Abstract
Hepatitis B virus (HBV) is a small DNA virus with a compact genomic organization. All HBV proteins identified to date have been encoded by unspliced HBV RNAs. Spliced HBV RNAs have been described, but their functions are unknown. We show here that a singly spliced HBV RNA encodes a novel HBV protein in vivo. This HBV splice-generated protein (HBSP) corresponds to the fusion of a part of the viral polymerase and a new open reading frame that is created by the splicing event. In vivo, HBSP protein was found in HBV-infected liver samples, and anti-HBSP antibodies occurred in one-third of sera samples collected from chronic HBV carriers. In vitro, the ectopic expression of HBSP had no effect on viral DNA replication or transcription but induced cell apoptosis without a cell-cycle block. Overall, our results suggest that HBV has evolved a mechanism that directly modulates virus-cell interaction through RNA splicing.
Introduction
Hepatitis B virus (HBV) is a small, enveloped DNA virus that causes acute and chronic liver infections (1). The viral genome contains 4 overlapping open reading frames encoding for capsid, surface, polymerase, and X proteins (1). Three major unspliced transcripts, of 3.5 kb, 2.4 kb, and 2.1 kb, and a less abundant 0.8-kb transcript with a common polyadenylation site, all code for viral proteins identified to date. The 3.5-kb pregenomic RNA also serves as a template for viral replication through a reverse transcription mechanism. In addition, spliced HBV transcripts have been identified in human liver tissues and in HBV-transfected hepatoma cell lines (2–8). Two major HBV-spliced forms of the pregenomic RNA, each with a size of 2.2 kb, have been found in vitro to represent about 30% of the pregenomic RNAs (5). Spliced RNAs have also been detected in ducks and woodchucks infected with duck hepatitis B virus or woodchuck hepatitis virus, respectively. However, the splicing pattern differed among these hepadnaviruses (9–11). We and others have detected defective HBV particles containing a 2.2-kb subgenomic HBV DNA, complementary to a singly spliced HBV RNA, in most livers and sera from the chronically infected patients tested (4, 12, 13). These defective HBV particles are secreted through transcomplementation with a helper wild-type virus, which allows for the packaging and reverse transcription of the singly spliced RNA (4). This raises the possibility that these defective HBV particles and their expressed proteins may be implicated in viral persistence.
The singly spliced RNA had previously been shown to encode for capsid and X proteins (12). In addition, it contains a novel open reading frame overlapping the splice junction, which potentially encodes for an HBV splice-generated protein (HBSP). This protein, with 93 amino acids, begins at the polymerase start codon and includes the first 46 amino acids of the polymerase, followed by the 47 amino acids corresponding to a new HBV amino acid sequence generated by the frameshift (Figure 1a). In this report we demonstrate the in vivo expression of this new protein.
Figure 1.
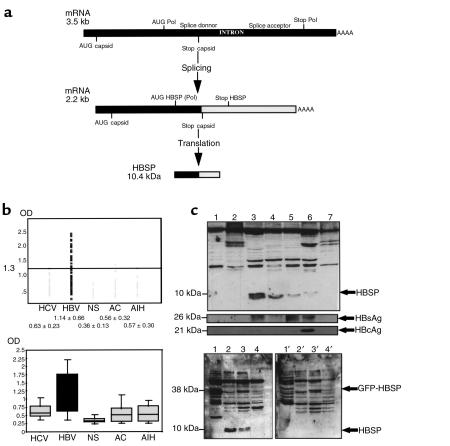
(a) Schematic representation of singly spliced HBV RNA and HBSP translation product. (b) Top: Indirect ELISA for the detection of anti-HBSP antibodies in sera from chronic HBV carriers (n = 45), in normal serum controls (NS, n = 45), and in sera from patients with HCV (n = 45), autoimmune hepatitis (AIH, n = 17), and chronic alcoholic cirrhosis (AC, n = 25). Bottom: Box plot representation of ELISA data. (c) Western blot analysis of liver biopsies. Top: HBSP was detected (using polyclonal chicken anti-HBSP antibodies raised against HBSP bacterial recombinant protein) in 4 HBV-infected livers (lanes 3–6), 2 HCV-infected livers (lanes 2 and 7), and 1 normal liver (lane 1). Middle: HBV envelope (HBsAg) and capsid (HBcAg) detection in the same samples using polyclonal HBs and HBc antibodies. Bottom: HBSP detection, without (left) or with (right) adsorption of anti-HBSP with GST-HBSP bacterial protein, in CCL13 GFP-HBSP–transfected cells (lanes 1 and 1′), HBV-infected livers (lanes 2, 2′, 3, and 3′), and normal liver (lanes 4 and 4′).
Methods
ELISA.
For indirect ELISA, 100 ng of HBSP-specific peptide was coated in a carbonate buffer and left overnight at 4°C in 96-well plates. After blocking, 100 μL each of control sera and sera from HBV-positive patients, diluted 1:20 in PBS, 0.1% Tween, and 10% FCS, was incubated for 1 hour at 37°C. This was washed in PBS and 0.1% Tween, and an anti-human antibody (1:2,000), labeling to the alkaline phosphatase, was added. This mixture was incubated for 30 minutes at 37°C. After revelation (30 minutes) with p-nitrophenylphosphate tablets (Sigma, Deinhoften, Germany), detection was undertaken using a Multiskan spectrophotometer set at an OD of 405 nm (Labsystems, Cergy-pontoise, France).
Plasmid constructs and antibody production.
For HBSP constructs, PCR was performed using a set of primers located at the 5′ (ATGCCCCTATCCTATCAACAC) or 3′ (AAGCCCAAGATGATGGGA) end of the HBSP gene on HBV DNA extracted from serum collected from a chronic HBV carrier. PCR product was cloned in eukaryotic (CMV promoter) or prokaryotic (pET and pGEX) vectors. To obtain the GFP-HBSP fusion protein, the HBSP gene was inserted in E-GFP vectors (CLONTECH Laboratories Inc., Palo Alto, California, USA). HBV 1.1 plasmid has been described previously (14). Anti-HBSP polyclonal antibodies were obtained after the immunization of rabbits and chickens with an HBSP recombinant protein produced in the pET bacterial system (Novagen, Madison, Wisconsin, USA), and were purified on a nickel column (QIAGEN Inc., Hilden, Germany). The pCluc and pXluc constructs (gifts of A. McLachan, The Scripps Research Institute, La Jolla, California, USA) corresponding to the luciferase gene under the control of the capsid or HBx gene promoters, respectively, have been previously described (15).
Cell culture and transfection.
HuH7 and CCL13 liver cells were grown in DMEM supplemented with 10% FBS, and were transfected with 10–20 μg of the indicated construct, using the calcium phosphate procedure. The efficiency of transfection was monitored by adding 1 μg of a β-galactosidase plasmid (pCH110; Pharmacia Biotech, Freiburg, Germany). For in situ β-galactosidase detection, cells were fixed in 4% paraformaldehyde (PFA) and then stained with X-Gal. Western blot analysis was performed on transfected cell extracts or tissue from liver biopsies pulverized in RIPA buffer (12). Blots were revealed using a chemiluminescence assay (ECL; Amersham Life Sciences Inc., Arlington Heights, Illinois, USA). For analysis of HBV RNAs, total RNA was extracted using Trizol (GIBCO BRL, Gaithersburg, Maryland, USA). RNAs were fractionated on a 1.5% agarose-formaldehyde gel and transblotted to Hybond N+ membranes. Southern blot analysis of the HBV replication intermediate was performed as described (12). A 32P-labelled HBV probe was used for Northern and Southern blot hybridization.
Luciferase assays.
HuH7 cells were transfected with 5 μg of pCluc or pXluc (10 μg of pNeo vector was added as carrier), or were cotransfected with 10 μg of pHBSP. One microgram of the β-galactosidase expression plasmid pCH110 was also transfected to monitor the efficiency of transfection. Two days after transfection, cells were lysed in NP-40 buffer and the extracts were clarified by brief centrifugation. The standard luciferase assay was performed on 50 μL of clarified supernatant using a Lumat LB 9501 Berthold luminometer (Analytical Luminescence Laboratory, San Diego, California, USA). Transfections were performed in duplicate.
Morphological cell analysis and cell death assays.
For immunofluorescence analysis, transfected cells grown on coverslips were washed in PBS and fixed in 4% PFA for 10 minutes at 4°C. After blocking with 10% normal goat serum, HBSP antibody was incubated at 37°C for 1 hour. The secondary antibody, FITC–conjugated anti-rabbit (Amersham Life Sciences Inc.), was added. After 30 minutes at 37°C, the nuclei were counterstained with the DNA dye 7-amino-actinomycin D (7-AAD) at 40 μg/mL for 5 minutes. Similarly, cells transfected with recombinant green fluorescent protein (GFP) vectors that were grown on coverslips were fixed with 4% PFA and then analyzed using an epifluorescence laser microscope (Zeiss North America, Thornwood, New York, USA) or a confocal laser (Leica Inc., Deerfield, Illinois, USA) microscope. For flow cytometric analysis for DNA quantification, cells were harvested at indicated timepoints after transfection with GFP-HBSP. Green fluorescent cells and control cells were sorted using a FACS Vantage cytometer (Becton Dickinson, Mountain View, California, USA) and then incubated for 15 minutes at 37°C in lysis buffer (1× PBS, 0.1% citrate, and 0.1% Triton) containing 50 μg/mL of propidium iodide. The DNA content was analyzed by FACScan (Becton Dickinson).
Results
In vivo detection of HBSP protein.
Using a peptide with 20 amino acids, synthetized from the HBSP sequence (amino acids 43–63), we set up an indirect ELISA assay for the detection of anti-HBSP antibodies in sera from patients with chronic HBV. The results showed a significant increase in the mean reactivity in sera from chronic HBV carriers (1.14 OD) compared with normal sera (0.36 OD) or sera from patients with HCV (0.63 OD), autoimmune hepatitis (0.57 OD), or chronic alcoholic hepatitis (0.56 OD) (Figure 1b). Seventeen of 45 (38%) sera samples from chronic HBV carriers showed a reactivity higher than the 1.3 OD cutoff, compared with 3 of 132 (2.3%) samples for the control sera. Statistical analysis performed on a box plot representation using the Student’s t test showed a highly significant difference (P = 0.0001) between these 2 groups. In the group of chronic HBV carriers, anti-HBSP antibodies were principally found in patients with detectable viremia (15 of 17 patients). Conversely, the majority of HBV DNA–negative patients (10 of 12) had negative anti-HBSP assay results. However, we observed no significant correlation between the level of HBV viral load and ELISA positivity (data not shown).
Direct detection of the HBSP protein was then investigated using Western blot analysis of liver biopsy tissue obtained from 5 patients with HBV-related cirrhosis. A 10-kDa band corresponding to the HBSP protein was observed with variable intensity in 4 of the 5 HBV-infected liver specimens tested (Figure 1c, upper panel, lanes 3–6 and data not shown). In contrast, this band was undetectable in liver biopsy tissue from 2 patients with HCV-related cirrhosis and 1 normal liver, which acted as controls (Figure 1c, top, lanes 1, 2, and 7). A competitive experiment using a glutathione-S-transferase–HBSP bacterial recombinant protein further demonstrated the specificity of HBSP detection. The adsorption of anti-HBSP antibodies with GST-HBSP abolished detection of the GFP-HBSP fusion protein expressed transiently in CCL13 cells (Figure 1c, bottom; compare lanes 1 and 1′). Similarly, adsorption abrogated detection of the 10-kDa band in the 2 HBSP-positive liver biopsy samples tested (Figure 1c, bottom; compare lane 2 to lane 2′ and lane 3 to lane 3′). Overall, our data clearly demonstrated the in vivo expression of the HBSP protein. We also investigated capsid and envelope expression in 4 of 5 HBV-infected liver samples (Figure 1c, middle panels). There was no obvious correlation between expression of HBSP and HBcAg or HBsAg. Indeed, HBSP was strongly expressed in specimens with little or no capsid or envelope expression (Figure 1c, middle panels, lanes 3–5).
Biological effects of HBSP ectopic expression.
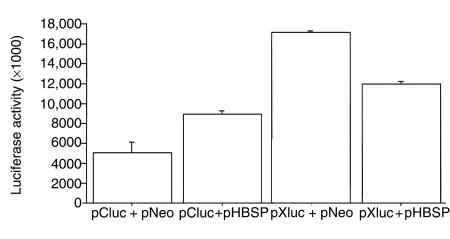
We investigated the influence of HBSP on HBV replication in HuH7 cells transiently transfected with HBV or cotransfected with vectors expressing HBV and HBSP. HBV genome transcription and replication were tested 40 hours after transfection. In cotransfection experiments, Northern blot analysis showed a 0.8-kb band (Figure 2a, lane 2) that corresponds to HBSP transcript and did not hybridize with an HBx-specific probe (data not shown). Thus, no variation in the level of the 3.5-kb and 2.1-kb viral RNA was observed upon HBSP expression (Figure 2a, lanes 1 and 2). Furthermore, in luciferase reporter assays, HBSP did not modify the activity of the capsid promoter regulating the production of pregenomic HBV RNA (Figure 3). Using the same assay, HBSP expression did not modify HBx gene promoter activity (Figure 3). Southern blot analysis did not demonstrate any modification to viral DNA replicative intermediates (Figure 2a, lanes 3 and 4). Thus the forced expression of HBSP had no detectable effect, in vitro, on HBV genome transcription and replication.
Figure 2.
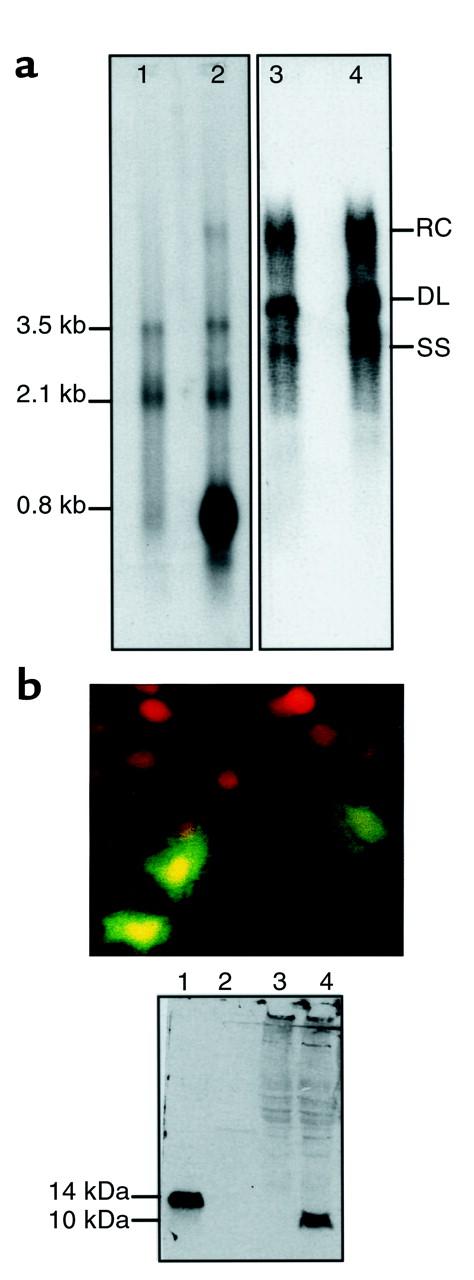
(a) Absence of an in vitro effect of HBSP expression on HBV transcription and replication. HuH7 cells were transiently transfected with HBV (lanes 1 and 3) or were cotransfected with HBV- and HBSP-expressing vectors (lanes 2 and 4). Forty hours after transfection, the HBV transcripts (left) and DNA replicative intermediates (right) were analyzed using Northern or Southern blots, respectively. RC, relax circular DNA; DL, double-stranded linear DNA; SS, single strand DNA. (b) Localization and in vitro expression of HBSP using a rabbit polyclonal anti-HBSP antibody. Top: Cellular distribution of HBSP in HuH7-transfected cells, revealed by immunofluorescence. Bottom: Western blot analysis of HuH7 cells transiently expressing HBSP (lane 4), and nontransfected cells (lane 3). Molecular weight (lane 2). Positive control was pET-HBSP bacterial protein (lane 1).
Figure 3.
Influence of HBSP protein on capsid and HBx promoters using luciferase assay. HuH7 cells were cotransfected either with the luciferase gene under the control of capsid or HBx promoters and pNeo (pCluc + pNeo; pXluc + pNeo), or with HBSP-expressing vector and pCluc (pCluc + pHBSP) or pXluc (pXluc + pHBSP). The values of luciferase activity were normalized by β-galactosidase activity to determine transfection efficiency.
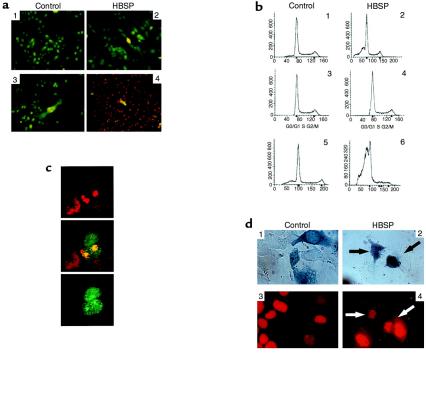
We also investigated the cellular localization of HBSP protein in HuH7 cells transiently transfected with an HBSP-expressing vector. Diffuse nuclear and cytoplasmic immunofluorescence staining was observed in transfected cells using rabbit polyclonal anti-HBSP antibodies (Figure 2b). The specificity of these antibodies was demonstrated using Western blot analysis of HuH7-transfected cells (Figure 2b). The effects of HBSP on cell growth were investigated using a cell viability assay on CCL13 cells transfected with GFP-HBSP or GFP vectors. We first verified that both GFP-HBSP and HBSP had the same cellular location (data not shown). There was no obvious effect of HBSP expression on cell viability 36 hours after transfection (Figure 4a, panels 1 and 2). In contrast, 96 hours after transfection, a major decrease in the number of GFP-HBSP expressing cells was observed when compared with that of GFP-expressing cells (Figure 4a, panels 3 and 4). To determine whether HBSP ectopic expression induces cell death, the cellular DNA content of CCL13 cells expressing GFP-HBSP or GFP was determined by FACS® analysis. Thirty-six hours after transfection, 30% of HBSP-expressing cells (Figure 4b, panel 2) showed a sub-G1 peak characteristic of hypoploid apoptotic cells, compared with 7% of nontransfected cells (Figure 4b, panel 1). The nonapoptotic cells expressing HBSP showed a normal cell-cycle distribution (Figure 4b, panels 3 and 4). Sixty-seven hours after transfection, 68% of HBSP-expressing cells were apoptotic (Figure 4b, panel 6), whereas apoptosis was detected in only 13% of control GFP-transfected cells (Figure 4b, panel 5). Confocal microscopy of GFP-HBSP–transfected cells revealed chromatin condensation and nuclear fragmentation (Figure 4c). The apoptotic effect of HBSP was also tested on transiently transfected HuH7 cells, in which the DNA content of the total cell population was determined using FACS® analysis. Eighteen percent of cells were found to be apoptotic after transfection with HBSP-specific vector, compared with 7% and 23% after transfection with salmon sperm DNA or p53 (acting as positive control), respectively (data not shown). HBSP-induced apoptosis in HuH7 cells was verified by nuclear fragmentation in β-galactosidase–positive transfected cells (Figure 4d). Overall, our data demonstrate that the ectopic expression of both native and GFP-fused HBSP induced apoptosis without a cell-cycle block.
Figure 4.
Effects of HBSP expression on viability. (a) CCL13 cells expressing GFP (panels 1 and 3) or GFP-HBSP (panels 2 and 4). In the same dish, transfected cells were viewed using GFP 36 hours (panels 1 and 2) and 96 hours (panels 3 and 4) after transfection. (b) DNA content of GFP-HBSP or GFP CCL13–transfected cells(FACS®). Sorted GFP-HBSP–expressing cells (panel 2) or nontransfected cells (panel 1) were analyzed 36 hours after transfection. Panels 3 and 4: Cell-cycle analysis on the same cell populations. Panels 5 and 6: Sorted GFP-HBSP–transfected cells (panel 6) or GFP-transfected cells (panel 5) were analyzed 67 hours after transfection. (c) HBSP-induced DNA condensation. Confocal micrograph of CCL13 cells (green) 40 hours after GFP-HBSP transfection. Nuclei were stained with 7-AAD (red). Using different-colored filters, red (top), green (bottom), or both fluorescences in the same field (middle) were detected. (d) HBSP-induced apoptosis in HuH7 cells. HuH7 cells were cotransfected with pCH110 β-galactosidase and pHBSP (panels 2 and 4) or salmon sperm DNA (panels 1 and 3). Transfected cells were revealed using β-galactosidase (blue) assay (panels 1 and 2) and nuclear fragmentation after staining with 7-AAD (red, panels 3 and 4).
Discussion
It is generally accepted that the encoding capacity of the HBV genome has been fully documented (16). In fact, our data demonstrate that, in vivo, a splice RNA encodes a new HBV protein that is a target for the immune response. There was no correlation between the prevalence of anti-HBSP antibodies and the HBV viral load in sera from chronic HBV carriers. Similarly, no correlation was shown between the level of HBSP expression and that of capsid and envelope protein expression in HBV-infected livers. This suggests a regulatory mechanism of singly spliced RNA synthesis that is independent of the level of pregenomic RNA expression. In vitro, the ectopic expression of HBSP demonstrated a lack of effect on HBV DNA replication and transcription. Clearly, based on data from this study we cannot exclude the possibility of an effect of HBSP on other steps of viral particle maturation and secretion. However, our results highlight the potentially interesting effects of HBSP on cell viability. We demonstrated that transient ectopic HBSP expression induced several hallmarks of cell apoptosis without associated cell-cycle block. We do not know as yet whether HBSP also induces liver cell apoptosis in the context of chronic HBV carriers. However, high levels of HBSP could be detected in liver biopsy tissue from HBV-infected patients. This contrasted with the barely detectable level of HBx protein (which also interferes with liver cell proliferation and viability) in liver specimens from patients with chronic hepatitis and hepatocellular carcinoma (17). There is increasing evidence that the regulation of apoptosis plays a key role in the life cycle of several DNA and RNA viruses and contributes directly to their cytopathogenic effects (18). In the case of HBV, it has been well established that the clearance of infected hepatocytes is mediated by the host immune response to various HBV proteins (19). In addition to this well-characterized mechanism, we now show evidence for direct modulation of liver-cell viability by HBSP. This might favor the dissemination of viral particles, as has been suggested for several other viruses (20). Alternatively, HBSP might participate in viral persistence by eliminating a subset of infected cells. Consistent with this hypothesis, we previously showed that defective HBV particles containing a 2.2-kb subgenomic HBV DNA were detected at a higher level in sera from patients with acute hepatitis progressing to chronic hepatitis than in sera from patients recovering from acute infection (12). Taken together, these data support the hypothesis that spliced HBV RNA and HBSP expression may be involved in the pathogenicity or persistence of HBV, or both.
Among DNA viruses, hepadnaviruses share with cauliflower mosaic virus the unique property of replicating their genomes using reverse transcription (21). Spliced RNAs have been detected for both hepadnaviruses and cauliflower mosaic virus (21, 22). It has been demonstrated in the case of cauliflower mosaic virus that spliced RNAs play an essential role in the viral life cycle (22). However, the precise involvement of spliced HBV RNAs in the pathogenesis of HBV has so far remained elusive. Alternative splicing is a well-documented mechanism that allows synthesis of viral proteins capable of modulating gene regulation and viral infectivity (23, 24). This study extends these data to human hepadnavirus.
Acknowledgments
This work was supported by INSERM, ARC, and LNC. We thank F. Demaugre and N. Radjef for helpful discussions and critical review of the manuscript, and M.-A. Petit and V. Maréchal for providing useful reagents.
References
- 1.Ganem D, Varmus HE. The molecular biology of the hepatitis B viruses. Annu Rev Biochem. 1987;56:651–693. doi: 10.1146/annurev.bi.56.070187.003251. [DOI] [PubMed] [Google Scholar]
- 2.Wu H-L, et al. Characterization and genetic analysis of alternatively spliced transcripts of hepatitis B virus in infected human liver tissues and transfected Hep G2 cells. J Virol. 1991;65:1680–1686. doi: 10.1128/jvi.65.4.1680-1686.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen P-J, Chen C-R, Sung J-L, Chen D-S. Identification of a doubly spliced viral transcript joining the separated domains for putative protease and reverse transcriptase of hepatitis B virus. J Virol. 1989;63:4165–4171. doi: 10.1128/jvi.63.10.4165-4171.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Terré S, Petit MA, Bréchot C. Defective hepatitis B virus particles are generated by packaging and reverse transcription of spliced viral RNAs in vivo. J Virol. 1991;65:5539–5543. doi: 10.1128/jvi.65.10.5539-5543.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu H-L, Chen P-J, Lin M-H, Chen D-S. Temporal aspects of major viral transcript expression in Hep G2 cells transfected with cloned hepatitis B virus DNA: with emphasis on the X transcript. Virology. 1991;185:644–651. doi: 10.1016/0042-6822(91)90535-j. [DOI] [PubMed] [Google Scholar]
- 6.Suzuki T, Kajino K, Masui N, Saito I, Miyamura T. Alternative splicing of hepatitis B virus RNAs in HepG2 cells transfected with the viral DNA. Virology. 1990;79:881–885. doi: 10.1016/0042-6822(90)90160-s. [DOI] [PubMed] [Google Scholar]
- 7.Susuki T, Masui N, Kajino K, Saito I, Miyamura T. Detection and mapping of spliced RNA from a human hepatoma cell line transfected with the hepatitis B virus genome. Proc Natl Acad Sci USA. 1989;86:8422–8426. doi: 10.1073/pnas.86.21.8422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Su TS, et al. Hepatitis B virus transcript produced by RNA splicing. J Virol. 1989;63:4011–4018. doi: 10.1128/jvi.63.9.4011-4018.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ogston CW, Razman DG. Spliced RNA of woodchuck hepatitis virus. Virology. 1992;189:245–252. doi: 10.1016/0042-6822(92)90700-y. [DOI] [PubMed] [Google Scholar]
- 10.Hantz O, Baginski I, Fourel I, Chemin I, Trepo C. Viral spliced RNA are produced, encapsidated and reverse transcribed during in vivo woodchuck hepatitis virus infection. Virology. 1992;190:193–200. doi: 10.1016/0042-6822(92)91205-9. [DOI] [PubMed] [Google Scholar]
- 11.Obert S, et al. A spliced hepadnavirus RNA that is essential for virus replication. EMBO J. 1996;15:2565–2574. [PMC free article] [PubMed] [Google Scholar]
- 12.Rosmorduc O, et al. In vivo and in vitro expression of defective hepatitis B virus particles generated by spliced hepatitis B virus RNA. Hepatology. 1995;22:10–19. [PubMed] [Google Scholar]
- 13.Günther S, Sommer G, Iwanska A, Will H. Heterogeneity and common features of defective hepatitis B virus genomes derived from spliced pregenomic RNA. Virology. 1997;238:363–371. doi: 10.1006/viro.1997.8863. [DOI] [PubMed] [Google Scholar]
- 14.Guidotti LG, Matzke B, Schaller H, Chisari FV. High-level hepatitis B virus replication in transgenic mice. J Virol. 1995;69:6158–6169. doi: 10.1128/jvi.69.10.6158-6169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang P, Raney AK, McLachlan A. Characterization of the hepatitis B virus X- and nucleocapsid gene transcriptional regulatory elements. Virology. 1992;191:31–41. doi: 10.1016/0042-6822(92)90163-j. [DOI] [PubMed] [Google Scholar]
- 16.Gerlich, W.H., and Heerman, K.H. 1991. Hepatitis B virus proteins and virus assembly. In Viral hepatitis and liver disease. F.B. Hollinger, S.M. Lemon, and H.S. Margolies, editors. Lippincott Williams & Wilkins. Baltimore, MD. 121–134.
- 17.Yen T. Hepadnaviral X protein: review of recent progress. J Biomed Sci. 1996;3:20–30. doi: 10.1007/BF02253575. [DOI] [PubMed] [Google Scholar]
- 18.O’Brien V. Viruses and apoptosis. J Gen Virol. 1998;79:1833–1845. doi: 10.1099/0022-1317-79-8-1833. [DOI] [PubMed] [Google Scholar]
- 19.Chisari FV. Hepatitis B virus transgenic mice: models of viral immunobiology and pathogenesis. Curr Top Microbiol Immunol. 1996;206:149–173. doi: 10.1007/978-3-642-85208-4_9. [DOI] [PubMed] [Google Scholar]
- 20.Teodoro JG, Branton PE. Regulation of apoptosis by viral gene products. J Virol. 1997;71:1739–1746. doi: 10.1128/jvi.71.3.1739-1746.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rothnie HM, Chapdelaine Y, Hohn T. Pararetroviruses and retroviruses: a comparative review of viral structure and gene expression strategies. Adv Virus Res. 1994;44:1–67. doi: 10.1016/s0065-3527(08)60327-9. [DOI] [PubMed] [Google Scholar]
- 22.Kiss-Laszlo Z, Blanc S, Hohn T. Splicing of cauliflower mosaic virus 35S RNA is essential for viral infectivity. EMBO J. 1995;14:3552–3562. doi: 10.1002/j.1460-2075.1995.tb07361.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Giron ML, de The H, Saib A. An evolutionarily conserved splice generates a secreted env-Bet fusion protein during human foamy virus infection. J Virol. 1998;72:4906–4910. doi: 10.1128/jvi.72.6.4906-4910.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kiss-Laszlo Z, Hohn T. Pararetro- and retrovirus RNA: splicing and the control of nuclear export. Trends Microbiol. 1996;4:480–485. doi: 10.1016/s0966-842x(97)82909-5. [DOI] [PubMed] [Google Scholar]