SUMMARY
Phagophore maturation is a key step in the macroautophagy pathway, which is critical in many important physiological and pathological processes. Here we identified Drosophila N-ethylmaleimide-sensitive fusion protein 2 (dNSF2) and soluble NSF attachment protein (Snap) as strong genetic modifiers of toxicity caused by mutant CHMP2B, an ESCRT-III component that causes frontotemporal dementia and autophagosome accumulation. Among several SNARE genes, Drosophila syntaxin 13 (syx13) exhibited a strong genetic interaction with mutant CHMP2B. Knockdown of syntaxin 13 (STX13) or its binding partner Vti1a in mammalian cells caused LC3-positive puncta to accumulate and blocks autophagic flux. STX13 was present on LC3-positive phagophores induced by rapamycin and was highly enriched on multilamellar structures induced by dysfunctional ESCRT-III. Loss of STX13 also caused the accumulation of Atg5-positive puncta and the formation of multilamellar structures. These results suggest STX13 is a genetic modifier of ESCRT-III dysfunction and participates in the maturation of phagophores into closed autophagosomes.
Keywords: autophagy, CHMP2B, Drosophila, ESCRT, frontotemporal dementia, SNARE, syntaxin 13
INTRODUCTION
Autophagy (i.e., macroautophagy) is a cytoplasmic bulk degradation pathway involved in many important physiological and pathological processes, including neurodegeneration (Wong and Cuervo, 2010; Ihara et al., 2012; Metcalf et al., 2012). The autophagy pathway involves a series of membrane fusion events that require the function of SNARE proteins. For instance, mutations in Vam3p (a yeast protein homologous to the syntaxin family of t-SNARE molecules) or Vti1p disrupt the fusion of autophagosomes with vacuoles (Darsow et al., 1997; Ishihara et al., 2001). VAMP7, VAMP8, Vti1b, and other v-SNARE proteins are required for the fusion between autophagosomes and endosomes/lysosomes (Fader et al., 2009; Furuta et al., 2010; Itakura et al., 2012). Moreover, multiple SNARE proteins are involved in the homotypic fusion of precursor membranes to form autophagic isolation membranes (phagophores) (Moreau et al., 2011; Nair et al., 2011). Once the isolation membrane is generated, the distal ends must fuse to form a closed autophagosome around the material destined for degradation. The molecular events required for this maturation step are poorly understood.
Endosomal sorting complexes required for transport (ESCRTs) were identified as several cytosolic protein complexes that participate in the biogenesis of multivesicular bodies and were later shown to play a key role in cytokinesis, HIV budding, autophagy, and other cellular processes (Henne et al., 2011). Dysfunctional ESCRT machinery, caused by ectopic expression of a mutant CHMP2B associated with frontotemporal dementia (FTD) or by loss of function of some subunits, leads to a dramatic accumulation of autophagosomes (Filimonenko et al., 2007; Lee et al., 2007; Rusten et al., 2007). However, it is still unclear how ESCRTs are involved in the autophagy pathway. In particular, dysfunctional ESCRT-III results in the formation of large LC3- and CD63-positive multilamellar structures (Lee et al., 2007, 2009; Urwin et al., 2010). The nature and formation of these abnormal membrane structures remain poorly understood.
Previously, we identified chromosomal deletions that enhance mutant CHMP2B toxicity in Drosophila (Ahmad et al., 2009). In this study, focusing on one chromosomal deletion, we set out to identify the gene responsible for the enhancement phenotype and investigate its role in autophagosome maturation.
RESULTS
A Genetic Screen in Drosophila Identifies dNSF2 as a Strong Enhancer of Mutant CHMP2B Toxicity
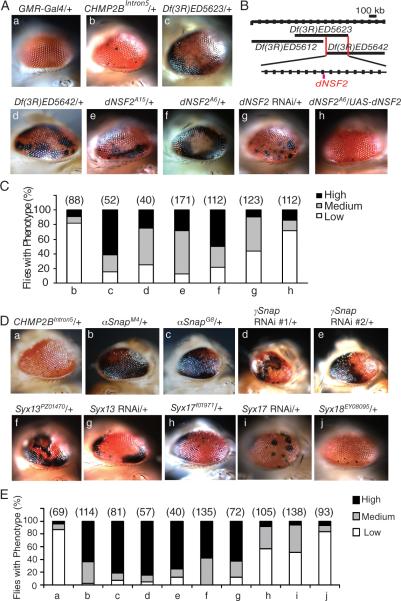
Previously, we established a fly model of FTD caused by the mutant ESCRT-III subunit CHMP2B (CHMP2BIntron5) (Ahmad et al., 2009). Using this model in an F1 deficiency genetic screen, we identified genetic modifiers whose partial loss of function enhanced the CHMP2BIntron5 eye phenotype (Ahmad et al., 2009). Expression of CHMP2BIntron5 in the eye by GMR-Gal4 (GMR-Intron5) leads to a modest phenotype with a few small melanin deposits in most flies (Ahmad et al., 2009 and Figure 1Ab). Df(3R)ED5623 heterozygosity greatly enhanced the eye phenotype of GMR-Intron5 flies (Figure 1Ac), suggesting that one or more genes in this deficiency region strongly modify CHMP2BIntron5 toxicity. A neighboring deficiency deletion, Df(3R)ED5642, also enhanced the phenotype, but a more distal deletion Df(3R)ED5612 did not (Figure 1Ad and 1B).
Figure 1. Identification of dNSF2 and Syx13 as Strong Genetic Modifiers of CHMP2BIntron5 Toxicity in Drosophila.
(A) Flies with mutant alleles of different genes in Df(3R)ED5623 were crossed with GMR-Intron5 flies to identify the gene responsible for the enhanced eye phenotype. All eye images are of 1-day old flies. a: Eyes expressing GMR-GAL4 have normal external morphology. b: UAS-CHMP2BIntron5 expression driven by GMR-Gal4 causes eye degeneration. c–h: All flies in these panels expressed GMR-GAL4 and UAS-CHMP2BIntron5. Genetic modifiers are listed above each panel.
(B) Schematic of the genomic region covered by deficiencies used in this study. The overlapping region between the two deficiencies is about 300 kb long and magnified with 20 kb divisions.
(C) Quantification of the eye phenotype in flies with different genotypes in panel A. Numbers in parentheses are numbers of flies of each genotype analyzed.
(D) Representative images of eye phenotypes in flies with different genotypes. All flies in a–j expressed UAS-CHMP2BIntron5 driven by GMR-Gal4. The modifier genes (e.g., Snap, Syx13, and Syx17) are listed above each image.
(E) Quantification of the eye phenotype in flies with different genotypes in panel D. Numbers in parentheses are numbers of flies of each genotype analyzed.
See also Tables S1 and S2.
The region of overlap between Df(3R)ED5623 and Df(3R)ED5642 contains 34 predicted protein-coding genes. To identify the gene(s) responsible for the genetic interaction, we tested all the available mutant lines that disrupt individual genes within this region. Two point mutations in the gene encoding the Drosophila NEM-sensitive fusion protein 2 (dNSF2, CG9931), dNSF2A6, and dNSF2A15 significantly enhanced the CHMP2BIntron5 phenotype (Figure 1Ae,f). Knockdown of dNSF2 by RNA interference (RNAi) produced a similar eye phenotype (Figure 1Ag). More importantly, the enhanced phenotype caused by the dNSF2A6 allele in GMR-Intron5 flies (Figure 1Ae) was rescued by ectopic expression of UAS-dNSF2 (Figure 1Ah). To exclude the possibility that this rescue was not due to the presence of an extra copy of the UAS element, we expressed UAS-GFP instead. It did not affect the genetic interaction between dNSF2A6 and CHMP2BIntron5 (data not shown). Thus, dNSF2 and CHMP2BIntron5 may regulate the same cellular pathway.
Syx13 Is a Strong Genetic Modifier of the CHMP2BIntron5 Eye Phenotype
Since NSF physically interacts with soluble trimeric NSF attachment protein (SNAP) in many membrane fusion events (Jahn and Scheller, 2006), we examined whether Snap is also a genetic modifier of CHMP2BIntron5 toxicity. Indeed, two mutant alleles of αSnap—αSnapM4 and αSnapG8—strongly enhanced the CHMP2BIntron5 eye phenotype (Figure 1Db,c), as did two RNAi lines specific for γSnap (Figure 1Dd,e).
SNAP proteins interact with SNAP receptors (SNAREs), an evolutionarily conserved protein superfamily with dozens of members involved in diverse vesicular fusions (Jahn and Scheller, 2006). To identify SNARE genes that genetically interact with CHMP2BIntron5 in Drosophila, we tested available identified SNARE mutant lines. Syntaxin13 (Syx13) strongly enhanced the CHMP2BIntron5 eye phenotype (Figure 1Df and Table S1), and the enhancement was confirmed by RNAi knockdown of Syx13 (Figure 1Dg). Other SNARE genes, such as Syx17 (Figure 1Dh,i), enhanced the phenotype to a lesser extent (Figure 1E); Syx18 (Figure 1Dj) and Syx8 (data not shown) had no effect.
Human syntaxin 12/13 (STX13) is highly homologous to Drosophila Syx13. To further test whether the genetic interaction between Syx13 and CHMP2BIntron5 is conserved in human cells, we cotransfected HEK293 cells with STX13 siRNA and CHMP2BIntron5. We screened for short, interfering RNAs (siRNAs) that could knock down the expression of STX13 (Figure S1A) and confirmed that siRNA #3 was very effective (Figure 2A), which we used here and for other experiments below. Knockdown of STX13 further increased the cellular toxicity caused by CHMP2BIntron5 expression (Figure S1B) or by knockdown of the ESCRT-III subunit Snf7-2 (Figure S1C) with Snf7-2-specific siRNA (Figure S1A) in HEK293 cells. Thus, STX13 and CHMP2BIntron5 seem to interact genetically to affect the same cellular pathway in human cells as well.
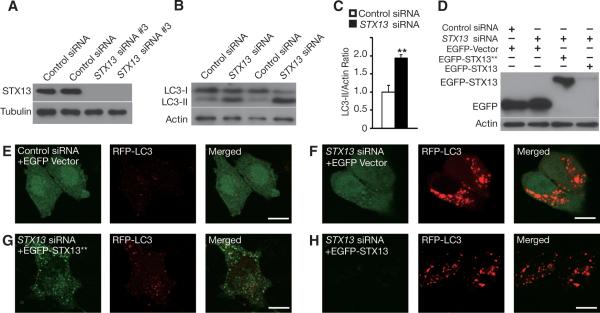
Figure 2. SiRNA Knockdown of STX13 Increases LC3-II Levels and Autophagosome Accumulation in Mammalian Cells.
(A) Western blot analysis confirmed the effectiveness of STX13 siRNA #3 in knocking down the expression of endogenous STX13 in HEK293 cells.
(B) SiRNA knockdown of STX13 significantly increased LC3-II level in HEK293 cells as shown by western blot analysis.
(C) Quantification of LC3-II/actin ratio. ** p < 0.01 by Student's t test. Values are mean ± SEM of three independent experiments.
(D) Western blot analysis confirmed that an siRNA-resistant STX13 (EGFP-STX13**) could be expressed in the presence of STX13 siRNA in HEK293 cells.
(E–H) RFP-LC3 was cotransfected into HeLa cells together with EGFP, EGFP-STX13**, or EGFP-STX13 as well as control siRNA or STX13 siRNA. STX13 knockdown (F) but not control siRNA (E) caused extensive accumulation of LC-3-positive puncta. The siRNA-resistant STX13 expression construct rescued the autophagy defects caused by siRNA knockdown of STX13 (G), but the normal STX13 expression construct failed to do so. Scale bar = 10 μm.
See also Figure S1 and Tables S2 and S4.
STX13 Is Involved in the Autophagy Pathway
Dysfunctional ESCRTs result in the accumulation of autophagosomes (Filimonenko et al., 2007; Lee et al., 2007; Rusten et al., 2007), so we examined whether STX13 is also involved in the autophagy pathway. Knockdown of STX13 with STX13 siRNA in HEK293 cells significantly increased the level of LC3-II relative to actin (Figure 2B,C), suggesting increased autophagy induction or a blockage in autophagic flux. Moreover, loss of STX13 led to extensive accumulation of LC3-positive puncta (Figure 2E, F). To confirm that this effect reflects the loss of STX13 and not other off-target effects of the siRNA, we used site-directed mutagenesis to generate an EGFP-STX13 construct that is insensitive to siRNA inhibition (EGFP-STX13**). This construct has four silent mutations in the coding region of STX13 that are targeted by STX13 siRNA; however, this siRNA still suppressed normal EGFP-STX13 expression (Figure 2D). Coexpression of EGFP-STX13** but not EGFP-STX13 prevented the massive accumulation of LC3-postive puncta in HeLa cells caused by STX13 siRNA (Figure 2G, H). These findings reveal a role for STX13 in the autophagy pathway and show that loss of its activity results in the accumulation of LC3-positive puncta.
STX13 Is Required for the Maturation of Early Autophagosomes
The increased accumulation of LC3-positive puncta could reflect increased induction of autophagy or defects in autophagic flux. To investigate whether STX13 directly affects the autophagy pathway, we examined whether STX13 localizes to LC3-II-positive puncta, in addition to its known localization on tubular early endosomes when the basal autophagy level is low (Prekeris et al., 1998). The expression patterns of endogenous and transfected STX13 were indistinguishable (Figure S1D). Thus, tagged STX13 was used in the following experiments. EGFP-tagged STX13 was localized in vesicular structures in DMSO-treated control HeLa cells, while endogenous LC3 was expressed in a mostly diffuse pattern (Figure 3A).
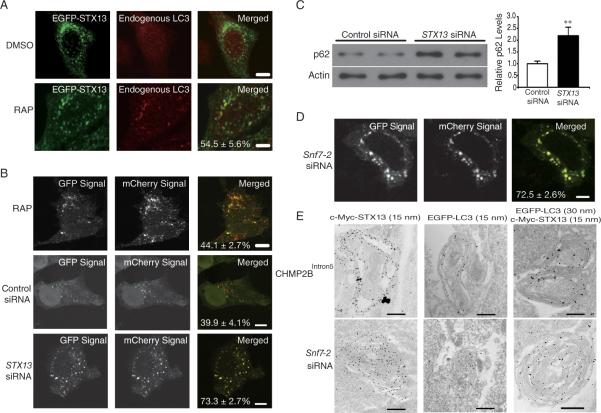
Figure 3. Depletion of STX13 Impairs Autophagic Flux in Mammalian Cells.
(A) EGFP-tagged STX13 was transfected into HeLa cells and LC3-positive puncta were induced by treatment with 1 μM rapamycin for 24 hr. The percentage of LC3-positive, STX13-positive puncta among all LC3-positive puncta is shown. Values are mean ± SEM. Bar = 10 μm.
(B) Double-tagged LC3 protein (mCherry-GFP-LC3) was used to indicate autophagic flux 48–72 hours after STX13 knockdown. The percentages of yellow dots (which maintain both the mCherry and the GFP signal) among total red dots are listed in each merged image. Values are mean ± SEM from three independent experiments. Bar = 10 μm.
(C) Western blot analysis showed that STX13 knockdown resulted in the accumulation of p62 in HEK293 cells. Values are mean ± SEM. ** p < 0.01 (Student's t test).
(D) SiRNA knockdown of Snf7-2 in HeLa cells caused a defect in autophagic flux. Bar = 10 μm.
(E) EGFP-LC3 and c-Myc-STX13 were transfected into HEK293 cells. After expression of CHMP2BIntron5 expression or siRNA knockdown of Snf7-2, C-Myc and EGFP antibodies were used for imuno-EM analysis. Bar = 0.5 μm.
See also Figures S1, S2, and S3, Tables S3 and S4.
In HeLa cells treated with 1 μM rapamycin to enhance induction of autophagy, about 55% of LC3-positive puncta are also positive for EGFP-STX13 (Figures 3A and S1E). Similarly, using c-Myc-tagged STX13 after autophagy induction, we found that a subset of vesicles was labeled by both GFP-LC3 and c-Myc-STX13 (Figure S1F). These results indicate that STX13 is localized on some LC3-positive puncta and that loss of STX13 activity results in autophagosome accumulation. Loss of STX13 did not seem to affect some aspects of general trafficking pathways, since transferrin uptake, as measured by fluorescence intensity (Figure S1G) and the quantity and maturation of the lysosomal protease cathepsin L (CathL) (Figure S1H), appeared to be normal. Transferrin uptake assays are often used to study early endocytosis (Mayle et al., 2012). CathL is produced as a pre-pro-protein in the endoplasmic reticulum that transits from the trans-Golgi network and is processed by proteolytic cleavage in the presence of decreasing pH to a mature form (of lower molecular weight) as it moves through the late endosome to the lysosome. Correct processing of CathL indicates normal endosomal function (Progida et al., 2010).
To further understand the precise role of STX13 in the autophagy pathway, we used LC3 with two fluorescent tags (mCherry-GFP-LC3) as an indicator of autophagic flux to differentiate early autophagosomes from amphisomes/autolysosomes in HeLa cells (Klionsky et al., 2012). In this assay, early autophagosomes were yellow and became red once they matured into autolysosomes, whose acidic environment quenches the GFP signal. In rapamycin-treated cells, the autophagy pathway was activated and more LC3-positive puncta became visible but autophagic flux was normal, as 44.1 ± 2.7% of the dots were yellow (and thus had lost the GFP signal) (Figure 3B). SiRNA knockdown of STX13 also blocked autophagic flux in HeLa cells (Figure 3B). With control siRNA, the yellow dot/total red dot ratio was 39.9 ± 4.1%, similar to that in rapamycin-treated cells. However, in STX13 siRNA-treated cells, the ratio increased to 73.3 ± 2.7% (p < 0.001). Furthermore, loss of STX13 activity resulted in the accumulation of p62, a substrate of autophagy (Figure 3C). Thus, STX13 is directly required for the maturation of autophagosomes, and defects caused by loss of STX13 occur before fusion with lysosomes. Consistent with this notion, ATG5 knockdown partially suppressed cellular toxicity caused by STX13 knockdown (Figure S1C), suggesting that the accumulation of immature autophagosomes that results from loss of STX13 activity is detrimental to cell survival, similar to the effects of dysfunctional ESCRT-III in rodent neurons (Lee et al., 2007; 2009).
STX13 Is Required for Maturation of Phagophores into Functional Autophagosomes
Our genetic interaction studies in Drosophila (Figure 1) and our finding that both ESCRT-III and STX13 are involved in the autophagy pathway (Lee et al., 2007 and Figure 2) prompted us to examine more closely how ESCRT-III might influence STX13 function. Our biochemical analyses in Drosophila (Figure S2A) and HEK293 cells (Figure S2B) did not reveal a direct physical interaction between the two protein complexes.
Since dysfunctional ESCRT-III also causes autophagosome accumulation (Lee et al., 2007), we used the mCherry-GFP-LC3 reporter to demonstrate that siRNA knockdown of the ESCRT-III subunit Snf7-2 also blocks autophagic flux (Figure 3D) without affecting the number and distribution of LAMP1-positive lysosomes (data not shown). Remarkably, when ESCRT-III function was compromised by ectopic expression of the FTD disease protein CHMP2BIntron5 or by siRNA knockdown of Snf7-2, immuno-electron microscopy (EM) revealed many LC3-positive multilamellar structures that were highly enriched with STX13 (Figure 3E). Immuno-EM with gold particles of two different sizes clearly showed colocalization of LC3 and STX13 on the same multilamellar structures in HEK293 cells (Figure 3E). Additional immune-EM images of multilamellar structures are shown in Figure S3. These abnormal structures are of autophagic origin, since no multilamellar structures in rodent cortical neurons isolated from atg5 knockout mice could be found when Snf7-2 was knocked down by siRNA (Lee et al., 2009). Moreover, siRNA knock-down of Snf7-2 in HeLa cells also resulted in the accumulation of STX13-positive puncta that were partially labeled with Atg5 (Figure S4A, B), a protein present in phagophores but normally absent in closed autophagosomes (Xie and Klionsky, 2007). These results demonstrate that STX13 is present on LC3-positive immature autophagosomes resulting from dysfunctional ESCRT-III.
In HEK293 cells treated with rapamycin, immuno-EM revealed STX13 on autophagic double membranes and surprisingly, in some cases, on elongating double membranes (Figure 4A), providing further evidence that STX13 is present on phagophores before sealing of the double membrane. Double immuno-EM confirmed that STX13 and LC3 colocalize on the same immature autophagosomes (Figure 4A). Thus, it is likely that multilamellar structures originate from phagophores positive for both STX13 and LC3. To further investigate the function of STX13 on immature autophagosomes, we used siRNA to knock down STX13 expression in HEK293 cells, which caused numerous multilamellar structures to form as revealed by Transmission EM (Figure 4B), which may not be identical but similar to that formed by dysfunctional ESCRT-III. The resulting LC3-positive puncta were also positive for Atg5 (Figure S4D). Thus, STX13 seems to be required for phagophore maturation into closed autophagosomes in HeLa cells.
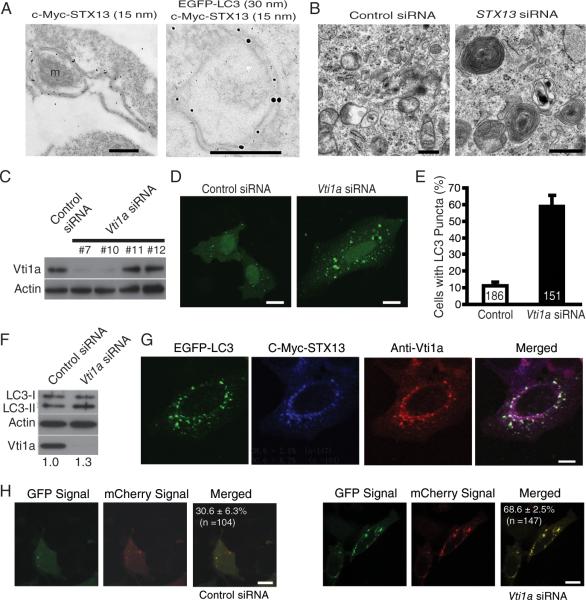
Figure 4. STX13 Is Present on Phagophores and Is Required for Their Maturation Into Closed Autophagosomes.
(A) HEK293 cells were cotransfected with EGFP-LC3 and c-Myc-STX13 and treated with rapamycin. c-Myc and EGFP antibodies were then used for immuno-EM to demonstrate STX13 localization on unclosed phagophores. M: a mitochondrion.
(B) SiRNA knockdown of STX13 resulted in the formation of numerous multilamellar structures as revealed by Transmission EM in HEK293 cells. Bar = 0.5 μm.
(C) Western blot analysis of the effectiveness of different Vti1a siRNA oligonucleotides on expression of endogenous Vti1a in HEK293 cells.
(D) SiRNA knockdown of Vti1a caused LC3-positive puncta to accumulate in HeLa cells. Bar = 10 μm.
(E) Quantification of cells with accumulated LC3-positive puncta. Values are mean ± SEM from three independent experiments. The number of cells for each condition is indicated in the column.
(F) SiRNA knockdown of Vti1a increased the LC3-II/actin ratio. The mean ratio 1.3 was based on three independent western blot experiments.
(G) After siRNA knockdown of Sn7-2, c-Myc-STX13 and Vti1a were immunostained to show colocalization with LC3. Bar = 10 μm.
(H) mCherry-GFP-LC3 was transfected into HeLa cells, and the percentage of yellow dots was quantified after treatment with control or Vti1a siRNA to indicate a defect in autophagic flux. Values are mean ± SEM. Bar = 10 μm.
See also Figure S2, Figure S4, and Table S4.
The accumulation of multilamellar structures positive for STX13 and LC3 in cells with dysfunctional ESCRT-III (Figure 3) suggests that ESCRT-III functions downstream of STX13. Indeed, in the presence of CHMP2BIntron5, which compromises ESCRT-III function through enhanced binding to other ESCRT-III components (Lee et al., 2007), the association between Syx13 and its SNARE partner Vti1 increased significantly (Figure S2A). Furthermore, knockdown of endogenous Snf7-2 with siRNA in HEK293 cells increased binding between STX13 and Vti1a (Figure S2B). Loss of Vti1a also resulted in autophagosome accumulation (Figure 4D, E) and an increased LC3-II/actin ratio (Figure 4F). Vti1a colocalized on autophagosomes induced by dysfunctional ESCRT-III either through expression of CHMP2BIntron5 (Figure S2D) or siRNA knockdown of Snf7-2 (Figure 4G). Loss of Vti1a also blocked autophagic flux as measured by the LC3 dual color assay (Figure 4H). Thus, STX13 acts together with its SNARE partner Vti1a upstream of ESCRT-III to control the maturation of autophagosomes.
DISCUSSION
Through an unbiased genetic screen in Drosophila, we identified several SNARE genes that interact with the gene encoding the FTD-associated mutant protein CHMP2BIntron5, which disrupts ESCRT-III function and causes an extensive accumulation of multilamellar autophagosomes. These SNARE proteins include Syx5, which is implicated in the late stage of autophagy (Renna et al., 2011), and Syx17, which is involved in autophagosome-lysosome fusion (Itakura et al., 2012) and in the recruitment of ATG14 to endoplasmic reticulum–mitochondria contact sites (Hamasaki et al., 2013). In this study, we focused on STX13, which showed the strongest genetic interaction with CHMP2BIntron5.
We identified STX13 as a key molecule required for the maturation of phagophores into closed autophagosomes. In contrast, three recent studies showed that SNARE complexes participate in very early homotypic membrane fusion events, before the formation of LC3-positive phagophores or fusion of autophagosomes and endosomes/lysosomes (Moreau et al., 2011; Nair et al., 2011; Itakura et al., 2012). Our findings suggest that STX13 is involved in the closure of phagophore membranes. First, we found that STX13 is present on phagophores and autophagosomes. Second, loss of STX13 resulted in autophagosome accumulation. Third, STX13 was required for autophagic flux, as shown by a dual-color reporter assay. Fourth, loss of STX13 caused the autophagy substrate p62 and Atg5-positive puncta to accumulate, indicating a failure in autophagosome maturation. Fifth, the effect of loss of STX13 was not due to defects in general trafficking pathways. Sixth, STX13 was present on LC3-positive phagophore membranes, as shown by EM analysis of cells in which autophagosomes were induced to accumulate by treatment with rapamycin. Finally, STX13 was highly localized on multilamellar structures formed by dysfunctional ESCRT-III, and loss of STX13 activity also led to the formation of multilamellar structures. It will be interesting to perform more sophisticated imaging and biochemical experiments to examine in detail the molecular events involving STX13.
Our results also indicate that ESCRT-III functions downstream of STX13 in autophagosome maturation. How ESCRT-III acts in this process is an open question. Our observation that STX13- and LC3-positive multilamellar structures accumulate in cells with dysfunctional ESCRT-III supports the notion that ESCRT-III is also required for the sealing of unclosed phagophores, a process topologically similar to vesicle budding during the formation of multivesicular bodies. Because the association of ESCRTs with phagophore membranes is thought to be transient (Hurley and Hanson, 2010) and because our immuno-EM analysis did not detect this association, other approaches are needed to determine whether ESCRTs act on autophagosomes directly or indirectly.
EXPERIMENTAL PROCEDURES
Fly Strains and Maintenance
Flies were raised at 25°C on a standard diet. UAS-CHMP2BIntron5 flies were described previously (Ahmad et al., 2009). GMR-Gal4, Df(3R)5263, Dr(3R)5612, Df(3R)5642, dNSF2A6, dNSF2A15, aSnapM4, aSnapG8, Syx13PZ01470, Syx17f01971, and Syx18EY08095 were from the Bloomington Drosophila Stock Center. UAS-dNsf2-RNAi, UAS-γSnap RNAi, UAS-Syx13 RNAi, UAS-Vti1 RNAi fly stocks were from the Vienna Drosophila RNAi Center. UAS-dNSF2 flies were kindly provided by Dr. Leo Pallanck (Golby et al., 2001). For genetic interaction studies, the recombined stock, GMR-GAL4, UAS-CHMP2BIntron5/CyO, was crossed with the deletion stocks or individual mutants. To quantify the CHMP2BIntron5 eye phenotype, we arbitrarily classified the eye phenotype with or without enhancers into three groups: strong (+++), medium (++), and weak (+). This classification was based on the relative abundance of black spots on the eye, ranging from a dozen or so scattered spots (+) to spots covering approximately 50–70% or more of the eye surface (+++).
Generation of Transgenic Stocks
To create UAS-Syx13 or UAS-Syx13-HA and c-Myc-Vti1 constructs, we used Pfu-polymerase from cDNA plasmids LD20781 and LD27967, respectively (Drosophila Genomics Resource Center), to amplify the coding region. The resulting fragments were digested and subcloned into pUAST vector, which was sequenced and microinjected into wildtype (w1118) flies to generate transgenic lines. The specific primers are listed in Table S2.
Immunoblots and Immunoprecipitation
Adult flies were frozen with dry ice and vortexed to remove the heads. Approximately 20 heads from each genotype were homogenized in lysis buffer (50 mM Tris-HCl, pH7.5, 150 mM sodium chloride, 1% Nonidet P40, 0.5% sodium deoxycholate, 1 tablet of complete Mini protein inhibitor cocktail/10 mL). Transfected mammalian cells were collected and homogenized in the same cold lysis buffer. Homogenates were centrifuged at 4°C for 20 minutes at 13,000 rpm. Protein concentrations were determined with the Bradford assay (Bio-Rad).
Supernatants containing 10–20 μg of protein were mixed with 4X sample buffer (1M Tris-HCL, pH 6.8, 8% SDS, 40% glycerol, 0.1% Bromophenol blue) and boiled for 5 minutes, then separated on a 10% polyacrylamide-SDS gel and transferred to a PVDF membrane (Bio-Rad) in a wet transfer system at 4°C for 60 minutes at 100 V. The membrane was incubated in the blocking solution (25 mM Tris-HCl, 137 mM NaCl, 3 mM KCl, pH 7.4, and 0.1% Tween-20) containing 5% milk at room temperature for 1 hour. The membrane was then incubated with primary antibody at room temperature for 3 hours and then with anti-rabbit or mouse horseradish peroxidase–conjugated secondary antibody (Jackson ImmunoResearch) for 1 hour. The signal was visualized with chemiluminescent substrate (SuperSignal West Pico, Pierce). The primary antibodies were anti-hemagglutinin (Roche), anti-c-Myc (EMD Millipore), anti-STX13 (Proteintech), anti-Vti1a (BD Transduction Laboratories), anti-LC3 (Novus), anti-Flag (Sigma), anti-CHMP2B (from Covance) (Ahmad et al., 2009), anti-Snf7-2 (Lee et al., 2011), anti-P62 (BD Transduction Laboratories), anti-ATG5 (Proteintech), anti-Cathepsin L (R&D systems), anti-actin antibody (A2228, Sigma), anti-tubulin antibody (Sigma), and β-actin antibody (4967, Cell Signaling).
For coimmunoprecipitation, the supernatants were pre-absorbed with protein A/G agarose overnight at 4°C. The specific antibody was added to the sample and incubated for 1 hour at 4°C. Then the homogeneous protein A/G agarose suspension was added to each tube, incubated for 1 hour at 4°C, and centrifuged to collect agarose beads. The beads were incubated and washed twice for 20 minutes each with washing buffer (50 mM Tris-HCl, pH7.5, 500 mM sodium chloride, 0.1% Nonidet p40, 0.05% sodium deoxycholate) and then with the washing buffer without sodium chloride. The agarose beads were then suspended in the gel loading buffer and boiled for 3 minutes. Aliquots of the final supernatant were analyzed by polyacrylamide-SDS gel.
Immunofluorescence
HeLa cells were fixed with 4% PFA at room temperature for 10 minutes, washed 3 times with phosphate-buffered saline (PBS) and blocked for 1 hour at room temperature with 10% normal goat serum (Sigma) diluted in PBS containing 0.1% Triton X-100 (PBS-T) and 2% bovine serum albumin (Sigma). Cells were then incubated overnight at 4°C with primary antibodies. These antibodies were rabbit anti-STX13 (Proteintech), mouse anti-Vti1a (BD Transduction Laboratories), rabbit anti-LC3 (Novus). After three washes in PBS (10 minutes each), cells were incubated for 1 hour at room temperature with secondary antibodies (goat anti-rabbit Alexa Fluor 568, goat anti-mouse Alexa Fluor 488, both 1:1000; Invitrogen) diluted in PBS. Cells were washed three times in PBS and mounted with Vectashield (Vector Laboratories). Immunostaining signals were examined with an Olympus IX-70 microscope or a Leica TCS-SP inverted scanning confocal microscope.
Immuno-Electron Microscopy
HEK293 cells were fixed with 1 ml of 4% paraformaldehyde in sodium phosphate buffer (pH 7.2) for 2 hours at room temperature. The next day, the fixed cells were washed three times in 0.75 M sodium phosphate buffer (pH 7.2), collected into a microfuge tube, pelleted, and briefly rinsed in distilled water. The fixed cells were then dehydrated through a graded ethanol series (20% increments) culminating in two changes of 100% ethanol. Samples were then infiltrated with a mixture of 50% ethanol 100%/50% LR White hard-grade acrylic resin and left overnight. The next morning, the cell pellets were transferred through three changes of fresh LR White hard-grade acrylic resin, embedded in closed-beam capsules, and polymerized for 48 hours at 68°C. Ultrathin sections (~80 nm) were cut on a Reichart-Jung ultramicrotome, mounted on gold support grids, immunolabeled with anti-GFP (Chemicon) and anti c-Myc antibodies (EMD Millipore), stained with lead citrate and uranyl acetate, and examined on a Philips CM 10 and a FEI Tecani 12 BT transmission electron microscopes at 80-kV accelerating voltage. Images were captured with a Gatan TEM CCD camera.
Mammalian Cell Culture, siRNAs, Constructs, and Transfection
Please see Supplementary Materials.
Supplementary Material
HIGHLIGHTS.
A genetic screen identified dNSF2 as a modifier of mutant CHMP2B in Drosophila.
Of SNAREs tested, Syx13/STX13 is a strong genetic modifier of mutant ESCRT-III.
Loss of STX13 causes LC3-positive puncta to accumulate and blocks autophagic flux.
STX13 is present on phagophores and required for their membrane closure.
ACKNOWLEDGMENTS
We thank S. Ordway and Gao lab members for comments, and several colleagues for reagents (please see details in the Methods Section). We also thank the University of Massachusetts Medical School (UMMS) Electron Microscopy and Digital Imaging Cores for assistance. This work was supported by a UMMS startup fund and the National Institutes of Health (R01 NS057553 and RO1 NS066586, F.-B.G.). Work in the STS lab is supported by the Biotechnology and Biological Sciences Research Council UK (BB/I012273/1) and the Alzheimer's Society UK.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Ahmad ST, Sweeney ST, Lee JA, Sweeney NT, Gao FB. Genetic screen identifies serpin5 as a regulator of the toll pathway and CHMP2B toxicity associated with frontotemporal dementia. Proc. Nat. Acad. Sci. USA. 2009;106:12168–12173. doi: 10.1073/pnas.0903134106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darsow T, Rieder SE, Emr SD. A multispecificity syntaxin homologue, Vam3p, essential for autophagic and biosynthetic protein transport to the vacuole. J. Cell Biol. 1997;138:517–529. doi: 10.1083/jcb.138.3.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fader CM, Sánchez DG, Mestre MB, Colombo MI. TI-VAMP/VAMP7 and VAMP3/cellubrevin: two v-SNARE proteins involved in specific steps of the autophagy/multivesicular body pathways. Biochim. Biophys. Acta. 1999;1793:1901–1916. doi: 10.1016/j.bbamcr.2009.09.011. [DOI] [PubMed] [Google Scholar]
- Filimonenko M, Stuffers S, Raiborg C, Yamamoto A, Malerød L, Fisher EM, Isaacs A, Brech A, Stenmark H, Simonsen A. Functional multivesicular bodies are required for autophagic clearance of protein aggregates associated with neurodegenerative disease. J. Cell Biol. 2007;179:485–500. doi: 10.1083/jcb.200702115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuta N, Fujita N, Noda T, Yoshimori T, Amano A. Combinational soluble N-ethylmaleimide-sensitive factor attachment protein receptor proteins VAMP8 and Vti1b mediate fusion of antimicrobial and canonical autophagosomes with lysosomes. Mol. Biol. Cell. 2010;21:1001–1010. doi: 10.1091/mbc.E09-08-0693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golby JA, Tolar LA, Pallanck L. Partitioning of N-ethylmaleimide-sensitive fusion (NSF) protein function in Drosophila melanogaster: dNSF1 is required in the nervous system, and dNSF2 is required in mesoderm. Genetics. 2001;158:265–278. doi: 10.1093/genetics/158.1.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamasaki M, Furuta N, Matsuda A, Nezu A, Yamamoto A, Fujita N, Oomori H, Noda T, Haraguchi T, Hiraoka Y, et al. Autophagosomes form at ER-mitochondria contact sites. Nature. 2013;495:389–393. doi: 10.1038/nature11910. [DOI] [PubMed] [Google Scholar]
- Henne WM, Buchkovich NJ, Emr SD. The ESCRT pathway. Dev. Cell. 2011;21:77–91. doi: 10.1016/j.devcel.2011.05.015. [DOI] [PubMed] [Google Scholar]
- Hurley JH, Hanson PI. Membrane budding and scission by the ESCRT machinery: it's all in the neck. Nat. Rev. Mol. Cell. Biol. 2010;11:556–566. doi: 10.1038/nrm2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihara Y, Morishima-Kawashima M, Nixon R. The ubiquitin-proteasome system and the autophagic-lysosomal system in Alzheimer disease. Cold Spring Harb. Perspect. Med. 2012;2(8) doi: 10.1101/cshperspect.a006361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihara N, Hamasaki M, Yokota S, Suzuki K, Kamada Y, Kihara A, Yoshimori T, Noda T, Ohsumi Y. Autophagosome requires specific early Sec proteins for its formation and NSF/SNARE for vacuolar fusion. Mol. Biol. Cell. 2011;12:3690–3702. doi: 10.1091/mbc.12.11.3690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itakura E, Kishi-Itakura C, Mizushima N. The hairpin-type tail-anchored SNARE syntaxin 17 targets to autophagosomes for fusion with endosomes/lysosomes. Cell. 2012;151:1256–1269. doi: 10.1016/j.cell.2012.11.001. [DOI] [PubMed] [Google Scholar]
- Jahn R, Scheller RH. SNAREs—engines for membrane fusion. Nat. Rev. Mol. Cell. Biol. 2006;7:631–643. doi: 10.1038/nrm2002. [DOI] [PubMed] [Google Scholar]
- Klionsky DJ, Abdalla FC, Abeliovich H, Abraham RT, Acevedo-Arozena A, Adeli K, Agholme L, Agnello M, Agostinis P, Aguirre-Ghiso JA, et al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 2012;8:445–544. doi: 10.4161/auto.19496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JA, Beigneux A, Ahmad ST, Young SG, Gao FB. ESCRT-III dysfunction causes autophagosome accumulation and neurodegeneration. Curr. Biol. 2007;17:1561–1567. doi: 10.1016/j.cub.2007.07.029. [DOI] [PubMed] [Google Scholar]
- Lee J-A, Gao F-B. Inhibition of autophagy induction delays neuronal cell loss caused by dysfunctional ESCRT-III in frontotemporal dementia. J. Neurosci. 2009;29:8506–8511. doi: 10.1523/JNEUROSCI.0924-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JA, Liu L, Javier R, Kreitzer AC, Delaloy C, Gao FB. ESCRT-III subunits Snf7-1 and Snf7-2 differentially regulate transmembrane cargos in hESC-derived human neurons. Mol. Brain. 2011;4:37. doi: 10.1186/1756-6606-4-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayle KM, Le AM, Kamei DT. The intracellular trafficking pathway of transferrin. Biochim. Biophys. Acta. 2012;1820:264–281. doi: 10.1016/j.bbagen.2011.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metcalf DJ, García-Arencibia M, Hochfeld WE, Rubinsztein DC. Autophagy and misfolded proteins in neurodegeneration. Exp. Neurol. 2012;238:22–28. doi: 10.1016/j.expneurol.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau K, Ravikumar B, Renna M, Puri C, Rubinsztein DC. Autophagosome precursor maturation requires homotypic fusion. Cell. 2011;146:303–317. doi: 10.1016/j.cell.2011.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair U, Jotwani A, Geng J, Gammoh N, Richerson D, Yen WL, Griffith J, Nag S, Wang K, Moss T, et al. SNARE proteins are required for macroautophagy. Cell. 2011;146:290–302. doi: 10.1016/j.cell.2011.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prekeris R, Klumperman J, Chen YA, Scheller RH. Syntaxin 13 mediates cycling of plasma membrane proteins via tubulovesicular recycling endosomes. J. Cell Biol. 1998;143:957–971. doi: 10.1083/jcb.143.4.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Progida C, Cogli L, Piro F, De Luca A, Bakke O, Bucci C. Rab7b controls trafficking from endosomes to the TGN. J. Cell Sci. 2010;123:1480–1491. doi: 10.1242/jcs.051474. [DOI] [PubMed] [Google Scholar]
- Renna M, Schaffner C, Winslow AR, Menzies FM, Peden AA, Floto RA, Rubinsztein DC. Autophagic substrate clearance requires activity of the syntaxin-5 SNARE complex. J. Cell Sci. 2011;124:469–482. doi: 10.1242/jcs.076489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusten TE, Vaccari T, Lindmo K, Rodahl LM, Nezis IP, Sem-Jacobsen C, Wendler F, Vincent JP, Brech A, Bilder D, et al. ESCRTs and Fab1 regulate distinct steps of autophagy. Curr. Biol. 2007;17:1817–1825. doi: 10.1016/j.cub.2007.09.032. [DOI] [PubMed] [Google Scholar]
- Urwin H, Authier A, Nielsen JE, Metcalf D, Powell C, Froud K, Malcolm DS, Holm I, Johannsen P, Brown J, et al. Disruption of endocytic trafficking in frontotemporal dementia with CHMP2B mutations. Hum. Mol. Genet. 2010;19:2228–2238. doi: 10.1093/hmg/ddq100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong E, Cuervo AM. Autophagy gone awry in neurodegenerative diseases. Nat. Neurosci. 2010;13:805–811. doi: 10.1038/nn.2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Klionsky DJ. Autophagosome formation: core machinery and adaptations. Nat. Cell Biol. 2007;9:1102–1109. doi: 10.1038/ncb1007-1102. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.