Abstract
Although not previously known to cause human infections, Kocuria species have now emerged as human pathogens, mostly in compromised hosts with severe underlying disease. Recently, there has been an increasing incidence of different types of Kocuria infections reported, most likely due to the adoption of better identification methods. Here, we report a case of peritonitis caused by Kocuria rosea in a diabetic nephropathy patient who was on continuous ambulatory peritoneal dialysis. Sepsis and peritonitis caused by K. rosea in our case yielded two identical Kocuria isolates from the peritoneal dialysate fluid within a period of three days. The infection was subsequently resolved by antibiotic treatment and catheter removal. In addition to reporting this case, we herein review the literature concerning the emergence of Kocuria as a significant human pathogen. The majority of cases were device-related, acquired in the hospital or endogenous, and different Kocuria species appear to share a common etiology of peritonitis. The overall disease burden associated with Kocuria appears to be high, and the treatment guidelines for diseases associated with Kocuria have not yet been clearly defined.
Keywords: CAPD, Kocuria rosea, peritonitis
INTRODUCTION
Organisms of the genus Kocuria (family Micrococcaceae, order Actinomycetales, class Actinobacteria) are gram-positive coccoid bacteria often found as tetrads and irregular clusters that are catalase-positive and coagulase-negative. These bacteria are responsible for different types of infection, mostly in immunocompromised hosts with serious underlying conditions.1,2 However, the prevalence of human infections caused by Kocuria species is underestimated, as commonly used phenotypic assays are known to misidentify Kocuria isolates as Staphylococci.2 Accordingly, a number of presumed staphylococcal pathologies might have been caused by Kocuria species, although it is plausible that a variety of presumed Kocuria infections might have actually been due to coagulase-negative Staphylococci (CoNS). Here, we report a case of peritonitis caused by Kocuria rosea that was initially assumed to be due to CoNS. The present article reviews all the cases of Kocuria infections reported in the English literature and discusses important issues pertaining to the diagnosis, etiology, identification, drug resistance, epidemiology, clinical presentation and management of such infections.
CASE PRESENTATION
A 57-year-old man, a previously known case of diabetic nephropathy with end-stage renal disease undergoing continuous ambulatory peritoneal dialysis (CAPD) for the last four years, was admitted to a tertiary-care hospital in Puducherry, with complaints of abdominal pain, pedal edema and loose stool for three days. Upon initial physical examination, he was not febrile and had a normal-appearing catheter exit site. However, the peritoneal dialysate fluid was turbid. Subsequently, he became febrile, and a peripheral blood examination showed a total cell count of white blood cells 8700 cells/cu mm, neutrophils 84%, lymphocytes 12% and eosinophils 4%. The hemoglobin concentration was 9.0 g/dL, and the blood urea and creatinine levels were 161 mg/dL and 8.4 mg/dL, respectively. Random blood sugar was 170 mg/dL, total protein 5.9 mg/dL, serum albumin 1.8 mg/dL and globulin 4.1 mg/dL; the C-reactive protein level was raised to 10 mg/dL.
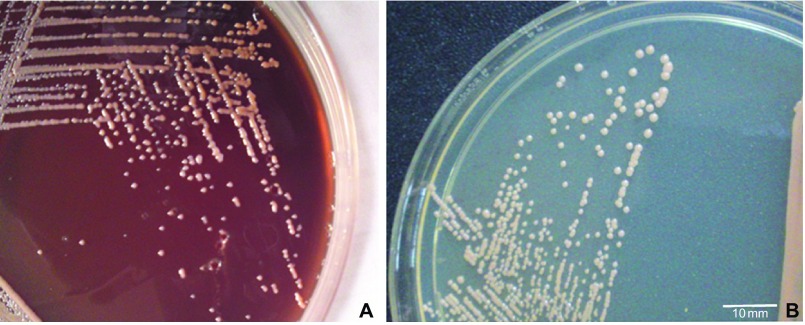
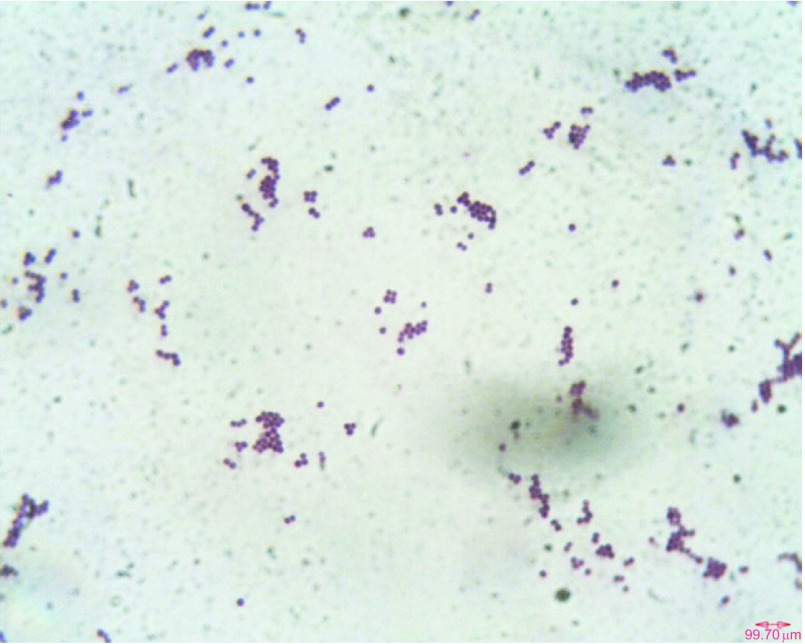
The peritoneal dialysis fluid (7 mL) inoculated into a BacT-alert FA bottle showed growth within 12 h (BacT-alert 240; bioMérieux, France). Direct microscopy of the sample showed gram-positive cocci in pairs and clusters. We performed all culturing procedures according to the 2005 update of the International Society for Peritoneal Dialysis recommendations and guidelines using specimens collected prior to antibiotic treatment.3 Subcultures of the peritoneal fluid were performed using sheep blood agar, MacConkey agar and chocolate agar; the plates were incubated at 35 °C for 48 h. After incubation, sheep blood agar yielded the pure luxuriant growth of pale-pink non-hemolytic colonies that were 1–2 mm size (Figure 1A). Subculture on nutrient agar yielded smooth, small, pale-cream to pale-pink colonies (Figure 1B). Gram staining of the culture revealed the cells to be gram-positive cocci in pairs or clusters (Figure 2). The organism was preliminarily identified as Kocuria based on phenotypic test results, such as positive reactions for catalase, oxidase, nitrate reduction and growth in 5% NaCl and motility test negativity.
Figure 1.
(A) Smooth, small, moist, slightly convex, pale-pink colonies of Kocuria observed on sheep blood agar. (B) Smooth, small, pale-cream to pale-pink colonies grown on nutrient agar.
Figure 2.
Gram-stain micrograph of Kocuria rosea PKS1409 obtained from a blood agar culture plate showing gram-positive cocci in pairs or clusters observed under oil immersion (×100).
Subsequent additional tests, such as bacitracin susceptibility, lysozyme sensitivity at 200 μg, resistance to furazolidone and lysostaphin, helped to discriminate micrococci from Staphylococci.4,5 The culture (designated as PKS1409) was later identified with a 97% probability as Kocuria rosea using a Vitek-2 system (bioMérieux) of 64 tests; the ID-GPC card panel tested positive only for α-glucosidase, leucine arylamidase, α-galactosidase, alanine arylamidase and tyrosine arylamidase. To verify the culture results, a second sample from the peritoneal fluid was evaluated, which also grew the same organism. The second isolate obtained from the second peritoneal dialysate fluid taken after 3 days showed a colony morphology, growth characteristics, biochemical test results and antibiogram identical to that of first isolate. However, an examination of the removed CAPD catheter and subsequent blood culture results showed no growth. Additionally, repeat blood samples collected from the patient who was already on antibiotics showed no growth.
We performed 16S rRNA gene sequencing, as previously described and compared the sequence results using the basic local alignment search tool with the EzTaxon database.6,7 The 16S rRNA gene amplified using a polymerase chain reaction Mastermix (Fermentas, Thermo Scientific, MA, USA) with the conserved primers PKRF (5′-ATC CTG GCT CAG AGC GAA CG-3′) and PKRR (5′-CCC TAC GGC TAC CTT GTT ACG-3′) generated a nearly complete sequence of the 16S rRNA gene (∼1365 bp) (Veriti-PCR; Applied Biosystems Inc., CA, USA). The polymerase chain reaction product was purified (Hi-Media, Mumbai, India) and sequenced using ABI technology (Eurofins, Bangalore, India). The basic local alignment search tool nucleotide sequence analysis revealed a high degree of homology (99.78%) with K. rosea (GenBank accession number JN084143); and the second closest match was K. polaris, with 99.56% homology (accession number AJ278868). This isolate was precisely identified and confirmed as K. rosea by a phylogenetic analysis of the 16S rRNA gene sequences by EzTaxon, a web-based tool used for the identification, using 16S rRNA gene sequences.7 Similar results were obtained when we constructed a neighbor-joining phylogenetic tree with the 16S rRNA gene sequences of all Kocuria species using MEGA v5.1 program (Figure 3).
Figure 3.
Phylogenetic analysis using MEGA V-5.1 based on the 16S rRNA gene sequences of all the representative Kocuria species, along with strain K. rosea PKS1409 obtained in this study. Bar=0.005 nucleotide substitutions per position.
Susceptibility testing through a modified Kirby-Bauer disc-diffusion technique and minimum inhibitory concentration (MIC) by microbroth dilution method was performed according to the Clinical and Laboratory Standards Institute guidelines for Staphylococcus.8 The disc-diffusion method revealed that PKS1409 was susceptible to ampicillin, cefotaxime, ciprofloxacin, cloxacillin, gentamicin, erythromycin, amikacin, imipenem, linezolid, teicoplanin and vancomycin, but showed intermediate resistance to ceftazidime. The MIC values for co-trimoxazole (10 mg/L), tetracycline (2 mg/L) and ceftazidime (2 mg/L) were relatively high when compared to other antimicrobials, such as benzylpenicillin (0.06 mg/L), clindamycin (0.5 mg/L) and levofloxacin (0.5 mg/L). The MIC value was less than 0.5 mg/L MIC for all the other antibiotics tested, including ciprofloxacin, erythromycin, gentamicin, linezolid, rifampicin, vancomycin and quinupristin–dalfopristin. The disc-diffusion results correlated well with the MIC values for all the common antibiotics evaluated. Biofilm production was assessed by the microtitre plate assay, and PKS1409 did not produce a biofilm.9 Biofilm production was further checked by growing the bacterium under a different set of conditions with different media, namely, brain–heart infusion, nutrient and Luria-Bertani broth, for different incubation time intervals of 6, 12 and 24 h. American Type Culture Collection reference strain Staphylococcus epidermidis 35984 was used as a positive control. All the experimental conditions reconfirmed that PKS1409 was not able to produce biofilm.
A chest X-ray of the patient was normal, and an abdominal ultrasound showed findings suggestive of chronic renal failure. The intraperitoneal administration of amikacin and cefazolin was started for the empirical treatment of CAPD peritonitis. In addition, intravenous injections of vancomycin (1 g) and amikacin were given for 5 days. Improvement of the patient was visible after the initiation of antibiotic treatment, with fever subsiding and a decrease in the C-reactive protein level. Two methods of susceptibility testing showed that PKS1409 was susceptible to vancomycin. The patient responded well to the vancomycin treatment and did not develop any other complications. The patient was symptomless after 14 days of antibiotic therapy and catheter removal and was discharged.
Nucleotide sequence accession number
The nearly complete sequence of the 16S rRNA gene of the Kocuria rosea isolate from this study has been deposited in GenBank under accession number HQ830206.
DISCUSSION AND LITERATURE REVIEW
Kocuria is a member of the Micrococcaceae family and presently comprises 18 species.1,2,10 This group was previously classified into the Micrococcus genus, but was later removed based on the phylogenetic and chemotaxonomic analyses of Stackebrant et al.1,10 Kocuria is ubiquitous in nature and is frequently found as normal skin flora in humans and other mammals. Only five of the 18 species in this genus are known to be opportunistic pathogens.2 Indeed, the documented cases of Kocuria infections in humans are very limited; however, many cases might have been missed owing to their misidentification as Staphylococci due to the limited biochemical tests and also due to automated identification systems.2,11,12,13,14 A search of the English-language research literature revealed nineteen other reports on Kocuria infections (Table 1).
Table 1. Details of all the significant cases of Kocuria infections reported to date.
| Reference(s) | Age (years)/sex | Site of infection | Type of infection and associated comorbidities | Susceptibility patterna | Treatment (antibiotics prescribed)c | Change in antibiotics after inappropriate therapy | Species | Removal of catheter/clinical prognosis | ||
|---|---|---|---|---|---|---|---|---|---|---|
| S | R | Ib | ||||||||
| Basaglia et al.15 | 51/F | Bloodstream | Catheter-related bacteremia, ovarian cancer | CD, E, CF, P, OX, CN, VA | — | — | Meropenem, ciprofloxacin and clindamycin | Yes | K. kristinae | Yes/recovered |
| Ben-Ami et al.11 | 47/F | Kidney | Ventriculo-peritoneal (VP) shunt infection | OX, VA, R, CD, TR, C | — | — | Vancomycin | No | K. varians | NMd/edema resolved |
| Altuntas et al.16 | 39/M | Bloodstream | Catheter-related bacteremia | AS, E, VA | — | — | Imipenem/cilastatin, amikacin, vancomycin | Yes | K. rosea | No/recovered |
| Ma et al.17 | 56/M | Gall bladder | Acute cholecystitis | P, CLX, E, CD, LZ, LE, TS, VA | — | — | Levofloxacin | No | K. kristinae | NM/recovered |
| Becker et al.13 | 8/M | Bloodstream | Sepsis | NX | — | Cefuroxime, vancomycin and amphotericin B | Yes | K. rhizophila | NM/multiple febrile episodes; subsequent recovery | |
| Lee et al.14 | 57/M | Peritoneum | CAPD-related peritonitis | P, A, G, CN, MO, TR, E, CD, VA, C, T, R | — | — | Ceftazidime and clindamycin | Yes | K. marina | Yes/recovered |
| Lee et al.14 | 73/M | Peritoneum | CAPD-related peritonitis | P, A, G, CN, MO, E, CD, VA, C, T, R | TR | — | Netilmicin | No | K. marina | NM/recovered |
| Lai et al.18 | 89/F | Bloodstream | Infective endocarditis/ischemic bowel disease | P, A, E, CD, VA, AC, TE, CF, LE, F, QD, LZ, R | OX, CZ | CE | Vancomycin, teicoplanin, oxacillin | No | K. kristinae | Yes/improved |
| 37/F | Bloodstream | Catheter-related bacteremia, gastric cancer | P, A, E, CD, VA, AC, TE, CF, LE, F, QD, LZ, R | OX, CZ | CE | Piperacillin-tazobactam, ciprofloxacin | No | K. kristinae | Yes/improved | |
| 2/M | Bloodstream | Catheter-related bacteremia, congenital short bowel syndrome, hypogammaglobulemia | P, A, E, CD, VA, AC, TE, CF, LE, F, QD, LZ, R | OX, CZ | CE | Oxacillin, vancomycin | Yes | K. kristinae | Yes/improved | |
| 68/F | Bloodstream | Catheter-related bacteremia, gastric cancer | P, A, E, CD, VA, AC, TE, CF, LE, F, QD, LZ, R | OX, CZ | CE | Oxacillin | Yes | K. kristinae | Yes/improved | |
| Tsai et al.19 | 52/M | Brain | Brain abscess | P, CLX, CF, CD, MR, AS, PT | — | — | Ceftazidime and ceftibuten | No | K. varians | NM/recovered |
| Carlini et al.20 | 78/M | Peritoneum | CAPD-related peritonitis | — | — | — | Cefotaxime, tobramycin, tazobactam, ciprofloxacin, teicoplanin, amoxicillin/clavulanic acid | Yes | K. kristinae | No/recovered |
| Cheung et al.21 | 69/M | Peritoneum | CAPD-related peritonitis | CLX, G, E, CD, F | — | — | Cefazolin and cefepime | No | K. kristinae | No/recovered |
| Dunn et al.22 | 29/F | Catheter-related bacteremia with pulmonary septic emboli | OX, VA, CZ, CD, R, TS | — | — | Ceftriaxone, azithromycin, vancomycin, clindamycin, oxacillin | Yes | K. kristinae | Yes/improved | |
| Moissenet et al.23 | 3/F | Bloodstream | Catheter-related bacteremia, hirschsprung's disease | P, G, AK, TOB, T, VA, TE | CF | E | Vancomycin, gentamicin | No | K. rhizophila | NM/septic episodes resolved |
| Meletis et al.24 | 70/M | Peritoneum | CAPD-related peritonitis | G, E, CD, T, LZ, glycopeptides | LE | — | Vancomycin, aztreonam | No | K. varians | Yes/improved |
| Oncel et al.25 | 4 months/F | Bloodstream | Black hairy tongue | - | — | — | Ceftriaxone, vancomycin | No | K. kristinae | NM/recovered |
| Citro et al.26 | 74/M | Bloodstream | Endocarditis, sepsis | Sulbactam/ampicillin, gentamicin | No | K. kristinae | NM/severe sepsis, MOFe, expired | |||
| Our study | 57/M | Peritoneum | Peritonitis, diabetic nephropathy | AC, CE, CF, CLX, G, E, MR, AK, LZ, TE, VA | CAZ | Vancomycin, cefazolin, amikacin | No | K. rosea | Yes/recovered | |
According to Clinical and Laboratory Standards Institute guidelines for Staphylococcus.
A, ampicillin; AC, amoxycillin/clavulanic acid; AK, amikacin; AS, ampicillin/sulbactam; C, chloramphenicol; CE, cephotaxime; CD, clindamycin; CLX, cloxacillin; CF, ciprofloxacin; CMZ, cefmetazole; CN, cefalothin; CZ, cefozolin; E, erythromycin; F, fusidic acid; G, gentamicin; LE, levofloxacin; LZ, lenezolid; MR, meropenem; MO, moxifloxacin; NX, norfloxacin; OX, oxacillin; P, penicillin; PT, piperacillin/tazobactam; QD, quinupristin/dalfopristin; R, rifampin; S, sulbactum; TE, teicoplanin; T, tetracycline; TOB, tobramycin; TR, trimethoprim; TS, trimethoprim/sulphamethoxazole; VA, vancomycin.
‘and' in between two antibiotics indicates simultaneous administration.
NM, not mentioned in the report.
MOF, multiple organ failure.
HUMAN INFECTIONS CAUSED BY Kocuria SPP.
The number of cases reported has increased over the past five years, most likely due to the improved identification systems for the diagnosis of this infective agent. Recovery in many of the cases of Kocuria infection required catheter removal, which resolved the infection. Among the members of this genus, K. kristinae, first described in 1974 (previously Micrococcus kristinae), is known to cause catheter-related bacteremia and infective endocarditis. These infections are associated with different cancers, acute cholecystitis and other metabolic disorders.15,17,18,27 The recently introduced Vitek-2 GP-card supplemented with an additional database allows for the identification of K. kristinae.28 Bacteremia caused by K. kristinae in a patient suffering from ovarian cancer who had multiple febrile episodes for a prolonged period of >six months has been described15 (Table 1). The strains yielded from all the blood and central venous catheter (CVC) cultures of this patient were indistinguishable and clonal in nature, strongly suggesting recurrent infection and the possibility of a CVC line as a port of entry.
On one occasion, a three-year screening of blood cultures in the National Taiwan University hospital led to the isolation of 21 Kocuria isolates from five patients, with 20 being K. kristinae and responsible for either catheter-related bacteremia or infective endocarditis.18 Subsequent random amplification of polymorphic DNA genotyping of these isolates revealed that there was no nosocomial transmission. Furthermore, multiple isolates from individual patients with persistent blood-stream infection (BSI) revealed identical random amplification of polymorphic DNA profiles, implying that there was either incomplete treatment of an occult focus or persistent carriage of the same bacterium. K. kristinae as the cause of acute peritonitis in a patient with end-stage renal failure undergoing CAPD for two years has also been reported, and bacterial entry into the peritoneal cavity through touch contamination of the catheter was suspected.20,21 The continued monitoring of bacterial growth for up to 72 h without any clinical indication after the onset of acute peritonitis was proven to be beneficial, as this led to an interruption and change in the antibiotic treatment, leading to improvement in the patient's condition.
Surprisingly, K. kristinae was also reported to cause severe intravascular infections in an immunocompetent host, an infection that was complicated by septic pulmonary emboli that were secondary to suppurative thrombosis. A problematic catheter-related BSI leading to such complications was striking in this case.22 A recent report of a female infant with a history of prolonged diarrhea showed the blood culture growth of K. kristinae, and this organism was considered responsible for bacteremia with black hairy tongue symptoms.25 Unlike the initial empirical ceftriaxone therapy, the infant showed a dramatic response to vancomycin treatment, which completely resolved the infection and symptoms within a week. A case of K. kristinae infection was recently documented in an elderly patient affected by endocarditis who was suffering from diabetes mellitus and hypertension who additionally had minor amputation for a left forefoot ulcer.26 This was a rare and unusual case, whereby a central venous catheter was not involved, but the disease was associated with a diabetic foot infection. Catheter removal prior to the clearance of infection was noted in many investigations, implying that the removal of the device should be considered and recommended for the treatment of K. kristinae infection. It is also evident that any repeat isolation of K. kristinae from blood cultures can no longer be ignored by either the microbiologist or clinician.
A brain abscess caused by Kocuria varians has been described, affecting the right occipital region, and was successfully treated with surgical excision and antimicrobial therapy.19 The potential source of the organism was suspected to be the hematogenous spread to brain parenchyma. The K. varians identification was based on Vitek-2, which showed a relatively reduced probability level (93%); however, the main drawback in this case was the lack of a 16S rRNA gene sequence analysis for species confirmation. CVC placement was suspected to be a possible risk factor for K. varians peritonitis relapse in patients undergoing dialysis, and the treatment of such infections might be a complicated task, as one study demonstrated that K. varians isolated from the environment was capable of producing biofilms.24,29 Interestingly, one investigation used Vitek-2 for the detection of a ventriculoatrial shunt infection by phenotypically variable Staphylococcus epidermidis that was masquerading as polymicrobial bacteremia caused by CoNS and K. varians.11 Therefore, the erroneous identification of CoNS as Kocuria or vice-versa is possible and should be excluded with certainty only through a genome-based analysis. Clearly, it is now more important than earlier to confirm whether K. varians indeed can cause different types of infection or not.
K. marina and K. rhizophila have also joined the emerging spectrum of Kocuria species causing human infections.13,14 Cases of K. marina peritonitis in patients undergoing CAPD revealed that dialysis fluid from these patients becoming turbid and straw colored and an increase in white blood cell and platelet counts was noticed;14 empirical therapy failed in these cases, and only catheter removal resolved the infection.14 The first case of K. rhizophila infection was reported in a boy with methylmalonic aciduria where the boy suffered multiple episodes of sepsis for more than two years, and the Port-A-Cath was shown to be the focus of infection. Indeed, the Port-A-Cath device might have provided a favorable niche for prolonged survival of the organism and subsequent recurrence.13 K. rhizophila has been widely used as a quality-control strain (American Type Culture Collection-9341) for sterility testing,30 and a recent case of K. rhizophila infection presented as persistent BSI associated with a damaged CVC in a girl with Hirschsprung's disease; advanced molecular methods were used for the identification in this case.23
K. rosea is yet another species capable of causing human infections, and the first report of K. rosea infection describes multiple episodes of febrile neutropenia in a patient with relapsed Hodgkin disease undergoing peripheral blood stem cell transplantation.31 Although the isolate was susceptible to vancomycin, its administration did not alter the clinical picture of the infection until the catheter was removed. It was previously reported that K. rosea can cause catheter-related bacteremia and peritonitis; however, no genotypic identification methods were employed to confirm this, thus the etiology is uncertain.16,31,32 Only an abstract was available as the reference in two of three cases because the original publication was not in English; thus, precise information could not be obtained. Slightly more information was available for the third case: the bacteremic patient was human immunodeficiency virus positive and was successfully treated with vancomycin and catheter removal.32 Such scanty information forced us to rely on a single existing report.16 However, the present study conclusively confirms K. rosea as an opportunistic pathogen that is able to cause peritonitis and BSI.
Catheter removal was one of the treatment options followed, and the possibility of a CVC line as the portal of entry was suspected. The possibility of biofilm formation on the catheter was attributed to treatment failure in one of the cases, although it was not investigated.16 Conversely, the K. rosea isolated in our case did not produce any biofilm, and hence the hypothesis of biofilm formation appears to be premature. Biofilm assays involving a large number of isolates may provide definite answers with regard to the biofilm-forming capability of K. rosea. Furthermore, K. rosea strain PKS1409 investigated in our study grew well and faster with enhanced pigment production when maintained at 4 °C. Interestingly, contrary to the earlier observations, our isolate is very close to K. polaris, a known psychrophile isolated from Antarctica, as revealed by 16S rRNA gene analysis.2,33 To sum up, human infections caused by only a few species of Kocuria affirm the presence of certain species-specific virulence characteristics that enable these bacteria to cause infections. Moreover, several new cases have expanded the clinical spectrum of disease caused by this unusual pathogen.
IDENTIFICATION OF Kocuria SPECIES
Phenotypic identification may fail to recognize all the members of Kocuria species because the results of biochemical and carbon assimilation tests are shown to be heterogeneous in different species of Kocuria.2 In reality, these organisms are often not correctly identified and are many times hastily discarded as contaminants. Furthermore, accurate identification is difficult because commercially available databases do not include all the classified Kocuria species. Even cellular fatty acid profile analyses were not much of use in identification.1,34 However, Kocuria classification is now typically confirmed by 16S rRNA gene sequencing, and one study even used matrix-assisted laser desorption/ionization time-of-flight mass spectrometry for species identification, which appears to be an efficient method.2,26
A series of 12 Kocuria isolates obtained from a patient (subsequently identified as K. rhizophila by 16S rRNA gene sequencing) in a study were ambiguously identified as either belonging to any one of the four different bacterial species, namely, K. varians, K. rosea, Dermacoccus nishinomiyaensis and Micrococcus luteus, by the Vitek 2 ID-GPC-card phenotypic system.14 The ID-32 Staph system also misidentified approximately four isolates in this collection as either Staphylococcus auricularis or S. capitis. Another study employing the Vitek-2 system initially identified two K. marina isolates as K. varians and Staphylococcus hominis; upon re-examination, they were once again misidentified as K. kristinae and Staphylococcus chromogenes,14 whereas subsequent 16S rRNA gene sequencing confirmed these two isolates as K. marina. When subjected to the Vitek ID-GPC-card panel test, Kocuria isolates are positive for only the alanine arylamidase reaction and alkalinization of L-lactate;13 strain PKS1409 from our case was positive for only five tests in the Vitek array. Such instances of only a few positive reactions observed in an array of 64 biochemical tests (Vitek-2-Compact; bioMérieux) with an automated system questions the validity of this identification process.
Previously, Ben-Ami et al.11 reported on the misidentification of CoNS as Kocuria species by the Vitek-2 assay and warned that a clinical specimen yielding Kocuria could easily raise suspicion of CoNS infection. However, recent studies, including ours, correctly identified Kocuria using the Vitek-2 ID-GPC gram-positive identification card, perhaps due to the recently introduced larger database that allows the identification of additional taxa.12 However, it is not clear whether the Vitek-2 GP-system will reduce the number of CoNS falsely identified as Kocuria spp., although there are claims of improvement in specificity after the addendum of new database.12 However, misidentification among members of the Kocuria genus cannot be ruled out altogether, as recent studies have reported such occurrences.13,14,17 Nonetheless, in a recent screening of 21 Kocuria isolates, only one was misidentified, suggesting that the newer Vitek-2 has improved efficiency.18
Furthermore, both Micrococci and Staphylococci are known to show phenotypic variability (PV) that complicates their identification.11 PV among isolates might be misidentified by phenotypic assays, as biochemical activities may change during growth to allow the microbes to adapt to changing environmental conditions.5,11,14,35 Moreover, PV becomes more common under stress conditions and likely contributes to virulence.36,37 It has been shown that the pathogenic clinical isolates cultured from blood are more phenotypically variable than the saprophytic strains cultured from skin or mucosal surfaces.38 Because PV is common in S. epidermidis isolates, misidentification of these isolates as Kocuria by commercial systems may be frequent and widespread. More troubling is the fact that such errors are likely to occur more frequently with the highly virulent strains of Staphylococcus that are known to show high PV.11 Thus, the biochemical analysis-based misidentification of CoNS as Kocuria and vice-versa due to PV appears to be a major problem faced by diagnostic laboratories. A summary of the reports on the misidentification of Kocuria by phenotypic systems is given in Table 2.
Table 2. Misidentification of Kocuria species and members of the Kocuria genus by different automated phenotypic identification systems.
| Study | Vitek2 | Bactec/BactAlert | Phoenix | API-STAPH/ATB | 16S rRNA gene sequencing (a genome-based assay) |
|---|---|---|---|---|---|
| Ma et al.17 | K. kristinae | — | K. varians | K. kristinae. | K. marina |
| Becker et al.13 | K. varians/K. rosea/Dermacoccus nishinomiyaensis/Micrococcus luteus/K. rhizophila | Micrococcus sp. | — | K. varians/K. rosea/Dermacoccus nishinomiyaensis/Micrococcus luteus/K. rhizophila | K. rhizophila |
| Lee et al.14 | Kocuria varians/Kocuria kristinae | — | — | K. kristinae | K. marina |
| Staphylococcus hominis/K. kristinae | — | — | Staphylococcus Chromogenes | K. marina | |
| Tsai et al.19 | K. varians | — | — | — | — |
| Cheung et al.21 | K. kristinae | Staphylococcus lugdunensis/Staphylococcus hominis | — | ||
| Moissenet et al.23 | K. rhizophila | — | — | Staphylococcus auricularis | K. rhizophila |
Despite its limitations, simple tests, such as susceptibility to bacitracin and lysozyme and resistance to nitrofurantoin/furazolidone and lysostaphin, appear to be very useful in differentiating Kocuria from Staphylococci. In addition, the typical pigmentation of Kocuria colonies generally becomes more distinct with age.2 Hence, the prolonged incubation of a culture for >48 h is recommended to appreciate the pigmentation better, which can also discriminate the organism from Staphylococci. Importantly, in most of the previous reports of misidentification, there was no mention of incubation time, and it can be assumed that the plates were kept only up to 24 h, thereby missing the characteristic pigmentation, which may appear some time later. We suggest that clinical laboratories worldwide prolong the incubation time to >48 h to avoid any chance of misidentification. In particular, our observations revealed massive growth and pigment enhancement of K. rosea at refrigeration temperatures. Finally, the identification of Kocuria spp. by any number of phenotypic tests should be always placed under suspicion and should be confirmed explicitly by performing molecular assays.
PATHOPHYSIOLOGY, VIRULENCE FACTORS AND EPIDEMIOLOGY
Multiple episodes of sepsis, febrile neutropenia, and increased platelets, white blood cell counts and C-reactive protein levels are the common manifestations of Kocuria infection.2,22 Although very little is known about the epidemiology and virulence properties of Kocuria, the involvement of biofilm in adhesion, colonization and subsequent infection has been suggested.24 Surprisingly, K. rosea PKS1409 characterized by us did not produce any biofilm. Furthermore, there are no reports of biofilm production by Kocuria in the literature, yet it has been implicated in device-related bacteremia, implying that the colonization of Kocuria on a device will likely result in chronic recurrent bacteremia in a compromised host.15,16,18 Hence, clinicians should not overlook and underestimate the isolation of Kocuria from implanted devices.
K. kristinae is shown to be a component of the skin and oral cavity flora and has been implicated as a common source of contamination in clinical specimens.5,17,39 Interestingly, K. rhizophila is a predominant bacterium isolated from chicken meat treated with oxalic acid.13,40 The most likely suspected source of K. rhizophila is patient contact with contaminated meat and dust from the environment, which may result in colonization. The possibility of a CVC line as a portal of entry should also be evaluated. A recent study emphasized the adherence of bacteria to the silastic tube as a possibility for the failure of antibiotic treatment.16 Biofilm production in any pathogen makes them more resistant and difficult to eradicate; nonetheless, it remains unclear whether Kocuria species produce biofilm. The lack of genotypic studies has greatly impacted epidemiological investigations and the understanding of the basic differences between the saprophytic and pathogenic traits of Kocuria strains. Indeed, an understanding of these aspects may avoid the hasty classification of Kocuria as contaminating micrococci in a clinical setup.
ANTIBIOTIC RESISTANCE AND TREATMENT
The lack of proper guidelines and breakpoints for Kocuria has forced many studies, including ours, to follow the susceptibility breakpoints used for Staphylococcus. As a result, the numerous reports referring to the interpretive values of Staphylococcus may cause the misdiagnosis of either sensitivity or resistance. We therefore emphasize the urgent need for the formulation of specific criteria for interpreting susceptibility in Kocuria species. A recent review analyzed the antibiotic resistance patterns of various Kocuria species and their clinical impact.2 Most Kocuria isolates were reported to be susceptible to many of the first- and second-line drugs, with the exception of ampicillin and norfloxacin.2,13,41
Decreased cell wall permeability and the presence of efflux pumps are implicated in the resistance of Kocuria species.2 The recently sequenced genome of K. rhizophila has revealed the presence of many proteins that are predicted to be involved in multidrug efflux mechanisms, although their role in imparting resistance is not clear.42 Pathogenic Kocuria species are highly susceptible to broad-spectrum antibiotics. Szczerba proposed amoxicillin/clavulanate along with such drugs as ceftriaxone, cefuroxime, doxycycline and amikacin as a first-line therapy against micrococcal pathologies.41 In addition, it has been suggested that the treatment duration should depend on the type of infection.2,13,14,15 The infusion of antibiotics in a catheter lock or catheter removal appears to be the best option for the treatment of implant-related sepsis.2,17,18,41 As there are no evidence-based guidelines for the management of Kocuria infections, previously reported successfully treated cases can only become the guiding principle for treating Kocuria infections.
Unfortunately, the skill required and high cost involved in molecular assays may limit their use in routine practice, and consequently, Kocuria species misidentification in many laboratories is likely to continue for some time. However, it is probable that the prevalence of Kocuria infections in coming years may increase as genome-based identification becomes routinely available. The first indication of such a scenario becoming true has been observed through the numerous new cases of Kocuria infections being reported in a short span of time.20,22,23,32 Owing to misidentification over the years, the diseases caused by Kocuria species are erroneously believed to be rare. Hence, it is suggested that physicians should not underestimate the importance of Kocuria spp. when isolated from clinical samples, particularly from blood and inert medical implants. Furthermore, cause for concern remains, as therapy guidelines for illnesses involving Kocuria are lacking, mainly due to the absence of established criteria for evaluating the killing and/or growth inhibition of Kocuria in the presence of antibiotics. Although a comprehensive view of Kocuria infections will have to await the documentation of many more cases, several recent reports of Kocuria species as the cause of different diseases clearly reveal the expanding clinical spectrum of infections caused by Kocuria.
Acknowledgments
This work was supported by University Grants Commission, Government of India, through a University Grants Commission MRP project grant (F. NO 36-190\2008 (SR) to K Prashanth). Ethics committee approval for this study was obtained from an institutional review board.
References
- Stackebrandt E, Koch C, Gvozdiak O, Schumann P. Taxonomic dissection of the genus Micrococcus: Kocuria gen. nov., Nesterenkonia gen. nov., Kytococcus gen. nov., Dermacoccus gen. nov., and Micrococcus Cohn 1872 gen. emend. Int J Syst Bacteriol. 1995;45:682–692. doi: 10.1099/00207713-45-4-682. [DOI] [PubMed] [Google Scholar]
- Savini V, Catavitello C, Masciarelli G, et al. Drug sensitivity and clinical impact of members of the genus Kocuria. J Med Microbiol. 2010;59:1395–402. doi: 10.1099/jmm.0.021709-0. [DOI] [PubMed] [Google Scholar]
- Piraino B, Bailie G, Bernardini J, et al. for the ISPD Ad Hoc Advisory Committee. Peritoneal dialysis-related infections recommendations2005 update Perit Dial Int 200525107–131. [PubMed] [Google Scholar]
- Baker JS. Comparison of various methods for differentiation of Staphylococci and Micrococci. J Clin Microbiol. 1984;19:875–879. doi: 10.1128/jcm.19.6.875-879.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Ami R, Navon-Venezia S, Schwartz D, Schlezinger Y, Mekuzas Y, Carmeli Y. Erroneous reporting of coagulase-negative Staphylococci as Kocuria spp. by the Vitek 2 system. J Clin Microbiol. 2005;43:1448–1450. doi: 10.1128/JCM.43.3.1448-1450.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wauters G, Charlier J, Janssens M, Delme'e M. Brevibacterium paucivorans sp. nov., from human clinical specimens. Int J Syst Evol Microbiol. 2001;51:1703–1707. doi: 10.1099/00207713-51-5-1703. [DOI] [PubMed] [Google Scholar]
- Chun J, Lee JH, Jung Y, et al. EzTaxon: a web-based tool for the identification of prokaryotes based on 16S ribosomal RNA gene sequences. Int J Syst Evol Microbiol. 2007;57:2259–2261. doi: 10.1099/ijs.0.64915-0. [DOI] [PubMed] [Google Scholar]
- Clinical and Laboratory Standards Institute . Performance standards for antimicrobial susceptibility testing; 19th informational supplement. CLSI document M100-S19. Wayne, PA; Clinical and Laboratory Standards Institute; 2009. [Google Scholar]
- Duggirala A, Kenchappa P, Sharma S, Peeters JK, Das T, Hasnain SE. High-resolution genome profiling differentiated Staphylococcus epidermidis strains isolated from patients with ocular infections and normal individuals. Invest Ophthalmol Vis Sci. 2007;48:3239–3245. doi: 10.1167/iovs.06-1365. [DOI] [PubMed] [Google Scholar]
- Stackebrandt E, Frederiksen W, Garrity GM, et al. Report of the ad hoc committee for the re-evaluation of the species definition in bacteriology. Int J Syst Evol Microbiol. 2002;52:1043–1047. doi: 10.1099/00207713-52-3-1043. [DOI] [PubMed] [Google Scholar]
- Ben-Ami R, Navon-Venezia S, Schwartz D, Carmeli Y. Infection of a ventriculoatrial shunt with phenotypically variable Staphylococcus epidermidis masquerading as polymicrobial bacteremia due to various coagulase-negative Staphylococci and Kocuria varians. . J Clin Microbiol. 2003;41:2444–2447. doi: 10.1128/JCM.41.6.2444-2447.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudewijns M, Vandeven J, Verhaegen J, Ben-Ami R, Carmeli Y. Vitek 2 automated identification system and Kocuria kristinae. J Clin Microbiol. 2005;43:5832. doi: 10.1128/JCM.43.11.5832.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker K, Rutsch F, Uekötter A, et al. Kocuria rhizophila adds to the emerging spectrum of micrococcal species involved in human infections. J Clin Microbiol. 2008;46:3537–3539. doi: 10.1128/JCM.00823-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JY, Kim SH, Jeong HS, et al. Two cases of peritonitis caused by Kocuria marina in patients undergoing continuous ambulatory peritoneal dialysis. J Clin Microbiol. 2009;47:3376–3378. doi: 10.1128/JCM.00847-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basaglia G, Carretto E, Barbarini D, et al. Catheter-related bacteremia due to Kocuria kristinae in a patient with ovarian cancer. J Clin Microbiol. 2002;40:311–313. doi: 10.1128/JCM.40.1.311-313.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altuntas F, Yildiz O, Eser B, Gündogan K, Sumerkan B, Cetin M. Catheter-related bacteremia due to Kocuria rosea in a patient undergoing peripheral blood stem cell transplantation. BMC Infect Dis. 2004;4:62. doi: 10.1186/1471-2334-4-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma ESK, Wong CL, Lai KT, Chan EC, Yam WC, Chan AC. Kocuria kristinae infection associated with acute cholecystitis. BMC Infect Dis. 2005;5:60. doi: 10.1186/1471-2334-5-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai CC, Wang JY, Lin SH, et al. Catheter-related bacteraemia and infective endocarditis caused by Kocuria species. Clin Microbiol Infect. 2011;17:190–192. doi: 10.1111/j.1469-0691.2010.03211.x. [DOI] [PubMed] [Google Scholar]
- Tsai CY, Su SH, Cheng YH, Chou YL, Tsai TH, Lieu AS. Kocuria varians infection associated with brain abscess: a case report. BMC Infect Dis. 2010;10:102. doi: 10.1186/1471-2334-10-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlini A, Mattei R, Lucarotti I, Bartelloni A, Rosati A. Kocuria kristinae: an unusual cause of acute peritoneal dialysis-related infection. Perit Dial Int. 2011;31:105–107. doi: 10.3747/pdi.2010.00132. [DOI] [PubMed] [Google Scholar]
- Cheung CY, Cheng NH, Chau KF, Li CS. An unusual organism for CAPD-related peritonitis: Kocuria kristinae. Perit Dial Int. 2011;31:107–108. doi: 10.3747/pdi.2010.00125. [DOI] [PubMed] [Google Scholar]
- Dunn R, Bares S, David MZ. Central venous catheter-related bacteremia caused by Kocuria kristinae: case report and review of the literature. Ann Clin Microbiol Antimicrob. 2011;10:31. doi: 10.1186/1476-0711-10-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moissenet D, Becker K, Mérens A, Ferroni A, Dubern B, Vu-Thien H. Persistent bloodstream infection with Kocuria rhizophila related to a damaged central catheter. J Clin Microbiol. 2012;50:1495–1498. doi: 10.1128/JCM.06038-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meletis G, Gogou V, Palamouti M, et al. Catheter-related relapsing peritonitis due to Kocuria varians in a patient undergoing continuous ambulatory peritoneal dialysis. Nefrologia. 2012;32:541–542. doi: 10.3265/Nefrologia.pre2012.Apr.11471. [DOI] [PubMed] [Google Scholar]
- Karadag Oncel E, Boyraz MS, Kara A. Black tongue associated with Kocuria (Micrococcus) kristinae bacteremia in a 4-month-old infant. Eur J Pediatr. 2012;171:593. doi: 10.1007/s00431-011-1573-8. [DOI] [PubMed] [Google Scholar]
- Citro R, Prota C, Greco L, et al. Kocuria kristinae endocarditis related to diabetic foot infection. J Med Microbiol. 2013;62:932–934. doi: 10.1099/jmm.0.054536-0. [DOI] [PubMed] [Google Scholar]
- Martinaud C, Gisserot O, Gaillard T, Brisou P, de Jaureguiberry JP. Bacteremia caused by Kocuria kristinae in a patient with acute leukaemia. Med Mal Infect. 2008;38:334–335. doi: 10.1016/j.medmal.2008.02.006. [DOI] [PubMed] [Google Scholar]
- Funke G, Funke-Kissling P. Performance of the new Vitek 2 GP card for identification of medically relevant Gram-positive cocci in a routine clinical laboratory. J Clin Microbiol. 2005;43:84–88. doi: 10.1128/JCM.43.1.84-88.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Midelet G, Kobilinsky A, Carpentier B. Construction and analysis of fractional multifactorial designs to study attachment strength and transfer of Listeria monocytogenes from pure or mixed biofilms after contact with a solid model food. Appl Environ Microbiol. 2006;72:2313–2321. doi: 10.1128/AEM.72.4.2313-2321.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang JS, Gillevet PM. Reclassification of ATCC 9341 from Micrococcus luteus to Kocuria rhizophila. Int J Syst Evol Microbiol. 2003;53:995–997. doi: 10.1099/ijs.0.02372-0. [DOI] [PubMed] [Google Scholar]
- Kaya KE, Kurtoğlu Y, Cesur S, et al. Peritonitis due to Kocuria rosea in a continuous ambulatory peritoneal dialysis case Mikrobiyol Bul 200943335–337.Turkish. [PubMed] [Google Scholar]
- Corti M, Villafañe MF, Soto I, Palmieri O, Callejo R.Bacteremia by Kocuria rosea in an AIDS patient Rev Chilena Infectol 201229355, 356Spanish. [DOI] [PubMed] [Google Scholar]
- Reddy GS, Prakash JS, Prabahar V, Matsumoto GI, Stackebrandt E, Shivaji S. Kocuria polaris sp.nov., an orange-pigmented psychrophilic bacterium isolated from an Antarctic cyanobacterial mat sample. Int J Syst Evol Microbiol. 2003;53:183–187. doi: 10.1099/ijs.0.02336-0. [DOI] [PubMed] [Google Scholar]
- Stead DE, Sellwood JE, Wilson J, Viney I. Evaluation of a commercial microbial identification system based on fatty acid profiles for rapid, accurate identification of plant pathogenic bacteria. J Appl Bacteriol. 1992;72:315–321. [Google Scholar]
- Savini V, Catavitello C, Bianco A, Balbinot A, D'Antonio D. Epidemiology, pathogenicity and emerging resistances in Staphylococcus pasteuri: from mammals and lampreys, to man. Recent Pat Anti infect Drug Discov. 2009;4:123–129. doi: 10.2174/157489109788490352. [DOI] [PubMed] [Google Scholar]
- Christensen GD, Baddour LM, Madison BM, et al. Colonial morphology of Staphylococci on Memphis agar: phase variation of slime production, resistance to β-lactam antibiotics, and virulence. J Infect Dis. 1990;161:1153–1169. doi: 10.1093/infdis/161.6.1153. [DOI] [PubMed] [Google Scholar]
- Deighton M, Pearson S, Capstick J, Spelman D, Borland R. Phenotypic variation of Staphylococcus epidermidis isolated from a patient with native valve endocarditis. J Clin Microbiol. 1992;30:2385–2390. doi: 10.1128/jcm.30.9.2385-2390.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziebuhr W, Heilmann C, Gotz F, et al. Detection of the intercellular adhesion gene cluster (ica) and phase variation in Staphylococcus epidermidis blood culture strains and mucosal isolates. Infect Immun. 1997;65:890–896. doi: 10.1128/iai.65.3.890-896.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloos WE, Tornabene TG, Schleifer KH. Isolation and characterization of micrococci from human skin, including two new species: Micrococcus lylae and Micrococcus kristinae. Int J Syst Bacteriol. 1974;24:79–101. [Google Scholar]
- Anang DM, Rusul G, Radu S, Bakar J, Beuchat LR. Inhibitory effect of oxalic acid on bacterial spoilage of raw chilled chicken. J Food Prot. 2006;69:1913–1919. doi: 10.4315/0362-028x-69.8.1913. [DOI] [PubMed] [Google Scholar]
- Szczerba I.[Susceptibility to antibiotics of bacteria from genera Micrococcus, Kocuria, Nesternkonia, Kytococcus and Dermacoccus Med Dosw Mikrobiol 20035575, 80Polish. [PubMed] [Google Scholar]
- Takarada H, Semine M, Kosugi H, et al. Complete genome sequence of the soil actinomycete Kocuria rhizophila. J Bacteriol. 2008;190:4139–4146. doi: 10.1128/JB.01853-07. [DOI] [PMC free article] [PubMed] [Google Scholar]