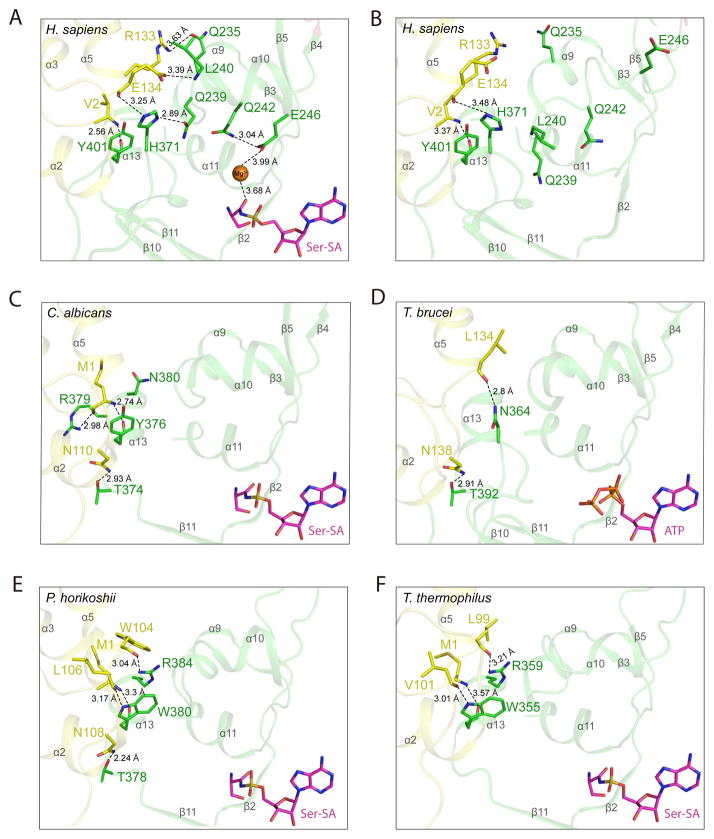
Figure 2. Ser-SA induced conformational change in human SerRS is caused by shifting the TBD-AD interactions from α13 in lower organisms to α9–α10 in human SerRS (see also Figure S5).
The hydrogen bonding interactions in the “hinge” region mediating TBD (yellow) and AD (green) in crystal structures of (A) human SerRS bound with Ser-SA (PDB 4L87) (B) ligand-free human SerRS (PDB 3VBB) (C) C. albicans SerRS bound with Ser-SA (PDB 3QO8) (D) T. brucei SerRS bound with ATP (PDB 3LSS) (E) P. horikoshii SerRS bound with Ser-SA (PDB 2DQ0) and (F) T. thermophiles SerRS bound with Ser-SA (PDB 1SET). The secondary structures are labeled in gray.