Abstract
Ibalizumab is a humanized monoclonal antibody that binds human CD4—a key receptor for HIV—and blocks HIV-1 infection. However, HIV-1 strains with mutations resulting in loss of an N-linked glycan from the V5 loop of the envelope protein gp120 are resistant to ibalizumab. Previous structural analysis suggests that this glycan fills a void between the gp120 V5 loop and the ibalizumab L chain, perhaps causing steric hindrance that disrupts viral entry. If this void contributes to HIV-1 resistance to ibalizumab, we reasoned that ‘refilling’ it by engineering an N-linked glycan into the ibalizumab L chain at a position spatially proximal to gp120 V5 may restore susceptibility to ibalizumab. Indeed, one such ibalizumab variant neutralized 100% of 118 tested diverse HIV-1 strains in vitro, including ten strains resistant to parental ibalizumab. These findings demonstrate that the strategic placement of a glycan in the variable region of a monoclonal antibody can substantially enhance its activity.
INTRODUCTION
The human immunodeficiency virus type 1 (HIV-1) epidemic continues to spread at an alarming rate of approximately 2.5 million new cases per year, and a safe and effective HIV vaccine is still elusive. Currently used strategies to fight the spread of HIV-1 include pre-exposure prophylaxis (PrEP) using small-molecule antiretroviral drugs1-3 and virus-inhibiting immunoglobulins4. However, compared to antiviral drugs, HIV-1 neutralizing antibodies tend to have better in vivo pharmacokinetic profiles in humans. HIV-1-neutralizing antibodies have shown efficacy in several animal models. For example, passive administration of anti-envelope (gp120 or anti-gp41) monoclonal antibodies (mAb) such as b12, 2G12, 2F5, and 4E10 protects rhesus macaques against challenge with simian-human immunodeficiency virus (SHIV)5, 6. However, mAb-based passive immunization therapy was considered infeasible for a long time due to the relatively weak potency and/or narrow breadth of the available HIV-1-neutralizing mAbs. However, recently identified human anti-HIV mAbs including VRC017, PG98, 3BNC1179, PGT antibodies10, 11, NIH45-46G54W12, and 10E813 with much greater breadth and potency increase enthusiasm about the prospect of using mAb for PrEP or passive immunization. Indeed, compared to first-generation HIV-1-neutralizing mAb, much lower concentrations of one such next-generation antibody protected of monkeys from virus challenge11. In addition, AAV-based expression of VRC01 in a humanized mice model led to successful prophylaxis against HIV-1 infection14. Nevertheless, with the exception of 10E8, most of these next-generation mAbs only neutralize around 70% to 90% of circulating HIV-1 strains, even at concentrations as high as 50 μg/mL.
PrEP strategies may also use mAbs specific for the HIV-1 receptors CCR515 and CD416-19, as such mAbs also show potent and broad inhibitory activity against HIV-1. For example, ibalizumab (formerly TNX-355) is a humanized IgG4 mAb that blocks HIV-1 entry by binding to human CD4 with high affinity17-21. Ibalizumab inhibits entry of a diverse spectrum of clinical and laboratory-adapted HIV-1 isolates, including CCR5-tropic and CXCR4-tropic strains from multiple subtypes. Mutagenesis22 and structural studies23 demonstrated that ibalizumab binds CD4 mainly by direct contacts with the BC-loop (AA 121-125) in domain 2 (D2) of CD4. Additional contacts include those between residues 164-165 (the short FG loop in D2) of CD4 and the ibalizumab H chain, as well as between the Ser79 and Glu77 (in the EF loop in D1) of CD4 and the ibalizumab L chain. Located at the interface between D1 and D2 of CD4, the ibalizumab epitope is positioned on the opposite side from the region of CD4 that engages HIV-1 gp120 or major histocompatibility complex class II (MHCII) (Fig. 1). Consistent with these findings, ibalizumab does not inhibit binding of CD4 to monomeric gp12016. Thus, ibalizumab is thought to inhibit a post–HIV-1 attachment step required for virus entry. In Phase 1, Phase 2a, and Phase 2b clinical trials in HIV-1 patients, ibalizumab treatment resulted in substantial reductions (~1 log) in viral load and significant increases in CD4+ T-cell counts, without serious immunologic impairments or adverse effects17, 19. Ibalizumab is now awaiting a Phase 3 clinical trial to examine its efficacy in HIV-1 patients with multi-drug resistant viruses in need of salvage antiretroviral therapy. We are also exploring the feasibility of using ibalizumab and ibalizumab variants for the purpose of HIV-1 prevention.
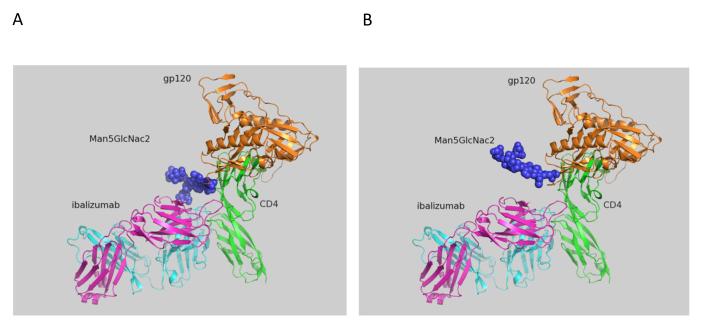
Figure 1.
Model of glycosylation in V5 of HIV-1 gp120, in the context of both CD4 and ibalizumab (using PyMOL). The complex was modeled by superimposing the structure of D1 and D2 of CD4 in complex with gp120 (Protein Data Bank accession number 2NXY) onto the same domains of CD4 in complex with ibalizumab (PDB 3O2D). The glycan (blue) was introduced at the relevant asparagine by superimposing the asparagine with that of a glycan-bound asparagine from PDB 3TYG. The heavy and L chains of ibalizumab are shown as cyan and magenta ribbons, respectively. The first two domains of human CD4 are green, while HIV-1 gp120 is tan. (A) Man5GlcNac2 at the position 459 of gp120 in the V5 loop (N-terminal). (B) Man5GlcNac2 at the position of 463 of gp120 in the V5 loop (C-terminal).
Unfortunately, HIV-1 strains with reduced susceptibility to ibalizumab (in terms of ibalizumab effects on virus infectivity) were isolated from HIV-1 patients who experienced a rebound in viral load after the addition of ibalizumab to failing antiretroviral drug regimens24. In most of these cases, a much reduced plateau of maximum percentage of inhibition (MPI) in the dose-response curve was observed17,24. In other words, complete virus inhibition cannot be achieved despite increasing antibody concentrations. Such flattening of the virus-inhibition curve is characteristic of the resistance profile for a noncompetitive inhibitor of HIV-1 entry. Further evaluation of ibalizumab-resistant viruses indicated that reduced susceptibility to ibalizumab is conferred by the loss of one or two potential N-linked glycosylation sites (PNGS) in the V5 loop of HIV-1 gp12024. Indeed, among unselected wild-type HIV-1 isolates, there is a strong correlation between the number of V5 glycosylation sites and ibalizumab susceptibility25. Moreover, site-directed mutagenesis confirmed that the loss of glycan at the V5 N-terminus was the major contributor to ibalizumab resistance25.
Here, based on structural modeling we propose that a glycan on the N-terminus of gp120 fills a vacant space between the L chain of ibalizumab and gp120 V5, and that ibalizumab’s effect on HIV-1 entry is sterically mediated by the mass effect of this glycan. We confirm this hypothesis by creating a panel of ibalizumab mutants with PNGS at various positions in the L chain and measuring their ability to neutralize HIV-1 infectivity in vitro. Indeed, ibalizumab mutants bearing a glycan located in close proximity to V5 in the ibalizumab-CD4-gp120 complex efficiently neutralized HIV-1. These ibalizumab mutants also neutralized HIV-1 strains that are resistant to wild-type ibalizumab. One particular ibalizumab mutant, LM52, neutralized 100% of the tested 118 HIV-1 isolates at a potency more than 10-fold higher than wild-type ibalizumab judging by the geometric mean IC80, the antibody concentration required to neutralize 80% of infection. The results of this study suggest that ibalizumab blocks HIV-1 entry through a steric hindrance mechanism, and provide an example of how a strategic placement of an N-linked glycosylation site can be used to improve the activity of a mAb. In addition, to our knowledge there is no previous report of a monospecific mAb of natural architecture reported to neutralize the infectivity of 100% of the HIV-1 strains tested.
RESULTS
Design of ibalizumab variants
Studies of ibalizumab-resistant HIV-1 strains revealed that resistance is mainly conferred by the loss of glycan(s) from the V5 loop of HIV-1 Env gp12024, 25. To further explore the role of V5 glycosylation in ibalizumab susceptibility, we modeled the interactions between gp120, CD4 and ibalizumab using the structures reported to the Protein Data Bank (accession number 2NXY and 3O2D). The V5 N-terminal glycan is situated closest to the ibalizumab L chain (Fig. 1A), while the V5 C-terminal glycan is further away from ibalizumab (Fig. 1B). This model raises the possibility that the fit of the N-terminal glycan into the space between gp120 and ibalizumab exerts a mass effect on gp120, thereby disrupting its conformational changes (twists and turns) that are essential for HIV-1 entry into the target cell. This model also suggests that introduction of a similarly sized glycan into the ibalizumab L chain may boost its ability to inhibit entry of HIV-1 strains that have lost their V5 N-terminal glycan.
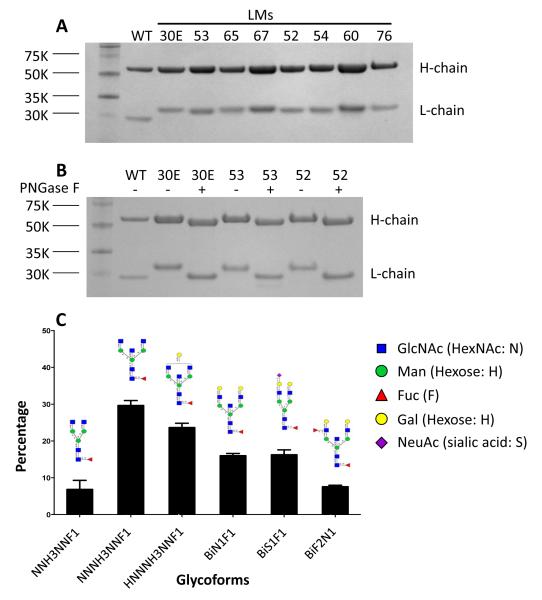
We therefore introduced PNGS into residues (30E, 52, 53, 54, 60, 65, 67, and 76) in the variable region of the ibalizumab L chain that are proximal to the V5 loop of gp120. These ibalizumab L chain mutants (LMs) were constructed, sequenced, and transfected into 293A cells to produce mutant proteins. We purified the variant mAbs by protein-A agarose chromatography, and analyzed them by SDS-PAGE (Fig. 2A). Yields of LMs from transient expression in 293A cells were comparable to those of wild-type ibalizumab (data not shown). A slightly bigger L chain was observed in each of the LMs compared to the wild-type L chain. To confirm that the larger sizes are due to glycosylation, LMs 30E, 52, and 53 were treated with PNGase F under denaturing conditions and analyzed by SDS-PAGE (Fig. 2B). As expected, the sizes of the L chains of these variants post-PNGase F treatment were decreased to the size of the wild-type. We next used mass spectrometry to identify the form of N-glycans introduced to the L chain of LM52 routinely produced in 293A cells. Over six forms of complex type N-glycans with 8-12 rings were identified on the L chain, with 80% of these forms having 9-11 rings (Fig. 2C). Taken together, these data confirmed that N-linked glycans were introduced into the desired residues of the ibalizumab L chain.
Figure 2.
N-linked glycosylation in the L chain of ibalizumab. (A) Ibalizumab L chain mutants (LMs) were constructed, co-transfected into 293A cells with the WT ibalizumab H chain plasmid, purified on a protein-A agarose column, and analyzed by SDS-PAGE. WT ibalizumab was analyzed the same way. (B) Purified WT, LM30E, LM53, and LM52 antibodies were treated with or without PNGase F at denaturing conditions and analyzed by SDS-PAGE. (C) N-linked glycoforms on the L chain of LM52 produced in 293A cells were analyzed by mass spectrometry.
CD4 binding and HIV-1 neutralization by LMs
We next evaluated the kinetics of binding of these ibalizumab LMs to human soluble CD4 (sCD4) by surface plasmon resonance. In a Biacore assay using sCD4 as the analyte, WT ibalizumab and LMs bound sCD4 with similar binding kinetics. The KD of six of these LMs bound to sCD4 was in the range of 0.19 nM to 0.8 nM (Table S1), and these numbers were within 2-fold of the KD of wild-type ibalizumab (0.43 nM). These data showed that the addition of an N-linked glycan at these select locations in the L chain of ibalizumab did not markedly affect its ability to bind CD4.
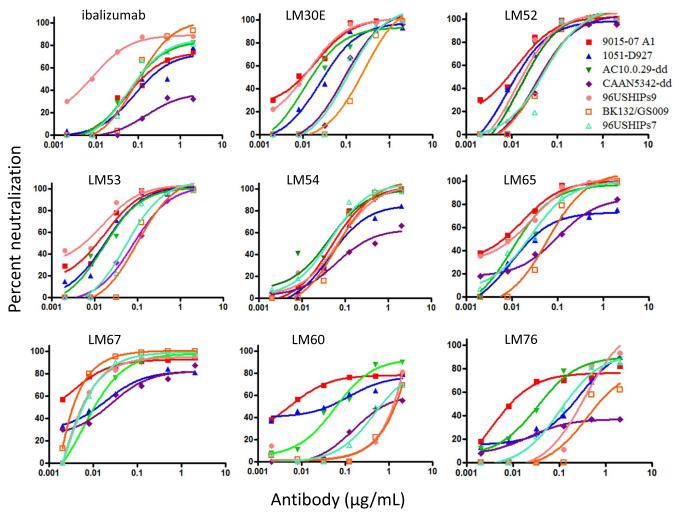
We next explored the HIV-1 neutralizing capacity of the LMs compared to WT ibalizumab. To this end, we tested the LMs against a panel of HIV-1 viruses that are resistant or partially resistant to ibalizumab, including 3 replication-competent HIV-1 strains (Fig. 3). Using this panel of ibalizumab-resistant viruses, an average MPI of 75% and IC80 of 1.43 μg/mL were observed when WT ibalizumab was tested up to 2 μg/mL. Markedly improved neutralization activities were observed for four LMs (LM30E, LM52, LM53, and LM67), with an average MPI of 91-99% and IC80 of 0.05-0.14 μg/mL (Fig. 3 and Table S2). Among these, LM52 appears to have the best HIV-1 neutralization potency. Two other variants (LM54 and LM65) yielded more modest improvement in virus-neutralization activities compared to ibalizumab. Interestingly, the PNGS in the six LMs that showed improved HIV-1 neutralizing activity are also closer to V5 (459) of gp120 (Table S2 and Figure S1) than the PNGS in LM60 and LM76, which showed HIV-1 neutralizing activity comparable to those of WT ibalizumab. Therefore, it seems that positioning the N-glycan closer to V5 improves the neutralization profile of ibalizumab variants. We note, however, that the orientation of the glycan, in addition to the distance to V5, may also play a critical role.
Figure 3.
Neutralization activities of WT ibalizumab and its LMs. Neutralization against a panel of ibalizumab-resistant or partially ibalizumab-resistant pseudovirus or replication-competent HIV-1 strains was measured by TZM-bl assay. 96USHIPs9, BK132/GS009, and 96USHIPs7 are replication-competent. CAAN5342.A2-dd and AC10.0.29-dd are site-directed Env mutants without any PNGS in V5 and are resistant or partially resistant to neutralization by wild-type ibalizumab. 9015-07 A1 and 1051-D927 are clade B transmitted founder viruses. The data represent three independent experiments.
Glycan size influences LM52 activity
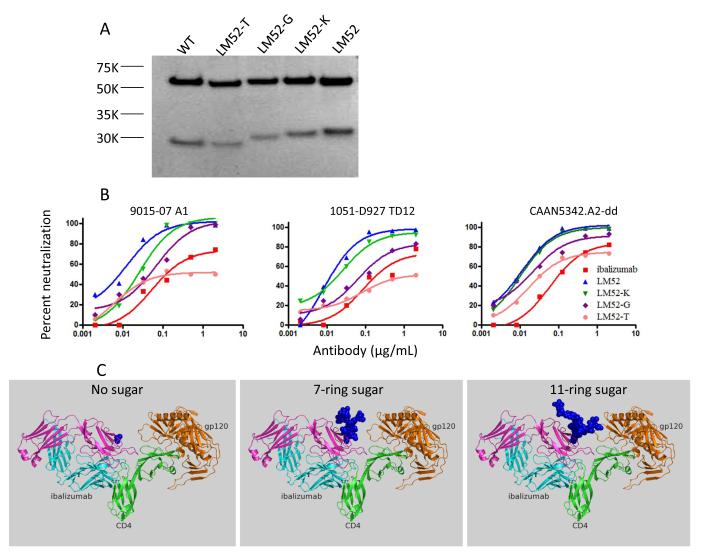
We next studied whether glycan size influences the improved HIV-1 neutralization activity of the LMs. We focused on LM52 because of its superior HIV-1 neutralization profile (Fig. 3). To produce a version of LM52 lacking any N-linked glycan, we added tunicamycin26, which inhibits GlcNAc phosphotransferase, to 293A cells immediately after transient transfection with the LM52 construct. Similarly, to produce a version of LM52 tagged with glycans of 10-11 rings (the high Man-type N-glycan Man8GlcNAc2 (Man8) or Man9GlcNAc2 (Man9)) we added kifunensine27 to 293A cells. Lastly, to produce a version of LM52 bearing a 7-ring glycan (high mannose (Man)-type N-glycan Man5GlcNAc2 (Man5)), we used N-acetylglucosaminyltransferase I-negative GnT1(−) HEK293S cells28. As shown by SDS-PAGE, although the addition of kifunensine did not noticeably change the size of the L chain, addition of tunicamycin or growth in GnT1(−) cells noticeably reduced the size of the L chain (Fig. 4a). Similarly, the neutralization activities against three HIV-1 strains were more severely reduced for antibodies produced in the presence of tunicamycin or in GnT1(−) cells than for the antibody produced in the presence of kifunensine (Fig. 4B). Thus, stronger HIV-1-neutralizing activity was observed when larger glycans were present in the L chain at residue 52. Modeling suggests that larger glycans better fill the space between the L chain and gp120 (Fig. 4C), but the nature of the branching of the glycan may also affect the HIV-1 neutralizing activity of LM52.
Figure 4.
The influence of glycan size on the HIV-1 neutralization activity of LM52. (A) LM52 was produced in HEK293A cells with or without tunicamycin (LM52-T) or kifunensine (LM52-K). Alternatively, LM52 was produced in the N-acetylglucosaminyltransferase I-negative GnT1(−) HEK293S cells (LM52-G). The purified LM52 proteins, together with unmodified LM52 and WT ibalizumab, were analyzed by SDS-PAGE. (B) Neutralization activities of ibalizumab and different glycan variants of LM52 against three ibalizumab-resistant pseudoviruses, as measured in in TZM-bl cells. (C) Depiction (using PyMOL) of the space filled by glycans of representative conformations and sizes, when tagged on residue 52 of ibalizumab. Depiction is based on the model generated in Figure 1 and colors are the same. The 7-ring N-glycan, Man5GlcNac2, was extracted from PDB entry 3TYG. An 11-ring N-glycan, Man3GlcNac5Fuc, was extracted from PDB entry 3QUM. These results represent three independent experiments.
Neutralization breadth and potency of LM52
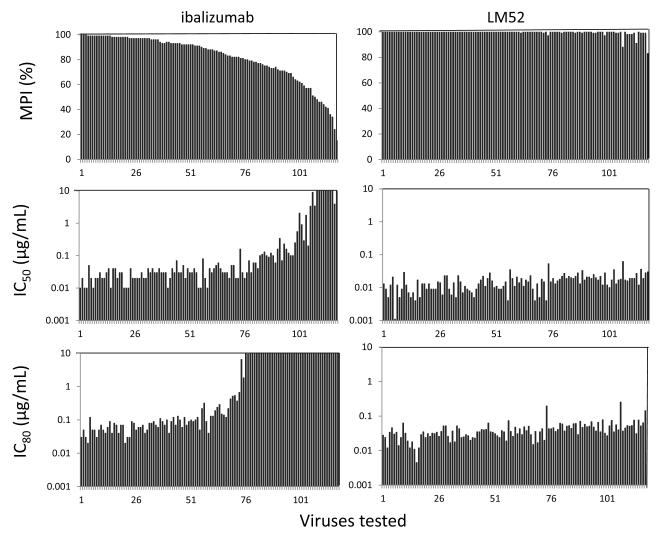
We then tested the HIV-1 neutralization activity of LM52 against a panel of 118 diverse HIV-1 viral strains covering 11 clades. LM52 showed significantly improved neutralization breadth and potency compared to WT ibalizumab in this single-cycle TZM-bl assay (Figure 5 and Table S3). LM52 had no neutralizing activity against the negative control virus (murine leukemia virus) (Table S3), but it neutralized (as defined by ≥50% inhibition) all HIV-1 strains tested, compared to 92% of strains for WT ibalizumab. In fact, LM52 neutralized 97% of viruses to ≥95% inhibition, compared to only 31% for WT ibalizumab (Fig. 5, upper panels). LM52 also exhibited IC50 values of <0.1 μg/mL for all 118 viruses, compared to 75% of viruses for WT ibalizumab (Fig. 5, middle panels). Indeed, all of the viruses were neutralized by LM52 with an IC80 <0.3 μg/mL, whereas 36% of the viruses were not neutralized by 80% with WT ibalizumab even at a concentration of 10 μg/mL (Fig. 5, lower panels).
Figure 5.
Neutralization of a panel of 118 HIV-1 Env pseudoviruses. Neutralization by LM52 and WT ibalizumab was measured in a TZM-bl assay. For each virus, black bars indicate maximum percent inhibition (MPI) when tested at Ab concentrations up to 10 μg/mL, and the corresponding IC50 (μg/mL) or IC80 (μg/mL). Viruses are ordered by descending MPI for ibalizumab. Given the large of number of viruses being tested, this experiment was done only once.
Overall, the geometric mean IC50 value for LM52 was 14 ng/mL, compared to 74 ng/mL for WT ibalizumab, and the geometric mean IC80 value for LM52 was 37 ng/mL, compared to 510 ng/mL for WT ibalizumab (Fig. S2). When the viruses were separated into ibalizumab-sensitive (MPI>80%) and ibalizumab-resistant strains (MPI≤80%), it was evident that the enhanced potency of LM52 was most obvious in ibalizumab-resistant viruses (Fig. S3). In ibalizumab-resistant viruses, the geometric mean IC80 value for LM52 was 50 ng/mL, compared to almost 10,000 ng/mL for WT ibalizumab. The potency of LM52 was enhanced by about 3-fold even in ibalizumab-sensitive viruses.
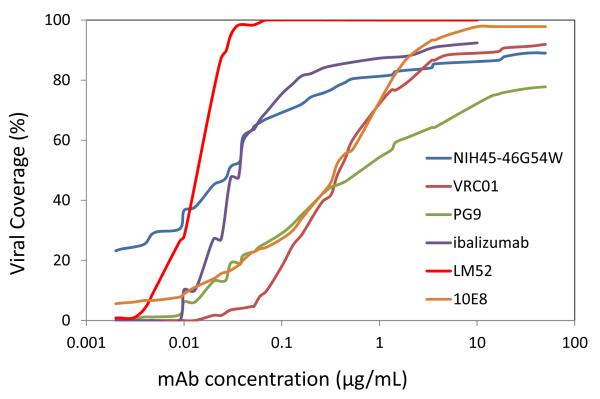
The improved activity of LM52 was also evident in a plot comparing its HIV-1 strain coverage across increasing antibody concentrations (Fig. 6). The improvement over WT ibalizumab was readily apparent, as was its superiority over mAbs (PG9, VRC01, 10E8, and NIH45-46G54W) known for their broad and potent HIV-1 neutralizing activity. LM52 had the greatest viral coverage at all concentrations >0.01 μg/mL and it reached 100% at a concentration less than 0.1 μg/mL. Only at concentrations below 0.01 μg/mL was LM52 coverage inferior to NIH45-46G54W12, a modified version of a human mAb directed to the CD4-binding site on gp120. It should be noted, however, that ibalizumab inhibits HIV-1 infection by binding to CD4, while VRC01, PG9, 10E8, and NIH45-46 G54W inhibit HIV-1 infection by binding directly to the virus. Nonetheless, regardless of whether these mAbs bind viral envelope or CD4, they all neutralize HIV-1 infection by blocking viral entry.
Figure 6.
HIV-1 strain coverage of LM52. Viral coverage of WT ibalizumab, LM52, and PG9, 10E8, VRC01, and NIH45-46G54W HIV-1 bnAb. LM52 and ibalizumab were tested up to 10 μg/mL, while the other monoclonal antibodies were tested up to 50 μg/mL. The data for PG9, 10E8, VRC01, and NIH45-46G54W were obtained from the published literature7, 8, 12, 13.
Effects of multiple glycans
We next examined the effect of placing two or three glycans in the region of interest in the ibalizumab L chain. We produced LM30E-52, LM30E-53, and LM52-67 double mutants, as well as LM30E-52-67 and LM30E-53-67 triple mutants in 293A cells and then purified and analyzed the antibodies by SDS-PAGE (Fig. S4A). Compared to WT ibalizumab or single mutant LMs, slightly bigger L chain was observed in each of the double mutants, and an even bigger L chain was observed in each of the triple mutants. We next explored the HIV-1 neutralization profile of these variant mAbs against a panel of HIV-1 strains known to be fully or partially resistant to ibalizumab. In general, the double and triple mutants showed neutralizing activity comparable to that of LM52 (Table S4; Fig. S4B left panel). In fact, the geometric mean IC80 values suggest that the triple mutants may be slightly less potent compared to LM52. The only exception noted was for SHIVsf162P3N, a virus more sensitive to the double and triple mutants than to LM52 (Table S4; Fig. S4B right panel). Overall, we found no evidence that adding more than one glycan further improves the HIV-1 neutralization profile of LM52 against ibalizumab-sensitive viruses. The improvement to LM52 by the addition of a second glycan may only be apparent with viruses resistant or partially resistant to LM52.
Analysis of LM52 polyreactivity
A property common to some HIV-1-neutralizing mAbs is their cross-reactivity with self-antigens29, 30. Neither LM52 nor ibalizumab bound to HEp-2 epithelial cell extracts (Fig. S5A) even at concentrations of 10 μg/mL. In addition, neither LM52 nor ibalizumab showed reactivity with single-stranded DNA, double-stranded DNA, insulin, lipopolysaccharide, or keyhole limpet hemocyanin (KLH) (Fig. S5B). Human CD4 protein, the intended target of ibalizumab, was the only self antigen tested that was recognized by LM52. Thus, we find no evidence that LM52 is polyreactive with self-antigens.
DISCUSSION
Here we created a superior HIV-1 entry-blocking mAb product in the ibalizumab variant LM52, which at relatively low concentrations could neutralize all tested HIV-1 strains tested. To our knowledge, no previous study has reported a mAb demonstrating 100% viral coverage against a large panel of HIV-1 isolates. The virus-neutralizing properties of LM52 are also evidently superior to those of the WT ibalizumab as well as of a number of the best anti-Env mAbs reported to date, for example VRC0131, PG98, 10E813, and NIH45-46G54W12. Together with the established safety record of the parental ibalizumab in humans, these observations suggest that LM52 may be a good candidate for clinical development for the treatment or prevention of HIV-1. This modified antibody may be particularly suitable as a long-acting PrEP agent given its expected (based on the known pharmacokinetic properties of the parental ibalizumab) schedule of monthly administration.
These findings also provide insight into the mechanism of action of ibalizumab. That HIV-1 loses a glycan in the N-terminus of gp120 V5 to become resistant to ibalizumab24, 25 already suggested the possibility that the ibalizumab mechanism of action is mediated by the glycan resulting in a steric clash between the antibody L chain and the viral envelope glycoprotein, thereby sterically disrupting a step in HIV-1 entry. Our observations indicating that a glycan on the L chain restores the antiviral activity of ibalizumab against ibalizumab-resistant strains, that positioning this glycan on key residues spatially closest to V5 resulted in greatest antiviral effect, and that larger glycans conferred larger effects on virus neutralization collectively support, but do not prove, that ibalizumab blocks HIV-1 infection via a steric hindrance mechanism.
This study also expands our knowledge about the influence of glycosylation on antibody activity. Under normal circumstances, an N-linked glycosylation site exists at amino acid 297 in the H chain of all IgG subclasses32, 33. The effect of this glycan has been studied extensively and found to be essential in mediating proinflammatory activity by maintaining the H chain in an open conformation required for FcγR binding. It has also been noted that Fc molecules with fucosylated and sialylated glycans have reduced affinities for FcRs compared with afucosylated or asialylated Fc molecules32, 34. In addition, approximately 20% of human IgGs have PNGS motifs within the variable region34, but the role of N-glycan in this region has not been thoroughly investigated. Although two studies reported that the introduction of an N-linked carbohydrate in the variable region resulted in improved solubility35, 36, no previous study has shown that a glycan strategically placed in the variable region of an antibody can markedly improve its activity.
MAbs have become a major class of drugs in our therapeutic arsenal, particularly for cancers and autoimmune disorders. While some antibody drugs mediate their clinical effect by acting as competitive inhibitors, others act by steric hindrance. For select mAbs in the latter category, the glycan-addition approach described herein may be adapted to enhance their functional activity. Guided by structural information on the antigen-antibody complex, it is conceivable that increasing the bulk at key positions on the antibody could lead to better activity. Our study provides an example of how structural-activity relationship (SAR) could be exploited to generate a superior mAb product. It also demonstrates the power of using SAR to develop improved biological products, as chemists have done for small-molecule drugs for decades.
METHODS
Cell lines, reagents, and pseudotyped viruses
TZM-bl cells (catalog no. 8129) were obtained through the AIDS Research and Reference Reagent Program (ARRRP), Division of AIDS, NIAID, NIH. This is a genetically engineered HeLa cell line that expresses CD4, CXCR4, and CCR5 and contains Tat-responsive reporter genes for luciferase and β-galactosidase under the control of an HIV-1 long terminal repeat. The Standard Reference Panels of Subtype B HIV-1 Env clones from acute and early infections and Env-deficient backbone plasmid (SG3ΔEnv) were also obtained through the NIH ARRRP. HIV-1 env pseudotyped viruses were prepared by co-transfection of 293A cells (Invitrogen) with an Env-expression plasmid and SG3ΔEnv. Recombinant sCD4 comprising the full-length extracellular domain of human CD4 was obtained from Progenics Pharmaceuticals, Inc. (Tarrytown, NY). Ibalizumab protein was provided by TaiMed Biologics (Irvine, CA). Plasmids pMV1 and pLC, which encode for ibalizumab H chain and L chain, respectively, were amplified from cDNA and cloned into pCDNA3.1 (+) (Invitrogen). N-acetylglucosaminyltransferase I-negative GnT1(−) human embryonic kidney (HEK) 293S cells were obtained from ATCC (catalog no. CRL-3022).
Addition of N-linked glycosylation sites to the ibalizumab L chain
The Asn-Ala-Thr (LM30E, LM53, LM54, LM60, LM65, LM67, and LM76) or Asn-Ser-Thr (LM52) sequences were introduced by mutagenesis to create ibalizumab L chain mutants. Mutagenesis was carried out with Quikchange mutagenesis kit (Stratagene, Santa Clara, VA). LM constructs were sequenced and transiently transfected into HEK293A cells (1:1 ratio of heavy and L chain plasmids) with polyethylenimine (PEI)-DNA complex. Supernatants were harvested on day 5 post transfection and LM proteins were purified with a protein-A agarose (Thermo Scientific, Rockford, IL) column. Ibalizumab and LM30E, LM52, and LM53 were treated with PNGase F (New England Biolabs, Ipswich, MA) under denaturing conditions, before analysis by SDS-PAGE.
Virus neutralization assay using TZM-bl cells
Neutralization assay was performed based on the method of Wei et al.37 with modifications38. Briefly, 10,000 cells per well were seeded in a 96-well plate in 100 μL/well of DMEM supplemented with 10% fetal bovine serum (D10) and incubated overnight. The next day, serially diluted ibalizumab or LM proteins were added to the cells and incubated for 1 h. Then, 200X 50%-tissue-culture-infective-doses (TCID50) of replication-competent or pseudotyped HIV-1 were prepared in D10 containing DEAE-Dextran (Sigma, St. Louis, MO) and added to the cells. The cells were incubated for 48 h and β-galactosidase activity was measured using the Galacto-Star System (Applied Biosystems, Cedarville, OH). The percentage of inhibition of viral infectivity was calculated as 1 minus the ratio of antibody-treated wells versus untreated-infected wells multiplied by 100. The IC50 and IC80 values (the antibody concentrations that confer 50% and 80% neutralization, respectively) were calculated by a non-linear regression analysis.
Surface plasmon resonance
Binding affinity analyses were performed with a Biacore T3000 optical biosensor (GE Healthcare, Piscataway, NJ). Immobilization of ibalizumab and all the glycan variants were performed following the standard amine coupling procedure. Briefly, carboxyl groups on the sensor chip surface were activated by injection of 35 μL of a solution containing 0.2 M N-(3-dimethylaminopropyl)-N-ethylcarbodiimide and 0.05 M Nhydroxysuccinimide at a flow rate of 5 μL/minute. Next, ibalizumab or its mutant variant, at a concentration of 2 μg/mL in 10 mM sodium-acetate buffer, pH 4.5, was allowed to flow over the chip surface at a rate of 10 μL/minute until the desired level of response units of reacted protein (150-200 RU) was achieved. After unreacted protein was washed out, excess active ester groups on the sensor surface were capped by the injection of 35 μL of 1 M ethanolamine, pH 8.0, at a flow rate of 5 μL/minute. As background to correct instrument and buffer artifacts, a reference was generated under the same conditions with omission of the protein ligand. Binding experiments were performed at 25°C in HBS-EP buffer (0.01 M HEPES, 0.15M NaCl, 3 mM EDTA, 0.005% vol/vol surfactant P20 (GE Healthcare). Binding kinetics were measured by passing various concentrations of analyte (human sCD4 protein) over the chip surface at a flow rate of 30 μL/minute for 3 min. Dissociation of bound analytes was monitored while the surface was washed for 10 min. Remaining analytes were removed at a flow rate of 50 μL/minute with two 30-sec injections of 10 mM glycine-HCl, pH 2.0. For kinetics data analysis, the kinetic parameters were determined by collectively fitting the overlaid sensograms locally using the BIAevaluation 4.1 software to the 1:1 Langmuir binding model.
Identification of N-linked glycosylation on LM52
Three micrograms of LM52 protein was dissolved in 50 mM ammonium bicarbonate (ABC)/50% tetrafluoroethylene and reduced by adding 40 mM dithiothreitol (DTT). After incubation at 65°C for 1 h, the protein samples were processed for alkylation by adding 40 mM iodoacetamide and incubating at room temperature for 1 h in the dark. The reaction was quenched by adding 40 mM DTT followed with 1h incubation. Then 25 mM ABC was added before trypsin digestion. Protein samples were then treated with 0.2 μg trypsin (Promega) for overnight. The digested protein samples were dried and re-dissolved with 20 μL of water before LC-MS/MS analysis. For the assignment for N-glycans on antibodies, the measured masses of trypsin-digested antibody were compared to a database that combined predicted tryptic peptides and N-linked glycans. The assigned glycopeptides were confirmed by the appearance of glycan fragments in MS/MS spectra. For PNGase digestion, LM52 was treated with PNGase F (New England Biolabs) overnight.
Statistical analyses
Differences in antibody potencies shown in Figures S2 and S3 were assessed by parametric (Students paired t-test) analyses of 50% and 80% inhibitory concentrations, using GraphPad Prism v5.03 software. Statistical significance was achieved if P ≤ 0.05.
Supplementary Material
Figure S1. Two views of locations of asparagine-linked glycan mutants in the L chain of ibalizumab in the context of glycosylation at residue 460 in V5 of HIV-1 gp120 (using PyMOL). The model and the color scheme are as in Figure 1. Mutants with PNGS at AAs 30E, 52, 53, 54, 65, and 67 (yellow) showed improved HIV-1 neutralizing activities, whereas mutants with PNGS at AAs 60 and 76 (white) showed HIV-1 neutralizing activity comparable to that of WT ibalizumab.
Figure S2. Summary of the neutralization activity of LM52 and ibalizumab against a diverse panel of 118 HIV-1 Env pseudoviruses, as reflected by their IC50 (μg/mL) and IC80 (μg/mL). Red lines show the geometric mean values and the 95% confidence interval. Each dot represents an individual viral strain.
Figure S3. The IC80 values of LM52 and ibalizumab against ibalizumab-sensitive or ibalizumab-resistant viruses (resistance defined as IC80 > 10 μg/mL).
Figure S4. (A) Sizes of H chain and L chain of ibalizumab and its single, double and triple glycan variants as analyzed by SDS-PAGE. (B) Neutralization activities of ibalizumab, LM52, and several double or triple glycan mutants against two ibalizumab-resistant pseudoviruses in TZM-bl cells.
Figure S5. Analysis of LM52 polyreactivity. (A) HEp-2 reactivity of LM52 and ibalizumab as measured by ELISA using Quanta Lite ANA ELISA kit (INOVA Diagnostics). Negative control is a human serum with no antibodies to nuclear and cytoplasmic antigens. Strong and weak positive controls are human sera known to have abundant and small quantities of antibodies to nuclear and cytoplasmic antigens. (B) LM52 and ibalizumab reactivity to single-stranded DNA, double-stranded DNA, insulin, lipopolysaccharide, KLH, and CD4, as measured by ELISA assay. These results were derived from three independent experiments.
ACKNOWLEDGEMENTS
The authors thank N. Padte for project management; Y. Huang, M. Tsuji, M-W. Chen, C. Andrews, M. Sun, and J. Yu for helpful discussions; and M.C. Nussenzweig for reagents. We also thank T-L. Hsu, P-C. Chen and the MS Core Facility at the Genomics Research Center, Academia Sinica (Taiwan), for glycoform profiling. Funding support for this study was provided by the Bill and Melinda Gates Foundation (OPP50714 and OPP1040731) via the Collaboration for AIDS Vaccine Discovery. Additional funding support was provided by National Institutes of Health (NIH) grant number 1DP1DA033263-01.
Footnotes
AUTHOR CONTRIBUTIONS R.S. and D.D.H. conceived the study and designed the experiments. R.S., D.F., and M.S.S. performed the experiments. D.A.O., D.F., and R.S. carried out the structural analyses. R.S. and D.D.H. analyzed the data and wrote the manuscript.
COMPETING FINANCIAL INTERESTS DDH is the scientific founder of TaiMed Biologics, Inc., which owns the commercial rights to ibalizumab. In this capacity, DDH has equity in the company. R.S and D.D.H. are inventors on a patent describing glycan-modified anti-CD4 antibodies for HIV prevention and therapy.
REFERENCE
- 1.Baeten JM, et al. Antiretroviral prophylaxis for HIV prevention in heterosexual men and women. The New England journal of medicine. 2012;367:399–410. doi: 10.1056/NEJMoa1108524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thigpen MC, et al. Antiretroviral preexposure prophylaxis for heterosexual HIV transmission in Botswana. The New England journal of medicine. 2013;367:423–434. doi: 10.1056/NEJMoa1110711. [DOI] [PubMed] [Google Scholar]
- 3.Van Damme L, et al. Preexposure prophylaxis for HIV infection among African women. The New England journal of medicine. 2012;367:411–422. doi: 10.1056/NEJMoa1202614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huber M, Olson WC, Trkola A. Antibodies for HIV treatment and prevention: window of opportunity? Current topics in microbiology and immunology. 2008;317:39–66. doi: 10.1007/978-3-540-72146-8_2. [DOI] [PubMed] [Google Scholar]
- 5.Baba TW, et al. Human neutralizing monoclonal antibodies of the IgG1 subtype protect against mucosal simian-human immunodeficiency virus infection. Nature medicine. 2000;6:200–206. doi: 10.1038/72309. [DOI] [PubMed] [Google Scholar]
- 6.Mascola JR, et al. Protection of Macaques against pathogenic simian/human immunodeficiency virus 89.6PD by passive transfer of neutralizing antibodies. Journal of virology. 1999;73:4009–4018. doi: 10.1128/jvi.73.5.4009-4018.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu X, et al. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science. 2010;329:856–861. doi: 10.1126/science.1187659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Walker LM, et al. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science. 2009;326:285–289. doi: 10.1126/science.1178746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scheid JF, et al. Sequence and structural convergence of broad and potent HIV antibodies that mimic CD4 binding. Science. 2011;333:1633–1637. doi: 10.1126/science.1207227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Walker LM, et al. Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature. 2011;477:466–470. doi: 10.1038/nature10373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moldt B, et al. Highly potent HIV-specific antibody neutralization in vitro translates into effective protection against mucosal SHIV challenge in vivo. Proc Natl Acad Sci U S A. 2012;109:18921–18925. doi: 10.1073/pnas.1214785109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Diskin R, et al. Increasing the potency and breadth of an HIV antibody by using structure-based rational design. Science. 2011;334:1289–1293. doi: 10.1126/science.1213782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang J, et al. Broad and potent neutralization of HIV-1 by a gp41-specific human antibody. Nature. 2012;491:406–412. doi: 10.1038/nature11544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Balazs AB, et al. Antibody-based protection against HIV infection by vectored immunoprophylaxis. Nature. 2011;481:81–84. doi: 10.1038/nature10660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jacobson JM, et al. Antiviral activity of single-dose PRO 140, a CCR5 monoclonal antibody, in HIV-infected adults. The Journal of infectious diseases. 2008;198:1345–1352. doi: 10.1086/592169. [DOI] [PubMed] [Google Scholar]
- 16.Burkly LC, et al. Inhibition of HIV infection by a novel CD4 domain 2-specific monoclonal antibody. Dissecting the basis for its inhibitory effect on HIV-induced cell fusion. J Immunol. 1992;149:1779–1787. [PubMed] [Google Scholar]
- 17.Jacobson JM, et al. Safety, pharmacokinetics, and antiretroviral activity of multiple doses of ibalizumab (formerly TNX-355), an anti-CD4 monoclonal antibody, in human immunodeficiency virus type 1-infected adults. Antimicrobial agents and chemotherapy. 2009;53:450–457. doi: 10.1128/AAC.00942-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dimitrov A. Ibalizumab, a CD4-specific mAb to inhibit HIV-1 infection. Curr Opin Investig Drugs. 2007;8:653–661. [PubMed] [Google Scholar]
- 19.Kuritzkes DR, et al. Antiretroviral activity of the anti-CD4 monoclonal antibody TNX-355 in patients infected with HIV type 1. The Journal of infectious diseases. 2004;189:286–291. doi: 10.1086/380802. [DOI] [PubMed] [Google Scholar]
- 20.Zhang XQ, Sorensen M, Fung M, Schooley RT. Synergistic in vitro antiretroviral activity of a humanized monoclonal anti-CD4 antibody (TNX-355) and enfuvirtide (T-20) Antimicrobial agents and chemotherapy. 2006;50:2231–2233. doi: 10.1128/AAC.00761-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boon L, et al. Development of anti-CD4 MAb hu5A8 for treatment of HIV-1 infection: preclinical assessment in non-human primates. Toxicology. 2002;172:191–203. doi: 10.1016/s0300-483x(02)00002-1. [DOI] [PubMed] [Google Scholar]
- 22.Song R, et al. Epitope mapping of ibalizumab, a humanized anti-CD4 monoclonal antibody with anti-HIV-1 activity in infected patients. Journal of virology. 2010;84:6935–6942. doi: 10.1128/JVI.00453-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Freeman MM, et al. Crystal structure of HIV-1 primary receptor CD4 in complex with a potent antiviral antibody. Structure. 2010;18:1632–1641. doi: 10.1016/j.str.2010.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Toma J, et al. Loss of asparagine-linked glycosylation sites in variable region 5 of human immunodeficiency virus type 1 envelope is associated with resistance to CD4 antibody ibalizumab. Journal of virology. 2011;85:3872–3880. doi: 10.1128/JVI.02237-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pace CS, et al. Anti-CD4 Monoclonal Antibody Ibalizumab Exhibits Breadth and Potency Against HIV-1, With Natural Resistance Mediated by the Loss of a V5 Glycan in Envelope. J Acquir Immune Defic Syndr. 2013;62:1–9. doi: 10.1097/QAI.0b013e3182732746. [DOI] [PubMed] [Google Scholar]
- 26.Olden K, Pratt RM, Yamada KM. Role of carbohydrates in protein secretion and turnover: effects of tunicamycin on the major cell surface glycoprotein of chick embryo fibroblasts. Cell. 1978;13:461–473. doi: 10.1016/0092-8674(78)90320-3. [DOI] [PubMed] [Google Scholar]
- 27.Vallee F, Karaveg K, Herscovics A, Moremen KW, Howell PL. Structural basis for catalysis and inhibition of N-glycan processing class I alpha 1,2-mannosidases. The Journal of biological chemistry. 2000;275:41287–41298. doi: 10.1074/jbc.M006927200. [DOI] [PubMed] [Google Scholar]
- 28.Reeves PJ, Callewaert N, Contreras R, Khorana HG. Structure and function in rhodopsin: high-level expression of rhodopsin with restricted and homogeneous N-glycosylation by a tetracycline-inducible N-acetylglucosaminyltransferase I-negative HEK293S stable mammalian cell line. Proc Natl Acad Sci U S A. 2002;99:13419–13424. doi: 10.1073/pnas.212519299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Haynes BF, et al. Cardiolipin polyspecific autoreactivity in two broadly neutralizing HIV-1 antibodies. Science. 2005;308:1906–1908. doi: 10.1126/science.1111781. [DOI] [PubMed] [Google Scholar]
- 30.Mouquet H, et al. Polyreactivity increases the apparent affinity of anti-HIV antibodies by heteroligation. Nature. 2010;467:591–595. doi: 10.1038/nature09385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu X, et al. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science. 329:856–861. doi: 10.1126/science.1187659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Arnold JN, Wormald MR, Sim RB, Rudd PM, Dwek RA. The impact of glycosylation on the biological function and structure of human immunoglobulins. Annual review of immunology. 2007;25:21–50. doi: 10.1146/annurev.immunol.25.022106.141702. [DOI] [PubMed] [Google Scholar]
- 33.Anthony RM, Ravetch JV. A novel role for the IgG Fc glycan: the anti-inflammatory activity of sialylated IgG Fcs. Journal of clinical immunology. 2010;30(Suppl 1):S9–14. doi: 10.1007/s10875-010-9405-6. [DOI] [PubMed] [Google Scholar]
- 34.Nimmerjahn F, Ravetch JV. Divergent immunoglobulin g subclass activity through selective Fc receptor binding. Science. 2005;310:1510–1512. doi: 10.1126/science.1118948. [DOI] [PubMed] [Google Scholar]
- 35.Pepinsky RB, et al. Improving the solubility of anti-LINGO-1 monoclonal antibody Li33 by isotype switching and targeted mutagenesis. Protein Sci. 2010;19:954–966. doi: 10.1002/pro.372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu SJ, et al. Structure-based engineering of a monoclonal antibody for improved solubility. Protein Eng Des Sel. 2010;23:643–651. doi: 10.1093/protein/gzq037. [DOI] [PubMed] [Google Scholar]
- 37.Wei X, et al. Antibody neutralization and escape by HIV-1. Nature. 2003;422:307–312. doi: 10.1038/nature01470. [DOI] [PubMed] [Google Scholar]
- 38.Seaman MS, et al. Standardized assessment of NAb responses elicited in rhesus monkeys immunized with single- or multi-clade HIV-1 envelope immunogens. Virology. 2007;367:175–186. doi: 10.1016/j.virol.2007.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Two views of locations of asparagine-linked glycan mutants in the L chain of ibalizumab in the context of glycosylation at residue 460 in V5 of HIV-1 gp120 (using PyMOL). The model and the color scheme are as in Figure 1. Mutants with PNGS at AAs 30E, 52, 53, 54, 65, and 67 (yellow) showed improved HIV-1 neutralizing activities, whereas mutants with PNGS at AAs 60 and 76 (white) showed HIV-1 neutralizing activity comparable to that of WT ibalizumab.
Figure S2. Summary of the neutralization activity of LM52 and ibalizumab against a diverse panel of 118 HIV-1 Env pseudoviruses, as reflected by their IC50 (μg/mL) and IC80 (μg/mL). Red lines show the geometric mean values and the 95% confidence interval. Each dot represents an individual viral strain.
Figure S3. The IC80 values of LM52 and ibalizumab against ibalizumab-sensitive or ibalizumab-resistant viruses (resistance defined as IC80 > 10 μg/mL).
Figure S4. (A) Sizes of H chain and L chain of ibalizumab and its single, double and triple glycan variants as analyzed by SDS-PAGE. (B) Neutralization activities of ibalizumab, LM52, and several double or triple glycan mutants against two ibalizumab-resistant pseudoviruses in TZM-bl cells.
Figure S5. Analysis of LM52 polyreactivity. (A) HEp-2 reactivity of LM52 and ibalizumab as measured by ELISA using Quanta Lite ANA ELISA kit (INOVA Diagnostics). Negative control is a human serum with no antibodies to nuclear and cytoplasmic antigens. Strong and weak positive controls are human sera known to have abundant and small quantities of antibodies to nuclear and cytoplasmic antigens. (B) LM52 and ibalizumab reactivity to single-stranded DNA, double-stranded DNA, insulin, lipopolysaccharide, KLH, and CD4, as measured by ELISA assay. These results were derived from three independent experiments.