SUMMARY
Seryl-tRNA synthetase (SerRS), an essential enzyme for translation, also regulates vascular development. This “gain-of-function” has been linked to the UNE-S domain added to vertebrate SerRS during evolution. However, the significance of two insertions also specific to higher eukaryotic SerRS remains elusive. Here we determined the crystal structure of human SerRS in complex with Ser-SA, an aminoacylation reaction intermediate analog, at 2.9-Å resolution. Despite a 70-Å distance, binding of Ser-SA in the catalytic domain dramatically leverages the position of Insertion I in the tRNA binding domain. Importantly, this leverage is unique to higher eukaryotes and not seen in bacterial, archaeal and lower eukaryotic SerRSs. Deletion of Insertion I does not affect tRNA binding but instead reduce the catalytic efficiency of the synthetase. Thus, a long-range conformational and functional communication specific to higher eukaryotes is found in human SerRS, possibly to coordinate translation with vasculogenesis.
INTRODUCTION
As a member of the aminoacyl-tRNA synthetase family, seryl-tRNA synthetase (SerRS) catalyzes the aminoacylation reaction that charges serine onto its cognate tRNA for protein synthesis (Schimmel, 1987). This evolutionarily conserved essential reaction happens in two steps: in the first step, serine is activated by ATP to form serine-adenylate (Ser-AMP) as the enzyme-bound reaction intermediate; in the second step, the seryl moiety on Ser-AMP is transferred to the 3′ of the cognate tRNA to generate the final product Ser-tRNASer to be delivered to the ribosome. The dimeric SerRS belongs to class II tRNA synthetases (Cusack et al., 1991; Ribas de Pouplana and Schimmel, 2001; Schimmel and Ribas de Pouplana, 2001; Wu and Gross, 1993), whose catalytic domain contains a seven-stranded anti-parallel β-sheet and three conserved sequence motifs: motif 1 forms the dimer interface while motifs 2 and 3 contains active site residues critical for aminoacylation (Belrhali et al., 1994; Chimnaronk et al., 2005; Cusack et al., 1990; Cusack et al., 1996; Itoh et al., 2008; Leberman et al., 1991). Unlike most other tRNA synthetases, SerRS does not identify the anticodon, but instead recognizes the long variable arm that is unique to tRNASer (Asahara et al., 1994; Heckl et al., 1998; Normanly et al., 1992; Sampson and Saks, 1993; Soma and Himeno, 1998; Wu and Gross, 1993). The recognition is achieved by the N-terminal tRNA binding domain (TBD) which, in all organisms except for methanogens, is composed of two long α-helices protruding away from the catalytic/aminoacylation domain (AD). The TBD from one subunit of the SerRS dimer interacts with tRNASer at the variable arm and the TψC loop to direct the 3′-CCA end to enter the active site of the other subunit (Biou et al., 1994; Cusack et al., 1996). In addition to charging tRNASer, SerRS also serylates the selenocysteine-specific tRNA (tRNASec) to participate in the translational incorporation of selenocysteine—the 21st amino acid—into selenoproteins in all domains of life (Amberg et al., 1996; Commans and Bock, 1999).
While preserving its classic role in protein synthesis, vertebrate SerRS developed a second essential function in vascular development through acquisition of a C-terminal UNE-S domain that is dispensable for aminoacylation (Amsterdam et al., 2004; Fukui et al., 2009; Herzog et al., 2009; Xu et al., 2012). From fish to humans, the UNE-S domain harbors a nuclear localization signal (NLS) sequence that directs SerRS from the cytoplasm into the nucleus to control the expression of VEGFA (Xu et al., 2012) Mutations that disrupted SerRS nuclear localization caused abnormal vasculature and premature death in zebrafish. Prior to the acquisition of the UNE-S domain is the insertion of two sequence motifs that are present in all vertebrates as well as in some invertebrates. The first (Insertion I) of 23 residues is inserted between the two long α-helices in the TBD and is distal to the active site. The second (Insertion II) is only 9 residues located between motifs 1 and 2 in the AD. No functional annotation has been reported for these insertions. Interestingly, both insertions were completely or partially disordered in the crystal structure of the free human SerRS (Xu et al., 2012).
To gain more insight on human SerRS, we set out to determine the crystal structure of human SerRS in complex with its aminoacylation reaction intermediate analog. Remarkably, binding of the adenylate induced a conformational change of the TBD not seen in bacterial, archaeal and lower eukaryotic SerRSs. The binding also induced structural ordering of both insertions, including Insertion I that is 70 Å away from the adenylate binding site. Further structural and functional analyses suggest that Insertion II, albeit being close to the active site, plays a minimal role on aminoacylation, whereas Insertion I mediates a long-range conformational and functional communication with the active site.
RESULTS
Ser-SA induces conformational change of TBD unique to human SerRS
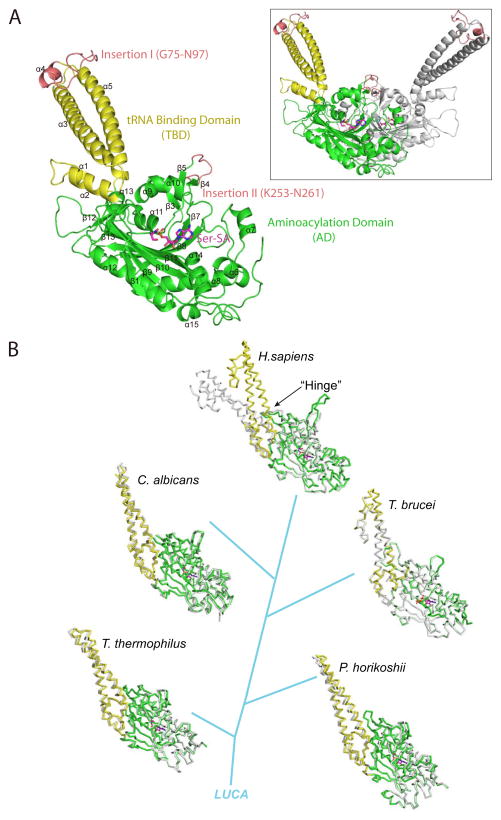
The crystal structure of human SerRS in complex with Ser-AMP analog 5′-O-(N-(L-Seryl)-Sulfamoyl)Adenosine (Ser-SA) is solved at 2.9-Å resolution (Table 1A). The P321 trigonal crystal contains one subunit of SerRS in the asymmetric unit and the structure of the dimeric protein can be generated through crystallographic symmetry operation (Figure 1A). Except for the C-terminal UNE-S domain, conformation of the 514-residue protein, including Insertions I (G75-N97) and II (K253-N261), is completely resolved (Figure 1A). Compared with the crystal structures of the free human SerRS we determined previously (Xu et al., 2012), binding of Ser-SA in the active site induces a prominent conformational change and a tightening of the TBD (Figure 1B; top). The conformational change acts as a lever that dramatically affected the position of Insertion I in the complex versus in the free protein (Figure S1). In contrast, no obvious conformational change of the TBD resulting from Ser-SA or ATP binding was observed in any previous structures of SerRS from organisms including Thermus thermophilus (bacterium) (Biou et al., 1994; Cusack et al., 1996; Fujinaga et al., 1993), Pyrococcus horikoshii (archaeon) (Itoh et al., 2008), Candida albicans (O’Sullivan et al., 2001) and Trypanosoma brucei (lower eukaryotes) (Figure 1B; bottom). Thus, the Ser-SA induced conformational change of the TBD is unique to human SerRS.
Table 1A.
X-ray crystallography data collection and refinement statistics
| Data collection | |
|
| |
| Cell parameters (Å) |
a = 167.074, b = 167.074, c = 65.156 α = β = 90°, γ = 120° |
| Space group | P321 |
| Wavelength (Å) | 0.9794 |
| Resolution (Å) | 50 (2.95) – 2.9 |
| No. of all reflections | 233550 |
| No. of unique reflections | 23503 |
| Completeness (%) | 99.9 (100) |
| Redundacy | 9.9 (10.1) |
| I/σI | 6.2 (2.7) |
| Rmerge (%) | 9.4 (66.9) |
|
| |
| Refinement | |
|
| |
| Resolution (Å) | 50 – 2.9 |
| Total No. of reflections | 23501 |
| No. of reflections used | 22293 |
| Rwork/Rfree (%) | 25.5/26.8 |
| No. of atoms | 3875 |
| Protein | 3836 |
| Water | 3 |
| SSA | 1 |
| PO43− | 1 |
| Mg2+ | 2 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.007 |
| Bond angle (°) | 1.0 |
| Average B-factors (Å2) | 69.7 |
Figure 1. Overall structure of human SerRS in complex with Ser-SA and the conformational change of TBD (see also Figure S1, S2 and S3).
(A) Crystal structure of human SerRS bound with Ser-SA. The secondary structure elements are labeled in black. Inset: Structure of the dimeric SerRS with the second subunit generated by crystallographic symmetry operation shown in gray. (B) Ser-SA induced conformational change of the TBD is unique to human SerRS. The Ser-SA bound and free structures from human (PDB 4L87 and 3VBB), C. albicans (PDB 3QO8 and 3QNE), T. brucei (PDB 3LSS and 3LSQ), P. horikoshii (PDB 2DQ0 and 2ZR3) and T. thermophilus (PDB 1SET and 1SRY) are superimposed at the AD. Free SerRSs are shown in gray and the Ser-SA bound SerRSs in colors.
Free human SerRS has a unique open conformation
A closer examination of the structures indicated that the tight conformation of the TBD in Ser-SA bound human SerRS is not unique. In fact, ligand bound or not, the TBD in SerRS from T. thermophiles, P. horikoshii, C. albicans or T. brucei all adopt the similar tight conformation (Figure S2). What is unique is the open and flexible conformation of the TBD in the free human SerRS. Indeed, multiple conformations and varies degrees of openness of the TBD were observed in the crystal structures of the free human SerRS (Xu et al., 2012). The conformational flexibility of the TBD was captured as conformational variability due to the differences in crystal lattice interactions for each TBD (Figure S3). Importantly, upon Ser-SA binding, the TBD adopts a conformation that is more closed than any one of the TBD conformations found in the free SerRS (Figure S1), suggesting that the open and flexible conformations of TBD can be tightened and leveraged upon Ser-SA binding.
The communication between AD and TBD in human SerRS involves residues specific to higher eukaryotes
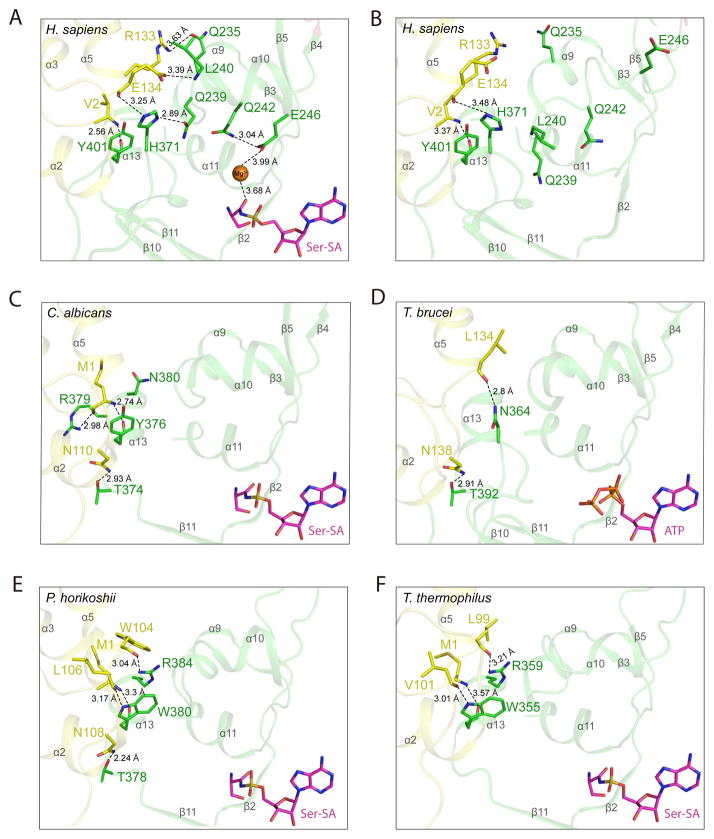
An important question is how the active site, upon Ser-SA binding, is communicated with TBD in human SerRS. In fact, Ser-SA is bound in the active site of human SerRS via residues that are highly conserved throughout evolution (Figure S4). Interestingly, extra electron densities are present in the active site, which most likely correspond to two Mg2+ ions mediating interactions between Ser-SA and the protein (Figure S4). The first Mg2+ is located in a conserved position that was also seen in the co-crystal structure of the SerRS/Ser-SA complex from C. albicans. The Mg2+ coordinates a sulfamoyl oxygen of Ser-SA with the carboxyl side chain of E391 and the hydroxyl side chain of S394. (As seen in the C. albicans structure, the rest of the octahedral coordination for the Mg2+ ion is through water molecules that do not resolve at this resolution of 2.9 Å.) The second Mg2+ ions is, however, in a unique position not seen in the C. albicans structure and coordinates the amide oxygen of Ser-SA with the carboxyl side chain of E246, which in turn forms a hydrogen bond with the carboxamide side chain of Q242. This Mg2+-mediated interaction induces the helical formation of α10, where E246 and Q242 are located (Figure 2A, B). Also located on α10 are two residues—Q239 and L240—that are important for engaging the TBD (Figure 2A and see below).
Figure 2. Ser-SA induced conformational change in human SerRS is caused by shifting the TBD-AD interactions from α13 in lower organisms to α9–α10 in human SerRS (see also Figure S5).
The hydrogen bonding interactions in the “hinge” region mediating TBD (yellow) and AD (green) in crystal structures of (A) human SerRS bound with Ser-SA (PDB 4L87) (B) ligand-free human SerRS (PDB 3VBB) (C) C. albicans SerRS bound with Ser-SA (PDB 3QO8) (D) T. brucei SerRS bound with ATP (PDB 3LSS) (E) P. horikoshii SerRS bound with Ser-SA (PDB 2DQ0) and (F) T. thermophiles SerRS bound with Ser-SA (PDB 1SET). The secondary structures are labeled in gray.
To understand which residues in human SerRS are involved in mediating the conformational change of TBD in respond to Ser-SA binding, we examined the “hinge” region where the TBD is connected to the AD (Figure 1B). We found two residues, R133 and E134, from the TBD domain, whose side chains interacts with residues from the AD only when Ser-SA is bound (Figure 2A, B). In particular, the guanidine side chain of R133 forms a hydrogen bond with the carboxamide side chain of Q235 on helix α9; and the carboxyl side chain of E134 form a hydrogen bond with the main chain N of L240 on α10. The only hydrogen-bonding interactions found in the free SerRS to bridge the TBD with the AD is between the main chain O of E134 and the imidazole side chain of H371 (preceding β10) and between the main chain N of V2 and the main chain O of Y401 (preceding α13) (Figure 2B). Upon Ser-SA binding, these two interactions are strengthened, as evidenced by shortened hydrogen bonding distances, and are used to engage another residue Q239 on α10 to further enhance the domain-domain interaction (Figure 2A). Therefore, Ser-SA binding induces the helical formation of α10 that allows its residues Q239 and L240 and residue Q235 on α9 to engage the TBD to rotate towards the active site (Figure 2A, B). Importantly, the residues involved in mediating the conformational change of TBD (i.e. R133, E134, Q235, Q239, L240, Q242, E246 and H371) are strictly conserved in all vertebrate SerRSs (Figure 3).
Figure 3. Conservations of key residues involved in the TBD-AD interaction and of the Insertions I and II in higher eukaryotes.
Key residues involved in communication of TBD and AD are highlighted in yellow (TBD) or in green (AD). The vertebrate specific Insertions I and II are shown in salmon.
TBD interactions shift from α13 in lower organisms to α9–α10 in human SerRS
To understand how the tight conformation of the TBD is achieved in bacterial, archaeal and lower eukaryotic SerRSs, we examined the crystal structures of SerRS from T. thermophiles, P. horikoshii, C. albicans and T. brucei to identify residues that mediate the TBD-AD interaction. In each structure, we found multiple hydrogen-bonding interactions between the two domains and, in contrast to the situation in human SerRS, the interactions are essentially unaffected by ligand binding (Figure 2C–F and Figure S5). Interestingly, in all non-human SerRS structures, a α–helix that is equivalent to α13 in human SerRS is key for the domain-domain interaction: in C. albicans, Y376 on α13 and T374 immediately preceding α13 interact with M1 and N110 of the TBD, respectively (Figure 2C). The M1-Y376 interaction also involves R379 (as well as N380 in the ligand-free structure) on α13 (Figure 2C); in T. brucei, T392 preceding α13 interacts with N138 of the TBD, while N364, the residue equivalent to H371 in human SerRS, interacts with L134 (Figure 2D); in P. horikoshii, T378 preceding α13 interacts with N108 of the TBD, while W380 and R384 on α13 interact with TBD residues M1, W104 and L106 (Figure 2E); similarly in T. thermophiles, W355 and R359 on α13 interact with TBD residues M1, L99 and V101 (Figure 2F). The only inter-domain hydrogen-bonding interaction found in human SerRS that is also conserved in other SerRSs is the V2-Y401 backbone interaction, which is equivalent to M1-Y376, M1-W380 and M1-W355 interaction in C. albicans, P. horikoshii and T. thermophiles SerRS, respectively (Figure 2A, C, E, F). (This interaction is not found in T. brucei, instead, the L134-N364 interaction similar to the E134-H371 interaction in human SerRS is found in T. brucei (Figure 2A, D).) Remarkably, except for the V2-Y401 backbone interaction, α13 is no longer used by human SerRS to interact with the TBD. Instead, α9 and especially α10 that are closer to the active site are used to create the communication between the active site and the TBD in human SerRS (Figure 2A).
Ser-SA binding stabilized the higher eukaryote-specific insertions in human SerRS
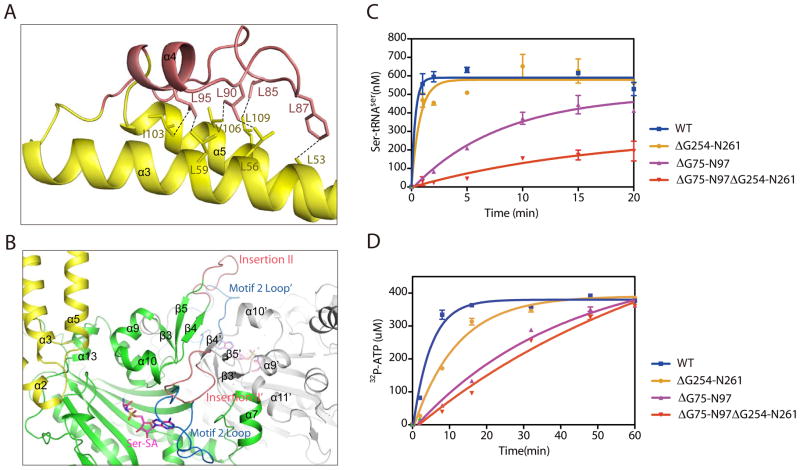
Ser-SA binding not only engages the TBD but also stabilizes the conformation of Insertion I, which is located at the far end of the TBD approximately 70 Å away from the active site (Figure 1A). This suggests that the communication between the active site and the TBD is long-range and not limited to the “hinge” region. Insertion I connects the two long α helices (α3 and α5) of the TBD with a long loop (G75-T91) followed by a short α-helix (α4; A92-N97). Interestingly, part of the Insertion I sequence contains one leucine residue at every five amino acids (L85SFDDL90TADAL95AN). This leucine-rich motif provides extensive hydrophobic interactions with residues from both α3 and α5 (Figure 4A). For example, L85 interacts with L109 of α5; L90 interacts with both L56 of α3 and V106 of α5; and L95 interacts with L59 of α3 and L103 of α5 (Figure 4A). These hydrophobic interactions not only stabilize the conformation of Insertion I but also that of the two long helices of the TBD, which is connected to the active site via the opposite ends (Figure 2A).
Figure 4. Structural and functional analyses of Insertions I and II.
(A) Leucine-rich motif of Insertion I (salmon) forms extensive hydrophobic interactions with residues from α3 and α5 (yellow). (B) Structural environment of Insertion II (salmon) near the dimer interface and the active site. (C) Aminoacylation assay indicating that deletion of Insertion I has a larger effect on SerRS aminoacylation activity than deletion of Insertion II. (D) ATP PPi-exchange assay confirming the effect of Insertion I on the active site.
Insertion II in the aminoacylation domain is also resolved as a result of Ser-SA binding (Figure 1A). This insertion forms a loop structure connecting the antiparallel β4 and β5 strands. It protrudes out of the aminoacylation domain to cross over the dimer interface and sits on top of the motif 2 loop of the other subunit (Figure 4B). Therefore, Insertion II is close to the active site of the other subunit and seems to stabilize the dimerization interface. Indeed, the dimer interface is increased from 1126 ± 36 Å2 to 1615 Å2 as a result of Ser-SA binding and the structural ordering of Insertion II.
Insertion I functionally communicates with the active site
The long-range conformational communication between the active site and Insertion I suggested that Insertion I may affect the enzymatic activity of SerRS. The proximity of Insertion II to the active site across the dimer also raised the same possibility. To test the effect of the insertions on aminoacylation, we created deletion mutants that remove Insertion I or II or both. Interestingly, despite that Insertion II is closer to the active site, removal of Insertion I (ΔG75-N97) has a greater impact than removal of Insertion II (ΔG254-S261) on the overall aminoacylation activity of human SerRS as measured by the incorporation of serine onto tRNASer (Figure 4C). Kinetics analysis indicated that ΔG75-N97 SerRS had a 19-fold decrease in maximum catalytic rate (kcat) and a 2.3-fold enhancement in tRNA binding (decrease in Km) to give a 8-fold decrease in the overall catalytic efficiency (kcat/Km). In contrast, ΔG254-S261 SerRS only had a 2-fold decrease in kcat and a 1.5-fold decrease in Km to give a negligible 1.4-fold decrease in kcat/Km (Table 1B). Simultaneous removal of both insertions had a more pronounced effect (Figure 4C and Table 1B).
Table 1B.
Kinetic parameters for tRNASer of human wild-type and mutant SerRS proteins.
| Enzyme | Km (μM)a | kcat (s−1)a | Relative kcat/Km a | Kd (μM)b |
|---|---|---|---|---|
| WT | 0.77±0.25 | 0.43±0.040 | 1 | 0.37±0.010 |
| ΔG254-N261 | 0.51±0.23 | 0.20±0.025 | 0.72 | 0.24±0.028 |
| ΔG75-N97 | 0.33±0.40 | 0.025±0.004 | 0.13 | 0.19±0.016 |
| ΔG75-N97ΔG254-N261 | 0.13±0.10 | 0.005±0.001 | 0.07 | 0.099±0.019 |
Determined by aminoacylation assay.
Determined by nitrocellulose filter binding assay.
The kinetic parameters from the aminoacylation assay suggested that Insertion I and Insertion II are unnecessary, and perhaps even slightly cumbersome, for tRNA binding. To confirm this interpretation, we performed the nitrocellulose filter binding assay to directly measure the tRNA binding constants (Kds) of SerRS and its deletion mutants. The results are highly consistent with those from the aminoacylation assay and show that removal of either or both insertions would enhance tRNA binding (Table 1B).
To confirm that Insertion I does remotely impact the active site, we also measured the serine-dependent ATP-PPi exchange activity of SerRS (and its deletion mutants) in the absence of tRNA. Consistent with the result from the tRNA aminoacylation assay, removal of Insertion I more dramatically impacted the catalytic activity than removal of Insertion II (Figure 4D). Therefore, both a structural and a functional interplay exist between Insertion I and the active site.
DISCUSSION
It has been well established that vertebrate SerRS has expanded its function beyond aminoacylation to also play an essential role in vascular development (Amsterdam et al., 2004; Fukui et al., 2009; Herzog et al., 2009; Xu et al., 2012). The functional expansion is dependent on the UNE-S domain that was added to SerRS during the invertebrate-to-vertebrate transition when the closed circulatory system was developed. Using its embedded NLS, the UNE-S domain functions to facilitate the translocation of SerRS from the cytoplasm into the nucleus to regulate VEGF expression. Based on our sequence analysis (Figure 3), the open and flexible conformation of the TBD found in human SerRS should be conserved in all vertebrates, and perhaps even in some insects such as the yellow fever mosquito Aedes aegypti, as they contain all the key residues for engaging the TBD in respond to ligand binding and therefore do not need to stay in the closed conformation to support catalysis. Although the mechanism of how SerRS regulates VEGF in the nucleus is not yet clear, it is tempting to speculate that the open conformation of SerRS specific to higher eukaryotes is developed or is used to accommodate the nuclear function of SerRS.
Also not known are the critical domain(s) or site(s) in SerRS that are used for supporting its nuclear function. In this regard, both Insertion I and Insertion II are newly added motifs that are concomitantly present in all vertebrates and in some but not all invertebrates; both insertions are rich in charged residues and have surface locations ideal for providing protein-protein or protein-nucleic acid interactions and, from our charging kinetic and tRNA binding analyses, it is clear that the insertions are not meant for enhancing tRNA binding. It would be interesting to test in future works if the two insertions are important for the nuclear function of SerRS in regulating vasculogenesis.
One of the advantages to expand the “functionome” of tRNA synthetases was thought to provide coordination between protein synthesis and other cellular processes important for higher organisms. For example, the nuclear import of human tyrosine tRNA synthetase is controlled by the cytoplasmic level of tRNATyr to link the role of TyrRS in the nucleus with the demands of protein synthesis in the cytoplasm (Fu et al., 2012). Our observation of a higher eukaryote-specific, long-range conformational and functional communication between the enzymatic core and a potential functional site in SerRS for regulating vasculogenesis raises the possibility that this communication is important for coordinating translation with vasculogenesis.
CONCLUSIONS
Crystal structure analyses of human SerRS revealed that the TBD adopts open and flexible conformations that are unique to SerRSs from higher eukaryotes. Upon Ser-SA binding, the TBD is tightened through interacting with a newly formed α-helix (α10) in the AD near the active site. The conformational change of the TBD is leveraged to dramatically change the position of a higher eukaryote-specific motif—Insertion I—that may be involved in the function of SerRS in vascular development. Inversely, deletion of Insertion I allosterically affected the enzymatic activity of SerRS indicating a long-range conformational and functional communication between the active site and Insertion I, which may serve to coordinate the dual roles of SerRS in protein synthesis and in vascular development.
EXPERIMENTAL PROCEDURES
Protein expression and purification
Human SerRS gene was cloned into pET20b vector (Novagen) with a C-terminal 6×His tag and overexpressed in E. coli BL21 (DE3) cells. The cells were lysed by M-110L Microfluidizer Processor (Microfluidics) in 25 mM Hepes-Na pH 7.5, 300 mM NaCl and 20 mM imidazole and 5 % glycerol. The cell lysates were centrifuged at 25,000 rpm for 40 min. The soluble fractions were applied to Ni-NTA column (Qiagen), and the bound SerRS was eluted by 250 mM imidazole. After buffer exchange into 25 mM Hepes-Na pH 7.5, SerRS was further purified by ion exchange chromatography using a Resource Q column (GE Healthcare) with a NaCl gradient from 0 to 1 M (SerRS was eluted around 250 mM NaCl), and by size exclusion chromatography using a HiLoad 16/60 Superdex 200 prep grade column (GE Healthcare). The purified human SerRS was collected and concentrated for crystallization. The Insertion I and Insertion II deletion mutants of SerRS were constructed by site-directed mutagenesis and purified using the same methods.
Crystallization
High-throughput crystallization screen was performed using Mosquito liquid transfer robot (TTP Labtech) and the sitting-drop vapor diffusion method. Crystals were obtained at room temperature by mixing 0.1 μL human SerRS (10 mg mL−1, pre-incubated with 250 μM Ser-SA in buffer containing 25 mM Hepes-Na pH 7.5, 100 mM KCl, 10 mM MgCl2 and 5 mM DTT) with 0.1 μL reservoir solution (20% PEG 3350, 0.2 M Ammonium Fluoride or Ammonium Formate, 0.1 M Hepes pH 7.0) and by equilibrating the mixture against 70 μL reservoir solution. The final crystals used for data collection were further optimized by adding 0.5% w/v n-Dodecyl-β-D-maltoside into the mixture.
Crystal data collection, structure determination and refinement
Crystals were cryoprotected with 15% glycerol added to the reservoir solution and flash-frozen with liquid nitrogen. A 2.9-Å resolution data set was collected at 100 K on beamline BL7-1 at Stanford Synchrotron Radiation Lightsource with an ADSC Q315 detector. The crystal belongs to space group P321 with unit cell dimensions a = b = 167.074 Å and c = 65.156 Å, α = β = 90°, γ = 120°. Diffraction data were processed, integrated and scaled with HKL2000 (Otwinowski. Z and Minor. W, 1997). The structure of human SerRS and Ser-SA complex was solved by molecular replacement using the human SerRS apo-structure (PDB ID 3VBB(Xu et al., 2012)) as a search model and the PHASER program (McCoy et al., 2007) from the CCP4 package (1994). Iterative model building and refinement were performed using Coot (Emsley and Cowtan, 2004) and Refmac5 (Murshudov et al., 1997) to obtain the final model with Rwork of 25.5% and Rfree of 26.8% at 2.9-Å resolution.
Active site titration assay
Active site titration assay was performed in 100 mM HEPES pH 7.5, 20 mM KCl, 10 mM MgCl2, 50 μM ATP, 22.2 nM [γ-32P]-ATP, 20 mM L-serine, 2 μg mL−1 pyrophosphatase (Roche) and 2 mM DTT as previously described (Beebe et al., 2007; Xu et al., 2012) to determine the concentration of active enzymes.
ATP-PPi exchange assay
ATP-PPi exchange assay was performed with 0.5 μM SerRS (WT or mutants) in 100 mM Hepes pH 7.5, 20 mM KCl, 10 mM MgCl2, 2 mM ATP, 500 μM L-serine, 1 mM Na-PPi and 0.07 mCi Na-32PPi and 2 mM DTT as described (Xu et al., 2012).
Aminoacylation assay
Human tRNASer was in vitro transcribed as previous described (Xu et al., 2012). Aminoacylation assays were performed with 50 nM enzyme in 50 mM HEPES pH 7.5, 20 mM KCl, 10 mM MgCl2, 4 mM ATP, 2 μM [3H]-L-serine, 20 μM L-serine, 2 mM DTT, 0.1mg mL−1 BSA, 4 μg mL−1 pyrophosphatase (Roche) and 1 μM tRNASer transcript.
Filter binding assay
In vitro transcribed human tRNASer was diluted to 25 pmol μL−1, and dephosphorylated by calf intestine alkaline phosphatase at 37 °C for 1 hr. The dephosphorylated tRNASer was labeled with 3′-32P by T4 polynucleotide kinase at 37 °C for 1hr (Ambion’s Kinasemax kit), and spun through a G-25 column to remove the unlabeled nucleotides. The labeled tRNA was re-annealed and maintained in buffer containing 100 mM Hepes, 2 mM MgCl2. The filter binding assay was performed by incubating different concentrations of SerRS (WT or mutants) with 32P-labeled tRNASer mixed with cold tRNASer in Assay buffer (50 mM Hepes, 20 mM KCl, 4 mM MgCl2, 2 mM DTT), after incubation for 30 min, the mixtures were spotted on nitrocellulose filter membrane (Millopore). The membranes were washed three times with 0.5 mL Wash buffer (25 mM Hepes, 10 mM KCl, 4 mM MgCl2) before dried. The amount of bound tRNASer on the membranes was measured using a multipurpose scintillation counter LS-6500 (Beckman Coulter).
Supplementary Material
Highlights.
Crystal structure of human SerRS in complex with Ser-SA solved at 2.9-Å resolution
Ser-SA binding induces conformational change of SerRS unique to higher eukaryotes
Ser-SA binding stabilizes conformation of higher eukaryote-specific insertions
Long-range structural and functional communication between active site and Insertion I
Acknowledgments
We thank Professor Christopher Francklyn for valuable discussion on SerRS structures. The work is supported by grant GM 088278 from National Institutes of Health and a fellowship from the National Foundation for Cancer Research. X-ray diffraction data was collected at Stanford Synchrotron Radiation Lightsource, a Directorate of SLAC National Accelerator Laboratory and an Office of Science User Facility operated for the U.S. Department of Energy Office of Science by Stanford University. The SSRL Structural Molecular Biology Program is supported by the DOE Office of Biological and Environmental Research, and by the National Institutes of Health, National Institute of General Medical Sciences (including P41GM103393).
Footnotes
ACCESSION NUMBERS
The atomic coordinates and structure factors of human SerRS in complex with Ser-SA have been deposited in the Protein Data Bank with the accession code 4L87.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Amberg R, Mizutani T, Wu XQ, Gross HJ. Selenocysteine synthesis in mammalia: an identity switch from tRNASer to tRNASec. J Mol Biol. 1996;263:8–19. doi: 10.1006/jmbi.1996.0552. [DOI] [PubMed] [Google Scholar]
- Amsterdam A, Nissen RM, Sun Z, Swindell EC, Farrington S, Hopkins N. Identification of 315 genes essential for early zebrafish development. Proc Natl Acad Sci U S A. 2004;101:12792–12797. doi: 10.1073/pnas.0403929101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asahara H, Himeno H, Tamura K, Nameki N, Hasegawa T, Shimizu M. Escherichia coli seryl-tRNA synthetase recognizes tRNASer by its characteristic tertiary structure. J Mol Biol. 1994;236:738–748. doi: 10.1006/jmbi.1994.1186. [DOI] [PubMed] [Google Scholar]
- Beebe K, Waas W, Druzina Z, Guo M, Schimmel P. A universal plate format for increased throughput of assays that monitor multiple aminoacyl transfer RNA synthetase activities. Anal Biochem. 2007;368:111–121. doi: 10.1016/j.ab.2007.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belrhali H, Yaremchuk A, Tukalo M, Larsen K, Berthet-Colominas C, Leberman R, Beijer B, Sproat B, Als-Nielsen J, Grubel G, et al. Crystal structures at 2.5 angstrom resolution of seryl-tRNA synthetase complexed with two analogs of seryl adenylate. Science. 1994;263:1432–1436. doi: 10.1126/science.8128224. [DOI] [PubMed] [Google Scholar]
- Biou V, Yaremchuk A, Tukalo M, Cusack S. The 2.9 A crystal structure of T. thermophilus seryl-tRNA synthetase complexed with tRNASer. Science. 1994;263:1404–1410. doi: 10.1126/science.8128220. [DOI] [PubMed] [Google Scholar]
- Chimnaronk S, Gravers Jeppesen M, Suzuki T, Nyborg J, Watanabe K. Dual-mode recognition of noncanonical tRNAsSer by seryl-tRNA synthetase in mammalian mitochondria. EMBO J. 2005;24:3369–3379. doi: 10.1038/sj.emboj.7600811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Commans S, Bock A. Selenocysteine inserting tRNAs: an overview. FEMS Microbiol Rev. 1999;23:335–351. doi: 10.1111/j.1574-6976.1999.tb00403.x. [DOI] [PubMed] [Google Scholar]
- Cusack S, Berthet-Colominas C, Hartlein M, Nassar N, Leberman R. A second class of synthetase structure revealed by X-ray analysis of Escherichia coli seryl-tRNA synthetase at 2.5 A. Nature. 1990;347:249–255. doi: 10.1038/347249a0. [DOI] [PubMed] [Google Scholar]
- Cusack S, Hartlein M, Leberman R. Sequence, structural and evolutionary relationships between class 2 aminoacyl-tRNA synthetases. Nucleic Acids Res. 1991;19:3489–3498. doi: 10.1093/nar/19.13.3489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cusack S, Yaremchuk A, Tukalo M. The crystal structure of the ternary complex of T.thermophilus seryl-tRNA synthetase with tRNA(Ser) and a seryl-adenylate analogue reveals a conformational switch in the active site. EMBO J. 1996;15:2834–2842. [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Fu G, Xu T, Shi Y, Wei N, Yang XL. tRNA-controlled nuclear import of a human tRNA synthetase. J Biol Chem. 2012;287:9330–9334. doi: 10.1074/jbc.C111.325902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujinaga M, Berthet-Colominas C, Yaremchuk AD, Tukalo MA, Cusack S. Refined crystal structure of the seryl-tRNA synthetase from Thermus thermophilus at 2.5 A resolution. J Mol Biol. 1993;234:222–233. doi: 10.1006/jmbi.1993.1576. [DOI] [PubMed] [Google Scholar]
- Fukui H, Hanaoka R, Kawahara A. Noncanonical activity of seryl-tRNA synthetase is involved in vascular development. Circ Res. 2009;104:1253–1259. doi: 10.1161/CIRCRESAHA.108.191189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckl M, Busch K, Gross HJ. Minimal tRNASer and tRNASec substrates for human seryl-tRNA synthetase: contribution of tRNA domains to serylation and tertiary structure. FEBS Lett. 1998;427:315–319. doi: 10.1016/s0014-5793(98)00435-9. [DOI] [PubMed] [Google Scholar]
- Herzog W, Muller K, Huisken J, Stainier DY. Genetic evidence for a noncanonical function of seryl-tRNA synthetase in vascular development. Circ Res. 2009;104:1260–1266. doi: 10.1161/CIRCRESAHA.108.191718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh Y, Sekine S, Kuroishi C, Terada T, Shirouzu M, Kuramitsu S, Yokoyama S. Crystallographic and mutational studies of seryl-tRNA synthetase from the archaeon Pyrococcus horikoshii. RNA Biol. 2008;5:169–177. doi: 10.4161/rna.5.3.6876. [DOI] [PubMed] [Google Scholar]
- Leberman R, Hartlein M, Cusack S. Escherichia coli seryl-tRNA synthetase: the structure of a class 2 aminoacyl-tRNA synthetase. Biochim Biophys Acta. 1991;1089:287–298. doi: 10.1016/0167-4781(91)90168-l. [DOI] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- Normanly J, Ollick T, Abelson J. Eight base changes are sufficient to convert a leucine-inserting tRNA into a serine-inserting tRNA. Proc Natl Acad Sci U S A. 1992;89:5680–5684. doi: 10.1073/pnas.89.12.5680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Sullivan JM, Mihr MJ, Santos MA, Tuite MF. Seryl-tRNA synthetase is not responsible for the evolution of CUG codon reassignment in Candida albicans. Yeast. 2001;18:313–322. doi: 10.1002/1097-0061(20010315)18:4<313::AID-YEA673>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray Diffraction Data Collected in Oscillation Mode Methods in Enzymology. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Ribas de Pouplana L, Schimmel P. Two classes of tRNA synthetases suggested by sterically compatible dockings on tRNA acceptor stem. Cell. 2001;104:191–193. doi: 10.1016/s0092-8674(01)00204-5. [DOI] [PubMed] [Google Scholar]
- Sampson JR, Saks ME. Contributions of discrete tRNASer domains to aminoacylation by E.coli seryl-tRNA synthetase: a kinetic analysis using model RNA substrates. Nucleic Acids Res. 1993;21:4467–4475. doi: 10.1093/nar/21.19.4467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schimmel P. Aminoacyl tRNA synthetases: general scheme of structure-function relationships in the polypeptides and recognition of transfer RNAs. Annu Rev Biochem. 1987;56:125–158. doi: 10.1146/annurev.bi.56.070187.001013. [DOI] [PubMed] [Google Scholar]
- Schimmel P, Ribas de Pouplana L. Formation of two classes of tRNA synthetases in relation to editing functions and genetic code. Cold Spring Harb Symp Quant Biol. 2001;66:161–166. doi: 10.1101/sqb.2001.66.161. [DOI] [PubMed] [Google Scholar]
- Soma A, Himeno H. Cross-species aminoacylation of tRNA with a long variable arm between Escherichia coli and Saccharomyces cerevisiae. Nucleic Acids Res. 1998;26:4374–4381. doi: 10.1093/nar/26.19.4374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu XQ, Gross HJ. The long extra arms of human tRNA(Ser)Sec and tRNASer function as major identify elements for serylation in an orientation-dependent, but not sequence-specific manner. Nucleic Acids Res. 1993;21:5589–5594. doi: 10.1093/nar/21.24.5589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Shi Y, Zhang HM, Swindell EC, Marshall AG, Guo M, Kishi S, Yang XL. Unique domain appended to vertebrate tRNA synthetase is essential for vascular development. Nat Commun. 2012;3:681. doi: 10.1038/ncomms1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.