Abstract
Objectives
This study sought to investigate the role of secretory phospholipase A2 (sPLA2)-IIA in cardiovascular disease.
Background
Higher circulating levels of sPLA2-IIA mass or sPLA2 enzyme activity have been associated with increased risk of cardiovascular events. However, it is not clear if this association is causal. A recent phase III clinical trial of an sPLA2 inhibitor (varespladib) was stopped prematurely for lack of efficacy.
Methods
We conducted a Mendelian randomization meta-analysis of 19 general population studies (8,021 incident, 7,513 prevalent major vascular events [MVE] in 74,683 individuals) and 10 acute coronary syndrome (ACS) cohorts (2,520 recurrent MVE in 18,355 individuals) using rs11573156, a variant in PLA2G2A encoding the sPLA2-IIA isoenzyme, as an instrumental variable.
Results
PLA2G2A rs11573156 C allele associated with lower circulating sPLA2-IIA mass (38% to 44%) and sPLA2 enzyme activity (3% to 23%) per C allele. The odds ratio (OR) for MVE per rs11573156 C allele was 1.02 (95% confidence interval [CI]: 0.98 to 1.06) in general populations and 0.96 (95% CI: 0.90 to 1.03) in ACS cohorts. In the general population studies, the OR derived from the genetic instrumental variable analysis for MVE for a 1-log unit lower sPLA2-IIA mass was 1.04 (95% CI: 0.96 to 1.13), and differed from the non-genetic observational estimate (OR: 0.69; 95% CI: 0.61 to 0.79). In the ACS cohorts, both the genetic instrumental variable and observational ORs showed a null association with MVE. Instrumental variable analysis failed to show associations between sPLA2 enzyme activity and MVE.
Conclusions
Reducing sPLA2-IIA mass is unlikely to be a useful therapeutic goal for preventing cardiovascular events.
Key Words: cardiovascular diseases, drug development, epidemiology, genetics, Mendelian randomization
Abbreviations and Acronyms: ACS, acute coronary syndrome(s); CI, confidence interval; LDL-C, low-density lipoprotein cholesterol; MI, myocardial infarction; MVE, major vascular events; OR, odds ratio; RCT, randomized clinical trial; SNP, single-nucleotide polymorphism; sPLA2, secretory phospholipase A2
The secretory phospholipase A2 (sPLA2) enzymes, mostly comprising sPLA2-IIA, sPLA2-III, sPLA2-V, and sPLA2-X, hydrolyze phospholipids from the cell membrane surface and lipoproteins, producing pro-inflammatory lysophospholipids and eicosanoids (1). This activity may also modify low-density lipoprotein cholesterol (LDL-C) particles in the circulation increasing the binding of LDL-C onto blood vessel wall proteoglycans, promoting foam cell formation and the development of atherosclerosis (2). sPLA2-IIA is thought to be the most highly expressed of the sPLA2 enzymes (2) and its mass can be quantified specifically in plasma by enzyme-linked immunosorbent assay (3). In contrast, the assay for sPLA2 enzyme activity does not distinguish between the secretory isoenzymes IIA, III, V, and X (2).
Observational studies have indicated that higher circulating sPLA2-IIA mass and sPLA2 enzyme activity are associated with an increased risk of incident and recurrent MVE (comprising cardiovascular death, myocardial infarction [MI], and stroke) 3, 4, 5, 6, with the evidence being more compelling in primary prevention (4) than in patients with ACS (7). This suggests that sPLA2 isoenzymes, in particular IIA, may represent a novel therapeutic target for cardiovascular disease prevention. This hypothesis is supported by studies in mouse models that show over-expression of sPLA2-IIA associates with increased atherosclerotic lesion size (8).
Despite these encouraging findings, mechanistic studies in animals may not faithfully model the disease process in humans, and observational studies in humans cannot provide definitive evidence on causation. This is because higher sPLA2-IIA mass or sPLA2 enzyme activity may be a consequence not a cause of atherosclerosis or reflect an association with causal risk factors. An evaluation of sPLA2 as a therapeutic target is timely to help put into context the recent announcement that a phase III randomized trial (VISTA-16 [Vascular Inflammation Suppression to Treat Acute Coronary Syndrome for 16 Weeks] trial) (10) of varespladib, a first-in-class sPLA2 inhibitor (9) for major vascular event (MVE) prevention in patients with acute coronary syndrome (ACS), was stopped prematurely for lack of efficacy (11). Varespladib was designed to selectively block sPLA2-IIA, however, it also has minor inhibitory effects on other sPLA2 isoenzymes (Online Fig. 1) 12, 13.
Methods
A total of 109,179 individuals of European descent from 36 studies were included in the analysis (Online Table 1), comprising 19 in general populations and 10 studies in patients with ACS. In addition, we included 4 case control studies of coronary artery disease, 1 cohort of patients with established arterial vascular disease or risk factors for cardiovascular disease (SMART [Second Manifestations of ARTerial disease] study), and 1 nested case-control study of coronary artery restenosis in patients with ACS undergoing percutaneous coronary intervention (GENDER [GENetic Determinants of Restenosis] study). These additional studies did not contribute toward the analyses set in general population or ACS studies, and were analyzed and reported separately (Online Appendix). Finally, tissue samples from 1 cohort of patients undergoing aortic valve surgery (ASAP [Advanced Study of Aortic Pathology]) were used to investigate the association of single-nucleotide polymorphisms (SNPs) in PLA2G2A with mRNA expression in liver, mammary artery, aorta, and heart with an external data source comprising 206 transplant donor liver samples used for replication (14). Approval from relevant ethical committees was obtained for collaborating studies. All analyses, unless otherwise stated, were performed using Stata 12.1 (StataCorp, College Station, Texas).
Measurement of sPLA2-IIA mass and sPLA2 enzyme activity
sPLA2-IIA mass and sPLA2 enzyme activity were measured in 7 and 6 of the collaborating studies, respectively (Online Table 2). Assay methods are reported in Online Table 3. Owing to the time of blood sampling being greater than 1 month after the acute coronary event, samples for the KAROLA (Langzeiterfolge der KARdiOLogischen Anschlussheilbehandlung) study were not included in the analysis. The distributions of both sPLA2-IIA mass and sPLA2 enzyme activity were skewed, hence the traits were natural log(e) transformed prior to analysis.
Observational analysis
We investigated correlations between log sPLA2-IIA mass and log sPLA2 enzyme activity in studies that measured both traits (Online Appendix, Online Fig. 2). To investigate the association between circulating sPLA2-IIA mass and sPLA2 enzyme activity with incident major vascular events in general populations, we used the European Prospective Investigation of Cancer (EPIC)-Norfolk study and to investigate the association with recurrent events in patients with ACS, we used 4 ACS cohorts (FAST-MI [French Registry of Acute ST-Elevation or Non–ST-elevation Myocardial Infarction], GRACE [Global Registry of Acute Coronary Events]-France, GRACE-Scotland, and MIRACL [Myocardial Ischemia Reduction with Acute Cholesterol Lowering] trial). For EPIC-Norfolk, the outcome was a composite of fatal and nonfatal MI, whereas for the 4 ACS cohorts, it was a composite of all-cause mortality or MI.
First, in the EPIC-Norfolk study we evaluated the cross-sectional correlates of sPLA2-IIA and sPLA2 enzyme activity with established and emerging risk factors using linear or logistic regression as appropriate. Second, we evaluated the shape of the association between sPLA2-IIA mass and sPLA2 enzyme activity with MI in the general population study, and with MI/all-cause mortality in the ACS cohorts. Third, we estimated the magnitude of the association per 1 log unit lower sPLA2-IIA mass and sPLA2 enzyme activity with MI or MI/all-cause mortality after statistical adjustment for potential confounders using logistic regression. Finally, we explored the independent effect of log sPLA2-IIA mass and log sPLA2 enzyme activity with MI or MI/all-cause mortality by fitting a logistic regression model that included both log sPLA2-IIA mass, and log sPLA2 enzyme activity in addition to potential confounders. The summary estimates were pooled across studies using fixed-effects meta-analysis. For full details of the observational analyses, please see the Online Appendix.
Genetic analysis
All studies apart from the MIRACL trial contributed toward the genetic analysis (Online Table 2). Genotype coding was arranged to be directionally consistent with the effect of varespladib on sPLA2-IIA mass and sPLA2 enzyme activity. Traits that were nonnormally distributed (sPLA2-IIA mass, sPLA2 enzyme activity, C-reactive protein, triglycerides, and interleukin-6) were log(e) transformed, and differences between genotype groups were reported as a percentage difference.
Selection of the genetic instrument and evaluation of its specificity and effect size
SNP Selection
Six tagging SNPs (15) that captured >90% of genetic variation in PLA2G2A in Europeans were evaluated in 3 studies (EPIC-Norfolk, GRACE-France, and UDACS [University College London Diabetes and Cardiovascular Study]). The rs11573156 variant that showed the lowest p value with sPLA2-IIA mass and sPLA2 enzyme activity (Online Fig. 3) was chosen for Mendelian randomization analysis. Rs11573156 was directly genotyped in all studies except 2 that imputed it and 1 that used a proxy SNP. Genotype frequencies were consistent across studies (Online Fig. 4) and did not deviate from Hardy-Weinberg equilibrium (at p < 0.001) (Online Table 4).
Specificity of Genetic Instrument for PLA2G2A
Affymetrix GeneChip Human Exon 1.0 ST expression arrays were used to quantify mRNA expression in the ASAP study, in which participants were genotyped using Illumina Human 610W-Quad Beadarray (including 101 SNPs in the region 200 kb upstream and downstream from the PLA2G2A locus). Please see the Online Appendix for further details of estimation of genotype association with mRNA expression.
Strength of Genetic Instrument (rs11573156 C>G) on sPLA2
We estimated the per C allele association between PLA2G2A rs11573156 and sPLA2 measures, as well as the proportion of variance (R2) of these measures explained by the rs11573156 variant.
Association Between Genetic Instrument and Putative and Established Cardiovascular Risk Factors
Twenty studies of individuals in which blood sampling occurred prior to the cardiovascular event were used to test the association of PLA2G2A rs11573156 (per C allele) with cardiovascular risk factors within each study using linear regression. Results were pooled using fixed (default) and random effects meta-analysis.
Cardiovascular Outcomes Examined
For the general population studies, MVE were separated into prevalent and incident, whereas for ACS cohorts, all events after recruitment were included and labeled as recurrent. Prevalent MVE were a composite of nonfatal MI and nonfatal stroke, and incident MVE were a composite of fatal/nonfatal MI and fatal/nonfatal stroke. For ACS cohorts, recurrent MVE were a composite of nonfatal MI, nonfatal stroke, and all-cause mortality. Individual components of MVE were also reported separately. See the Online Appendix for outcomes definitions per study and Online Table 5 for study contribution to the composite outcome.
Association Between Genetic Instrument and MVE
We conducted 2 genetic approaches: first, a genetic association analysis of the PLA2G2A rs11573156 variant with MVE, and; second, an instrumental variable analysis that quantified a causal effect per 1 log unit lower sPLA2-IIA mass and sPLA2 enzyme activity on MVE, under the assumptions of instrumental variable analysis (16). A total of 26 studies contributed to these 2 approaches, comprising 17 in general populations and 9 studies in patients with ACS.
For the genetic association approach, we estimated the within-study odds ratio (OR) per C allele of PLA2G2A rs11573156 with MVE using logistic regression and the results were pooled using fixed (default) and random-effects meta-analysis and used I2 to measure heterogeneity. All meta-analyses were stratified by clinical setting (general population or patients with ACS).
For the instrumental variable analysis, we first applied the pooled estimate of the gene variant on log sPLA2-IIA mass and log sPLA2 enzyme activity to each study, including studies that did not have measures of sPLA2-IIA mass or sPLA2 enzyme activity (17). An instrumental variable estimate was then generated (taking into account the uncertainty in both the gene-sPLA2 and gene-outcome associations) (18) for each study. The study-specific instrumental variable estimates were pooled using fixed-effects meta-analysis. Full details of the methodology are provided in the Online Appendix. We compared the instrumental variable estimates to the expected estimates based on the observational association between sPLA2-IIA mass, sPLA2 enzyme activity, and cardiovascular events.
Treatment trials of varespladib
In order to contextualize the effect of the genetic instrument with the sPLA2 inhibitor (varespladib), we conducted a systematic review of RCTs (following PRISMA guidance) (19) to evaluate the effects of varespladib on sPLA2-IIA mass and other cardiovascular traits. To investigate the dose response between varespladib and sPLA2-IIA mass, we conducted a meta-regression analysis (for details, see the Online Appendix).
Results
Observational analysis of sPLA2-IIA mass and sPLA2 enzyme activity
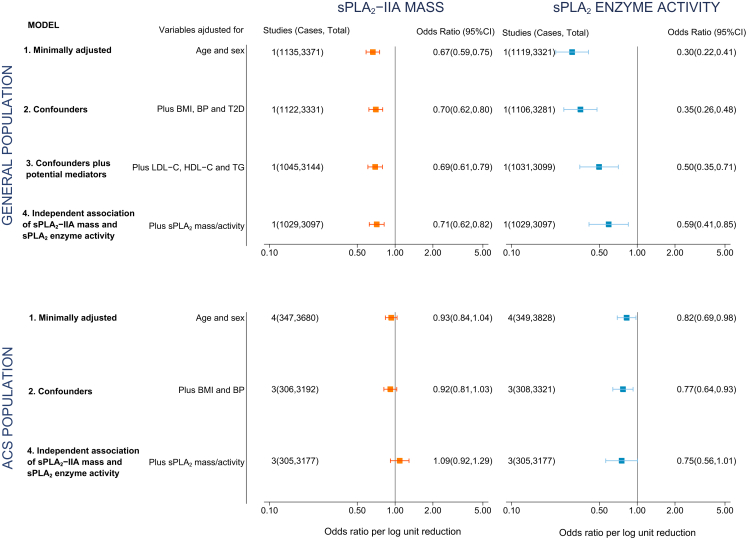
Lower levels of sPLA2-IIA mass and lower levels of sPLA2 enzyme activity each were associated with a reduced risk of cardiovascular events in the general population with an OR for fatal/nonfatal MI of 0.67 (95% confidence interval [CI]: 0.59 to 0.75) and 0.30 (95% CI: 0.22 to 0.41) per 1 log unit lower sPLA2-IIA mass and sPLA2 enzyme activity, respectively, after adjustment for age and sex (Fig. 1). For studies set in ACS, the corresponding summary ORs for all-cause mortality or MI were 0.93 (95% CI: 0.84 to 1.04) and 0.82 (95% CI: 0.69 to 0.98), respectively (Fig. 1). The log-linear model provided the best fit (p ≥ 0.1 for a quadratic model in all comparisons) (Online Table 6), indicating a constant proportional decrease in the relative odds per 1 log unit lower sPLA2-IIA mass or sPLA2 enzyme activity.
Figure 1.
Association of a 1 Log Unit Lower sPLA2-IIA Mass or sPLA2 Enzyme Activity With Fatal/Nonfatal Myocardial Infarction in General Population Studies and All-Cause Mortality/Myocardial Infarction in Acute Coronary Syndrome Studies
The general population study was EPIC-Norfolk and the 4 acute coronary syndrome cohorts were FAST-MI (French Registry of Acute ST-Elevation or Non–ST-elevation Myocardial Infarction), GRACE (Global Registry of Acute Coronary Events)-France, GRACE-Scotland, and MIRACL (Myocardial Ischemia Reduction with Acute Cholesterol Lowering). In Model 1, only age and gender were introduced as covariates. We then additionally adjusted for covariates (blood pressure [BP], body mass index [BMI], type 2 diabetes [T2D]) that could confound the association between secretory phospholipase A2 (sPLA2) and coronary heart disease (CHD; Model 2). Because lipids may mediate the association between sPLA2-IIA and CHD, we did not include lipids in Model 2, but included them in Model 3 (only available in the general population cohort). Finally, to investigate whether there was an independent association between sPLA2-IIA mass (orange), sPLA2 enzyme activity (blue), and CHD, we additionally included sPLA2 enzyme activity where sPLA2-IIA mass was the explanatory variable (and vice-versa; Model 4). CI = confidence interval; HDL-C = high-density lipoprotein cholesterol; LDL-C = low-density lipoprotein cholesterol; OR = odds ratio; TG = triglycerides.
sPLA2-IIA mass and sPLA2 enzyme activity associated with several established and emerging cardiovascular risk factors in the general population (Online Tables 7 and 8). In general, adjustment for cardiovascular risk factors diminished the association between sPLA2-IIA mass and sPLA2 enzyme activity with incident MI in the general population, though the association persisted following multivariate adjustment (Fig. 1). Interestingly, both associations (sPLA2-IIA mass and sPLA2 enzyme activity with MI) remained after adjustment for one another. For ACS cohorts, only sPLA2 enzyme activity was associated with recurrent events (Fig. 1).
Selection and validation of the genetic instrument for sPLA2-IIA
rs11573156 C>G showed the lowest p value with sPLA2-IIA mass (p = 5.49 × 10–180) and sPLA2 enzyme activity (p = 3.29 × 10–5) and was prioritized for analysis in the remaining studies (Online Fig. 3).
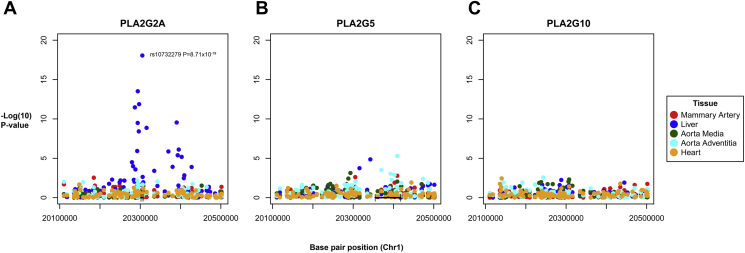
To evaluate the specificity of the rs11573156 variant for sPLA2-IIA, we analyzed the association of SNPs in PLA2G2A with mRNA expression of 3 different sPLA2 isoenzymes, encoded by distinct genes (PLA2G2A for sPLA2-IIA and PLA2G5 for sPLA2-V in close proximity on chromosome 1, and PLA2G10 for sPLA2-X on chromosome 10). PLA2G2A was mainly expressed in the liver, aortic adventitia and heart (Online Fig. 5). The SNP showing strongest association with PLA2G2A mRNA expression in liver was rs10732279A>G (p = 8.71 × 10–19) (Fig. 2A), in strong linkage disequilibrium with rs11573156 (R2 = 0.91 in Europeans, HapMap release 21) and explained 31% of the variance of PLA2G2A mRNA. These findings were replicated in an external data source comprising 206 transplant donor liver samples (p = 4.76 × 10–8) (14). rs10732279 showed no association with either PLA2G5 or PLA2G10 mRNA expression confirming the specificity of the genetic instrument for sPLA2-IIA (Figs. 2B and 2C).
Figure 2.
Association of SNPs in the PLA2G2A Region With mRNA Expression
Manhattan plots of single-nucleotide polymorphisms (SNPs) in the PLA2G2A region and association with mRNA expression of (A)PLA2G2A, (B)PLA2G5, and (C)PLA2G10, color-coded by tissue type. The p values for the association between rs10732279 and mRNA expression of PLA2G5 or PLA2G10 were 0.04 and 0.88, respectively.
Association of rs11573156 with sPLA2-IIA mass and sPLA2 enzyme activity
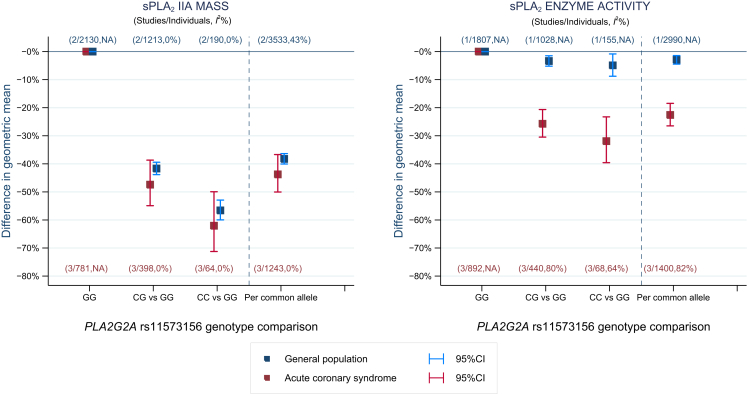
In 3 studies of 1,400 individuals with ACS and 2 general population studies of 3,533 individuals, an allele dose-dependent association was observed between rs11573156 and sPLA2-IIA mass and sPLA2 enzyme activity (Fig. 3). For each additional C allele of rs11573156, sPLA2-IIA mass was lower by 38% (95% CI: 36% to 40%) in studies of general populations and 44% (95% CI: 37% to 50%) in studies of ACS patients, compared with individuals homozygous for the G allele. The proportion of variance of sPLA2-IIA mass explained by rs11573156 in general population and ACS studies was 21% (95% CI: 18% to 23%) and 6% (95% CI: 3% to 9%), respectively.
Figure 3.
Meta-Analysis Pooled Estimates of the Association Between PLA2G2A rs11573156 With sPLA2-IIA Mass and sPLA2 Enzyme Activity
The analysis is separated by study setting into general populations (EPIC [European Prospective Investigation of Cancer]-Norfolk, UDACS [University College London Diabetes and Cardiovascular Study]; blue) and acute coronary syndrome (FAST-MI [French Registry of Acute ST-Elevation or Non–ST-elevation Myocardial Infarction], GRACE [Global Registry of Acute Coronary Events]-France, GRACE-Scotland; red). The percentage estimate was obtained by back-transforming the PLA2G2A rs11573156 log sPLA2 association to obtain the relative difference, which was converted to a percentage by subtracting 1 from the relative difference and multiplying the fraction by 100. NA = not applicable either because there were too few studies (<3 studies) to synthesize an I2 estimate, or the value could not be calculated for the reference genotype group (GG). sPLA2 = secretory phospholipase A2.The 3 genotype groups for the rs11573156 SNP are: 1) GG = reference group; 2) CG: 1 copy of the sPLA2-lowering (common) C-allele; 3) CC: 2 copies of the sPLA2-lowering C-allele.
In contrast, the effect of the rs11573156 C allele on sPLA2 enzyme activity was smaller and varied considerably by study setting, with a 3% reduction (95% CI: 1% to 5%) in studies of general populations and 23% reduction (95% CI: 19% to 27%) for studies of ACS patients. The proportion of variance of sPLA2 enzyme activity explained by rs11573156 was 0.5% (95% CI: 0.0% to 1.0%) and 3% (95% CI: 1% to 5%) in the general population and ACS cohorts, respectively.
We identified no major associations between rs11573156 and blood pressure, lipid fractions, inflammation markers, or carotid intima-media thickness (Online Tables 9 to 11).
Comparison of pharmacological modification of sPLA2 in randomized clinical trials and carriage of the PLA2G2A variant in populations
We identified 4 randomized clinical trials (RCTs) of the sPLA2 inhibitor varespladib in a total of 1,300 individuals (Online Fig. 6, Online Table 12) 9, 20, 21, 22. A meta-regression suggested varespladib treatment produced a dose-dependent reduction in sPLA2-IIA mass (p for meta-regression = 0.06) (Online Fig. 7). The most frequently studied dose of varespladib (500 mg/day) reduced sPLA2-IIA mass by 78% (95% CI: 62% to 94%). The effect of varespladib on sPLA2 enzyme activity was not reported in RCTs because activity was reported to be beneath the lower limit of quantification of the assay 20, 21, 22.
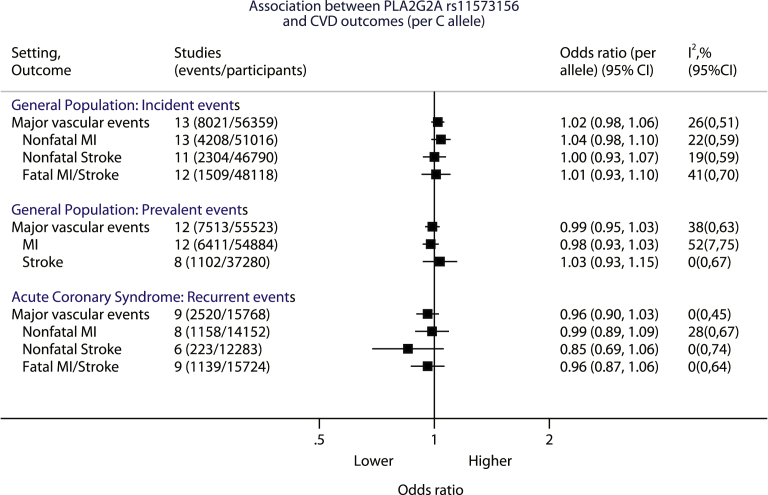
Association between rs11573156 and MVE
In a meta-analysis across 13 population studies (8,021 incident events in 56,359 individuals), there was no association between the C allele of rs11573156 and incident MVE (OR: 1.02; 95% CI: 0.98 to 1.06), nor with any of the individual components (Fig. 4, Online Fig. 8). Similarly, in 12 studies with 7,513 prevalent events in 55,523 individuals, there was no association between the rs11573156 C allele with prevalent MVE (OR: 0.99; 95% CI: 0.95 to 1.03), or with any of the individual components (Fig. 4, Online Fig. 9). For the 9 ACS studies with 2,520 recurrent events in 15,768 patients, there was also no association between the C allele of rs11573156 and recurrent MVE (OR: 0.96; 95% CI: 0.90 to 1.03) (Fig. 4, Online Fig. 10). Similar findings were obtained using a random-effects model for meta-analysis (Online Table 13).
Figure 4.
Meta-Analysis Pooled Estimates of the Association Between PLA2G2A rs11573156 and MVE (Including Individual Components) Stratified by Clinical Setting Into General Population and ACS Patients
Each plot represents the PLA2G2A rs11573156 per C allele odds ratio, with genotype grouping arranged to mimic the effects of pharmacological lowering of secretory phospholipase A2 (sPLA2)-IIA (i.e., if lowering of sPLA2-IIA mass were to reduce risk of cardiovascular events, the odds ratio in the plot should be <1). Major vascular events (MVE) comprise fatal/nonfatal MI or stroke in general population studies and fatal/nonfatal MI or stroke or all-cause mortality in ACS studies. CI = confidence interval. Fatal myocardial infarction (MI)/stroke included all-cause mortality for some acute coronary syndrome (ACS) studies (see Online Table 5 for further details).
Extreme genotype comparison
Individuals homozygous for the rs11573156 C allele had a 57% to 62% lower sPLA2-IIA mass compared with those homozygous for the G allele (Fig. 3), which was similar in magnitude to the 78% reduction seen with 500 mg/day varespladib dose used in VISTA-16. Using this genotype comparison, a null effect was again observed for MVE in all clinical settings: incident (5,175 cases; OR: 0.99; 95% CI: 0.89 to 1.10), prevalent (3,545 cases; OR: 1.00; 95% CI: 0.88 to 1.13), and recurrent (1,626 cases; OR 0.89; 95% CI: 0.74 to 1.06).
Instrumental variable analysis of sPLA2 on MVE
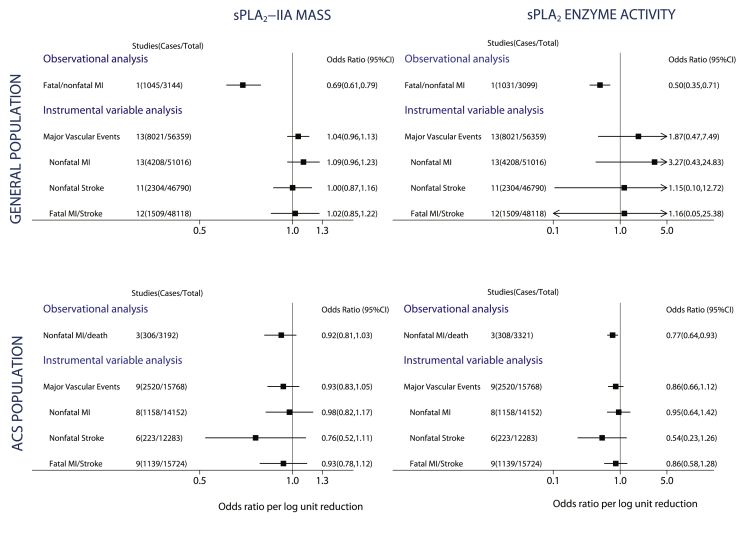
For the general population studies, instrumental variable analysis showed a null effect between sPLA2-IIA mass and incident MVE (OR per 1 log unit lower sPLA2-IIA mass: 1.04; 95% CI: 0.96 to 1.13) that was in contrast to the expected association based on observational analysis (OR: 0.69; 95% CI: 0.61 to 0.79). Similarly, for sPLA2 enzyme activity, observational studies showed an OR of 0.50 (95% CI: 0.35 to 0.71), yet null associations were obtained for the instrumental variable estimates for sPLA2 enzyme activity and incident MVE (OR: 1.87; 95% CI: 0.47 to 7.49), although the CIs were wide due to the weak effect of the rs11573156 variant on sPLA2 enzyme activity in the general population.
For the ACS studies, the instrumental variable estimate for sPLA2-IIA mass and recurrent MVE was also null (OR per 1 log unit lower sPLA2-IIA mass: 0.93; 95% CI: 0.83 to 1.05) and consistent with the observational estimate (OR: 0.92; 95% CI: 0.81 to 1.03). For sPLA2 enzyme activity, no association was identified for the instrumental variable estimate with MVE (OR: 0.86, 95% CI: 0.66 to 1.12), which was similar to the observational estimate (OR: 0.77, 95% CI: 0.64 to 0.93) (Fig. 5).
Figure 5.
Observational and Instrumental Variable Estimates Per 1 Log Unit Lower sPLA2-IIA Mass and sPLA2 Enzyme Activity With Major Vascular Events
The observational analyses were adjusted for age, sex, BMI, BP, T2D, LDL-C, HDL-C, and TG in general population cohorts (corresponding to Model 3 in Fig. 1) and for age, sex, BMI, and BP in ACS cohorts (corresponding to Model 2 in Fig. 1). Abbreviations as in Figure 1.
Discussion
We used a genetic approach to judge the causal role of sPLA2-IIA on MVE and, by extension, to evaluate if inhibition of sPLA2-IIA represents a valid therapeutic target for cardiovascular prevention. We identified a SNP in PLA2G2A (rs11573156) that had a large and specific effect on circulating sPLA2-IIA mass and a small-to-modest effect on sPLA2 enzyme activity, but found no association between rs11573156 and incident, prevalent or recurrent MVE. This study provides evidence that the observational association between sPLA2-IIA mass and MVE is likely due to residual confounding or reverse causality.
Our Mendelian randomization analysis used a single genetic instrument that had a remarkable effect on sPLA2-IIA mass, explaining between 6% and 21% of its variance, a value several times higher than that observed for all genome wide association studies hits on blood pressure (1% for 29 SNPs combined) (23) and similar to that for LDL-C (∼12% for 49 SNPs combined) (24). The strength of our genetic instrument together with the large sample size analyzed strongly support the instrumental variable estimates that indicate a null effect of sPLA2-IIA mass with cardiovascular events.
The key SNP in our study (rs11573156) had a smaller impact on sPLA2 enzyme activity than sPLA2-IIA mass, in particular for general population studies. Because sPLA2 enzyme activity is a composite of several sPLA2 isoenzymes (2), it is not surprising that a PLA2G2A SNP (specific for sPLA2-IIA) explained a smaller proportion of variance of sPLA2 enzyme activity compared with sPLA2-IIA mass.
While this manuscript was being prepared, a phase III RCT of varespladib (VISTA-16) (10) was halted for lack of efficacy (11). VISTA-16 was to enroll up to 6,500 patients with ACS and randomize them to 500 mg/day varespladib or placebo for 16 weeks with a primary combined endpoint of cardiovascular death, nonfatal MI, nonfatal stroke, or documented unstable angina. We hypothesize that the null findings from our Mendelian randomization analysis may provide an eventual explanation for the lack of efficacy of varespladib in VISTA-16.
We did not find an association between lower sPLA2-IIA mass and lower rates of recurrent MVE in ACS patients, unlike earlier reports (10). With CIs that span ORs from 0.81 to 1.03, we cannot rule out a false negative finding due to a limited number of events. Alternatively, initial studies often overestimate the effect of a biomarker on a health outcome, and as more evidence accrues, the magnitude of the effect may diminish and in some cases disappears, known as the “winner's curse.”
Study limitations
First, we did not have data from a common set of participants with all 3 key variables: sPLA2 measures, genetic information, and cardiovascular events. This is a common scenario with large-scale meta-analyses of Mendelian randomization studies that include novel biomarkers (25), but the instrumental variable approach helps overcome this problem. Second, given the impact of the SNP on sPLA2 enzyme activity was more modest than its effect on sPLA2-IIA mass, our genetic analyses do not exclude a possible causal role of other isoforms such as sPLA2-III, -V, and -X in cardiovascular disease. However, our genetic data do provide clear evidence against a causal role of sPLA2-IIA mass in incident MVE in the general population. With regard to an ACS population, our analysis includes 2,520 recurrent MVE in patients with ACS, which is 6-fold greater than the 385 primary events that VISTA-16 intended to accrue (10). Furthermore, comparing individuals homozygous for the rs11573156 C allele to those homozygous for the G allele resulted in a reduction in sPLA2-IIA mass similar to the effect of varespladib 500 mg/day and also showed no association with MVE.
Conclusions
Our large-scale Mendelian randomization analysis suggests that sPLA2-IIA is unlikely to be a valid therapeutic target for prevention of cardiovascular events. The concordance of our genetic findings with the lack of efficacy of the phase III varespladib trial supports the wider use of this type of genetic approach earlier in drug development to prioritize which drug targets to take through to RCTs.
Footnotes
The UCLEB consortium is funded by a BHG programme grant (ref RG/10/12/28456). This work was funded by the UK Medical Research Council (Population Health Scientist Fellowship G0802432: to M.V.H.), British Heart FoundationRG 10/12/28456 (A.D.H., J.P.C., S.E.H., and Meena Kumari), RG008/014 (S.E.H., A.D.H., and P.J.T.), RG/10/001/27643 (Z.M.), PG07/133/24260 (S.E.H., A.D.H., P.J.T., and Meena Kumari), and FS 08/048/25628 (P.J.T. and A.D.H.). Inserm A.T. and I.M. Arron D. Hingorani has provided non-remunerated advice on the Roche Actemra (tocilizumab) cardiovascular advisory board; Aroon D. Hingorani and Juan P. Casas are supported by the National Institute of Health Research University College London Hospitals Biomedical Research Centre. PREVEND genetics is supported by EU-LSHM-CT-2006-037697 and the Dutch Kidney Foundation (grant E033). P. van der Harst is supported by the Netherlands Organisation for Health Research and Development (NWO VENI grant 916.761.70), and the Dutch Inter University Cardiology Institute Netherlands (ICIN). GP is supported by the Canada Research Chair in Genetic and Molecular Epidemiology. The EPIC-NL study was funded by the Europe Against Cancer Programme of the European Commission (SANCO), Dutch Ministry of Public Health, Welfare and Sports (VWS), Netherlands Cancer Registry (NKR), LK Research Funds, Dutch Prevention Funds, Dutch Cancer Society; ZonMW the Netherlands Organisation for Health Research and Development, World Cancer Research Fund (WCRF) (the Netherlands). Genotyping was funded by IOP Genomics grant IGE05012 from Agentschap NL. Folkert W. Asselbergs is supported by a clinical fellowship from the Netherlands Organisation for Health Research and Development (ZonMw grant 90700342). The FAST-MI study is sponsored by the French Society of Cardiology and has been supported by a grant from La Caisse Nationale de l'Assurance Maladie (CNAM) and unrestricted grants from Pfizer and Servier. The WHII study has been supported by grants from the Medical Research Council; British Heart Foundation; Health and Safety Executive; Department of Health; National Heart, Lung, and Blood Institute (NHLBI: HL36310) and National Institute on Aging (AG13196), U.S., NIH; Agency for Health Care Policy Research (HS06516); and the John D. and Catherine T. MacArthur Foundation Research Networks on Successful Midlife Development and Socio-economic Status and Health. The ASAP study was supported by the Swedish Research Council (12660: P.E.), the Swedish Heart-Lung Foundation (20090541: P.E.), the European Commission (FAD, Health F2 2008 200647: P.E.), a donation by Fredrik Lundberg (A.F.C. and P.E.). Dr. Swerdlow is supported by a Medical Research Council Doctoral Training Award. M. Scholz was funded by LIFE–Leipzig Research Center for Civilization Diseases (LIFE Center, Universität Leipzig). LIFE–Leipzig Research Center for Civilization Diseases is funded by means of the European Union, by the European Regional Development Fund (ERFD) and by means of the Free State of Saxony within the framework of the excellence initiative. This publication is supported by LIFE–Leipzig Research Center for Civilization Diseases, Universität Leipzig). LIFE is funded by means of the European Union, by the European Regional Development Fund (ERFD) and by means of the Free State of Saxony within the framework of the excellence initiative. The British Regional Heart Study is a British Heart Foundation research group (grant number RG/08/013/25942). DNA extraction was supported in part by British Heart Foundation Senior Research Fellowship FS05/125. The British Women's Heart and Health Study is supported by funding from the British Heart Foundation (BHF) and the Department of Health Policy Research Programme (England). Human CVD genotyping of the BWHHS was funded by the BHF (PG/07/131/24254). D.A.L., T.M.P., T.R.G., and I.N.M.D. work in a center that receives funds from the UK Medical Research Council (G0600705) and University of Bristol. The EAS has been supported by grants from the British Heart Foundation. Recruitment of the CAD cases for the BHF-FHS Study was funded by the British Heart Foundation. Controls were collected as part of the Wellcome Trust Case Control Consortium Study. Genotyping was funded by the British Heart Foundation and the European Union FP6 Cardiogenics Study. Recruitment and genotyping of the GRAPHIC Study was funded by the British Heart Foundation. N.J.S. holds a Chair funded by the British Heart Foundation and is an NIHR Senior Investigator. Z.M. holds a Chair funded by the British Heart Foundation. M.D.T. is an MRC Senior Clinical Fellow. The BHF-FHS and GRAPHIC studies are part of the portfolio of research supported by the Leicester NIHR Biomedical Research Unit in Cardiovascular Disease. The IMPROVE study was supported by the European Commission (Contract number: QLG1-CT-2002-00896), the Swedish Heart-Lung Foundation, the Swedish Research Council (projects 8691 and 0593), Knut and Alice Wallenberg Foundation, the Torsten and Ragnar Söderberg Foundation, the Foundation for Strategic Research, the Stockholm County Council (project 562183), the Strategic Cardiovascular and Diabetes Programmes of Karolinska Institutet and Stockholm County Council, Academy of Finland (Grant #110413), Ministry of Education and Culture of Finland, the City of Kuopio, the British Heart Foundation (RG2008/014), and the Italian Ministry of Health (Ricerca Corrente). The BWHHS is supported by the Department of Health (England) Policy Research Programme and British Heart Foundation (PG/09/036/26739). The Cyprus study has been supported by grants from the Cyprus Research Promotion Foundation (YGEIA/1104/17 and PENEK_05/04) and the Cyprus Cardiovascular Educational and Research Trust. EPIC-Norfolk study is supported by grants from the Medical Research Council UK and Cancer Research UK. G. Kees Hovingh is supported by the Netherlands Organisation for Health Research and Development (NWO VENI grant 91612122). The Rotterdam Study is supported by the Erasmus Medical Center and Erasmus University Rotterdam; the Netherlands Organization for Scientific Research (NWO); the Netherlands Organization for Health Research and Development (ZonMw); the Research Institute for Diseases in the Elderly (RIDE); the Netherlands Heart Foundation; the Ministry of Education, Culture and Science; the Ministry of Health Welfare and Sports; the European Commission; and the Municipality of Rotterdam. Support for genotyping was provided by the Netherlands Organisation of Scientific Research NWO Investments (nr. 175.010.2005.011, 911-03-012), the Research Institute for Diseases in the Elderly (014-93-015; RIDE2), the Netherlands Genomics Initiative (NGI)/Netherlands Consortium for Healthy Aging (NCHA) project nr. 050-060-810. Jacqueline Witteman is supported by NWO grant (vici, 918-76-619). Abbas Dehghan is supported by NWO grant (veni, 916.12.154) and the EUR Fellowship. The research of the PROSPER study has been funded by the European Union's Seventh Framework Programme (FP7/2007-2013) under grant agreement n° HEALTH-F2-2009-223004. GRACE-Scotland was supported by an unrestricted educational grant from sanofi-aventis,. KAROLA: The study was supported in parts by the Association of German Pension Fund Agencies, the German Federal Ministry of Education and Research (#01GD9820/0), and the Pitzer Foundation (Bad Nauheim, Germany). UDACS was supported by a clinical training fellowship (JWS:BDA:RD 01 / 0001357) from Diabetes UK. GENDEMIP: supported by the project (Ministry of Health, Czech Republic) for development of research organization 00023001 (IKEM, Prague, Czech Republic)–Institutional support. UCP: The project was funded by Veni grant Organization for Scientific Research (NWO), Grant no. 2001.064Netherlands Heart Foundation (NHS), and TI Pharma Grant T6-101 Mondriaan.
S Yusuf holds the Heart and Stroke Foundation/ Marion W Burke Chair in cardiovascular diseases. PROCARDIS was supported by the British Heart Foundation; by the European Community Sixth Framework Programme (grant number LSHM-CT-2007-037273); and by AstraZeneca. A.B., M.F., and H.W. are supported by the British Heart Foundation Centre for Research Excellence in Oxford and the Wellcome Trust core award (090532/Z/09/Z). Anders Hamsten obtained support for this project from the Swedish Heart-Lung Foundation; from the Swedish Medical Research Council (grant number 8691); from the Knut and Alice Wallenberg Foundation; from the Karolinska Institute; and from the Stockholm County Council (grant number 560183). Dr. Schwartz, through his institution, has received research grants from Anthera, Resverlogix, Roche, and sanofi-aventis. The MIRACL study was supported by Pfizer. CCHS: Supported by The Danish Medical Research Council and The Research Fund at Rigshospitalet, Copenhagen University Hospital. The Whitehall II study is supported by the UK Medical Research Council (K013351), the British Heart Foundation, the ESRC, the U.S. NIH (R01 HL036310; R01 AG034454) Health and Safety Executive; the Department of Health; the National Heart, Lung, and Blood Institute (NHLBI; HL36310); the National Institute on Aging (AG13196); the Agency for Health Care Policy Research (HS06516); and the John D. and Catherine T. MacArthur Foundation Research Networks on Successful Midlife Development and Socio-economic Status and Health.
Ziad Mallat, Tabassome Simon, and Alain Tedgui are listed as coinventors on patents related to the use of sPLA2 activity and/or PLA2G2A rs11573156 SNP as a cardiovascular biomarker. Wolfgang Koenig has received a research grant from Anthera to measure sPLA2 in KAROLA, was the National Coordinator for the VISTA-16 clinical trial in Germany and has acted as consultant for Roche, BioInvent, Cerenis, Novartis, and Anthera. Gregory Schwartz, through his institution, has received research grants from Anthera, Resverlogix, Roche, and sanofi-aventis and was involved in the VISTA-16 clinical trial. Alain Tedgui has acted as a consultant for Aterovax. Damiano Baldassarre has acted as a consultant for Centro Cardiologico Monzino, IRCCS. J. W. Jukema has received research grants from and was speaker on (CME accredited) meetings sponsored by Astellas, Anthera, AstraZeneca, Biotronik, Boston Scientific, Daiichi Sankyo, Eli Lilly, Genzyme, Medtronic, Merck-Schering-Plough, Pfizer, Orbus Neich, Novartis, Roche, Servier, sanofi-aventis, the Netherlands Heart Foundation, the Interuniversity Cardiology Institute of the Netherlands, and the European Community Framework KP7 Programme. Martin Tobin has received funding from a Pfizer collaborative research project. Kathryn Carruthers has received an educational grant from sanofi-aventis. Ziad Mallat is a board member of AstraZeneca and acts as consultant for Aterovax. Tabassome Simon is a board member of Bayer; acts as consultant for Bayer, Eli Lilly, AstraZeneca, and sanofi-aventis; has received grants from Merck, AstraZeneca, sanofi-aventis, Eli Lilly, Daiichi Sankyo, Novartis, and GlaxoSmithKline; has received payment for lectures from sanofi-aventis and Eli Lilly. Sam Tsimikas has acted as consultant for Isis, Genzyme, and Quest; receives a grant from Pfizer; and has patents with and/receives royalties from UCSD. J.L.M. has received research grant support from AstraZeneca, Bayer, Bristol Myers Squibb, Daiichi Sankyo, Eli Lilly, Janssen, Sanofi-aventis and honoraria for consulting from: Boehringer Ingelheim, Janssen. M.S.S. received research grant support through Brigham and Women's Hospital from: Abbott Laboratories; Amgen; AstraZeneca; AstraZeneca/Bristol-Myers Squibb Alliance; Bristol-Myers Squibb/Sanofi-aventis Joint Venture; Critical Diagnostics; Daiichi-Sankyo; Eisai; Genzyme; GlaxoSmithKline; Intarcia; Merck; Nanosphere; Roche Diagnostics; Sanofi-aventis; Takeda. MSS received honoraria for consulting from: Aegerion; Amgen; AstraZeneca/Bristol-Myers Squibb Alliance; GlaxoSmithKline; Intarcia; Merck; Pfizer; Sanofi-aventis; Vertex. PROVE-IT TIMI 22 was by Bristol-Myers Squibb & Sankyo. MERLIN-TIMI 36 was supported by CV Therapeutics. All other authors have reported that they have no relationships relevant to the contents of this paper to disclose. Drs. Holmes, Simon, and Exeter are joint first authors. Drs. Hingorani, Sabatine, Mallat, Casas, and Talmud are joint last authors. Stephen Nicholls, MBBS, PhD, served as Guest Editor for this paper.
Appendix
For an expanded methods and results sections and supplemental figures and tables, please see the online version of this article.
Contributor Information
Michael V. Holmes, Email: mvholmes@gmail.com.
Juan P. Casas, Email: Juan.Pablo-Casas@lshtm.ac.uk.
Appendix
References
- 1.Rosenson R.S., Hurt-Camejo E. Phospholipase A2 enzymes and the risk of atherosclerosis. Eur Heart J. 2012;33:2899–2909. doi: 10.1093/eurheartj/ehs148. [DOI] [PubMed] [Google Scholar]
- 2.Mallat Z., Lambeau G., Tedgui A. Lipoprotein-associated and secreted phospholipases A in cardiovascular disease: roles as biological effectors and biomarkers. Circulation. 2010;122:2183–2200. doi: 10.1161/CIRCULATIONAHA.110.936393. [DOI] [PubMed] [Google Scholar]
- 3.Mallat Z., Steg P.G., Benessiano J. Circulating secretory phospholipase A2 activity predicts recurrent events in patients with severe acute coronary syndromes. J Am Coll Cardiol. 2005;46:1249–1257. doi: 10.1016/j.jacc.2005.06.056. [DOI] [PubMed] [Google Scholar]
- 4.Boekholdt S.M., Keller T.T., Wareham N.J. Serum levels of type II secretory phospholipase A2 and the risk of future coronary artery disease in apparently healthy men and women: the EPIC-Norfolk Prospective Population Study. Arterioscler Thromb Vasc Biol. 2005;25:839–846. doi: 10.1161/01.ATV.0000157933.19424.b7. [DOI] [PubMed] [Google Scholar]
- 5.Mallat Z., Benessiano J., Simon T. Circulating secretory phospholipase A2 activity and risk of incident coronary events in healthy men and women: the EPIC-Norfolk study. Arterioscler Thromb Vasc Biol. 2007;27:1177–1183. doi: 10.1161/ATVBAHA.107.139352. [DOI] [PubMed] [Google Scholar]
- 6.O'Donoghue M.L., Mallat Z., Morrow D.A. Prognostic utility of secretory phospholipase A(2) in patients with stable coronary artery disease. Clin Chem. 2011;57:1311–1317. doi: 10.1373/clinchem.2011.166520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ryu S.K., Mallat Z., Benessiano J. Phospholipase A2 enzymes, high-dose atorvastatin, and prediction of ischemic events after acute coronary syndromes. Circulation. 2012;125:757–766. doi: 10.1161/CIRCULATIONAHA.111.063487. [DOI] [PubMed] [Google Scholar]
- 8.Tietge U.J., Pratico D., Ding T. Macrophage-specific expression of group IIA sPLA2 results in accelerated atherogenesis by increasing oxidative stress. J Lipid Res. 2005;46:1604–1614. doi: 10.1194/jlr.M400469-JLR200. [DOI] [PubMed] [Google Scholar]
- 9.Dzavik V., Lavi S., Thorpe K. The sPLA2 Inhibition to Decrease Enzyme Release after Percutaneous Coronary Intervention (SPIDER-PCI) trial. Circulation. 2010;122:2411–2418. doi: 10.1161/CIRCULATIONAHA.110.950733. [DOI] [PubMed] [Google Scholar]
- 10.Nicholls S.J., Cavender M.A., Kastelein J.J. Inhibition of secretory phospholipase A(2) in patients with acute coronary syndromes: rationale and design of the Vascular Inflammation Suppression to Treat Acute Coronary Syndrome for 16 Weeks (VISTA-16) Trial. Cardiovasc Drugs Ther. 2012;26:71–75. doi: 10.1007/s10557-011-6358-9. [DOI] [PubMed] [Google Scholar]
- 11.Clinical Study Report for Study AN-CVD2233. Evaluation of the Safety and Efficacy of Short-Term A-002 Treatment in Subjects with Acute Coronary Syndromes. 2012. Available at: http://www.anthera.com/VISTA-16.pdf. Accessed September 2013.
- 12.Snyder D.W., Bach N.J., Dillard R.D. Pharmacology of LY315920/S-5920, [[3-(aminooxoacetyl)-2-ethyl-1- (phenylmethyl)-1H-indol-4-yl]oxy] acetate, a potent and selective secretory phospholipase A2 inhibitor: a new class of anti-inflammatory drugs, SPI. J Pharmacol Exp Ther. 1999;288:1117–1124. [PubMed] [Google Scholar]
- 13.Fraser H., Hislop C., Christie R.M. Varespladib (A-002), a secretory phospholipase A2 inhibitor, reduces atherosclerosis and aneurysm formation in ApoE-/- mice. J Cardiovasc Pharmacol. 2009;53:60–65. doi: 10.1097/FJC.0b013e318195bfbc. [DOI] [PubMed] [Google Scholar]
- 14.Innocenti F., Cooper G.M., Stanaway I.B. Identification, replication, and functional fine-mapping of expression quantitative trait loci in primary human liver tissue. PLoS Genet. 2011;7:e1002078. doi: 10.1371/journal.pgen.1002078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wootton P.T., Drenos F., Cooper J.A. Tagging-SNP haplotype analysis of the secretory PLA2IIa gene PLA2G2A shows strong association with serum levels of sPLA2IIa: results from the UDACS study. Hum Mol Genet. 2006;15:355–361. doi: 10.1093/hmg/ddi453. [DOI] [PubMed] [Google Scholar]
- 16.Lawlor D.A., Harbord R.M., Sterne J.A., Timpson N. Davey Smith G. Mendelian randomization: using genes as instruments for making causal inferences in epidemiology. Stat Med. 2008;27:1133–1163. doi: 10.1002/sim.3034. [DOI] [PubMed] [Google Scholar]
- 17.Thompson J.R., Minelli C., Abrams K.R., Tobin M.D., Riley R.D. Meta-analysis of genetic studies using Mendelian randomization—a multivariate approach. Stat Med. 2005;24:2241–2254. doi: 10.1002/sim.2100. [DOI] [PubMed] [Google Scholar]
- 18.Thomas D.C., Lawlor D.A., Thompson J.R. Re: estimation of bias in nongenetic observational studies using “Mendelian triangulation” by Bautista et al. Ann Epidemiol. 2007;17:511–513. doi: 10.1016/j.annepidem.2006.12.005. [DOI] [PubMed] [Google Scholar]
- 19.Moher D., Liberati A., Tetzlaff J., Altman D.G. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med. 2009;6:e1000097. doi: 10.1371/journal.pmed.1000097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rosenson R.S., Hislop C., Elliott M., Stasiv Y., Goulder M., Waters D. Effects of varespladib methyl on biomarkers and major cardiovascular events in acute coronary syndrome patients. J Am Coll Cardiol. 2010;56:1079–1088. doi: 10.1016/j.jacc.2010.06.015. [DOI] [PubMed] [Google Scholar]
- 21.Rosenson R.S., Hislop C., McConnell D. Effects of 1-H-indole-3-glyoxamide (A-002) on concentration of secretory phospholipase A2 (PLASMA study): a phase II double-blind, randomised, placebo-controlled trial. Lancet. 2009;373:649–658. doi: 10.1016/S0140-6736(09)60403-7. [DOI] [PubMed] [Google Scholar]
- 22.Rosenson R.S., Elliott M., Stasiv Y., Hislop C. Randomized trial of an inhibitor of secretory phospholipase A2 on atherogenic lipoprotein subclasses in statin-treated patients with coronary heart disease. Eur Heart J. 2011;32:999–1005. doi: 10.1093/eurheartj/ehq374. [DOI] [PubMed] [Google Scholar]
- 23.Padmanabhan S., Newton-Cheh C., Dominiczak A.F. Genetic basis of blood pressure and hypertension. Trends Genet. 2012;28:397–408. doi: 10.1016/j.tig.2012.04.001. [DOI] [PubMed] [Google Scholar]
- 24.Teslovich T.M., Musunuru K., Smith A.V. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466:707–713. doi: 10.1038/nature09270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Minelli C., Thompson J.R., Tobin M.D., Abrams K.R. An integrated approach to the meta-analysis of genetic association studies using Mendelian randomization. Am J Epidemiol. 2004;160:445–452. doi: 10.1093/aje/kwh228. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.