Abstract
Elucidation of sub-cellular signaling networks by multiparameter imaging is hindered by a lack of sensitive FRET pairs spectrally compatible with the classic CFP/YFP pair. Here we present a generic strategy to enhance the traditionally poor sensitivity of red FRET sensors by developing self-associating variants of mOrange and mCherry that allow sensors to switch between well-defined on- and off states. Requiring just a single mutation of the mFruit domain, this new FRET pair improved the dynamic range of protease sensors up to 10-fold and was essential to generate functional red variants of CFP-YFP-based Zn2+ sensors. The large dynamic range afforded by the new red FRET pair allowed simultaneous use of differently colored Zn2+ FRET sensors to image Zn2+ over a broad concentration range in the same cellular compartment.
Genetically-encoded FRET (Förster Resonance Energy Transfer) sensors allow ratiometric, noninvasive intracellular imaging of small molecules and enzyme activities in single living cells with high spatial and temporal resolution.1, 2 These sensors consist of donor and acceptor fluorescent domains that report a conformational change in a native receptor domain through a change in FRET. Elucidation of the spatial and temporal relationships between different biochemical pathways requires the use of spectrally distinct sensors, permitting two or more probes to be used in the same cell at the same time.3, 4 Several examples of orthogonal FRET pairs have recently been reported, but most of these are incompatible with the classical CFP/YFP pair.5, 6 Attempts to develop spectrally-distinct sensors by simply replacing CFP and YFP by the fully red-shifted mOrange/mCherry pair, resulted in FRET sensors with no or strongly attenuated responses,7-9 often precluding their simultaneous use with CFP-YFP-based sensors in the same cellular compartment.10 The development of new FRET sensors is often a process of trial-and-error that requires laborious optimization of linker lengths, linker positions and domain orientations. We recently introduced an alternative, rational design strategy for FRET sensor proteins based on the principle of mutually exclusive domain interactions. These sensors, which include probes for protease activity, Zn2+, bile acids, and antibodies, rely on CFP and YFP variants that interact in one state of the sensor, but whose interaction is readily disrupted upon a ligand-induced interaction at the input domains.11-15 This modular design not only ensures robust changes in energy transfer efficiency, but should also allow easy exchange of the traditional CFP- and YFP output domains by alternative pairs of self-associating FPs.
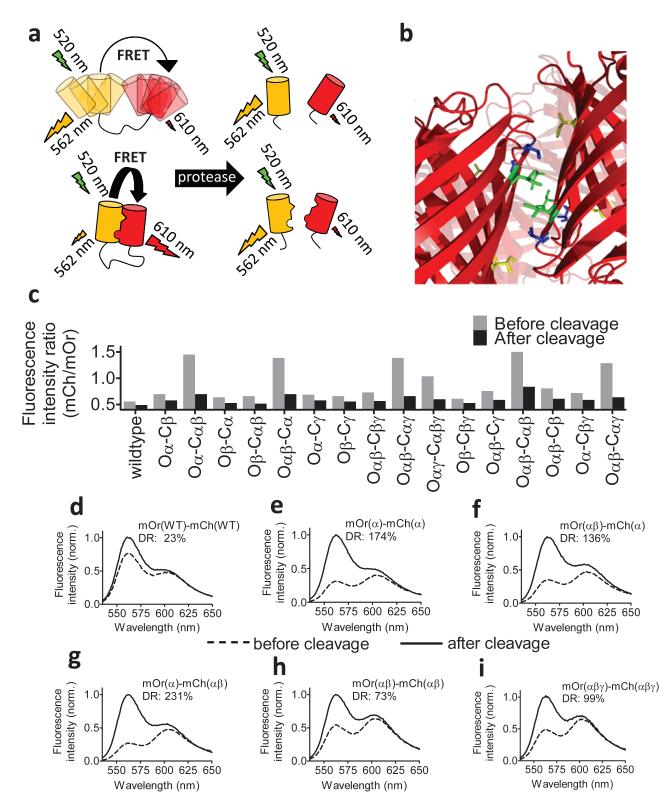
To identify mutations that promote intramolecular association of mOrange and mCherry, we constructed a test protein consisting of mOrange and mCherry fused by a long and flexible glycine-serine linker (Figure 1a). Cleavage of the linker by proteinase K resulted in a relatively modest reduction of emission ratio from 0.58 to 0.47 (Figure 1d). This poor dynamic range (ΔR/Rmin) of 23% is attributable to the low quantum yield of the acceptor, a critical factor in sensitized emission FRET. Since mOrange and mCherry are both derived from the obligate tetramer DsRed, we reasoned that reversion of one or more of the mutations originally carried out to break the hydrophobic A-B dimeric interface, I125R, V127T and I180T (Figure 1b),16 might be used to promote intramolecular self-association of these fluorescent domains (Figure 1a). Screening of a small library of sensor variants in which one or more of these mutations were reversed revealed a subset of mutants with a large increase in emission ratio (Figure 1c). Significantly, all these variants carried the R125I reversion on both fluorescent domains, suggesting that this mutation is crucial for promoting intramolecular association between the donor and acceptor domain. In all cases, protease addition reduced the emission ratio to that of the sensors with the monomeric fluorescent domains (Figure 1c). The emission spectra before and after proteolytic cleavage for several of these variants show that the increased dynamic range is caused by a large change of the mOrange emission, resulting in a three-to ten-fold increase in dynamic range (Figure 1e-i).
Figure 1.
Development of self-associating mOrange and mCherry variants for improved performance in red-shifted FRET sensors. (a) Promoting intramolecular domain interactions increases FRET before proteolytic cleavage. (b) Structure showing the key residues I125 (green), V127 (blue) and I180 (yellow) at the A-B hydrophobic dimer interface in DsRed (PDB 1G7K). (c) A library of mOrange(O)-mCherry(C) fusion proteins carrying reversions R125I (α), T127V (β) and/or T180I (γ) was screened for increased change in FRET after proteolytic cleavage of the linker. (d-i) Emission spectra of purified red protease sensor variants before and after cleavage. Dynamic range (DR), calculated as the difference in ratios before and after cleavage, divided by the ratio after proteinase K cleavage, is indicated for each variant.
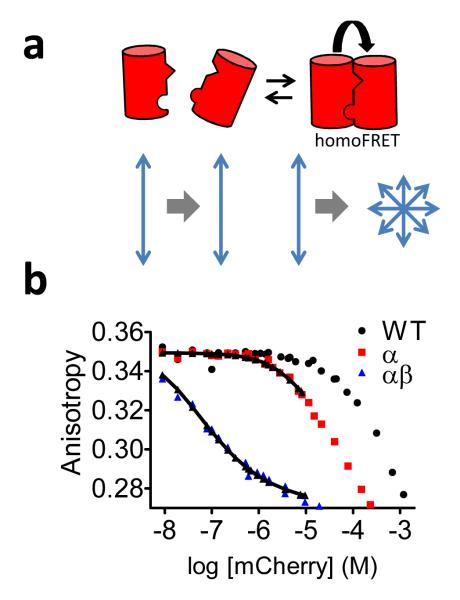
To quantify the strength of the protein-protein interaction brought about by the reversions, we used fluorescence polarization to monitor homodimerization of mCherry variants as a function of protein concentration (Figure 2a).17 Fitting these titration data yielded a Kd for dimerization of 35 ± 4 μM for the R125I reversion, while a 250-fold stronger interaction was observed for the R125I/T127V double mutation (Figure 2b). As the effective concentration provided by the linker is in the low mM range,18 the Kd of 35 μM obtained for the R125I mutation is sufficient to promote essentially complete intramolecular association between the FPs in these constructs. The T127V mutation, while not sufficient to induce dimerization on its own, further strengthens the interaction induced by the R125I mutation, providing an additional possibility to tune FRET sensor properties.11 However, the R125I/T127V mutation also increases the chance of additional intermolecular interactions, which could partially counteract the increase in dynamic range, as shown in Figure 1e-i.
Figure 2.
Fluorescence anisotropy was used to monitor fluorescent protein dimerization. (a) Schematic representation of the anisotropy assay. Upon homodimerization, the decreased interchromophore distance allows homoFRET to occur, which has a strong depolarizing effect. (b) Fluorescence anisotropy of mCherry variants as a function of mCherry concentration. Data was fitted to equation 2, yielding dimer dissociation constants of 35 ± 4 μM and 91 ± 7 nM for mCherry-R125I (α) and mCherry-R125I/T127V (β), respectively.
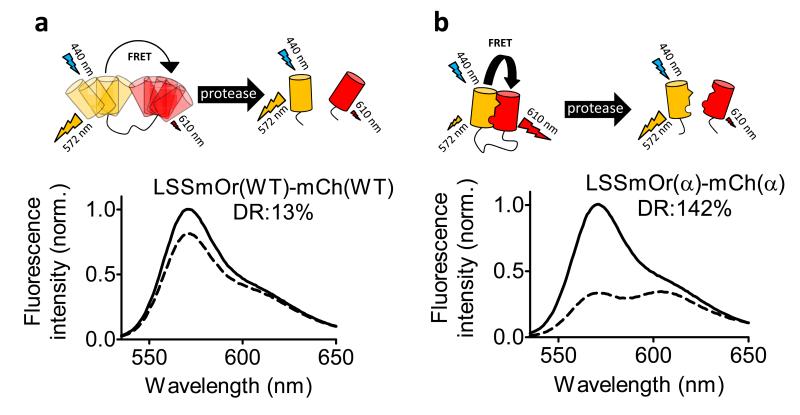
Since R125 and T127 are present in all mFruit variants, the results obtained here for mOrange and mCherry should be generally applicable to all monomeric derivatives of dsRed. We therefore tested whether R125I would also improve the performance of FRET sensors containing LSSmOrange, a recently described variant of mOrange that has a blue-shifted excitation peak (440 nm) and a long Stokes-shifted emission peak (572 nm).19 This variant is useful as a FRET donor to a red acceptor since it prevents direct acceptor excitation and allows simultaneous excitation of CFP-YFP-based FRET sensors. Replacement of mOrange by LSSmOrange resulted in a protease sensor that could be excited at 440 nm and detected in the red part of the spectrum, but with a low dynamic range (13%) (Figure 3a). Introduction of the R125I reversion on both fluorescent domains again resulted in a 10-fold improvement in dynamic range (Figure 3b). Interestingly, these results show that preventing direct acceptor excitation by itself does not significantly improve the sensor’s dynamic range, and that intramolecular interaction between the FPs is essential to enhance the sensor’s performance.
Figure 3.
Protease sensors employing LSSmOrange as donor in the absence (a) or presence (b) of the R125I mutation (α) on the fluorescent domains. Emission spectra of LSSmOrange-linker-mCherry sensors are shown before (dashed line) and after protease cleavage (solid line).
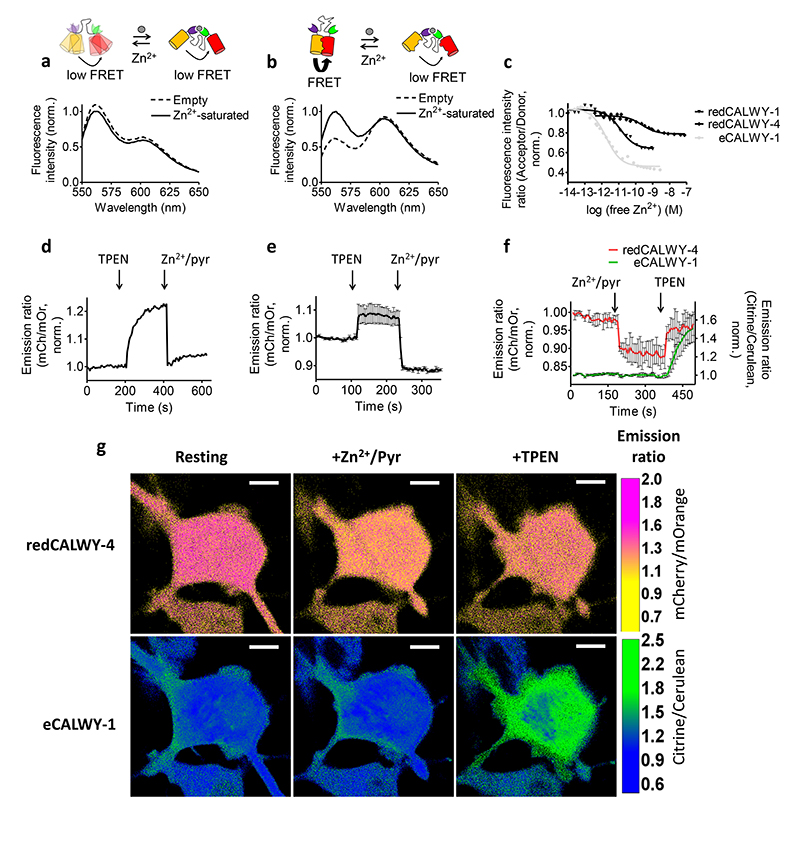
We next tested whether the use of self-associating FPs indeed allows straightforward replacement of the fluorescent domains in sensors based on mutually exclusive domain interactions. We previously developed the eCALWY series of Zn2+ sensors for intracellular imaging of Zn2+ homeostasis and signaling.13, 20 These sensors consist of a fusion of two cysteine-containing metal binding domains (ATOX1 and WD4) connected by a long, flexible glycine-serine linker. Binding of Zn2+ between ATOX1 and WD4 is mutually exclusive with the self-association of the FPs, translating to a decrease in emission ratio with increasing Zn2+free. As expected, replacement of the self-associating FPs in eCALWY-1 by mOrange and mCherry resulted in a sensor with a low emission ratio (0.55) that did not significantly change upon addition of a saturating concentration of Zn2+ (Figure 4a). Introduction of the R125I mutation in both fluorescent domains restored the switching capability however, yielding a genetically-encoded red Zn2+ FRET sensor (redCALWY-1) with a higher emission ratio in absence of Zn2+ (1.41), a large dynamic range of 62% and a Kd for Zn2+ of 12.3 ± 2 pM (Figure 4b, c). The same strategy could also be applied to obtain a red version of eCALWY-4, a variant bearing a mutation on the WD4 domain, yielding redCALWY-4 whose affinity of 234 ± 5 pM is better tuned to monitor increases in cytosolic Zn2+ concentrations (Figure 4c). In both cases, the Zn2+ affinities of the red variants mirrored those of their CFP-YFP counterparts,13 demonstrating that the fluorescent domains in these sensors can be replaced without affecting their ligand binding properties.
Figure 4.
In vitro characterization and intracellular application of red Zn2+ sensors. (a, b) Emission spectra measured in vitro in the absence and presence of 1.5 nM Zn2+free for redCALWY-1, either without (a) or with (b) the R125I reversions. (c) Emission ratio as a function of [Zn2+]free for redCALWY-1 (Kd = 12.3 ± 2 pM) and redCALWY-4 (Kd = 234 ± 5 pM). (d, e). Response of HeLa cells expressing redCALWY-1 (d) or redCALWY-4 (e) to the addition of TPEN followed by the addition of excess Zn2+/pyrithione. (f) Response of HeLa cells expressing both eCALWY-1 (green) and redCALWY-4 (red) to the addition of Zn2+/pyrithione followed by excess TPEN. All traces in d-f represent the average of multiple cells after normalization of the emission ratio at t=0. Error bars represent SEM. (g) False-colored ratiometric images of one of the cells measured in f in the resting state, at high Zn2+ concentration (+Zn2+/pyr) and at low Zn2+ concentration (+TPEN). White scale bars represent 10 μm.
To assess the performance of these new red variants for intracellular imaging, we replaced mOrange by the more photostable mOrange2 variant21 carrying the same R125I mutation and transfected HeLa cells with one or more CALWY variants for cytosolic expression. HeLa cells expressing the high affinity redCALWY-1 showed a clear increase in emission ratio upon addition of the membrane permeable Zn2+-chelator TPEN, which was completely reversed upon subsequent addition of the Zn2+-specific ionophore pyrithione together with excess Zn2+ (Figure 4d). This response is similar to that previously reported for the eCALWY-1 sensor13 and shows that these high affinity sensors are completely saturated with Zn2+ under normal physiological conditions. The lower-affinity redCALWY-4 is mostly empty at the start of the experiment, as only a small increase in emission ratio is observed in the presence of TPEN, whereas a large decrease is observed with Zn2+/pyrithione (Figure 4e). As expected, the same sensor lacking the R125I reversion on the mFruit domains did not show any response to perturbations in intracellular Zn2+free (Supplementary Figure 5a). Experiments with redCALWY-4 in HEK293 cells showed results similar to those in HeLa cells (Supplementary Figure 6). To provide a critical test for the orthogonality of the green and red FRET sensors, we co-expressed eCALWY-1 together with redCALWY-4 in the HeLa cytosol. Since their Zn2+ affinities differ by more than an order of magnitude, together these sensors allow measurement of a greatly extended range of cytosolic Zn2+ concentrations. First, increasing the intracellular Zn2+free led to a decrease in the redCALWY-4 emission ratio without the affecting eCALWY-1 emission ratio. The lack of a response observed for the green eCALWY-1 is consistent with our previous work that showed that this sensor is already fully saturated with Zn2+ under normal conditions.13 Subsequent addition of TPEN resulted in an immediate increase in emission ratio for redCALWY-4, followed by a slower increase in emission ratio for the high affinity green eCALWY-1 sensor (Figure 4f, g). This result establishes the feasibility of multiparameter imaging using a combination of our newly developed optimized mOrange-mCherry FRET pair with a CFP-YFP based sensor.
In conclusion, we have shown that promoting intramolecular interactions between donor and acceptor fluorescent domains can be an efficient strategy to improve the traditionally poor performance of red FRET sensors. The mutations identified here allowed precise tuning of the intramolecular interaction between mOrange and mCherry, yielding protease and Zn2+ sensors with dramatically improved dynamic ranges. It is important to realize that successful application of this new FRET pair in other sensors will critically depend on their design. FRET sensors based on mutually exclusive domain interactions are most likely to benefit, whereas improvement of sensor performance is not guaranteed for sensors based on more subtle allosteric mechanisms, where introduction of these self-associating domains could lead to an increase in FRET in both the on and off states of the sensor.11 In addition to being directly applicable to dsRed-derived fluorescent domains, these design principles should be readily extendable to fluorescent domains from other families, providing a generic method for engineering alternatively colored FRET sensors for use in multiparameter imaging.
METHODS
Cloning and mutagenesis
A detailed description of cloning strategies, primers used, as well as final nucleotide sequences for all constructs described in this paper, can be found in the Supplementary Information.
Protein expression and purification
Detailed descriptions of protein expression and purification may be found in the Supplementary Information.
Library screening
Initial expression, purification and screening of a small library of mOrange-linker-mCherry reversion bearing variants were carried out at small scale. Briefly, protein expression from 100 mL E. coli BL21 (DE3) cultures was induced with IPTG overnight at 25 °C. The His-tagged proteins were purified from the soluble fraction of the cell lysate by microfuge-based Ni2+-NTA affinity chromatography (GE). Proteins were salt exchanged to 50 mM Tris-HCl (pH 8), 100 mM NaCl and subsequently incubated at 37 °C overnight to allow complete maturation of the mOrange chromophore. Protein samples were prepared in 200 μL Tris buffer at a 10 μM concentration in a 96-well plate. Fluorescence emission spectra were recorded using a plate reader (TECAN Safire) before and after the addition of 0.006 U proteinase K (Sigma) using 525 nm excitation light. Emission ratios were calculated as emission at 610 nm divided by emission at 565 nm.
Fluorescence spectroscopy and Zn2+ titration experiments
Before any measurements, proteins were incubated at room temperature overnight to ensure complete maturation of the mOrange or LSSmOrange chromophore. Red protease sensor fluorescence emission spectra were measured with 1 μM sensor protein in 2 mL buffer (50 mM Tris-HCl (pH 8), 100 mM NaCl) at 25 °C in quartz cuvettes with a 1 cm path length (Hellma) and an excitation wavelength of 525 nm (mOrange) or 440 nm (LSSmOrange), using a Cary Eclipse fluorescence spectrometer (Varian). To cleave the flexible linker, a total of 0.005 units of proteinase K (Sigma) was added. Zn2+ titrations were carried out with 0.25 μM redCALWY protein, in buffer consisting of 50 mM Tris (pH 7.1), 100 mM NaCl, 10% (v/v) glycerol, 0.5 mM TCEP and 2 μM DTT, together with different Zn2+-chelators (EDTA, HEDTA, DHPTA, NTA and EGTA) and different concentrations of ZnCl2, as previously described13. The free Zn2+ concentration was calculated using the published critical stability constants of the chelators for Zn2+. To determine the redCALWY variants’ dissociation constants (Kd) for Zn2+, the emission ratio (R) was fitted as a function of [Zn2+] using equation 1.
| (1) |
Here R is the ratio of mCherry (at 605 nm) to mOrange (at 562 nm) emission, [Zn2+] the calculated Zn2+free concentration, P1 the difference in ratio between the Zn2+ saturated and Zn2+ depleted states and P2 the ratio in the Zn2+ depleted state of the sensor.
Fluorescence anisotropy
To determine the dissociation constant for dimerization of the mCherry mutants, fluorescence anisotropy was used to follow fluorescence emission depolarization induced by homoFRET. This technique was recently described in detail for variants of CFP17. Measurements were performed at 25 °C in 50 mM Tris-HCl (pH 8), 100 mM NaCl in a quartz cuvette with a 1×1 cm path length (Hellma), with an excitation wavelength of 587 nm, using a Cary Eclipse fluorescence spectrophotometer (Varian). At concentrations above 10 μM, a 3×3 mm pathlength cuvette was used instead. The decrease in anisotropy that is observed for mCherry WT at concentrations above 10 μM is most likely not due to formation of a homodimer, as mCherry has been reported to be monomeric up to at least 350 μM.22 Instead this decrease results from homoFRET between molecularly dissolved monomeric proteins and we therefore limited the analysis of the mCherry variants to concentrations no higher than 10 μM. We fitted the data in Figure 1b measured at mCherry concentrations below 10 μM to the dimerization model (equation 2) previously derived for CFP17 to arrive at the Kd’s reported here:
| (2) |
Here r is the observed anisotropy, Kd the dissociation constant for FP homodimerization, c the FP concentration, rm the FP anisotropy in the monomeric state and rd the anisotropy of the FP in the dimeric state. A global fit over both data sets allowed us to exploit the information present in both datasets, with mCherry (R125I/T127V) providing the value for anisotropy in the fully dimerized state and mCherry (R125I) providing the value for anisotropy in the completely monomeric state.
Mammalian cell culture and imaging
HeLa cells were grown in Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 25 mM glucose, 10% Fetal Bovine Serum (FBS), 100 U/mL penicillin and 100 μg/mL streptomycin (all from Life Technologies) but lacking sodium pyruvate. Chambered borosilicate cover glass wells (4.2 cm2 each, Lab-Tek, Nunc) were seeded with 200,000 cells one day before transfection. Transfection was carried out with Lipofectamine 2000 (Life Technologies) according to manufacturer’s instructions. Cells were imaged two days after transfection in a HEPES buffer (Live Cell Imaging Solution, Life Technologies) at 37 °C. Imaging was performed with a confocal microscope (Leica, TCS SP5X) equipped with a 40× water immersion objective, acousto-optical beamsplitters (AOBS), a white light laser and a 405 nm laser. To excite cerulean, a 405 nm laser was used, while a white light laser set to 550 nm (5% of full power) was used for excitation of mOrange2. Emission was monitored over the following windows using the AOBS and avalanche photo diode/photomultiplier tubes hybrid detectors (HyD, Leica): cerulean (460-490 nm), citrine (510-550 nm), mOrange2 (565-600 nm) and mCherry (600-630 nm). Images were acquired at either 8.21 second intervals (two sensor experiments) or at 5 second intervals (single sensor experiments). Cells were first imaged for several minutes with HEPES buffer alone, before HEPES buffer with N,N,N′,N′-Tetrakis(2-pyridylmethyl)ethylenediamine (TPEN, TCI Europe) was added to a final concentration of 50 μM TPEN, followed a few minutes later by HEPES buffer containing ZnCl2 and 2-Mercaptopyridine N-oxide (pyrithione, Sigma) with a final concentration of 36 μM TPEN, 100 μM ZnCl2 and 5 μM pyrithione. In other experiments, this order was reversed, and cells were imaged first in their resting state, followed by 10 μM ZnCl2 and 1 μM pyrithion, before the addition of 50 μM TPEN. For ease of comparison, all emission ratios have been normalized to ratio at t=0. To generate pseudocolored ratiometric images, images corresponding to channels (mOrange, mCherry, cerulean or citrine) obtained under three different conditions (in the cell’s resting state, at saturating Zn2+ levels or in the Zn2+-depleted state) were averaged, after which acceptor averaged images (mCherry or citrine) were divided by donor averaged images (mOrange or cerulean respectively), which were then thresholded, removing values above 2 (for mOrange/mCherry images) or 2.5 (for cerulean/citrine images).
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to members of the Laboratory of Soft Tissue Biomechanics & Engineering (Eindhoven University of Technology) for the use of the fluorescence confocal microscope and to M. van Turnhout for technical assistance. We thank M. van ‘t Erve for help in characterization of the effect of mFruit reversions and P. Goodarzy Fard for initial preparation and measurement of redCALWY protein samples. This work was supported by grants from the Netherlands Organization of Scientific Research (VIDI grant 700.56.428 and ECHO grant 700.59.013) and an ERC starting grant (ERC-2011-StG 280255).
Footnotes
Supporting Information Molecular cloning methods, primer sequences, protein purification methods, nucleotide sequences for expression constructs and additional redCALWY HeLa and HEK293 cell measurements may be found in the Supporting Information. This material is available free of charge at http://pubs.acs.org.
Notes The authors declare no competing financial interests.
REFERENCES
- 1.Campbell RE. Fluorescent-protein-based biosensors: modulation of energy transfer as a design principle. Anal. Chem. 2009;81:5972–5979. doi: 10.1021/ac802613w. [DOI] [PubMed] [Google Scholar]
- 2.Palmer AE, Qin Y, Park JG, McCombs JE. Design and application of genetically encoded biosensors. Trends Biotechnol. 2011;29:144–152. doi: 10.1016/j.tibtech.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carlson HJ, Campbell RE. Genetically encoded FRET-based biosensors for multiparameter fluorescence imaging. Curr. Opin. Biotechnol. 2009;20:19–27. doi: 10.1016/j.copbio.2009.01.003. [DOI] [PubMed] [Google Scholar]
- 4.Welch CM, Elliott H, Danuser G, Hahn KM. Imaging the coordination of multiple signalling activities in living cells. Nat. Rev. Mol. Cell Biol. 2011;12:749–756. doi: 10.1038/nrm3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ai HW, Hazelwood KL, Davidson MW, Campbell RE. Fluorescent protein FRET pairs for ratiometric imaging of dual biosensors. Nat. Methods. 2008;5:401–403. doi: 10.1038/nmeth.1207. [DOI] [PubMed] [Google Scholar]
- 6.Ding Y, Ai HW, Hoi H, Campbell RE. Forster resonance energy transfer-based biosensors for multiparameter ratiometric imaging of Ca2+ dynamics and caspase-3 activity in single cells. Anal. Chem. 2011;83:9687–9693. doi: 10.1021/ac202595g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Piljic A, Schultz C. Simultaneous recording of multiple cellular events by FRET. ACS Chem. Biol. 2008;3:156–160. doi: 10.1021/cb700247q. [DOI] [PubMed] [Google Scholar]
- 8.Ouyang M, Huang H, Shaner NC, Remacle AG, Shiryaev SA, Strongin AY, Tsien RY, Wang Y. Simultaneous visualization of protumorigenic Src and MT1-MMP activities with fluorescence resonance energy transfer. Cancer Res. 2010;70:2204–2212. doi: 10.1158/0008-5472.CAN-09-3698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jost CA, Reither G, Hoffmann C, Schultz C. Contribution of fluorophores to protein kinase C FRET probe performance. ChemBioChem. 2008;9:1379–1384. doi: 10.1002/cbic.200700728. [DOI] [PubMed] [Google Scholar]
- 10.Miranda JG, Weaver AL, Qin Y, Park JG, Stoddard CI, Lin MZ, Palmer AE. New alternately colored FRET sensors for simultaneous monitoring of Zn2+ in multiple cellular locations. PLoS One. 2012;7:e49371. doi: 10.1371/journal.pone.0049371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kotera I, Iwasaki T, Imamura H, Noji H, Nagai T. Reversible dimerization of Aequorea victoria fluorescent proteins increases the dynamic range of FRET-based indicators. ACS Chem. Biol. 2010;5:215–222. doi: 10.1021/cb900263z. [DOI] [PubMed] [Google Scholar]
- 12.Vinkenborg JL, Evers TH, Reulen SW, Meijer EW, Merkx M. Enhanced sensitivity of FRET-based protease sensors by redesign of the GFP dimerization interface. ChemBioChem. 2007;8:1119–1121. doi: 10.1002/cbic.200700109. [DOI] [PubMed] [Google Scholar]
- 13.Vinkenborg JL, Nicolson TJ, Bellomo EA, Koay MS, Rutter GA, Merkx M. Genetically encoded FRET sensors to monitor intracellular Zn2+ homeostasis. Nat. Methods. 2009;6:737–740. doi: 10.1038/nmeth.1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Golynskiy MV, Rurup WF, Merkx M. Antibody Detection by Using a FRET-Based Protein Conformational Switch. Chembiochem. 2010;11:2264–2267. doi: 10.1002/cbic.201000143. [DOI] [PubMed] [Google Scholar]
- 15.van der Velden LM, Golynskiy MV, Bijsmans IT, van Mil SW, Klomp LW, Merkx M, van de Graaf SF. Monitoring bile acid transport in single living cells using a genetically encoded Forster resonance energy transfer sensor. Hepatology. 2013;57:740–752. doi: 10.1002/hep.26012. [DOI] [PubMed] [Google Scholar]
- 16.Campbell RE, Tour O, Palmer AE, Steinbach PA, Baird GS, Zacharias DA, Tsien RY. A monomeric red fluorescent protein. Proc. Natl. Acad. Sci. USA. 2002;99:7877–7882. doi: 10.1073/pnas.082243699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Espagne A, Erard M, Madiona K, Derrien V, Jonasson G, Levy B, Pasquier H, Melki R, Merola F. Cyan fluorescent protein carries a constitutive mutation that prevents its dimerization. Biochemistry. 2011;50:437–439. doi: 10.1021/bi1015875. [DOI] [PubMed] [Google Scholar]
- 18.Evers TH, Appelhof MA, de Graaf-Heuvelmans PT, Meijer EW, Merkx M. Ratiometric detection of Zn(II) using chelating fluorescent protein chimeras. J. Mol. Biol. 2007;374:411–425. doi: 10.1016/j.jmb.2007.09.021. [DOI] [PubMed] [Google Scholar]
- 19.Shcherbakova DM, Hink MA, Joosen L, Gadella TW, Verkhusha VV. An orange fluorescent protein with a large Stokes shift for single-excitation multicolor FCCS and FRET imaging. J. Am. Chem. Soc. 2012;134:7913–7923. doi: 10.1021/ja3018972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Taylor KM, Hiscox S, Nicholson RI, Hogstrand C, Kille P. Protein kinase CK2 triggers cytosolic zinc signaling pathways by phosphorylation of zinc channel ZIP7. Sci. Signal. 2012;5:ra11. doi: 10.1126/scisignal.2002585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shaner NC, Lin MZ, McKeown MR, Steinbach PA, Hazelwood KL, Davidson MW, Tsien RY. Improving the photostability of bright monomeric orange and red fluorescent proteins. Nat. Methods. 2008;5:545–551. doi: 10.1038/nmeth.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shemiakina II, Ermakova GV, Cranfill PJ, Baird MA, Evans RA, Souslova EA, Staroverov DB, Gorokhovatsky AY, Putintseva EV, Gorodnicheva TV, Chepurnykh TV, Strukova L, Lukyanov S, Zaraisky AG, Davidson MW, Chudakov DM, Shcherbo D. A monomeric red fluorescent protein with low cytotoxicity. Nat. Commun. 2012;3:1204. doi: 10.1038/ncomms2208. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.