Abstract
Aneuploidy and chromosomal instability frequently co-exist, and aneuploidy is recognized as a direct outcome of chromosomal instability. However, chromosomal instability is widely viewed as a consequence of mutations in genes involved in DNA replication, chromosome segregation and cell cycle checkpoints. Telomere attrition and presence of extra centrosomes have also been recognized as causative for errors in genomic transmission. Here, we examine recent studies suggesting that aneuploidy itself can be responsible for the procreation of chromosomal instability. Evidence from both yeast and mammalian experimental models suggest that changes in chromosome copy number can cause changes in dosage of the products of many genes located on aneuploid chromosomes. These effects on gene expression can alter the balanced stoichiometry of various protein complexes, causing perturbations of their functions. Therefore, phenotypic consequences of aneuploidy will include chromosomal instability if the balanced stoichiometry of protein machineries responsible for accurate chromosome segregation is affected enough to perturb the function. The degree of chromosomal instability will depend on specific karyotypic changes, which may be due to dosage imbalances of specific genes or lack of scaling between chromosome segregation load and the capacity of the mitotic system. We propose that the relationship between aneuploidy and chromosomal instability can be envisioned as a “vicious cycle”, where aneuploidy potentiates chromosomal instability leading to further karyotype diversity in the affected population.
Introduction
The genome of each eukaryotic species is divided into a certain number of chromosomes. This number is specific for a given species, but varies widely among species. Even closely related species may have different numbers of chromosomes; for example, humans have 46 chromosomes while chimpanzees have 48, even though the genome sequences are ~98.8% identical on the DNA level [1]. Most species of the animal kingdom, with few exceptions, are diploid, which means each of their chromosomes has a pair, and this balanced chromosome number is called euploid. On a macro evolutionary scale, heritable and stable change in chromosome number (the “new euploid”) has long been thought to abet the emergence of new species [2].
Aneuploidy is an unbalanced change in chromosome number on a cellular or organismal level, when certain chromosomes no longer come in pairs. In an aneuploid cell the total chromosome number is increased or reduced compared to the normal euploid genome of a given biological species. Chromosome instability refers to the lack of capacity to maintain the same chromosome number from one cell generation to the next. While aneuploidy, especially in cancer, is often a product of chromosomal instability, these two concepts are not equivalent. That is, if a cell is aneuploid it does not necessarily imply that it is also chromosomally unstable and will not pass on its exact chromosome number to its offspring. However, aneuploidy and chromosomal instability frequently coexist, which may underlie very complex and diverse aneuploid karyotypes in high grade tumors.
Aneuploidy and chromosomal instability have both been recognized as hallmarks of cancer [3,4] and a source of genetic variation fueling tumor adaptation to stressful environments, the host’s immune response and cytotoxic effects of anti-cancer drugs. There is little doubt that continuous genetic change is one of the key factors that underlies the adaptive evolution of malignant cells. The continuous change in cancer genomes makes cancer a “moving target” for defensive responses of the host or attacks of anticancer treatments. In cancer research, both aneuploidy and chromosomal instability have received much attention. However, it has been difficult to dissect the exact relationship between aneuploidy and chromosomal instability, because in addition to complex aneuploid karyotypes, cancer cells typically carry a wide variety of genetic and epigenetic alterations compared to non-cancer cells. As such, it is difficult to determine the extent by which aneuploidy contributes to any specific phenotype or genome instability associated with cancer. Studies in experimental models such as yeast, plants and cultured non-malignant mammalian cells have begun to elucidate the alliance of aneuploidy and chromosomal instability.
This review is focused on the effect of aneuploidy on chromosomal instability. In other words, is the relationship between chromosomal instability and aneuploidy inadvertent or are they locked in a vicious cycle? A vicious cycle occurs when either problem, aneuploidy or chromosomal instability, has a high likelihood of exacerbating the other. We will discuss recent studies on the cause-effect relationship between aneuploidy and chromosome instability in the budding yeast and mammalian cells and speculate on the implication of these findings in cellular evolution.
Elevated chromosomal instability in aneuploid yeast and plants
In unicellular eukaryotic organisms such as the budding yeast Saccharomyces cerevisiae, aneuploidy is not uncommon and exists in natural populations. All budding yeast disomies are viable except disomy VI [5,6]. Although standard laboratory yeast strains are maintained as haploid or diploid, yeast strains isolated from the wild are often polyploid and/or aneuploid [7]. In some mutant strains, aneuploidy arises spontaneously and has been observed in a considerable portion of the yeast ORF deletion library. In laboratory settings, aneuploid strains of yeast can be artificially generated using various methods, such as sporulation of triploid or pentaploid strains, or by means of chromoduction, where a centromere on a specific chromosome is transiently turned off causing mitotic non-disjunction [7,8].
In the field of yeast genetics, the idea that aneuploid strains have higher rates of genomic instability than isogenic euploid counterparts has a long history. In 1970, Parry and Fox generated a series of aneuploid strains using triploid meiosis strategy. They noticed that extra chromosomes in these strains were prone to elimination [9]. Ten years later, Campbell and colleagues reported that aneuploid strains generated by triploid meiosis experienced repeated chromosome loss [10]. Two recent studies of strains obtained from triploid or pentaploid meiosis also found that many aneuploid strains displayed various degrees of chromosome instability [11,12]. In a recent study of disomic strains, the rate of chromosome missegregation, inferred from the loss of an artificial chromosome, was found to be elevated in 9 out of 13 disomic strains compared to the euploid control strain [13]. Monosomic yeast strains (referring to a diploid losing a single copy of a particular chromosome) have also been shown to exhibit increased chromosomal instability and tend to restore their diploid chromosome number, albeit at different rates with different monosomes. For instance, chromosome VIII monosomes restore their diploid karyotype after 22 generations, whereas monosomic V cells become diploid much more quickly [14,15].
A new study attempted to estimate the rate of chromosomal instability in aneuploid yeast obtained via triploid meiosis in order to identify genome-level and chromosome-specific determinants of chromosomal instability. This study showed that chromosome instability correlated with cellular ploidy, i.e. aneuploids with DNA content closer to the haploid state tend to be more stable than those between 1.5N and 2N. In addition, chromosomal instability was found to be associated with copy number imbalance between specific chromosome pairs. For instance, strains with balanced gains of Chromosome VII and Chromosome X were more likely to be stable than strains where these chromosomes have different copy numbers [16]. The mechanisms behind these observations are discussed further below.
In plants, unlike animals, whole chromosome aneuploidy seems to be tolerated on an organism level. For instance, in the model organism Arabidopsis thaliana all possible trisomies have been isolated [17], although chromosome stability has not been investigated extensively during vegetative growth in these plants. An example of increased chromosomal instability in an aneuploid plant came from a study in a trisomic tobacco line that was engineered to have kanamycin resistance maker gene on the trisomic chromosome [18]. When plants were regenerated from single cells of the trisomic tobacco, some of the clones were kanamycin sensitive, suggesting loss of the chromosome locus carrying the resistance gene. This was not observed in euploid plants carrying kanamycin resistance on the same chromosome, indicating that the trisome, but not its euploid counterpart, was genomically unstable.
The effect of aneuploidy on chromosome instability in mammalian organisms
Does aneuploidy also promote chromosomal instability in mammalian cells? Arguments for and against this idea are decidedly nontrivial, partly because in mammals whole chromosome aneuploidy is poorly tolerated on an organism level. In the most commonly used mammalian model organism –a mouse, every single whole chromosome gain or loss results in embryonic lethality [19]. In humans, the loss of any autosome is universally fatal, and so is the gain of any autosomes except for chromosomes 13, 19 and 21. Yet on a single cell level, wrong chromosome number can be tolerated as in tumors, where aneuploidy and chromosome instability are very typical. However, malignant cells frequently carry mutations and epigenetic alterations in genes important for the fidelity of genome transmission. In addition, cancer cells frequently have supernumerary centrosomes that interfere with accurate chromosome segregation in mitosis. Therefore, it is difficult to separate the impact of specific changes in chromosome number from other coexisting genetic and epigenetic alterations in cancer cells. One strategy to adequately assess effects of aneuploidy on genomic stability would be to use experimental systems with either naturally occurring or artificially created specific chromosome aneuploidies.
Investigating genomic instability in naturally occurring congenital aneuploidy of non-cancer origin is also challenging because most whole chromosome aneuploidies in humans are fatal, and all mouse trisomies are embryonic lethal. However, some human aneuploidies can be viable but cause congenital pathologies: trisomy 21 (Down syndrome), trisomy 18 (Edwards syndrome), trisomy 13 (Patau syndrome), and variation in the copy number of sex chromosomes. Few studies addressed the issue of genomic instability in these disorders. In one study, lymphocytes isolated from individuals with trisomies 13, 18 and 21 were treated with phytohaemagglutinin (PHA), which stimulates proliferation, and were then subjected to ploidy analysis by FISH using probes for three different autosomes - 8, 15 and 18. For each of these chromosomes, samples from trisomic individuals had a two-fold higher number of aneuploid cells compared to healthy euploid individuals [20]. Another study from the same group PHA-stimulated lymphocytes from patients with monosomy X (Turner syndrome). The number of aneuploid cells in this monosomy was almost twice the control [21]. Interestingly, these studies observed more frequently monosomies than polysomies.
Increased rates of chromosomal instability observed in these studies may be relevant to elevated risks of certain malignancies in congenital aneuploidy patients. For instance, children with Down syndrome have a high risk of acute myeloid leukemia, particularly the megakaryoblastic subtype [22]. Turner and Edwards syndromes are usually fatal, but a small number of children who survive with Edwards syndrome are at risk of developing Wilms’ tumor, a form of kidney cancer. Women with Turner syndrome have an increased risk of gonadoblastoma, childhood brain tumors, and possibly other malignancies [23].
In the instance of sex chromosome aneuploidy – triple X, multiple Y and XXY (Klinefelter’s syndrome), there have been no studies purposely addressing chromosomal instability. However, men with Klinefelter syndrome reportedly have elevated risks of several malignancies –such as lung cancer, breast cancer and non-Hodgkin’s lymphoma [24], whereas the rates of cancer incidence among men with Y polysomy were reported to be not different from the general population[25]. Although the number of studies exploring genomic instability in human trisomies is limited, existing data suggest that cells from patients with Down, Edward, Turner and Patau syndromes may be karyotypically less stable than cells from normal diploid individuals.
It is possible to introduce specific chromosome gain in cultured mammalian cells, but this approach has its own challenges. Primary cells that are diploid and karyotypically stable can be difficult to work with because of their limited proliferative ability. Historically, most of the immortal human tissue culture cell lines were derived from various cancers. Unlike primary cells from normal tissues, cancer-derived cell lines typically carry multiple chromosomal aberrations and display various degrees of genomic instability possibly as a result of the malignant transformation. There are, however, some cancer-derived cell lines that have relatively stable and near-diploid chromosomal number, such as HCT116 and DLD-1 (both derived from colon cancer). Non-malignant human cells immortalized by the expression of telomerase (hTERT), such as RPE1, are another source of genetically stable, diploid cell lines that can be used as the starting point for studying whole chromosomal aneuploidy in vitro.
Introducing extra copies of specific chromosomes without bringing in other chromosomes and extra centrosomes is possible by classical technique termed “microcell - mediated chromosome transfer”. This technique allows transferring a whole chromosome from a donor cell line into a recipient cell line in a form of membrane-covered micronucleus [26]. Several studies implemented this technique to incorporate extra chromosomes in established cell lines. For instance, an extra copy of chromosome 8 was introduced in diploid human fibroblasts using this technique. With passages, these originally trisomic cells developed numeral chromosomal changes and structural aberrations such as gaps, breaks and diplochromosomes (chromosomes consisting of four chromatids instead of the normal two) [27]. In a similar study, an extra copy of chromosome 3 was introduced into the genome of KH39 cell line derived from renal cancer. This cell line is aneuploid with a modal number of 51 chromosomes (and 2 copies of chromosome 3), but was shown to maintain its characteristic karyotype throughout multiple passages. With introduction of an extra copy of chromosome 3, these cells acquired chromosomal copy number instability and structural aberrations such as breaks in pericentromeric regions [28].
Another approach to evoke aneuploidy is through induction of chromosome missegregation during mitosis. In one study, aneuploid HCT116 cells were generated by transient treatment of mitotic cells with the microtubule depolymerizing drug nocodazole, the mitotic kinesin Eg5 inhibitor monastrol, or by depletion of microtubule-depolymerizing motor MCAK [29]. This approach presumably works through high rate of chromosome missegregations resulting from merotelic kinetochore/microtubule attachments, when a single kinetochore is erroneously attached to microtubules from both poles of the mitotic spindle. Merotelically attached chromosomes tend to escape the mitotic checkpoint surveillance and either segregate toward the wrong spindle pole or lag behind the rest of segregating chromatids during anaphase. The latter frequently results in chromosomes being trapped in the cleavage furrow during cytokinesis, leading to abscission failure and tetraploidy. Subsequent mitosis in tetraploid cells is highly error-prone and produces grossly aneuploid cells. Aneuploid HCT116 cells generated by creating merotelic attachments in this study exhibited high-level chromosome instability, which lead to karyotype diversity. In this study, chromosome instability was assayed by counting chromosome spreads and number of LacO-GFP spots incorporated in a certain chromosome [29].
Although the results discussed above suggest that aneuploid mammalian genomes are unstable, other studies that utilized aneuploid human cells generated by microcell-mediated chromosome transfer or trisomic mouse embryonic fibroblasts derived from early embryos did not report increased chromosomal instability. For instance, trisomy 3, 7 and 13 created in near-diploid colon carcinoma DLD1 cells by microcell mediated chromosome transfer, and trisomy 3 created in diploid human hTERT mammary cells, have stable chromosome numbers as assayed by spectral karyotyping (SKY) and array-based comparative genomic hybridization (aCGH) [30]. Similarly, HCT116 cells made tri- and tetrasomic for chromosomes 3 and 5, and RPE1 cells trisomic for chromosomes 5, 12 and 21, were not reported to be unstable as assayed by aCGH and chromosome-specific FISH probes [31]. Mouse embryonic fibroblast cell lines trisomic for chromosomes 1, 13, 16 and 19 also were not found to be unstable before immortalization (examined by SKY and aCGH) [32].
All in all, while there are indications that aneuploidy per se may evoke chromosomal instability, there is also evidence inconsistent with this notion. These discrepancies may have several causes. First, different chromosome aneuploidies may have distinct impacts on chromosome instability. This would be in line with findings in budding yeast, where chromosome instability is associated with certain genome level and chromosome-specific determinants. However, in the case of trisomy 3 and 21, their effect on karyotype stability was not the same in different studies [20],[28,31].
Another possible source of discrepancy may lie in the use of different analytical methodologies to assess karyotypes. For instance, aCGH examines karyotype in a population and may not adequately reflect variation on the single cell level. Counting chromosomes in metaphase spreads gives an idea of a total chromosome number and major structural aberrations, but does not identify which chromosome is altered. SKY, while being a definitive and high resolution single-cell technique, is labor intensive and costly, which makes it difficult to use for sufficient statistics.
It is also possible that the impact of an extra chromosome may vary in different tissues or in vitro cultured cell lines. All things considered, more comprehensive studies that examine karyotype dynamics in aneuploid cells on the single-cell level are needed. To accurately track genomic changes, the karyotype needs to be followed throughout multiple generations. Unless karyotype dynamics can be accurately tracked, measuring precise rates of genomic instability will continue to present a challenge.
Effect of aneuploidy on the gene expression
Understanding the link between aneuploidy and chromosome instability on a mechanistic level requires identifying the effects of changes in chromosome copy number on the mRNA and protein composition of the cell. Studies of the yeast transcriptome showed that genes on aneuploid chromosomes are expressed, and for the majority of genes their expression level scales proportionally to the chromosome copy number (cis-effect). That is, genes on a disomic chromosome usually showed a 2-fold increase in transcript abundance, while genes on a monosomic chromosome showed a 0.5-fold decrease, compared to their isogenic strains [6,12,33,34]. However, there were a small number of genes throughout the genome that showed a disproportionate increase or decrease in mRNA levels relative to the chromosome copy number change (trans-effect) [12,34]. Transcriptional regulatory network analysis of disproportionately expressed genes revealed enrichment for direct and indirect targets of transcription factors encoded on aneuploid chromosomes. This suggests that the trans-effect may be partially explained as the consequence of cis-mediated expression alteration of upstream transcription factors.
The question of whether or not the amount of mRNA from genes located on aneuploid chromosomes scales with chromosome dosage in mammalian cells has been baffling researchers and physicians for some time, in their quest to understand the molecular basis for congenital aneuploidy disorders such as Down syndrome. The advent of microarray technology at the beginning of the 21st century enabled genome-scale gene expression analysis, and because of the disease relevance, the mRNA analysis from patient studies preceded studies in model organisms. Early microarray studies of human fetal brain from trisomy 21 and primary astrocytes derived from trisomy 21 specimens showed a chromosome-wide increase in transcription for genes located on chromosome 21 compared to euploid cells [35,36], consistent with the cis-effect that was demonstrated in yeast. However, a later microarray analysis of lymphoblastoid cells derived from adult Down syndrome patients showed that in this experimental setup, only about 30% of the transcripts from chromosome 21 increased in abundance proportionally to the copy number, while other transcripts were either unchanged or varied among patients [37]. This finding implies that at least in the case of human trisomy 21, there may be tissue and/or age-specific mechanisms dampening gene expression changes caused by aneuploidy chromosome dosage.
Another early microarray study investigated gene expression in induced trisomic colon carcinoma DLD1 cells, where extra chromosomes 3, 7 or 13 were inserted by microcell mediated chromosome transfer [30]. The expression of genes located on trisomic chromosomes was overall proportional to the increase in chromosome copy number. However, this study also identified additional misregulated genes throughout genomes, located on different chromosomes. There was no overlap among genes deregulated in each of these trisomies, possibly consistent with the trans-effect seen in aneuploid yeast strains. More recent microarray-based measurements of gene expression in trisomic MEFs [32] and trisomic HCT116 cells [31] also agree that the level for most transcripts expressed from aneuploid chromosomes varies proportionally to the copy number of that chromosome. Nevertheless, as in DLD1 cells [30], few deregulated genes located on various chromosomes were also found. In mammalian cells, the basis for this transcriptional deregulation of a small number of genes that are not located on aneuploid chromosomes is entirely unclear.
Since the increase in chromosome copy number results in largely proportional buildup of transcripts from these chromosomes, the question arises whether the expression of proteins follows the pattern of DNA and mRNA dosage variation in aneuploid cells. Using budding yeast as a model system, this question was recently addressed directly by two quantitative proteomics studies. The study that utilized aneuploid yeast strains generated by triploid meiosis showed, by using the MudPIT (Multidimensional Protein Identification) technology that in these freshly generated aneuploid strains the relative expression level of proteins encoded on aneuploid chromosomes is, in general, proportional to the relative DNA and mRNA levels. However, there were also outliers whose expression changes are far beyond those predicted from chromosome copy number change [12]. Similarly, proteomic analysis of disomic yeast strains using the SILAC (Stable Isotope Labeling with Amino Acids in Cell Culture) technology also found that the relative level of the majority of proteins encoded on aneuploid chromosomes scales proportionally to the relative chromosome copy number. However, this study identified a higher fraction of proteins whose levels did not scale with ploidy. Bioinformatics analysis showed that many proteins in this group of outliers were annotated as components of protein complexes [38]. The difference between the two studies in the fraction of proteins whose concentration do not scale with chromosome dosage can be attributed to distinct proteomics methodologies (MudPIT vs. SILAC) or the different ways of generating and maintaining aneuploid strains [12] [38].
Does the expression of proteins follow the pattern of DNA and mRNA in mammalian aneuploid cells? So far, only one study has addressed this question in human trisomic and tetrasomic tissue culture cells, as mentioned above. Proteomic analysis by SILAC in HCT116 cells found that the majority of proteins encoded on aneuploid chromosomes in tri- and tetrasomic cell lines were more abundant than in isogenic diploid cell lines. However, about 25% of proteins encoded on aneuploid chromosomes were found in lower amounts than expected based on the chromosome copy number. Protein kinases and components of large macromolecular complexes were over-represented among the latter category [31]. This observation suggests that there may be mechanisms that correct for the altered component stoichiometry of protein complex. Explanations for the phenomenon of “correcting” protein concentrations may be quite complex. How might a cell detect that a certain protein may be in excess? Does this reflect the existence of a surveillance mechanism sensitive to the proteins’ catalytic properties, stoichiometric ratios in relation to their interaction partners, or both? It is also possible that overexpressed protein subunits are not fully incorporated in native complexes and thus are more prone to be degraded as free monomers via the ubiquitin-proteasome pathway.
Of note, this pioneering study explored the DNA-RNA-Protein relationship in the condition of “chronic” aneuploidy, where aneuploid cell lines were selected to survive and proliferate while maintaining a certain extra chromosome throughout multiple passages. It is likely that these cell lines may have evolved compensatory or stress-coping mechanisms to alleviate their persistent gene dosage disarray. It is also possible that similar compensatory mechanisms may help aneuploid cancer cells to thrive. However, chromosomally unstable aneuploid cells will always bear some acute changes in their genome, transcriptome and proteome with every erroneous cell division. Whether the same or different mechanisms operate in response to acute or chronic aneuploidy is an important question for future research.
How perturbations in gene dosages can cause chromosome instability
The impact of aneuploidy on gene expression implies that variation in chromosome copy number may lead to altered stoichiometry of proteins that interact physically or functionally. Stoichiometric perturbations of protein interaction networks involved in chromosome segregation machinery or mitotic spindle checkpoint can easily lead to chromosome missegregation and more aneuploidy (Figure 1). A profound example of this comes from a recent study in aneuploid budding yeast, which showed that changes in copy number of chromosome VII and X perturbed the ratio of essential mitotic checkpoint complexes Mad1 and Mad2. Consequentially, this weakened the mitotic checkpoint and lead to chromosomal instability [16]. In yeast, the MAD2 gene is located on chromosome X, and MAD1 on chromosome VII. Maintaining the 1:1 stoichiometry of Mad2:Mad1 proteins in the cell is essential for stringent mitotic checkpoint surveillance mechanism that monitors unattached or improperly attached kinetochores [39]. Aneuploid strains that gained copies of both chromosomes VII and X together were karyotypically more stable than when the copy number of these two chromosomes was nonequivalent. Measuring the Mad2:Mad1 ratio in various aneuploid strains derived from triploid meiosis revealed that in many highly unstable aneuploid strains Mad2:Mad1 ratio was 0.5, whereas aneuploid strains that were relatively stable had a Mad2:Mad1 ratio close to 1. This example illustrates how a dosage imbalance between specific chromosome pairs could alter the stoichiometry of a functional protein complex and evoke chromosome instability.
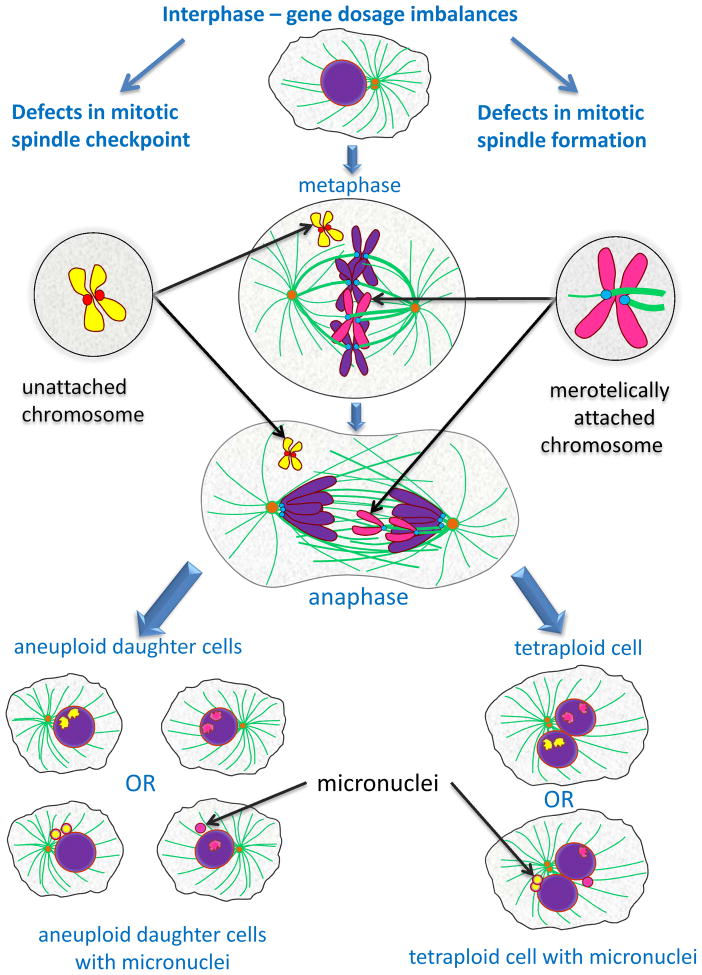
Figure 1. Perturbations of mitotic protein machinery lead to chromosome instability.
During mitosis condensed chromosomes (violet) form stable kinetochore (blue)-microtubule (green) attachments, align on the metaphase plate, and then segregate to opposite spindle poles in anaphase. Gene dosage imbalance can lead to defects in spindle assembly checkpoint and proper formation of the mitotic spindle. Spindle assembly checkpoint is a surveillance mechanism that generates the “delay anaphase” signal in response to unattached kinetochores. Defects in spindle assembly checkpoint allow anaphase to proceed despite the presence of a misaligned chromosome (yellow) whose kinetochores (red) are not stably attached to microtubules. Unattached sister chromatids cannot segregate to opposite spindle poles during anaphase and wind up in one of the daughter cells. Merotelically attached chromosome (magenta), where a single kinetochore is attached to microtubules from both spindle poles, can be a consequence of defects in mitotic spindle formation and microtubule dynamics. Merotelic attachments can evade mitotic spindle checkpoint surveillance but form lagging chromosomes that tend to missegregate and can disrupt the last step in cytokinesis - abscission. The failure to segregate properly leads to altered chromosome copy number in daughter cells: both copies of the unattached or lagging chromosomes end up in one cell while the other cell lacks a copy. Unattached and lagging chromosomes can form micronuclei – detached minute nuclei consisting of de-condensed DNA from missegregated chromosomes surrounded by the nuclear membrane. Micronuclei are prone to having defects in DNA replication, which adds to genomic instability. Chromosome missegregations, as well as protein dosage imbalances, can lead to cytokinesis failure and formation of tetraploid cells which are binucleate and contain twice the number of centrosomes (orange) as diploid cells. Tetraploid cells can also have micronuclei.
Notably, this study also found that aneuploid yeast cells with only few extra chromosomes (near-haploid) tended to be more karyotypically stable than those with many extra chromosomes (near-diploid). In other words, the more aneuploid the karyotype was, the higher was its propensity to be unstable in general. A possible explanation for this is that in yeast, the capacity of the mitotic machinery is insufficient for accurate segregation of too many extra chromosomes.
There is yet to be a study in a mammalian system showing that aneuploidy based gene dosage imbalances cause genomic instability. Nevertheless, there is ample evidence that changing stoichiometric ratios of components of protein complexes or proteins with structural or regulatory roles could have dramatic functional effects on biological processes. There are multiple cellular processes whose malfunctioning can manifest in chromosomal instability, all of which cannot be comprehensively covered in this review. Below we will discuss some instances where imbalance of gene dosages affects accuracy of chromosome segregation in mitosis.
The spindle assembly checkpoint is a complex biological mechanism that ensures faithful segregation of sister chromatids to opposite spindle poles during cell division [40]. Genes involved in mitotic checkpoint have been extensively studied in vitro and in mouse models. Mouse models expressing haploinsufficient and/or hypomophic alleles of mitotic checkpoint components such as Mad1 [41] and Mad2 [42], Cenp E [43], Bub1 [44], BubR1 [45], Rae1 [46] showed increased chromosome instability and aneuploidy in all cases. Interestingly, the spectrum of tumors induced in each case varied widely, and not all of these mouse models were tumorogenic, indicating that the relationship between chromosome instability and malignant growth is complicated.
Reduced expression of mitotic checkpoint components can partially mimic the effect of decreased gene dosage in an event of a chromosome loss. However, in cancer karyotypes chromosome gains are frequent, and many cancers are hyperploid – i.e., have a greater chromosome number than the normal euploid [47]. Therefore, cancer cells may be characterized by increased, not decreased gene dosages, which can equally perturb the ratio of components of functional protein complexes. Consistent with this, overexpression of some mitotic checkpoint gene products such as Mad1 [48], Mad2 [49], Bub1 [50] and their downstream targets separase [51] and securin [52] also leads to chromosomal instability and frequently escalates tumor formation. While these effects of protein overexpression can be attributed to the dominant gain of function, they can equally be impacted by the gene dosage imbalance.
In the case of Mad1 overexpression, increased dosage of this essential mitotic checkpoint component actually weakens the checkpoint by sequestration of its binding partner Mad2 [48]. Mad 2 overexpression, on the other hand, presumably caused over-active mitotic checkpoint and high frequency of lagging chromosomes [49,53]. These effects of Mad2 overexpression could be due to overly stable kinetochore-microtubule attachments brought about by partially displacing Aurora B from the inner centromere [54]. In cells overexpressing the mitotic kinase Bub1, spindle checkpoint was not defective. Yet, chromosomes were frequently misaligned during metaphase and lagging in anaphase, resulting in near-diploid aneuploidies. A possible mechanism for chromosomal instability here involves elevated activity of Aurora B, mitotic kinase regulated in part by Bub1 [55].
Changing dosages of other protein components of mitotic machinery that do not play a direct role in mitotic checkpoint can also cause chromosome segregation errors and subsequent aneuploidy. For instance, altering dosages of mitotic kinesins Kif2b and MCAK that regulate microtubule dynamics in the mitotic spindle increases frequency of merotelic kinetochore-microtubule attachments [56,57].
As mentioned earlier, merotelically attached chromosomes tend to be “invisible” to mitotic checkpoint and can either missegregate or form lagging chromosomes. Lagging chromosomes can be torn apart during cytokinesis, and resulting double-stranded DNA breaks leading to chromosome structural aberrations in daughter cells such as unbalanced translocations [58]. Lagging chromosomes frequently form micronuclei, which tend to have defects in DNA replication. Incompletely replicated chromosomes trapped in micronuclei can become “pulverized”-broken in pieces - during the subsequent mitosis [59]. However, “pulverized” chromosomes may also be a consequence of surviving chromosome fragmentation, which is an unusual type of mitotic cell death that can occur spontaneously or may be induced by chemotherapeutic agents [60]. This chromosome “pulverization” was proposed to be linked to chromotrypsis, - a form of genomic instability where a chromosome does not change its copy number but becomes rearranged as clusters of its own genetic material, get stitched together in a random order [61,62].
Cytokinesis failure and the tetraploidy route to chromosome instability
Another major source of chromosome instability is tetraploidy, which can result from cytokinesis failure, mitotic slippage and cell-cell fusion. In mammalian mitosis cytokinesis is, arguably, the most error-prone step. Cytokinesis is the final step in cell division when the mother cell separates into two daughter cells. It involves a concerted action of cytoskeleton machinery, membrane trafficking, protein dynamics and signaling pathways [63]. The complexity of this process makes it vulnerable to stoichiometric changes in the proteins involved, and the knockdown of numerous different proteins can cause cytokinesis to fail, leading to formation of binucleate cells. One interesting yet controversial study showed that chromosome missegregation per se (specifically missegregation of chromosomes 8 and 12) can cause cytokinesis to fail at the last stage – abscission [64]. The reasons for that were not entirely clear, but it did not involve chromatin bridges stuck inside the midbody. In light of the unbalanced stoichiometry idea, this phenomenon may be explained by proteomic changes in aneuploid daughter cells that compromise the abscission process. Abscission occurs in G1 stage of the cell cycle when gene transcription is resumed, and possibly requires newly synthesized proteins. Chromosome 8 contains a poorly characterized gene BIN3, which has the ability to localize F-actin and may be involved in cytokinesis and abscission [65]. Chromosome 12 contains a number of genes involved in cytokinesis: Rab 35 (a component of the endocytic pathway involved in abscission [66], Citron kinase (regulates cleavage furrow formation through binding a key GTPase RhoA and [67]) and MgcRacGAP – a GAP for RhoA [68]. Dosage perturbations of these genes may have predisposed cells to the high rate of abscission failure observed in that study.
Cytokinesis can also fail when a lagging chromosome gets stuck in the cleavage furrow. Nearly all chromosome segregation problems discussed above, either from mitotic spindle checkpoint defects or merotelic kinetochore-microtubule attachments, can produce lagging chromosomes in anaphase. Lagging chromosomes can easily become jammed in the cleavage furrow, resulting in chromatin bridges trapped inside the midbody. Structural chromosomal defects that are consequences of pre-existing genomic instability, such as dicentric chromosomes [69] or double-stranded DNA breaks repaired by non-homologous end joining [70] also lead to segregation defects manifested in chromatin bridges. Chromatin bridges trapped in the midbody frequently interfere with abscission, causing cleavage furrow to regress and giving rise to tetraploid cells [71].
Proliferation of tetraploid cells is a major source of genomic instability. Compared to diploid cells, tetraploid cells contain twice the number of chromosomes and centrosomes, which can lead to multipolar spindle formation during the next mitosis. Multipolar mitosis is very inaccurate and gives rise to daughter cells with drastically altered number of chromosomes and supernumerary centrosomes [72,73,74,75]. Sometimes, multipolar anaphases in cells with extra centrosomes are suppressed by centrosome clustering, restoring bipolar mitosis. However, even with centrosome clustering chromosomes often missegregate due to a high propensity for merotelic attachments [76].
Tetraploidy can be subdivided into acute (developed during most recent cell cycle) and chronic (propagated). Acutely tetraploid cells generated by cytokinesis failure are tumorigenic in immunodefficient mouse models while their diploid counterparts are not [77]. Chronic tetraploidy has been observed in many human cancers and is viewed as an early event in tumorogenesis [78]. However, a recent computational study of somatic DNA alterations in ovarian cancers revealed that genome-doubling events? are common throughout cancer progression and in fact frequently occur in cells that are already aneuploid [79], consistent with the idea discussed above that aneuploidy may promote tetraploidy. Therefore, tetraploidy can be the cause and the consequence of aneuploidy, carrying on the self-propagating cycle of genomic instability.
Interestingly, polyploidization is a normal physiological process in some tissues or cell types, such as liver or megakaryocytes. Polyploid hepatocytes frequently divide with multipolar spindles thereby reducing their ploidy to near-diploid aneuploidy [80]. In fact, the number of aneuploid cells in human liver is very high [81]. However, aneuploidy in this case does not seem to predispose hepatocytes to malignant transformation but instead promotes genetic variation that may be beneficial for the ability of liver to metabolize a diverse variety of substances [82].
Conclusions and perspective
The evidence discussed above strongly suggests that the relationship between aneuploidy and chromosomal instability can be envisioned as a “vicious cycle”, where one potentiates the other (Figure 2). This vicious cycle can lead to a rapid broadening of the karyotype diversity in a population, providing the substrate for natural selection. In laboratory strains of yeast, under normal conditions the fittest karyotype is euploid, and induction of aneuploidy generally leads to reduced fitness. However, under unfavorable conditions chromosomal instability can potentiate rapid adaptation via karyotypic diversity. For example, gain of chromosome XV in budding yeast confers resistance to the antibiotic radicicol, while loss of chromosome XVI confers resistance to another antibiotic, tunicamycin. Importantly, inducing chromosome instability surges the number of colonies resistant to these antibiotics [83]. Therefore, chromosomal instability, while being mostly detrimental, may produce karyotypes that are advantageous under certain environmental conditions. Future research may enable the prediction of specific phenotypic consequences for a given karyotype change, which may allow predicting trajectory of the adaptive evolution through chromosome instability.
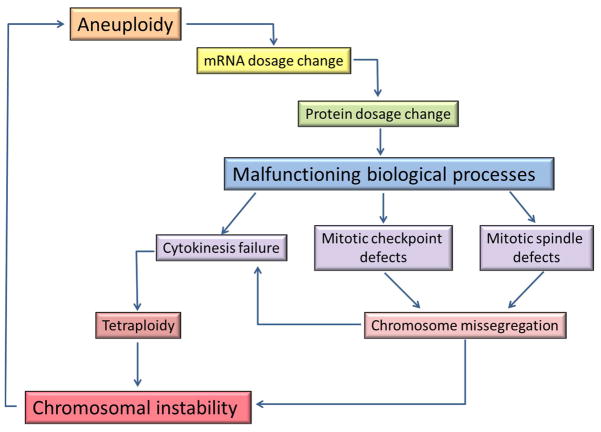
Figure 2. Vicious cycle of aneuploidy and chromosome instability.
Aneuploidy (presence of extra or missing chromosomes) is a consequence of chromosome instability. Alterations in chromosome copy number in aneuploid cells produce proportional changes in the level of transcripts of genes located on aneuploid chromosomes. This largely translates in dosage changes of protein products of these genes, which can alter the balanced stoichiometry of various protein complexes or pathways leading to malfunctioning of corresponding biological processes. Stoichiometric imbalances in protein complexes or regulatory activities involved in mitosis can cause various defects in mitotic spindle formation, mitotic checkpoint and cytokinesis. These defects can lead to errors in chromosome segregation and tetraploidy, thus elevating the level of chromosome instability and leading to more aneuploidy.
In cancer, the relationship between aneuploidy and chromosomal instability is much more complicated than in yeast because of the vast diversity and complexity of chromosome aberrations present in malignancies. Nonetheless, advanced cancer generally correlates with progressive karyotypic diversity [84]. Chromosomal instability is a common feature of nearly all types of solid tumors, and the level of the genetic instability within the tumor tends to be proportional to its level of aneuploidy [85,86]. It is likely that despite the overall complexity of human cancers, aneuploidy and chromosome instability can potentiate each other as they do in yeast, and this vicious cycle plays an important role in tumor evolution. However, it is important to bear in mind that many established cancer cell lines are aneuploid and yet have relatively stable karyotype. In fact, HeLa cells, one of the oldest and most widely used cancer cell lines, is grossly aneuploid yet has a very robust mitotic spindle checkpoint. It is unclear how gene dosages of mitotic checkpoint components are balanced in this case. Either abnormal gene dosages may be compensated on mRNA and/or protein level, or new gene dosage equilibrium emerged that makes this particular aneuploid karyotype have strong mitotic spindle checkpoint capability.
Understanding how aneuploid genomes can acquire and maintain karyotype stability may be an important insight in cancer biology. It is intuitive that karyotypes of aggressive cancers must retain a certain degree of stability to circumvent fitness costs of continuous karyotypic changes. Indeed, a mouse model study demonstrated that whereas heightened chromosome instability promotes tumor formation, exceedingly high levels of instability impairs tumor growth [87].
In yeast studies, the cycle of aneuploidy and chromosome instability has emerged as a powerful engine for adaptive evolution. Yeast can develop various aneuploid karyotypes as long as it presents an advantage in surviving and outcompeting less fit populations. In multicellular organisms, such unabated evolution of individual cells would be vastly deleterious if it was not for the tumor suppressor protein p53. In mammals, p53 is the central component of a signaling pathway that suppresses proliferation of aneuploid cells [88]. Yeast does not have the p53, and their karyotypic diversity is controlled only by relative fitness constraints. P53 pathway essentially prevents aneuploidy-driven evolution of single cells in a multicellular organism, thus acting as a tumor suppressor. Mutations of the p53 gene are detected in more than half of human malignancies [89]. Considering alterations in other p53 network components, p53 pathway is inactivated in most human cancers [90]. Evidently, inactivation of the p53 pathway unleashes tumor evolution, making cancer cells proliferate much like yeast - limited only by their relative fitness. In essence, p53 may be an anti-evolvability invention of multicellularity that breaks the vicious cycle of aneuploidy and chromosome instability by limiting the proliferation of aneuploid cells.
In spite of the enormous amount of research vested in p53 pathway over the years, it is still not entirely clear how p53 is activated by aneuploidy. Nailing it down has been difficult because of the very wide variety of external and internal stresses that can trigger activation of p53 [91,92,93]. DNA damage and oxidative stress – powerful p53 inducers-can sometimes accompany aneuploidy, but not always. Trisomic mouse embryonic fibroblasts were reported to show signs of proteotoxic stress that may activate p53 through p38 stress kinase [32,94]. However, aneuploid cells occur naturally in some tissues such as liver [81] and brain [95], and it is unknown how these aneuploidies evade p53 detection. Figuring out how p53 recognizes aneuploidy may teach us a great deal about how to break the vicious cycle of aneuploidy and chromosome instability to curb cancer evolution.
Ploidy terminology.
Ploidy is the number of sets of chromosomes in a cell or in an organism.
Haploid number refers to one set of chromosomes (1N), as in gametes or certain strains of budding yeast.
Diploid number means two sets of chromosomes (2N) that are homologous (one from each parent).
Euploid means an exact multiple of the haploid number normal for a particular species (i.e. human euploid genome contains 46 chromosomes – 23 pairs).
Polyploid indicates more than two sets of homologous chromosomes (triploid -3N, tetraploid - 4N, pentaploid – 5N, etc.).
Aneuploid denotes chromosome number that deviates from a multiple of the haploid, i.e. when there are extra or missing single chromosomes. Whole chromosomal aneuploidy is when entire chromosomes are gained or lost. Segmental aneuploidy is when pieces of chromosomes are amplified or deleted.
Disomy means haploid genome gained an additional chromosome (1N+1). Disomy 1 indicates an extra chromosome 1 in a haploid genome.
Trisomy means a diploid genome gained an additional chromosome (2N+1). Trisomy 21 indicates extra chromosome 21 in a diploid genome.
Monosomy means a diploid genome lost a chromosome (2N−1). Monosomy X in females indicates loss of one X chromosome.
Acknowledgments
We would like to thank Guangbo Chen and members of the Rong Li lab for their insightful discussions; Gary Gorbsky for comments on the manuscript; and Mary Toth for help with review preparation.
References
- 1.Initial sequence of the chimpanzee genome and comparison with the human genome. Nature. 2005;437:69–87. doi: 10.1038/nature04072. [DOI] [PubMed] [Google Scholar]
- 2.Dobzhansky Theodosius., editor. Genetics and the Origin of Species. 3. New York: Columbia University Press; 1951. pp. 73–118. [Google Scholar]
- 3.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 4.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 5.Anders K, Kudrna J, Keller K, Kinghorn B, Miller E, et al. A strategy for constructing aneuploid yeast strains by transient nondisjunction of a target chromosome. BMC Genetics. 2009;10:36. doi: 10.1186/1471-2156-10-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Torres EM, Sokolsky T, Tucker CM, Chan LY, Boselli M, et al. Effects of Aneuploidy on Cellular Physiology and Cell Division in Haploid Yeast. Science. 2007;317:916–924. doi: 10.1126/science.1142210. [DOI] [PubMed] [Google Scholar]
- 7.Ezov TK, Boger-Nadjar E, Frenkel Z, Katsperovski I, Kemeny S, et al. Molecular-Genetic Biodiversity in a Natural Population of the Yeast Saccharomyces cerevisiae From a “Evolution Canyona”: Microsatellite Polymorphism, Ploidy and Controversial Sexual Status. Genetics. 2006;174:1455–1468. doi: 10.1534/genetics.106.062745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carreto L, Eiriz M, Gomes A, Pereira P, Schuller D, et al. Comparative genomics of wild type yeast strains unveils important genome diversity. BMC Genomics. 2008;9:524. doi: 10.1186/1471-2164-9-524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Parry EM, Cox BS. The tolerance of aneuploidy in yeast. Genetical Research. 1970;16:333–340. doi: 10.1017/s0016672300002597. [DOI] [PubMed] [Google Scholar]
- 10.Campbell D, Doctor JS, Feuersanger JH, Doolittle MM. DIFFERENTIAL MITOTIC STABILITY OF YEAST DISOMES DERIVED FROM TRIPLOID MEIOSIS. Genetics. 1981;98:239–255. doi: 10.1093/genetics/98.2.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Charles JS, Hamilton ML, Petes TD. Meiotic Chromosome Segregation in Triploid Strains of Saccharomyces cerevisiae. Genetics. 2010;186:537–550. doi: 10.1534/genetics.110.121533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pavelka N, Rancati G, Zhu J, Bradford WD, Saraf A, et al. Aneuploidy confers quantitative proteome changes and phenotypic variation in budding yeast. Nature. 2010;468:321–325. doi: 10.1038/nature09529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sheltzer JM, Blank HM, Pfau SJ, Tange Y, George BM, et al. Aneuploidy Drives Genomic Instability in Yeast. Science. 2011;333:1026–1030. doi: 10.1126/science.1206412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Waghmare SK, Bruschi CV. Differential chromosome control of ploidy in the yeast Saccharomyces cerevisiae. Yeast. 2005;22:625–639. doi: 10.1002/yea.1226. [DOI] [PubMed] [Google Scholar]
- 15.Zang Y, Garrè M, Gjuracic K, Bruschi CV. Chromosome V loss due to centromere knockout or MAD2-deletion is immediately followed by restitution of homozygous diploidy in Saccharomyces cerevisiae. Yeast. 2002;19:553–564. doi: 10.1002/yea.859. [DOI] [PubMed] [Google Scholar]
- 16.Zhu J, Pavelka N, Bradford WD, Rancati G, Li R. Karyotypic Determinants of Chromosome Instability in Aneuploid Budding Yeast. PLoS Genet. 2012;8:e1002719. doi: 10.1371/journal.pgen.1002719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koncz C, Chua N-H, Schell J. Methods in Arabidopsis Research. World Scientific Publishing; 1992. p. 496. [Google Scholar]
- 18.Papp I, Iglesias VA, Moscone EA, Michalowski S, Spiker S, et al. Structural instability of a transgene locus in tobacco is associated with aneuploidy. The Plant Journal. 1996;10:469–478. doi: 10.1046/j.1365-313x.1996.10030469.x. [DOI] [PubMed] [Google Scholar]
- 19.Hernandez D, Fisher EM. Mouse autosomal trisomy: two’s company, three’s a crowd. Trends Genet. 1999;15:241–247. doi: 10.1016/s0168-9525(99)01743-6. [DOI] [PubMed] [Google Scholar]
- 20.Reish O, Regev M, Kanesky A, Girafi S, Mashevich M. Sporadic aneuploidy in PHA-stimulated lymphocytes of trisomies 21, 18, and 13. Cytogenet Genome Res. 2011;133:184–189. doi: 10.1159/000323504. [DOI] [PubMed] [Google Scholar]
- 21.Reish O, Brosh N, Gobazov R, Rosenblat M, Libman V, et al. Sporadic aneuploidy in PHA-stimulated lymphocytes of Turner’s syndrome patients. Chromosome Res. 2006;14:527–534. doi: 10.1007/s10577-006-1050-9. [DOI] [PubMed] [Google Scholar]
- 22.Khan I, Malinge S, Crispino J. Myeloid leukemia in Down syndrome. Crit Rev Oncog. 2011;16:25–36. doi: 10.1615/critrevoncog.v16.i1-2.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schoemaker MJ, Swerdlow AJ, Higgins CD, Wright AF, Jacobs PA. Cancer incidence in women with Turner syndrome in Great Britain: a national cohort study. Lancet Oncol. 2008;9:239–246. doi: 10.1016/S1470-2045(08)70033-0. [DOI] [PubMed] [Google Scholar]
- 24.Swerdlow AJ, Schoemaker MJ, Higgins CD, Wright AF, Jacobs PA. Cancer incidence and mortality in men with Klinefelter syndrome: a cohort study. J Natl Cancer Inst. 2005;97:1204–1210. doi: 10.1093/jnci/dji240. [DOI] [PubMed] [Google Scholar]
- 25.Higgins CD, Swerdlow AJ, Schoemaker MJ, Wright AF, Jacobs PA. Mortality and cancer incidence in males with Y polysomy in Britain: a cohort study. Hum Genet. 2007;121:691–696. doi: 10.1007/s00439-007-0365-8. [DOI] [PubMed] [Google Scholar]
- 26.Friedberg EC, Henning K, Lambert C, Saxon PJ, Schultz RA, et al. Microcell-mediated chromosome transfer: a strategy for studying the genetics and molecular pathology of human hereditary diseases with abnormal responses to DNA damage. Basic Life Sci. 1990;52:257–267. doi: 10.1007/978-1-4615-9561-8_21. [DOI] [PubMed] [Google Scholar]
- 27.Nawata H, Kashino G, Tano K, Daino K, Shimada Y, et al. Dysregulation of gene expression in the artificial human trisomy cells of chromosome 8 associated with transformed cell phenotypes. PLoS One. 2011;6:e25319. doi: 10.1371/journal.pone.0025319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kost-Alimova M, Fedorova L, Yang Y, Klein G, Imreh S. Microcell-mediated chromosome transfer provides evidence that polysomy promotes structural instability in tumor cell chromosomes through asynchronous replication and breakage within late-replicating regions. Genes Chromosomes Cancer. 2004;40:316–324. doi: 10.1002/gcc.20054. [DOI] [PubMed] [Google Scholar]
- 29.Thompson SL, Compton DA. Proliferation of aneuploid human cells is limited by a p53-dependent mechanism. J Cell Biol. 2010;188:369–381. doi: 10.1083/jcb.200905057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Upender MB, Habermann JK, McShane LM, Korn EL, Barrett JC, et al. Chromosome transfer induced aneuploidy results in complex dysregulation of the cellular transcriptome in immortalized and cancer cells. Cancer Res. 2004;64:6941–6949. doi: 10.1158/0008-5472.CAN-04-0474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stingele S, Stoehr G, Peplowska K, Cox J, Mann M, et al. Global analysis of genome, transcriptome and proteome reveals the response to aneuploidy in human cells. Mol Syst Biol. 2012;8:608. doi: 10.1038/msb.2012.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Williams BR, Prabhu VR, Hunter KE, Glazier CM, Whittaker CA, et al. Aneuploidy affects proliferation and spontaneous immortalization in mammalian cells. Science. 2008;322:703–709. doi: 10.1126/science.1160058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hughes TR, Roberts CJ, Dai H, Jones AR, Meyer MR, et al. Widespread aneuploidy revealed by DNA microarray expression profiling. Nat Genet. 2000;25:333–337. doi: 10.1038/77116. [DOI] [PubMed] [Google Scholar]
- 34.Rancati G, Pavelka N, Fleharty B, Noll A, Trimble R, et al. Aneuploidy Underlies Rapid Adaptive Evolution of Yeast Cells Deprived of a Conserved Cytokinesis Motor. Cell. 2008;135:879–893. doi: 10.1016/j.cell.2008.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mao R, Zielke CL, Zielke HR, Pevsner J. Global up-regulation of chromosome 21 gene expression in the developing Down syndrome brain. Genomics. 2003;81:457–467. doi: 10.1016/s0888-7543(03)00035-1. [DOI] [PubMed] [Google Scholar]
- 36.FitzPatrick DR, Ramsay J, McGill NI, Shade M, Carothers AD, et al. Transcriptome analysis of human autosomal trisomy. Hum Mol Genet. 2002;11:3249–3256. doi: 10.1093/hmg/11.26.3249. [DOI] [PubMed] [Google Scholar]
- 37.Ait Yahya-Graison E, Aubert J, Dauphinot L, Rivals I, Prieur M, et al. Classification of human chromosome 21 gene-expression variations in Down syndrome: impact on disease phenotypes. Am J Hum Genet. 2007;81:475–491. doi: 10.1086/520000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Torres EM, Dephoure N, Panneerselvam A, Tucker CM, Whittaker CA, et al. Identification of aneuploidy-tolerating mutations. Cell. 2010;143:71–83. doi: 10.1016/j.cell.2010.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Barnhart EL, Dorer RK, Murray AW, Schuyler SC. Reduced Mad2 expression keeps relaxed kinetochores from arresting budding yeast in mitosis. Mol Biol Cell. 2011;22:2448–2457. doi: 10.1091/mbc.E09-01-0029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Musacchio A, Salmon ED. The spindle-assembly checkpoint in space and time. Nat Rev Mol Cell Biol. 2007;8:379–393. doi: 10.1038/nrm2163. [DOI] [PubMed] [Google Scholar]
- 41.Iwanaga Y, Chi YH, Miyazato A, Sheleg S, Haller K, et al. Heterozygous deletion of mitotic arrest-deficient protein 1 (MAD1) increases the incidence of tumors in mice. Cancer Res. 2007;67:160–166. doi: 10.1158/0008-5472.CAN-06-3326. [DOI] [PubMed] [Google Scholar]
- 42.Sotillo R, Schvartzman JM, Socci ND, Benezra R. Mad2-induced chromosome instability leads to lung tumour relapse after oncogene withdrawal. Nature. 2010;464:436–440. doi: 10.1038/nature08803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weaver BA, Bonday ZQ, Putkey FR, Kops GJ, Silk AD, et al. Centromere-associated protein-E is essential for the mammalian mitotic checkpoint to prevent aneuploidy due to single chromosome loss. J Cell Biol. 2003;162:551–563. doi: 10.1083/jcb.200303167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Baker DJ, Jin F, Jeganathan KB, van Deursen JM. Whole chromosome instability caused by Bub1 insufficiency drives tumorigenesis through tumor suppressor gene loss of heterozygosity. Cancer Cell. 2009;16:475–486. doi: 10.1016/j.ccr.2009.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hartman TK, Wengenack TM, Poduslo JF, van Deursen JM. Mutant mice with small amounts of BubR1 display accelerated age-related gliosis. Neurobiol Aging. 2007;28:921–927. doi: 10.1016/j.neurobiolaging.2006.05.012. [DOI] [PubMed] [Google Scholar]
- 46.Baker DJ, Jeganathan KB, Malureanu L, Perez-Terzic C, Terzic A, et al. Early aging-associated phenotypes in Bub3/Rae1 haploinsufficient mice. J Cell Biol. 2006;172:529–540. doi: 10.1083/jcb.200507081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Storchova Z, Kuffer C. The consequences of tetraploidy and aneuploidy. J Cell Sci. 2008;121:3859–3866. doi: 10.1242/jcs.039537. [DOI] [PubMed] [Google Scholar]
- 48.Ryan SD, Britigan EM, Zasadil LM, Witte K, Audhya A, et al. Up-regulation of the mitotic checkpoint component Mad1 causes chromosomal instability and resistance to microtubule poisons. Proc Natl Acad Sci U S A. 2012;109:E2205–2214. doi: 10.1073/pnas.1201911109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sotillo R, Hernando E, Diaz-Rodriguez E, Teruya-Feldstein J, Cordon-Cardo C, et al. Mad2 overexpression promotes aneuploidy and tumorigenesis in mice. Cancer Cell. 2007;11:9–23. doi: 10.1016/j.ccr.2006.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ricke RM, Jeganathan KB, van Deursen JM. Bub1 overexpression induces aneuploidy and tumor formation through Aurora B kinase hyperactivation. J Cell Biol. 2011;193:1049–1064. doi: 10.1083/jcb.201012035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang N, Ge G, Meyer R, Sethi S, Basu D, et al. Overexpression of Separase induces aneuploidy and mammary tumorigenesis. Proc Natl Acad Sci U S A. 2008;105:13033–13038. doi: 10.1073/pnas.0801610105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yu R, Lu W, Chen J, McCabe CJ, Melmed S. Overexpressed pituitary tumor-transforming gene causes aneuploidy in live human cells. Endocrinology. 2003;144:4991–4998. doi: 10.1210/en.2003-0305. [DOI] [PubMed] [Google Scholar]
- 53.Schvartzman JM, Duijf PH, Sotillo R, Coker C, Benezra R. Mad2 is a critical mediator of the chromosome instability observed upon Rb and p53 pathway inhibition. Cancer Cell. 2011;19:701–714. doi: 10.1016/j.ccr.2011.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kabeche L, Compton DA. Checkpoint-independent stabilization of kinetochore-microtubule attachments by Mad2 in human cells. Curr Biol. 2012;22:638–644. doi: 10.1016/j.cub.2012.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ricke RM, van Deursen JM. Aurora B hyperactivation by Bub1 overexpression promotes chromosome missegregation. Cell Cycle. 2011;10:3645–3651. doi: 10.4161/cc.10.21.18156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bakhoum SF, Thompson SL, Manning AL, Compton DA. Genome stability is ensured by temporal control of kinetochore-microtubule dynamics. Nat Cell Biol. 2009;11:27–35. doi: 10.1038/ncb1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kline-Smith SL, Khodjakov A, Hergert P, Walczak CE. Depletion of centromeric MCAK leads to chromosome congression and segregation defects due to improper kinetochore attachments. Mol Biol Cell. 2004;15:1146–1159. doi: 10.1091/mbc.E03-08-0581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Janssen A, van der Burg M, Szuhai K, Kops GJ, Medema RH. Chromosome segregation errors as a cause of DNA damage and structural chromosome aberrations. Science. 2011;333:1895–1898. doi: 10.1126/science.1210214. [DOI] [PubMed] [Google Scholar]
- 59.Crasta K, Ganem NJ, Dagher R, Lantermann AB, Ivanova EV, et al. DNA breaks and chromosome pulverization from errors in mitosis. Nature. 2012;482:53–58. doi: 10.1038/nature10802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stevens JB, Liu G, Bremer SW, Ye KJ, Xu W, et al. Mitotic cell death by chromosome fragmentation. Cancer Res. 2007;67:7686–7694. doi: 10.1158/0008-5472.CAN-07-0472. [DOI] [PubMed] [Google Scholar]
- 61.Stephens PJ, Greenman CD, Fu B, Yang F, Bignell GR, et al. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell. 2011;144:27–40. doi: 10.1016/j.cell.2010.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Heng HH, Stevens JB, Bremer SW, Liu G, Abdallah BY, et al. Evolutionary mechanisms and diversity in cancer. Adv Cancer Res. 2011;112:217–253. doi: 10.1016/B978-0-12-387688-1.00008-9. [DOI] [PubMed] [Google Scholar]
- 63.Normand G, King RW. Understanding cytokinesis failure. Adv Exp Med Biol. 2010;676:27–55. doi: 10.1007/978-1-4419-6199-0_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shi Q, King RW. Chromosome nondisjunction yields tetraploid rather than aneuploid cells in human cell lines. Nature. 2005;437:1038–1042. doi: 10.1038/nature03958. [DOI] [PubMed] [Google Scholar]
- 65.Routhier EL, Burn TC, Abbaszade I, Summers M, Albright CF, et al. Human BIN3 complements the F-actin localization defects caused by loss of Hob3p, the fission yeast homolog of Rvs161p. J Biol Chem. 2001;276:21670–21677. doi: 10.1074/jbc.M101096200. [DOI] [PubMed] [Google Scholar]
- 66.Kouranti I, Sachse M, Arouche N, Goud B, Echard A. Rab35 regulates an endocytic recycling pathway essential for the terminal steps of cytokinesis. Curr Biol. 2006;16:1719–1725. doi: 10.1016/j.cub.2006.07.020. [DOI] [PubMed] [Google Scholar]
- 67.Madaule P, Eda M, Watanabe N, Fujisawa K, Matsuoka T, et al. Role of citron kinase as a target of the small GTPase Rho in cytokinesis. Nature. 1998;394:491–494. doi: 10.1038/28873. [DOI] [PubMed] [Google Scholar]
- 68.Yuce O, Piekny A, Glotzer M. An ECT2-centralspindlin complex regulates the localization and function of RhoA. J Cell Biol. 2005;170:571–582. doi: 10.1083/jcb.200501097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Therman E, Trunca C, Kuhn EM, Sarto GE. Dicentric chromosomes and the inactivation of the centromere. Hum Genet. 1986;72:191–195. doi: 10.1007/BF00291876. [DOI] [PubMed] [Google Scholar]
- 70.Acilan C, Potter DM, Saunders WS. DNA repair pathways involved in anaphase bridge formation. Genes Chromosomes Cancer. 2007;46:522–531. doi: 10.1002/gcc.20425. [DOI] [PubMed] [Google Scholar]
- 71.Steigemann P, Wurzenberger C, Schmitz MH, Held M, Guizetti J, et al. Aurora B-mediated abscission checkpoint protects against tetraploidization. Cell. 2009;136:473–484. doi: 10.1016/j.cell.2008.12.020. [DOI] [PubMed] [Google Scholar]
- 72.Nigg EA. Origins and consequences of centrosome aberrations in human cancers. Int J Cancer. 2006;119:2717–2723. doi: 10.1002/ijc.22245. [DOI] [PubMed] [Google Scholar]
- 73.Ganem NJ, Godinho SA, Pellman D. A mechanism linking extra centrosomes to chromosomal instability. Nature. 2009;460:278–282. doi: 10.1038/nature08136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Godinho SA, Kwon M, Pellman D. Centrosomes and cancer: how cancer cells divide with too many centrosomes. Cancer Metastasis Rev. 2009;28:85–98. doi: 10.1007/s10555-008-9163-6. [DOI] [PubMed] [Google Scholar]
- 75.Gisselsson D, Jin Y, Lindgren D, Persson J, Gisselsson L, et al. Generation of trisomies in cancer cells by multipolar mitosis and incomplete cytokinesis. Proc Natl Acad Sci U S A. 2010;107:20489–20493. doi: 10.1073/pnas.1006829107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kwon M, Godinho SA, Chandhok NS, Ganem NJ, Azioune A, et al. Mechanisms to suppress multipolar divisions in cancer cells with extra centrosomes. Genes Dev. 2008;22:2189–2203. doi: 10.1101/gad.1700908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fujiwara T, Bandi M, Nitta M, Ivanova EV, Bronson RT, et al. Cytokinesis failure generating tetraploids promotes tumorigenesis in p53-null cells. Nature. 2005;437:1043–1047. doi: 10.1038/nature04217. [DOI] [PubMed] [Google Scholar]
- 78.Ganem NJ, Storchova Z, Pellman D. Tetraploidy, aneuploidy and cancer. Curr Opin Genet Dev. 2007;17:157–162. doi: 10.1016/j.gde.2007.02.011. [DOI] [PubMed] [Google Scholar]
- 79.Carter SL, Cibulskis K, Helman E, McKenna A, Shen H, et al. Absolute quantification of somatic DNA alterations in human cancer. Nat Biotechnol. 2012;30:413–421. doi: 10.1038/nbt.2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Duncan AW, Taylor MH, Hickey RD, Hanlon Newell AE, Lenzi ML, et al. The ploidy conveyor of mature hepatocytes as a source of genetic variation. Nature. 2010;467:707–710. doi: 10.1038/nature09414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Duncan AW, Hanlon Newell AE, Smith L, Wilson EM, Olson SB, et al. Frequent aneuploidy among normal human hepatocytes. Gastroenterology. 2012;142:25–28. doi: 10.1053/j.gastro.2011.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Duncan AW, Hanlon Newell AE, Bi W, Finegold MJ, Olson SB, et al. Aneuploidy as a mechanism for stress-induced liver adaptation. J Clin Invest. 2012;122:3307–3315. doi: 10.1172/JCI64026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chen G, Bradford WD, Seidel CW, Li R. Hsp90 stress potentiates rapid cellular adaptation through induction of aneuploidy. Nature. 2012;482:246–250. doi: 10.1038/nature10795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Albertson DG, Collins C, McCormick F, Gray JW. Chromosome aberrations in solid tumors. Nat Genet. 2003;34:369–376. doi: 10.1038/ng1215. [DOI] [PubMed] [Google Scholar]
- 85.Duesberg P, Rausch C, Rasnick D, Hehlmann R. Genetic instability of cancer cells is proportional to their degree of aneuploidy. Proc Natl Acad Sci U S A. 1998;95:13692–13697. doi: 10.1073/pnas.95.23.13692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gordon DJ, Resio B, Pellman D. Causes and consequences of aneuploidy in cancer. Nat Rev Genet. 2012;13:189–203. doi: 10.1038/nrg3123. [DOI] [PubMed] [Google Scholar]
- 87.Weaver BA, Silk AD, Montagna C, Verdier-Pinard P, Cleveland DW. Aneuploidy acts both oncogenically and as a tumor suppressor. Cancer Cell. 2007;11:25–36. doi: 10.1016/j.ccr.2006.12.003. [DOI] [PubMed] [Google Scholar]
- 88.Aylon Y, Oren M. p53: guardian of ploidy. Mol Oncol. 2011;5:315–323. doi: 10.1016/j.molonc.2011.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Olivier M, Hollstein M, Hainaut P. TP53 mutations in human cancers: origins, consequences, and clinical use. Cold Spring Harb Perspect Biol. 2010;2:a001008. doi: 10.1101/cshperspect.a001008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Brown CJ, Lain S, Verma CS, Fersht AR, Lane DP. Awakening guardian angels: drugging the p53 pathway. Nat Rev Cancer. 2009;9:862–873. doi: 10.1038/nrc2763. [DOI] [PubMed] [Google Scholar]
- 91.Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408:307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- 92.Horn HF, Vousden KH. Coping with stress: multiple ways to activate p53. Oncogene. 2007;26:1306–1316. doi: 10.1038/sj.onc.1210263. [DOI] [PubMed] [Google Scholar]
- 93.Vousden KH, Lane DP. p53 in health and disease. Nat Rev Mol Cell Biol. 2007;8:275–283. doi: 10.1038/nrm2147. [DOI] [PubMed] [Google Scholar]
- 94.Tang YC, Williams BR, Siegel JJ, Amon A. Identification of aneuploidy-selective antiproliferation compounds. Cell. 2011;144:499–512. doi: 10.1016/j.cell.2011.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Rehen SK, Yung YC, McCreight MP, Kaushal D, Yang AH, et al. Constitutional aneuploidy in the normal human brain. J Neurosci. 2005;25:2176–2180. doi: 10.1523/JNEUROSCI.4560-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]