-specific conservation of expression of ncRNAs in equivalent genomic loci, but no conservation of the nucleotide sequences (Louro et al., 2008, Khaitovich et al., 2006). This shows that the evolutionary constraints that apply to lintRNA are different from mRNA, siRNA, snoRNA, etc. Expression of lintRNAs seems to be predominantly nuclear; however, some subsets were primarily detected in the cytoplasm, and only a few seem to be equally expressed in both compartments (Clark et al., 2012).
As the focus of ncRNA discovery has overlooked the intronic regions, the biogenesis and function of lintRNAs is poorly understood at this time. Nevertheless, because their expression tends to correlate with the expression of their corresponding protein genes, it is thought that lintRNA are transcribed by RNA polymerase II. However, when a specific inhibitor of this RNA polymerase is used, more than 10% of the lintRNA are upregulated, suggesting that a proportion of this ncRNA could be transcribed by other RNA polymerases. Currently, there are only two specific lintRNA validated in HGNC; KCNIP4-IT1 and SPRY4-IT1 (Michel et al., 2002). KCNIP4-IT is specifically expressed in adult cerebellum, but no further studies have been performed (Michel et al., 2002). SPRY4-IT1 is derived from an intron within the SPRY4 gene, a member of the Sprouty (SPRY) family of genes, which encode Ras/Erk inhibitor proteins. SPRY4-IT1 is upregulated in melanoma cells, and its knockdown resulted in defects in cell growth, invasion, and elevated rates of apoptosis in melanoma cells (Khaitan et al., 2011). While these represent the only lintRNAs that are currently validated, a set of lintRNAs have been identified whose expression correlates with Pancreatic ductal adenocarcinoma and metastasis in pancreatic cells, pointing to the potential relevance of this class of transcripts in biological processes related to malignant transformation and metastasis (Tahira et al., 2011). Also, at least a fraction of lintRNA may be regulated by common physiological signals such as hormones (Louro et al., 2007). In mouse models of inflammation, thousands of lintRNAs change in overall abundance (St Laurent et al., 2012). These data argue in favor of the cellular relevance and the function of this RNAs and further corroborate the notion that the intronic sequence play functional roles in the human gene-expression program in yet unknown mechanisms. So far, the focus of non-coding RNA discovery has avoided intronic regions as those were believed to simply encode parts of pre-mRNAs. The evidences collected here suggest a very different situation, where the sequences encoded in the introns appear to be a yet unexplored reservoir of novel, functional RNAs. As such, they should not be ignored in surveys of functional transcripts or other genomic studies.
While the general information about lintRNA is scant, in the oral biology field our understanding is far more limited. Serial analysis of gene expression studies performed using both exfoliated cells and biopsies of the oral cavity from normal oral mucosa and oral premalignant lesions showed the presence of 325 lncRNAs in normal mucosa. Additionally, approximately 60% of lncRNA expressed in oral mucosa had an aberrant expression in premalignant lesions. This suggests that lncRNA expression contributes significantly to the oral transcriptome and maybe also to the progression of the lesion (Gibb et al., 2011). A more detailed analysis is necessary to establish how many of the 325 lncRNAs described correspond to lintRNA. Oral squamous cell carcinoma (OSCC) represents ~3% of all malignancies and accounts for >274,000 newly diagnosed cancer cases every year worldwide (de Camargo Cancela et al., 2010). There is just one study in OSCC analyzing lncRNA, however in this study the analysis was performed by PCR looking for specific lncRNA all of them were NATs (Tang et al., 2012). The identity of the lintRNAs is recognized by HGNC and evidence suggests they may be important contributors in the control of the protein expression in development and pathology. Investigation of these signatures in future studies may give a new insight on normal oral physiology and oral pathology.
Long intergenic non-coding RNAs (lincRNAs)
LincRNAs are long non-coding RNAs sequences between protein-coding genes transcribed from DNA interspersed outside known protein-coding genome areas as independent transcriptional units (Sun et al., 2013). LincRNA are longer than 200 nucleotides and presently over 8,000 human lincRNA have been identified and catalogued (Sun et al., 2013). Most of lincRNA discovery is attributed to the exponential growth of deep sequencing experiments. LincRNAs are not only evolutionarily conserved but they seem to have specific spatial and temporal expression, similar to the expression of microRNAs and lintRNAs. A recent study of experimentally validated lincRNAs showed that there is a very high orthology between human and mouse, with 60-70% shared between those 2 species (Managadze et al., 2013). The large number of lincRNAs possibly suggests that they might be involved in several cellular functions and mechanisms. Also, in contrast to some of the classes of the short ncRNAs, lincRNAs have highly variable length and the structure their sequences form is more complex and variable. Some of the functions in which lincRNAs have been implicated include genomic programming (Amaral & Mattick, 2008), maintenance of pluripotency and lineage commitment control (Guttman et al., 2011) and control of epigenetic changes (Koziol & Rinn, 2010).
Short noncoding RNA
According to the HGNC nomenclature, short ncRNAs classes include: small cytoplasmic RNA (scRNA), small nuclear RNA (snRNA), small nucleolar RNA (snoRNA), microRNA (miRNA), PIWI interacting RNA (piRNA). In addition, 4 more classes (transfer RNA, Ro ribonucleo-particle RNA, ribosomal RNA, vault RNA) have been recognized, but will not discussed in this review as their functions are either well characterized or they only include a very small set of genes.
It is likely that as deep sequencing studies increase, the official classes of short ncRNA will increase. In this review we will also discuss 3 types on short ncRNA that are not yet included in the standard nomenclature system: the MicroRNA offset RNA (moRNA), the small interfering RNA (siRNA) and the Promoter-associated small RNAs (PASRS) (Table 1).
Small cytoplasmic RNA (scRNA)
Small cytoplasmic RNA (scRNA) is a small (7S; 129 nucleotides) RNA found in the cytosol and rough endoplasmic reticulum (ER) interacted with ribonucleoproteins (scRNP) that are involved in the specific selection and transport of other proteins. High abundance and evolutionarily conserved RNA sequences within these small RNP suggest some important but still relatively unclear biological roles (Selvakumar et al., 2012). The presence of Ro and La autoantibodies against scRNPs led to the discovery of scRNA termed Y RNAs in patients with Systemic Lupus Erythematosus (SLE) and with Sjögren's syndrome (Fabini et al., 2000). The Y RNAs are non-coding RNA components of either 60-kDa or 50-kDa of Ro or La RNPs, respectively (Verhagen & Pruijn, 2011).
Currently four different Y RNAs (69-112 nucleotides) have been identified in humans. The secondary structure of scRNAs molecules shows a long base-paired stem loop with pyrimidine-rich sequence (Perreault et al., 2007). Within this stem region, a conserved sequence contains a bulged cytosine that was suggested to be the site that interacted with Ro protein. The La protein was proposed to bind to 3′ oligo-uridylate sequences (Stein et al., 2005, Teunissen et al., 2000, Green et al., 1998). Several lines of evidence suggest that Y RNA either plays a role as a repressor of Ro (Reinisch & Wolin, 2007) or is functionally required for DNA replication (Christov et al., 2006). Indeed, a recent study has indicated that small RNAs derived from Y RNAs may be broadly involved in autoimmune diseases (Verhagen & Pruijn, 2011). Another type of scRNAs is reported found in rodent and primate cells. The Alu-related scRNAs are less abundant but accumulate steadily in cytoplasm. These scRNAs show Alu-like sequences at both 5′ and 3′ ends and bind to another type of scRNPs called signal recognition particles such as the SRP14/19 heterodimer. Although the function of these scRNP complexes is unclear, it might play a role in protein translation (Bovia & Strub, 1996). Besides the Sjogen's syndrome, where the scRNAs play a critical role in Ro and La autoantibody recognition, no other oral disease has been reported related to the effects of scRNAs.
Small nuclear RNA (snRNA)
Small nuclear RNAs (snRNAs) are a group of non-coding RNA, also known as U-RNA, due to their uridylate-rich sequence. SnRNAs are highly abundant, non-polyadenylated, and found within the nucleoplasm of eukaryotic cells (Busch et al., 1982). The size of snRNAs is about 150 nucleotides and they can be transcribed either by RNA polymerase II (for Sm-class RNAs, U1, U2, U4, U4atac, U5, U7, U11, U12) or by RNA polymerase III (Lsm-class RNAs, U6 and U6atac). snRNAs are associated with a group of small nuclear proteins (snRNPs), which play important roles in RNA biogenesis and processing. The Sm-class RNAs contain a 5′ trimethylguanosine cap and bind several Sm proteins. The Lsm-RNAs have a monomethylphosphate 5′ cap and an uridine rich 3′ end, and both are important for the binding of Lsm proteins. While Lsm-class snRNAs function within the nucleus, the Sm-class snRNAs are exported from the nucleus to the cytoplasm for maturation. The most common components of these snRNA complexed with snRNPs are U1, U2, U4, U5 and U6 snRNAs (Guo et al., 2009).
These five snRNAs together with more than 150 proteins form spliceosomes whose the primary function is in the processing of pre-mRNA splicing (Faustino & Cooper, 2003). The primary role of snRNAs (Busch et al., 1982) is to recognize the 5′ and 3′ intron/exon junction and recruit (Matera et al., 2007) the RNPs to facilitate the splicing. The precise removal of intron sequences is guided by the base-pairing interaction of the spliceosomal snRNAs at the intron-exon junctions. Therefore, any error that occurs in either step will lead to abnormal mRNA, and disrupt the open reading frame and result in human diseases, such as atypical cystic fibrosis, Frasier syndrome, spinal muscular atrophy, etc. Faustino and Cooper provided comprehensive documentation on abnormal splicing mechanisms that lead to human disorders (Faustino & Cooper, 2003). So far none of these have been examined in oral diseases. However, it is expected that alternatively spliced transcripts will be identified in oral pathologies. For example, a recent publication of deep sequencing results of buccal mucosa squamous cell carcinomas identified 11 novel splice junctions mainly originating from alternate 5′ splice sites (Shah et al., 2013).
Small nucleolar RNA (snoRNA)
Small nucleolar RNAs (snoRNAs) are a class of non-coding RNA whose primary functions are to guide chemical modification of pre-rRNA and facilitate RNA processing (Lafontaine & Tollervey, 1998) (Terns & Terns, 2002). Based on the sequence motifs and secondary structures, snoRNAs can be classified as the C/D box or H/ACA box snoRNAs, which are involved in methylation and pseudouridylaton of pre-rRNA molecules, respectively, to generate mature rRNA (Kiss, 2001, Lafontaine & Tollervey, 1998, Terns & Terns, 2002). Most snoRNAs are encoded in the introns of proteins that participate in ribosome synthesis or translation and are transcribed by RNA polymerase II.
There are currently more than 200 snoRNAs described in mammals (Brown et al., 2003, Lestrade & Weber, 2006) and they are among the most diverse trans-acting ncRNAs. To guide the modification on pre-rRNA, each snoRNA associates with at least four proteins and forms a snoRNP. These proteins include either methyltransferase (for methylation) or pseudouridine synthase (for pseudouridylation) in the complex. The conserved box of C/D and H/ACA motifs can serve as protein binding signals. The snoRNA contains a complementary sequence (antisense, around 10-21 nucleotides) which target the modification site of pre-rRNA. Human rRNA contains about 115 methyl group modifications and about 95 pseudouridine modification sites (Maden & Hughes, 1997). Besides modification of pre-rRNA, it has been reported that some snoRNAs are capable of processing pre-rRNA cleavage. These include C/D snoRNAs (U3, U8, U22) and H/ACA snoRNAs (snR10, snR30, E2 and E3) (Kressler et al., 1999, Venema & Tollervey, 1999). In addition, snoRNA can also modify the small nuclear RNA (snRNA) which mediates the splicing of mRNA.
SnoRNAs play an important role in imprinting several diseases in humans, especially in neurodevelopmental disorders such as Prader-Willi syndrome and Angelman syndrome, which are due to the loss of snoRNA expression. The Prader-Willi syndrome is linked to the deletion of region on chromosome 15 which contains a brain-specific C/D RNA (HBII-52) that processes serotonin-2c receptor (HTR2C) pre-mRNA. Absence of the C/D snoRNA and the reduced level of functional serotonin-2C receptor mRNA are found in these patients (Nicholls & Knepper, 2001). So far, no snoRNA abnormality has been reported for any related oral disease.
MicroRNA (miRNA)
MiRNA were first identified as regulators of larval development in C. elegans (Wightman et al., 1993) and are currently are the most widely studied class of ncRNA. MiRNAs are single-stranded RNAs of 17-25 nucleotides and function as guide molecules in post-transcriptional gene silencing by partially complementing with their target mRNAs, leading to translational repression or in some cases, transcript degradation. The biogenesis of miRNAs starts in the nucleus, where RNA polymerase II transcribes primary miRNAs (primiRNA). The primiRNA is edited and cleaved by the RNase Drosha and its co-factor DGCR8, generating a hairpin precursor (pre-miRNA) that is exported to the cytoplasm, where the RNase Dicer further processes the pre-miRNA to produce a miRNA duplex. This duplex is incorporated into the RNA-induced silencing complex (RISC) where it mediates gene repression. The RISC recognizes the duplex, unwinds it, selects the guide miRNA strand (while degrading the passenger strand) and mediates recognition of target RNAs. Human cells encode over 1,000 miRNA species, some of which regulate specific targets, but in most cases miRNAs are capable of regulating the expression of hundreds of genes at the same time, working as master regulators of the gene expression process (Kim et al., 2009).
MiRNAs represent the best studied of non-coding RNAs and there is a plethora of reviews about their biogenesis and function. They have also been studied in the field of oral research. More specifically, miRNA's role has been studied in Sjögren's syndrome (Alevizos et al., 2011), in oral cancer (Gorenchtein et al., 2012), oral immunology (Nahid et al., 2011), and salivary exosomes have been shown to contain miRNAs (Gallo et al., 2012, Michael et al., 2010). Still, there is a large number of oral diseases for which any role of miRNAs has not been examined. This is especially true in developmental abnormalities, as miRNAs are considered instrumental in controlling development by their temporal and tissue specific expression.
Micro-RNA offset RNAs (moRNA)
MoRNA were first discovered in Ciona intestinalis (Shi et al., 2009) and identified as 20-nt-long RNAs derived from the ends of pre-miRNAs. moRNAs can originate from either end of the pre-miRNA, but they are mainly derived from the 5′ arm, regardless of the major miRNA position. This suggested that their biogenesis is connected but, as in Ciona, the moRNa expression pattern is different than their miRNA and their abundance can exceed that of the corresponding mature miRNA. More recently, using short read sequencing from human prefrontal cortex, it was shown that moRNAs are also produced from human miRNA precursors, albeit at quite low expression levels. The expression levels of moRNAs suggest that despite this processing connection with miRNA, additional mechanisms exist downstream (Langenberger et al., 2009). Using deep sequencing analysis of miRNAs in 23 solid tumor specimens of breast, bladder, colon and lung cancer, seven types of moRNA were identified. All moRNAs sequenced in the human tumors were highly conserved, derived exclusively from the 5′ arm of the miRNA precursor directly upstream to the 5′ miRNA, and expressed at lower levels relative to the main miRNA product of the precursor (Meiri et al., 2010). More recently it was shown that in a cellular model for myeloproliferative neoplasms, mutant SET2 cells express at least 58 moRNA at moderate to high level and 95% of these moRNAs correspond to the 5′ arm of its miRNA (Bortoluzzi et al., 2012). The biological function of moRNAs is still not known, however, the possibility that these ncRNAs are also functional regulators of protein expression and cell function suggest they could have a role in human disease. In vivo experiments with rhesus adinovirus moRNA suggest that moRNAs might guide RISC to complementary target mRNAs, acting like a miRNA (Umbach et al., 2010). Nevertheless, moRNA enrichment observed in the nucleus (Taft et al., 2010) suggests that some moRNAs play a different role specifically related to nuclear processes, like transcript initiation and splice site regulation.
Both functional characterization of moRNAs and elucidation of their biogenesis are highly relevant for future research. In this respect, deep sequencing data analysis is a powerful tool that can increase our understanding of this process and will help to generate new hypotheses about their role within the context of functional gene regulation.
PIWI-interacting RNAs (piRNA)
This type of small RNA was first described in Drosophila (Sarot et al., 2004). They are between 25 to 33 nucleotides in length and bind the Piwi-class Argonaute proteins. PiRNAs are located in clusters at transposon loci in the genome (in association with Piwi family members). A cluster can encode between 10 to 1000 individual piRNAs. There is about 114 piRNA clusters in the human genome, and about 23,000 individual human piRNA sequences has been identified by HGNC (Wright & Bruford, 2011). piRNAs are produced independently of Dicer ribonuclease (Siomi et al., 2011) and no significant secondary structures in the stem-loop of their precursors have been detected in regions surrounding piRNAs (Mei et al., 2013). However, like miRNA, precursors of piRNAs need further post transcriptional processing to become fully mature (Siomi et al., 2011, Mei et al., 2013).
PiRNAs have been shown to act in the Piwi-dependent transposon silencing and heterochromatin modification in germinal cells to maintain the genomic integrity of these cells (Lin, 2007, Pillai & Chuma, 2012). However, only about 20% of known mammalian piRNAs map to transposons and other repeat genomic regions (Kim, 2006). Also piRNAs and piwi proteins have recently been identified outside of germinal cell lines, e.g., in human cancer cells (Yu & Pestell, 2012, Cheng et al., 2011, Cheng et al., 2012, Law et al., 2013, Huang et al., 2012), suggesting a role for this ncRNAs in the epigenetic regulation of aberrant cancer “stem cell like” cells that are proposed to be critical for tumor initiation and progression.
Small Interfering RNAs (siRNAs)
For this review, siRNAs will only refer to endogenous small interfering RNA, also referred to as endo-siRNAs in literature. This class of short ncRNAs has very recently been discovered in human cells (Xia et al., 2013), although it has been shown to have functional roles in eukaryotic cells of C. elegans (Ruby et al., 2006), D. melanogaster (Ghildiyal et al., 2008, Okamura et al., 2008, Kawamura et al., 2008), mouse oocytes (Tam et al., 2008) and mouse testicular cells (Song et al., 2011). Endo-siRNAs have been shown to arise from transposable elements (Okamura & Lai, 2008). This biogenesis mechanism is Dicer-dependent but does not require the involvement of the Drosha/Dgcr8 complex (Xia et al., 2013), i.e., different than the microRNA biogenesis. Endo-siRNAs regulate the post-transcriptional expression of mRNAs and have also been proposed to mediate and program chromatin modifications (Xia et al., 2013, Lejeune & Allshire, 2011). Endo-siRNAs require a very high complementarity with mRNA function and this high complementarity leads only to mRNA degradation, contrary to microRNAs that can either cleave their mRNA target or block its translation without degrading the transcript.
Promoter-associated small RNAs (PASRs)
PASRs represent one of the most abundant individual class of small RNAs of the non-annotated fraction the genome. They were initially defined as small RNAs mapped on a genome area found within 500 nucleotides of a transcription start site (TSS). Since then PASRs have also been mapped to the first exons of genes (Kapranov et al., 2007, 2009). Two possibilities regarding the origin of PASRs have been suggested. First, that they are either transcribed independently with a 5′ cap from promoters that also transcribe long RNAs or second, that they are the by-products of processed long RNAs. A large number of the PASRs possess a 5′ cap and this class of PASRs is termed 5-capped promoter-associated small RNA (Affymetrix ENCODE Transcriptome Project, 2009). Detailed bioinformatics analysis of the known PASRs revealed that there is a statistically significant enrichment of those RNAs on both the sense and antisense strands immediately next to the TSS, and that both of those strands had mirror-image profiles (Affymetrix ENCODE Transcriptome Project, 2009). Equally interesting was the finding that on the sense strand, PASRs were downstream of the TSS while on the antisense strand they were mainly upstream of the TSS. As with most of the non-coding RNA classes, their involvement in oral and maxillofacial diseases is totally unexplored.
Conclusion
Characterizion of the ncRNAs involved in oral diseases may be a key to our understanding of their pathogenesis, pathophysiology, and risk factors. As more and more deep sequencing data are generated and released for oral pathologies, extensive bioinformatics analysis can shed light on the genetic changes of the non-coding RNAs and map those changes to specific phenotypes, or answer questions about the role in oral biology and organ landscape of the very important class of ncRNAs.
Bibliography
- Affymetrix ENCODE Transcriptome Project CSHLETP Post-transcriptional processing generates a diversity of 5′-modified long and short RNAs. Nature. 2009;457:1028–32. doi: 10.1038/nature07759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alevizos I, Alexander S, Turner RJ, Illei GG. MicroRNA expression profiles as biomarkers of minor salivary gland inflammation and dysfunction in Sjogren's syndrome. Arthritis Rheum. 2011;63:535–44. doi: 10.1002/art.30131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaral PP, Mattick JS. Noncoding RNA in development. Mamm Genome. 2008;19:454–92. doi: 10.1007/s00335-008-9136-7. [DOI] [PubMed] [Google Scholar]
- Bortoluzzi S, Bisognin A, Biasiolo M, Guglielmelli P, Biamonte F, Norfo R, Manfredini R, Vannucchi AM. Characterization and discovery of novel miRNAs and moRNAs in JAK2V617F-mutated SET2 cells. Blood. 2012;119:e120–30. doi: 10.1182/blood-2011-07-368001. [DOI] [PubMed] [Google Scholar]
- Bovia F, Strub K. The signal recognition particle and related small cytoplasmic ribonucleoprotein particles. Journal of cell science. 1996;109(Pt 11):2601–8. doi: 10.1242/jcs.109.11.2601. [DOI] [PubMed] [Google Scholar]
- Brown JW, Echeverria M, Qu LH, Lowe TM, Bachellerie JP, Huttenhofer A, Kastenmayer JP, Green PJ, Shaw P, Marshall DF. Plant snoRNA database. Nucleic acids research. 2003;31:432–5. doi: 10.1093/nar/gkg009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch H, Reddy R, Rothblum L, Choi YC. SnRNAs, SnRNPs, and RNA processing. Annu Rev Biochem. 1982;51:617–54. doi: 10.1146/annurev.bi.51.070182.003153. [DOI] [PubMed] [Google Scholar]
- Cheng J, Deng H, Xiao B, Zhou H, Zhou F, Shen Z, Guo J. piR-823, a novel non-coding small RNA, demonstrates in vitro and in vivo tumor suppressive activity in human gastric cancer cells. Cancer letters. 2012;315:12–7. doi: 10.1016/j.canlet.2011.10.004. [DOI] [PubMed] [Google Scholar]
- Cheng J, Guo JM, Xiao BX, Miao Y, Jiang Z, Zhou H, Li QN. piRNA, the new non-coding RNA, is aberrantly expressed in human cancer cells. Clinica chimica acta; international journal of clinical chemistry. 2011;412:1621–5. doi: 10.1016/j.cca.2011.05.015. [DOI] [PubMed] [Google Scholar]
- Christov CP, Gardiner TJ, Szuts D, Krude T. Functional requirement of noncoding Y RNAs for human chromosomal DNA replication. Molecular and cellular biology. 2006;26:6993–7004. doi: 10.1128/MCB.01060-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark MB, Johnston RL, Inostroza-Ponta M, Fox AH, Fortini E, Moscato P, Dinger ME, Mattick JS. Genome-wide analysis of long noncoding RNA stability. Genome research. 2012;22:885–98. doi: 10.1101/gr.131037.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conley AB, Jordan IK. Epigenetic regulation of human cis-natural antisense transcripts. Nucleic acids research. 2012;40:1438–45. doi: 10.1093/nar/gkr1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crick F. Central dogma of molecular biology. Nature. 1970;227:561–3. doi: 10.1038/227561a0. [DOI] [PubMed] [Google Scholar]
- de Camargo Cancela M, Voti L, Guerra-Yi M, Chapuis F, Mazuir M, Curado MP. Oral cavity cancer in developed and in developing countries: population-based incidence. Head & neck. 2010;32:357–67. doi: 10.1002/hed.21193. [DOI] [PubMed] [Google Scholar]
- Fabini G, Rutjes SA, Zimmermann C, Pruijn GJ, Steiner G. Analysis of the molecular composition of Ro ribonucleoprotein complexes. Identification of novel Y RNA-binding proteins. European journal of biochemistry / FEBS. 2000;267:2778–89. doi: 10.1046/j.1432-1327.2000.01298.x. [DOI] [PubMed] [Google Scholar]
- Faustino NA, Cooper TA. Pre-mRNA splicing and human disease. Genes & development. 2003;17:419–37. doi: 10.1101/gad.1048803. [DOI] [PubMed] [Google Scholar]
- Gallo A, Tandon M, Alevizos I, Illei GG. The majority of microRNAs detectable in serum and saliva is concentrated in exosomes. PloS one. 2012;7:e30679. doi: 10.1371/journal.pone.0030679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghildiyal M, Seitz H, Horwich MD, Li C, Du T, Lee S, Xu J, Kittler EL, Zapp ML, Weng Z, Zamore PD. Endogenous siRNAs derived from transposons and mRNAs in Drosophila somatic cells. Science. 2008;320:1077–81. doi: 10.1126/science.1157396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibb EA, Enfield KS, Stewart GL, Lonergan KM, Chari R, Ng RT, Zhang L, MacAulay CE, Rosin MP, Lam WL. Long non-coding RNAs are expressed in oral mucosa and altered in oral premalignant lesions. Oral oncology. 2011;47:1055–61. doi: 10.1016/j.oraloncology.2011.07.008. [DOI] [PubMed] [Google Scholar]
- Gorenchtein M, Poh CF, Saini R, Garnis C. MicroRNAs in an oral cancer context - from basic biology to clinical utility. Journal of dental research. 2012;91:440–6. doi: 10.1177/0022034511431261. [DOI] [PubMed] [Google Scholar]
- Green CD, Long KS, Shi H, Wolin SL. Binding of the 60-kDa Ro autoantigen to Y RNAs: evidence for recognition in the major groove of a conserved helix. RNA. 1998;4:750–65. doi: 10.1017/s1355838298971667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Z, Karunatilaka KS, Rueda D. Single-molecule analysis of protein-free U2-U6 snRNAs. Nature structural & molecular biology. 2009;16:1154–9. doi: 10.1038/nsmb.1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, Munson G, Young G, Lucas AB, Ach R, Bruhn L, Yang X, Amit I, Meissner A, Regev A, Rinn JL, Root DE, Lander ES. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477:295–300. doi: 10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattori M. [Finishing the euchromatic sequence of the human genome]. Tanpakushitsu kakusan koso. Protein, nucleic acid, enzyme. 2005;50:162–8. [PubMed] [Google Scholar]
- Huang G, Hu H, Xue X, Shen S, Gao E, Guo G, Shen X, Zhang X. Altered expression of piRNAs and their relation with clinicopathologic features of breast cancer. Clinical & translational oncology : official publication of the Federation of Spanish Oncology Societies and of the National Cancer Institute of Mexico. 2012 doi: 10.1007/s12094-012-0966-0. [DOI] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931–45. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT, Stadler PF, Hertel J, Hackermuller J, Hofacker IL, Bell I, Cheung E, Drenkow J, Dumais E, Patel S, Helt G, Ganesh M, Ghosh S, Piccolboni A, Sementchenko V, Tammana H, Gingeras TR. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316:1484–8. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- Kapranov P, Drenkow J, Cheng J, Long J, Helt G, Dike S, Gingeras TR. Examples of the complex architecture of the human transcriptome revealed by RACE and high-density tiling arrays. Genome research. 2005;15:987–97. doi: 10.1101/gr.3455305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamura Y, Saito K, Kin T, Ono Y, Asai K, Sunohara T, Okada TN, Siomi MC, Siomi H. Drosophila endogenous small RNAs bind to Argonaute 2 in somatic cells. Nature. 2008;453:793–7. doi: 10.1038/nature06938. [DOI] [PubMed] [Google Scholar]
- Khaitan D, Dinger ME, Mazar J, Crawford J, Smith MA, Mattick JS, Perera RJ. The melanoma-upregulated long noncoding RNA SPRY4-IT1 modulates apoptosis and invasion. Cancer research. 2011;71:3852–62. doi: 10.1158/0008-5472.CAN-10-4460. [DOI] [PubMed] [Google Scholar]
- Khaitovich P, Kelso J, Franz H, Visagie J, Giger T, Joerchel S, Petzold E, Green RE, Lachmann M, Paabo S. Functionality of intergenic transcription: an evolutionary comparison. PLoS genetics. 2006;2:e171. doi: 10.1371/journal.pgen.0020171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim VN. Small RNAs just got bigger: Piwi-interacting RNAs (piRNAs) in mammalian testes. Genes & development. 2006;20:1993–7. doi: 10.1101/gad.1456106. [DOI] [PubMed] [Google Scholar]
- Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009;10:126–39. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- Kiss T. Small nucleolar RNA-guided post-transcriptional modification of cellular RNAs. The EMBO journal. 2001;20:3617–22. doi: 10.1093/emboj/20.14.3617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koziol MJ, Rinn JL. RNA traffic control of chromatin complexes. Current opinion in genetics & development. 2010;20:142–8. doi: 10.1016/j.gde.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kressler D, Linder P, de La Cruz J. Protein trans-acting factors involved in ribosome biogenesis in Saccharomyces cerevisiae. Molecular and cellular biology. 1999;19:7897–912. doi: 10.1128/mcb.19.12.7897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafontaine DL, Tollervey D. Birth of the snoRNPs: the evolution of the modification-guide snoRNAs. Trends in biochemical sciences. 1998;23:383–8. doi: 10.1016/s0968-0004(98)01260-2. [DOI] [PubMed] [Google Scholar]
- Langenberger D, Bermudez-Santana C, Hertel J, Hoffmann S, Khaitovich P, Stadler PF. Evidence for human microRNA-offset RNAs in small RNA sequencing data. Bioinformatics. 2009;25:2298–301. doi: 10.1093/bioinformatics/btp419. [DOI] [PubMed] [Google Scholar]
- Lapidot M, Pilpel Y. Genome-wide natural antisense transcription: coupling its regulation to its different regulatory mechanisms. EMBO reports. 2006;7:1216–22. doi: 10.1038/sj.embor.7400857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law PT, Qin H, Ching AK, Lai KP, Co NN, He M, Lung RW, Chan AW, Chan TF, Wong N. Deep sequencing of small RNA transcriptome reveals novel non-coding RNAs in hepatocellular carcinoma. Journal of hepatology. 2013 doi: 10.1016/j.jhep.2013.01.032. [DOI] [PubMed] [Google Scholar]
- Lejeune E, Allshire RC. Common ground: small RNA programming and chromatin modifications. Current opinion in cell biology. 2011;23:258–65. doi: 10.1016/j.ceb.2011.03.005. [DOI] [PubMed] [Google Scholar]
- Lestrade L, Weber MJ. snoRNA-LBME-db, a comprehensive database of human H/ACA and C/D box snoRNAs. Nucleic acids research. 2006;34:D158–62. doi: 10.1093/nar/gkj002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YY, Qin L, Guo ZM, Liu L, Xu H, Hao P, Su J, Shi Y, He WZ, Li YX. In silico discovery of human natural antisense transcripts. BMC bioinformatics. 2006;7:18. doi: 10.1186/1471-2105-7-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H. piRNAs in the germ line. Science. 2007;316:397. doi: 10.1126/science.1137543. [DOI] [PubMed] [Google Scholar]
- Louro R, El-Jundi T, Nakaya HI, Reis EM, Verjovski-Almeida S. Conserved tissue expression signatures of intronic noncoding RNAs transcribed from human and mouse loci. Genomics. 2008;92:18–25. doi: 10.1016/j.ygeno.2008.03.013. [DOI] [PubMed] [Google Scholar]
- Louro R, Nakaya HI, Amaral PP, Festa F, Sogayar MC, da Silva AM, Verjovski-Almeida S, Reis EM. Androgen responsive intronic non-coding RNAs. BMC biology. 2007;5:4. doi: 10.1186/1741-7007-5-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maden BE, Hughes JM. Eukaryotic ribosomal RNA: the recent excitement in the nucleotide modification problem. Chromosoma. 1997;105:391–400. doi: 10.1007/BF02510475. [DOI] [PubMed] [Google Scholar]
- Managadze D, Lobkovsky AE, Wolf YI, Shabalina SA, Rogozin IB, Koonin EV. The vast, conserved mammalian lincRNome. PLoS computational biology. 2013;9:e1002917. doi: 10.1371/journal.pcbi.1002917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matera AG, Terns RM, Terns MP. Non-coding RNAs: lessons from the small nuclear and small nucleolar RNAs. Nat Rev Mol Cell Biol. 2007;8:209–20. doi: 10.1038/nrm2124. [DOI] [PubMed] [Google Scholar]
- Mei Y, Clark D, Mao L. Novel dimensions of piRNAs in cancer. Cancer letters. 2013 doi: 10.1016/j.canlet.2013.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meiri E, Levy A, Benjamin H, Ben-David M, Cohen L, Dov A, Dromi N, Elyakim E, Yerushalmi N, Zion O, Lithwick-Yanai G, Sitbon E. Discovery of microRNAs and other small RNAs in solid tumors. Nucleic acids research. 2010;38:6234–46. doi: 10.1093/nar/gkq376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael A, Bajracharya SD, Yuen PS, Zhou H, Star RA, Illei GG, Alevizos I. Exosomes from human saliva as a source of microRNA biomarkers. Oral Dis. 2010;16:34–8. doi: 10.1111/j.1601-0825.2009.01604.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel U, Kallmann B, Rieckmann P, Isbrandt D. UM 9(5)h and UM 9(5)p, human and porcine noncoding transcripts with preferential expression in the cerebellum. RNA. 2002;8:1538–47. [PMC free article] [PubMed] [Google Scholar]
- Mihalich A, Reina M, Mangioni S, Ponti E, Alberti L, Vigano P, Vignali M, Di Blasio AM. Different basic fibroblast growth factor and fibroblast growth factor-antisense expression in eutopic endometrial stromal cells derived from women with and without endometriosis. The Journal of clinical endocrinology and metabolism. 2003;88:2853–9. doi: 10.1210/jc.2002-021434. [DOI] [PubMed] [Google Scholar]
- Modarresi F, Faghihi MA, Lopez-Toledano MA, Fatemi RP, Magistri M, Brothers SP, van der Brug MP, Wahlestedt C. Inhibition of natural antisense transcripts in vivo results in gene-specific transcriptional upregulation. Nature biotechnology. 2012;30:453–9. doi: 10.1038/nbt.2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nahid MA, Satoh M, Chan EK. MicroRNA in TLR signaling and endotoxin tolerance. Cell Mol Immunol. 2011;8:388–403. doi: 10.1038/cmi.2011.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakaya HI, Amaral PP, Louro R, Lopes A, Fachel AA, Moreira YB, El-Jundi TA, da Silva AM, Reis EM, Verjovski-Almeida S. Genome mapping and expression analyses of human intronic noncoding RNAs reveal tissue-specific patterns and enrichment in genes related to regulation of transcription. Genome biology. 2007;8:R43. doi: 10.1186/gb-2007-8-3-r43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholls RD, Knepper JL. Genome organization, function, and imprinting in Prader-Willi and Angelman syndromes. Annu Rev Genomics Hum Genet. 2001;2:153–75. doi: 10.1146/annurev.genom.2.1.153. [DOI] [PubMed] [Google Scholar]
- Okamura K, Balla S, Martin R, Liu N, Lai EC. Two distinct mechanisms generate endogenous siRNAs from bidirectional transcription in Drosophila melanogaster. Nature structural & molecular biology. 2008;15:998. doi: 10.1038/nsmb0908-998c. [DOI] [PubMed] [Google Scholar]
- Okamura K, Lai EC. Endogenous small interfering RNAs in animals. Nat Rev Mol Cell Biol. 2008;9:673–8. doi: 10.1038/nrm2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osato N, Suzuki Y, Ikeo K, Gojobori T. Transcriptional interferences in cis natural antisense transcripts of humans and mice. Genetics. 2007;176:1299–306. doi: 10.1534/genetics.106.069484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang KC, Frith MC, Mattick JS. Rapid evolution of noncoding RNAs: lack of conservation does not mean lack of function. Trends in genetics : TIG. 2006;22:1–5. doi: 10.1016/j.tig.2005.10.003. [DOI] [PubMed] [Google Scholar]
- Perreault J, Perreault JP, Boire G. Ro-associated Y RNAs in metazoans: evolution and diversification. Molecular biology and evolution. 2007;24:1678–89. doi: 10.1093/molbev/msm084. [DOI] [PubMed] [Google Scholar]
- Pillai RS, Chuma S. piRNAs and their involvement in male germline development in mice. Development, growth & differentiation. 2012 doi: 10.1111/j.1440-169X.2011.01320.x. [DOI] [PubMed] [Google Scholar]
- Project. AETPCSHLET Post-transcriptional processing generates a diversity of 5'-modified long and short RNAs. Nature. 2009;457:1028–32. doi: 10.1038/nature07759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinisch KM, Wolin SL. Emerging themes in non-coding RNA quality control. Curr Opin Struct Biol. 2007;17:209–14. doi: 10.1016/j.sbi.2007.03.012. [DOI] [PubMed] [Google Scholar]
- Ruby JG, Jan C, Player C, Axtell MJ, Lee W, Nusbaum C, Ge H, Bartel DP. Large-scale sequencing reveals 21U-RNAs and additional microRNAs and endogenous siRNAs in C. elegans. Cell. 2006;127:1193–207. doi: 10.1016/j.cell.2006.10.040. [DOI] [PubMed] [Google Scholar]
- Runte M, Huttenhofer A, Gross S, Kiefmann M, Horsthemke B, Buiting K. The IC-SNURF-SNRPN transcript serves as a host for multiple small nucleolar RNAspecies and as an antisense RNA for UBE3A. Human molecular genetics. 2001;10:2687–700. doi: 10.1093/hmg/10.23.2687. [DOI] [PubMed] [Google Scholar]
- Sarot E, Payen-Groschene G, Bucheton A, Pelisson A. Evidence for a piwi-dependent RNA silencing of the gypsy endogenous retrovirus by the Drosophila melanogaster flamenco gene. Genetics. 2004;166:1313–21. doi: 10.1534/genetics.166.3.1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwanhausser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, Selbach M. Global quantification of mammalian gene expression control. Nature. 2011;473:337–42. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- Selvakumar T, Gjidoda A, Hovde SL, Henry RW. Regulation of human RNA polymerase III transcription by DNMT1 and DNMT3a DNA methyltransferases. The Journal of biological chemistry. 2012;287:7039–50. doi: 10.1074/jbc.M111.285601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah TM, Patel AK, Bhatt VD, Tripathi AK, Shah S, Shankar V, Joshi CG. The landscape of alternative splicing in buccal mucosa squamous cell carcinoma. Oral oncology. 2013 doi: 10.1016/j.oraloncology.2013.03.431. [DOI] [PubMed] [Google Scholar]
- Shi W, Hendrix D, Levine M, Haley B. A distinct class of small RNAs arises from pre-miRNA-proximal regions in a simple chordate. Nature structural & molecular biology. 2009;16:183–9. doi: 10.1038/nsmb.1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siomi MC, Sato K, Pezic D, Aravin AA. PIWI-interacting small RNAs: the vanguard of genome defence. Nat Rev Mol Cell Biol. 2011;12:246–58. doi: 10.1038/nrm3089. [DOI] [PubMed] [Google Scholar]
- Song R, Hennig GW, Wu Q, Jose C, Zheng H, Yan W. Male germ cells express abundant endogenous siRNAs. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:13159–64. doi: 10.1073/pnas.1108567108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St Laurent G, Shtokalo D, Tackett MR, Yang Z, Eremina T, Wahlestedt C, Urcuqui-Inchima S, Seilheimer B, McCaffrey TA, Kapranov P. Intronic RNAs constitute the major fraction of the non-coding RNA in mammalian cells. BMC genomics. 2012;13:504. doi: 10.1186/1471-2164-13-504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein AJ, Fuchs G, Fu C, Wolin SL, Reinisch KM. Structural insights into RNA quality control: the Ro autoantigen binds misfolded RNAs via its central cavity. Cell. 2005;121:529–39. doi: 10.1016/j.cell.2005.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun K, Chen X, Jiang P, Song X, Wang H, Sun H. iSeeRNA: identification of long intergenic non-coding RNA transcripts from transcriptome sequencing data. BMC genomics. 2013;14(Suppl 2):S7. doi: 10.1186/1471-2164-14-S2-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taft RJ, Simons C, Nahkuri S, Oey H, Korbie DJ, Mercer TR, Holst J, Ritchie W, Wong JJ, Rasko JE, Rokhsar DS, Degnan BM, Mattick JS. Nuclear-localized tiny RNAs are associated with transcription initiation and splice sites in metazoans. Nature structural & molecular biology. 2010;17:1030–4. doi: 10.1038/nsmb.1841. [DOI] [PubMed] [Google Scholar]
- Tahira AC, Kubrusly MS, Faria MF, Dazzani B, Fonseca RS, Maracaja-Coutinho V, Verjovski-Almeida S, Machado MC, Reis EM. Long noncoding intronic RNAs are differentially expressed in primary and metastatic pancreatic cancer. Molecular cancer. 2011;10:141. doi: 10.1186/1476-4598-10-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam OH, Aravin AA, Stein P, Girard A, Murchison EP, Cheloufi S, Hodges E, Anger M, Sachidanandam R, Schultz RM, Hannon GJ. Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nature. 2008;453:534–8. doi: 10.1038/nature06904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H, Wu Z, Zhang J, Su B. Salivary lncRNA as a potential marker for oral squamous cell carcinoma diagnosis. Molecular medicine reports. 2012 doi: 10.3892/mmr.2012.1254. [DOI] [PubMed] [Google Scholar]
- Terns MP, Terns RM. Small nucleolar RNAs: versatile trans-acting molecules of ancient evolutionary origin. Gene Expr. 2002;10:17–39. [PMC free article] [PubMed] [Google Scholar]
- Teunissen SW, Kruithof MJ, Farris AD, Harley JB, Venrooij WJ, Pruijn GJ. Conserved features of Y RNAs: a comparison of experimentally derived secondary structures. Nucleic acids research. 2000;28:610–9. doi: 10.1093/nar/28.2.610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tufarelli C, Stanley JA, Garrick D, Sharpe JA, Ayyub H, Wood WG, Higgs DR. Transcription of antisense RNA leading to gene silencing and methylation as a novel cause of human genetic disease. Nature genetics. 2003;34:157–65. doi: 10.1038/ng1157. [DOI] [PubMed] [Google Scholar]
- Umbach JL, Strelow LI, Wong SW, Cullen BR. Analysis of rhesus rhadinovirus microRNAs expressed in virus-induced tumors from infected rhesus macaques. Virology. 2010;405:592–9. doi: 10.1016/j.virol.2010.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venema J, Tollervey D. Ribosome synthesis in Saccharomyces cerevisiae. Annu Rev Genet. 1999;33:261–311. doi: 10.1146/annurev.genet.33.1.261. [DOI] [PubMed] [Google Scholar]
- Verhagen AP, Pruijn GJ. Are the Ro RNP-associated Y RNAs concealing microRNAs? Y RNA-derived miRNAs may be involved in autoimmunity. BioEssays : news and reviews in molecular, cellular and developmental biology. 2011;33:674–82. doi: 10.1002/bies.201100048. [DOI] [PubMed] [Google Scholar]
- Watanabe T, Totoki Y, Toyoda A, Kaneda M, Kuramochi-Miyagawa S, Obata Y, Chiba H, Kohara Y, Kono T, Nakano T, Surani MA, Sakaki Y, Sasaki H. Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature. 2008;453:539–43. doi: 10.1038/nature06908. [DOI] [PubMed] [Google Scholar]
- Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–62. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- Wright MW, Bruford EA. Naming ‘junk’: human non-protein coding RNA (ncRNA) gene nomenclature. Human genomics. 2011;5:90–8. doi: 10.1186/1479-7364-5-2-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia J, Joyce CE, Bowcock AM, Zhang W. Noncanonical microRNAs and endogenous siRNAs in normal and psoriatic human skin. Human molecular genetics. 2013;22:737–48. doi: 10.1093/hmg/dds481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z, Pestell RG. Small non-coding RNAs govern mammary gland tumorigenesis. Journal of mammary gland biology and neoplasia. 2012;17:59–64. doi: 10.1007/s10911-012-9246-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu XS, Liu QR, Wei L. Genome-wide in silico identification and analysis of cis natural antisense transcripts (cis-NATs) in ten species. Nucleic acids research. 2006;34:3465–75. doi: 10.1093/nar/gkl473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Affymetrix ENCODE Transcriptome Project CSHLETP Post-transcriptional processing generates a diversity of 5′-modified long and short RNAs. Nature. 2009;457:1028–32. doi: 10.1038/nature07759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrieri C, Cimatti L, Biagioli M, Beugnet A, Zucchelli S, Fedele S, Pesce E, Ferrer I, Collavin L, Santoro C, Forrest AR, Carninci P, Biffo S, Stupka E, Gustincich S. Long non-coding antisense RNA controls Uchl1 translation through an embedded SINEB2 repeat. Nature. 2012;491:454–7. doi: 10.1038/nature11508. [DOI] [PubMed] [Google Scholar]
- Cavaille J, Bachellerie JP. SnoRNA-guided ribose methylation of rRNA: structural features of the guide RNA duplex influencing the extent of the reaction. Nucleic acids research. 1998;26:1576–87. doi: 10.1093/nar/26.7.1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong C, Maquat LE. lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3′ UTRs via Alu elements. Nature. 2011;470:284–8. doi: 10.1038/nature09701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, Thomas K, Presser A, Bernstein BE, van Oudenaarden A, Regev A, Lander ES, Rinn JL. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:11667–72. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009;10:126–39. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- Kishore S, Khanna A, Zhang Z, Hui J, Balwierz PJ, Stefan M, Beach C, Nicholls RD, Zavolan M, Stamm S. The snoRNA MBII-52 (SNORD 115) is processed into smaller RNAs and regulates alternative splicing. Human molecular genetics. 2010;19:1153–64. doi: 10.1093/hmg/ddp585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lejeune E, Allshire RC. Common ground: small RNA programming and chromatin modifications. Current opinion in cell biology. 2011;23:258–65. doi: 10.1016/j.ceb.2011.03.005. [DOI] [PubMed] [Google Scholar]
- Mercer TR, Mattick JS. Structure and function of long noncoding RNAs in epigenetic regulation. Nature structural & molecular biology. 2013;20:300–7. doi: 10.1038/nsmb.2480. [DOI] [PubMed] [Google Scholar]
- Perreault J, Perreault JP, Boire G. Ro-associated Y RNAs in metazoans: evolution and diversification. Molecular biology and evolution. 2007;24:1678–89. doi: 10.1093/molbev/msm084. [DOI] [PubMed] [Google Scholar]
- Pillai RS, Chuma S. piRNAs and their involvement in male germline development in mice. Development, growth & differentiation. 2012;54:78–92. doi: 10.1111/j.1440-169X.2011.01320.x. [DOI] [PubMed] [Google Scholar]
- Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, Chang HY. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–23. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selvakumar T, Gjidoda A, Hovde SL, Henry RW. Regulation of human RNA polymerase III transcription by DNMT1 and DNMT3a DNA methyltransferases. The Journal of biological chemistry. 2012;287:7039–50. doi: 10.1074/jbc.M111.285601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi W, Hendrix D, Levine M, Haley B. A distinct class of small RNAs arises from pre-miRNA-proximal regions in a simple chordate. Nature structural & molecular biology. 2009;16:183–9. doi: 10.1038/nsmb.1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siomi MC, Sato K, Pezic D, Aravin AA. PIWI-interacting small RNAs: the vanguard of genome defence. Nat Rev Mol Cell Biol. 2011;12:246–58. doi: 10.1038/nrm3089. [DOI] [PubMed] [Google Scholar]
- Terns MP, Terns RM. Small nucleolar RNAs: versatile trans-acting molecules of ancient evolutionary origin. Gene Expr. 2002;10:17–39. [PMC free article] [PubMed] [Google Scholar]
- Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, Shi Y, Segal E, Chang HY. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–93. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umbach JL, Strelow LI, Wong SW, Cullen BR. Analysis of rhesus rhadinovirus microRNAs expressed in virus-induced tumors from infected rhesus macaques. Virology. 2010;405:592–9. doi: 10.1016/j.virol.2010.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhagen AP, Pruijn GJ. Are the Ro RNP-associated Y RNAs concealing microRNAs? Y RNA-derived miRNAs may be involved in autoimmunity. BioEssays : news and reviews in molecular, cellular and developmental biology. 2011;33:674–82. doi: 10.1002/bies.201100048. [DOI] [PubMed] [Google Scholar]
- West S. The increasing functional repertoire of U1 snRNA. Biochemical Society transactions. 2012;40:846–9. doi: 10.1042/BST20120058. [DOI] [PubMed] [Google Scholar]
- Willingham AT, Orth AP, Batalov S, Peters EC, Wen BG, Aza-Blanc P, Hogenesch JB, Schultz PG. A strategy for probing the function of noncoding RNAs finds a repressor of NFAT. Science. 2005;309:1570–3. doi: 10.1126/science.1115901. [DOI] [PubMed] [Google Scholar]
- Xia J, Joyce CE, Bowcock AM, Zhang W. Noncanonical microRNAs and endogenous siRNAs in normal and psoriatic human skin. Human molecular genetics. 2013;22:737–48. doi: 10.1093/hmg/dds481. [DOI] [PMC free article] [PubMed] [Google Scholar]