Abstract
The entry of T cell progenitors to the thymus marks the beginning of a multistage developmental process that culminates in the generation of self-MHC-restricted CD4+ and CD8+ T cells. Although multiple factors including the chemokine receptors CCR7 and CCR9 are now defined as important mediators of progenitor recruitment and colonization in both the fetal and adult thymi, the heterogeneity of thymus-colonizing cells that contribute to development of the T cell pool is complex and poorly understood. In this study, in conjunction with lineage potential assays, we perform phenotypic and genetic analyses on thymus-settling progenitors (TSP) isolated from the embryonic mouse thymus anlagen and surrounding perithymic mesenchyme, including simultaneous gene expression analysis of 14 hemopoietic regulators using single-cell multiplex RT-PCR. We show that, despite the known importance of CCL25-CCR9 mediated thymic recruitment of T cell progenitors, embryonic PIR+c-Kit+ TSP can be subdivided into CCR9+ and CCR9− subsets that differ in their requirements for a functional thymic microenvironment for thymus homing. Despite these differences, lineage potential studies of purified CCR9+ and CCR9− TSP reveal a common bias toward T cell-committed progenitors, and clonal gene expression analysis reveals a genetic consensus that is evident between and within single CCR9+ and CCR9− TSP. Collectively, our data suggest that although the earliest T cell progenitors may display heterogeneity with regard to their requirements for thymus colonization, they represent a developmentally homogeneous progenitor pool that ensures the efficient generation of the first cohorts of T cells during thymus development.
A fundamental requirement for T cell development is the recruitment of progenitors produced in the hemopoietic tissues into the thymic microenvironment. This allows interactions that results in the selective maturation of CD4+ and CD8+ T cells that are tolerant to self-Ags yet are capable of participating in immune responses to foreign Ags. Many of the developmental checkpoints that occur during the intrathymic generation of mature T cells from immature progenitors are controlled by stromal cell components that collectively constitute the thymic microenvironment (1). Thus, molecular interactions involving direct cell–cell interactions (e.g., Notch-Notch ligand and abTCR-peptide/MHC), as well as soluble chemokines (e.g., CCR7-CCL19/CCL21 and CCR9-CCL25) and cytokines (e.g., IL-7 and c-Kit) acting between developing thymocytes and thymic epithelial and mesenchymal cells (2, 3), guide progenitors through subcapsular, cortical, and medullary thymic microenvironments and ensure an ordered program of differentiation (4). Importantly, because the thymus does not contain cells with long-term self-renewing potential (5), intrathymic T cell production throughout life requires the continued recruitment of T cell progenitors from sites of hemopoietic cell production such as yolk sac, fetal liver, and bone marrow (6, 7). Several studies have shown that multiple types of hemopoietic progenitors can develop into T lineage progeny (8, 9), including lymphoid-primed multipotent progenitors (MPP) (10), CCR9+ MPP (11), common lymphoid progenitors (CLP) (12) and BB20+ CLP (13) (CLP-2), and circulating T cell progenitors (14-17). Moreover, several key mediators of thymus colonization have been identified including the homeostatic chemokine receptors CCR7 and CCR9 (11, 12, 18-25), which bind CCL19/CCL21 and CCL25, respectively, the CDM family members DOCK2 and DOCK180 (26), the selectin/ligand pair of P-selectin/P-selectin glycoprotein ligand 1 (27, 28), as well as polysialic acid, a product of the polysialyltransferase ST8Sia IV (29). However, despite these advances and an increasingly refined definition of T cell progenitors at the molecular level (30, 31), the thymus-settling progenitors (TSP) (19) that colonize the thymus and give rise to early thymic progenitors as a first step along the T cell development pathway remain poorly defined. In addition, several studies analyzing the developmental potential of TSP, in particular in relation to T cell and myeloid lineage potential, have generated conflicting data (32-34, reviewed in Ref. 35).
In this study, we isolate progenitors from the embryonic stage 12 (E12) fetal thymus anlagen to investigate cellular and molecular heterogeneity in TSP. First, we show that although a dominant population of PIR+c-Kit+ progenitors is separable into two discrete subsets on the basis of expression of CCR9, limit dilution analysis of their developmental potential reveals that both CCR9+ and CCR9− subsets represent T cell progenitors with no B cell potential and residual myeloid potential. In addition to their differential expression of CCR9, we show that CCR9+, but not CCR9−, TSP are efficiently recruited to the nude thymus anlage, suggesting that thymic recruitment of CCR9− TSP is strictly dependent on the presence of FoxN1+ thymic epithelial cells. Finally, single-cell analysis of a panel of 14 genetic regulators of hemopoietic cell differentiation reveals striking homogeneity between CCR9+ and CCR9− TSP subsets in terms of their genetic profile, which also reveals a genetic consensus at the single-cell level. Collectively, although chemokine receptor heterogeneity in TSP and a differential requirement for FoxN1-expressing thymic epithelium are findings suggestive of multiple mechanisms of thymus colonization, our study identifies a common molecular profile of the first thymus-colonizing T cell progenitors that collectively give rise to the initial cohorts of intrathymically produced T cells.
Materials and Methods
Mice
For the isolation of wild-type (WT) and nude tissues, the following strains were used as indicated: BALB/c (H-2d), C57BL/6 (H-2b), and C57BL/6 Nude (H-2b). Both Ccr9−/− and Ccl25−/− mice (both H-2b) were obtained under exclusive use agreements from private collections within the European Mutant Mouse Archive through the Wellcome Trust mutant mouse resource. For the generation of timed embryos, the day of vaginal plug detection was designated day 0. All mice were bred and maintained at the Biomedical Services Unit at the University of Birmingham (Birmingham, U.K.).
Isolation of TSP
Freshly dissected E12 thymus lobes from the indicated strains were disaggregated by incubation in 10% FCS RPMI 1640 solution containing 3.2 mg/ml collagenase D (Roche) for 5–10 min at 37°C. To isolate progenitors from perithymic mesenchyme (36), whole E12 thymus lobes were incubated at room tempereature for 5 min in 2.4 mg/ml collagenase D in 10% FCS RPMI 1640 solution, placed under a dissecting microscope, and aspirated gently using a glass pipette to separate perithymic mesenchyme from the inner epithelial rudiment. Perithymic mesenchyme and intact epithelial rudiments were transferred into a fresh solution of 3.2 mg/ml collagenase D at 37°C, and enzymatic treatment continued until a single-cell suspension was produced. To obtain hematopoietic cells from fetal livers, organs from E12 C57BL/6 and C57BL/6 Nude embryos were disaggregated mechanically by gentle pipetting in 10% FCS RPMI 1640 solution. Blood was isolated from the umbilical cord, dorsal aorta, and heart of WT and nude C57/BL6 embryos at E11 of gestation. At E12, blood was obtained by mechanical disruption of hearts.
Abs and flow cytometry
The following Abs were purchased from eBioscience: anti–CCR9-PE or biotin (eBioCW-1.2), anti–F4/80-FITC or allophycocyanin (clone BM8), anti–CD45.2-FITC (clone 104), anti–CD45-PEcy7 or -PE or -allophycocyanin (clone 30-F11), anti–CD4-PEcy7 (clone GK1.5), anti–CD8α-allophycocyanin (clone 53-6.7), anti–TcRβ-PE (clone H57-597), anti–CD11b-biotin (clone M1/70), anti–c-Kit-allophycocyanin (clone 2B8), and streptavidin-PEcy7. The anti-paired IgR (PIR)-A/B-PE (clone 6C1; BD Pharmingen) and anti–CCR9-PE (clone 242503; R&D Systems) were also used. For FACS analysis, cell suspensions were stained in PBS containing 10% FCS, 10% mouse serum, and 10% rat serum solution, washed, and resuspended in PBS containing 250 μg/ml propidium iodide (Sigma-Aldrich) or 250 μg/ml DAPI (Invitrogen). Four-color flow cytometry analysis was performed by using CellQuest software and a duallaser LSR machine (BD Biosciences, San Jose, CA) or by using FACS-Diva software and a three-laser Fortessa LSR machine (BD Biosciences) with forward/side scatter gates set on propidium iodide (PI)-negative events or DAPI-negative events to study viable lymphocytes. Data were analyzed with FlowJo software (Tree Star). FACS sorting was performed on a MoFlo high-speed cell sorter (Beckman Coulter) using Summit software.
Limiting dilution assays
E12 thymic lobes were isolated from BALB/c embryos by microdissection, disaggregated, and then stained with PI, anti-CD45.2, anti-F4/80, anti-PIR, and anti-CCR9 (clone eBioCW-1.2). CD45+F4/80−CCR9− cells and CD45+F4/80−CCR9+ cells were MoFlo sorted and collected in an Eppendorf tube containing 400 μl culture medium (MEM-α, 10% FCS, 50 IU/ml penicillin and streptomycin, and 5 × 10−5 M 2-ME). The sorted samples were serially diluted according to the number of cells obtained. At least three different cell concentrations were plated per assay, with 48–96 wells plated for each cell concentration. The sorted cells were seeded on OP9-DL1 cell and OP9 cell monolayers (37) in the presence of IL-7, Flt3 ligand (Flt3-L), and c-Kit ligand (c-Kit–L) cytokines to assess the T cell and B cell potential, respectively. Myeloid potential was assessed by seeding sorted cells onto OP9 cell monolayers in the presence of M-CSF, GM-CSF, IL-3, Flt3-L, and c-Kit–L. Cultures were fed with fresh cytokines after 6 d. All cytokines were used at the final concentration of 10 ng/ml. IL-7, Flt3-L, M-CSF, GM-CSF, and IL-3 were purchased from R&D Systems, and c-Kit–L was purchased from AbD Serotec. Lymphoid and myeloid potential was assessed by flow cytometry after 14 and 10 d, respectively. According to the Poisson statistic law, the potential frequency is determined when 37% of the wells are negative.
Multiplex PCR
Cell suspensions of perithymic mesenchyme were stained with anti-CD45 and anti-CCR9 (clone 242503; R&D Systems) in PBS 3% FCS solution. CD45+CCR9− and CD45+CCR9+ cells were clonally sorted into a 0.2-ml thermo-strip (Thermo Scientific) containing 5 μl Nuclease-free water (Promega) and processed for multiplex RT-PCR, as described previously (38, 39). In brief, cells were lysed by freezing at −80°C, followed by heating to 65°C for 2 min. After cooling at 4°C, RNA was specifically reverse transcribed at 37°C for 60 min with inactivation of reverse transcriptase at 95°C for 3 min. cDNA was amplified by a seminested PCR. The first round of PCR consisted of a denaturation step at 95°C for 10 min, 15 amplification cycles (45 s at 94°C, 60s at 60°C, and 90s at 72°C), and a final step at 72°C for 10 min. This simultaneous amplification of all cDNAs was followed by a second round of specific PCRs (i.e., the first-round PCR products were separated and amplified with specific primers). The second round of PCR consisted of a denaturation step at 95°C for 10 min and then 48 amplification cycles (30 s at 94°C, 45 s at 70°C, and 60 s at 72°C, with the hybridization temperature decreased from 70 to 60°C every other cycle). The primer sequences have been reported previously (38). PCR products were detected on a 2.0% agarose GelRed (Biotium) gel.
Real-time PCR for expression of Dock2 and ST8siaIV
CCR9− and CCR9+ TSP were isolated from E12 fetal blood, and gene expression analysis was performed as described previously (40). Briefly, high-purity cDNA was obtained from purified mRNA by using μMacs One-step cDNA synthesis kit, according to the manufacturer’s instructions (Miltenyi Biotec). Real-time PCR was performed using SYBR Green with primers specific for the target genes. PCR were conducted in triplicate in 15-μl volumes in a reaction buffer containing 1 μl SensiMix qPCR SYBR Green Mix (Quantace) and 200 nM primers. After an initial denaturation step (95°C for 10 min), cycling was performed at 95°C for 15 s, 60–62°C (depending on primer set) for 20 s, and 72°C for 15 s (40 cycles). Specific amplification was verified by melt curve analysis. Reaction amplification efficiency and the Ct values were obtained from Rotor Gene 6.0 software (Corbett Research) by using standard curves generated from Mouse Universal cDNA(oligodT primed)Reference (Biochain). Calculation of the relative expression values for each sample normalized to β-actin was performed as described previously (25). Primer sequences, the National Center for Biotechnology Information Reference sequence, and amplicon sizes are as follows: ST8sia4 (NM_009183.2) forward, 5′-ACGCAACTCATCGGAGATGGTGA-3′, and reverse, 5′-GTGTCCGGCGTCTGTCCAGC-3′ (253 bp); and Dock2 (NM_033374.2) forward 5′-CCTTCCAGACCAGCTCAGAC-3′, and reverse, 5′-CGTCTCATGTCCCCGTACTT-3′ (319 bp); β-actin primers (149 bp) were predesigned and synthesized by Qiagen (QT00095242).
Analysis of apoptosis in CCR9− and CCR9+ TSP
First, freshly mechanically disaggregated E12 C57BL/6 fetal livers were FACS stained at 4°C by using Abs to F4/80, PIR, CCR9, and CD45 as described above. Cells were washed in a solution of 3% FCS in PBS and subsequently washed in an Annexin V binding buffer solution composed of 10 mM HEPES, 140 mM NaCl, and 2.5 mM CaCl2. Cells were resuspended in 100 μl of the Annexin V binding buffer solution containing Annexin V–FITC (Invitrogen). Before FACS analysis, 120 μl Annexin V binding buffer solution was added to the cell suspension.
Results
CCR9 expression defines heterogeneity within the T cell progenitor fraction of thymus-settling cells
Several lines of evidence show that chemokine–chemokine receptor interactions, including CCL25-CCR9, play a role in the recruitment of T cell progenitors to the thymus (11, 12, 18-24). However, although some of these subsets are detectable within the blood, it is unclear whether some or all of these cells represent the TSP that are recruited to the thymus under physiological conditions (8, 19). To provide a clearer definition of the developmental potential and molecular profile of TSP, we analyzed the hemopoietic progenitors recruited to the thymus anlage at E12 of development. As previously shown (36), the ability to isolate these cells from the vicinity of the early thymic rudiment allows direct analysis of the first cohort of TSP prior to their sustained contact with thymic epithelium and avoids possible confusion caused by the presence of mature lineage-restricted cells within the thymus or thymic blood volume.
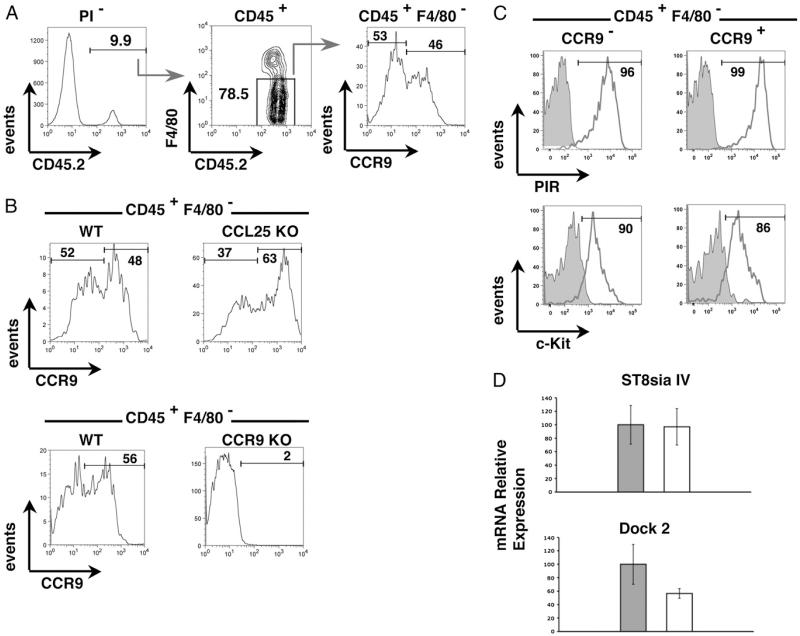
Intact thymus anlagen were dissected from E12 mouse embryos and enzymatically digested in isolation to form a single-cell suspension. Such preparations contain a small but detectable population of CD45+ hemopoietic cells that represent the first thymus-settling T cell progenitors (36, 41). Interestingly, when we performed flow cytometric analysis of CCR9 expression in conjunction with F4/80, enabling exclusion of myeloid-restricted cells colonizing the thymic anlage (42), we found that CD45+F4/80− TSP consisted of distinct CCR9+ and CCR9− subsets of approximately equal proportions (Fig. 1A). Importantly, the level of staining observed on CCR9− TSP using anti-CCR9 (eBioCW-1.2) is comparable to that seen in E12 TSP isolated from Ccr9−/− mice (Fig. 1B), providing a stringent control for the negative staining of these cells. Moreover, the lack of detectable cell surface CCR9 expression in some TSP is not due to ligand-mediated downregulation of CCR9 expression, because CCR9− and CCR9+ subsets are also detected in TSP isolated from Ccl25−/− thymus anlagen (Fig. 1B). Interestingly, however, when we next analyzed additional T cell progenitor markers, both CCR9+ and CCR9− TSP subsets were uniformly positive for c-Kit (CD117) and expressed high levels of PIR, a marker (43) previously shown to be specific to T/NK/dendritic cell progenitors (Fig. 1C). Interestingly, although heterogeneity in chemokine receptor expression is observed in TSP, we found that both CCR9− and CCR9+ TSP expressed Dock2 (Fig. 1D), a CDM family molecule linked to chemokine-mediated cytoskeletal reorganization. Given that Dock2 has been linked to fetal thymus colonization (26), this finding may suggest that additional undefined chemokine receptors are involved in the recruitment of CCR9− TSP. Moreover, we also found that the polysialyltransferase St8siaIV, shown to play a role in cell adhesion and migration in the nervous system and immune system (44, 45), as well as being linked to thymus colonization (29), is also expressed by CCR9− and CCR9+ TSP (Fig. 1D), suggesting that chemokine-independent mechanisms may also influence recruitment of TSP.
FIGURE 1. CCR9 expression defines heterogeneity in CD45+F4/80−c-Kit+PIRhi TSP recruited to the embryonic thymus anlage.
CD45+ haemopoietic cells from freshly digested E12 WT thymic anlagen were analyzed for expression of F4/80 and CCR9 (A). B shows an analysis of CCR9 expression by CD45+F4/80− progenitors from WT and either Ccl25−/− (upper panels) or Ccr9−/− (lower panels) E12 thymus. C shows expression of PIR (upper panels) and c-Kit (lower panels) in CCR9− and CCR9+ subsets of CD45+F4/80− cells. D shows quantitative PCR analysis of purified CCR9− (gray bars) and CCR9+ (open bars) TSP, isolated from E12 blood, for expression of ST8siaIV and Dock2 mRNA. Expression of the housekeeping gene β-actin was used as a reference.
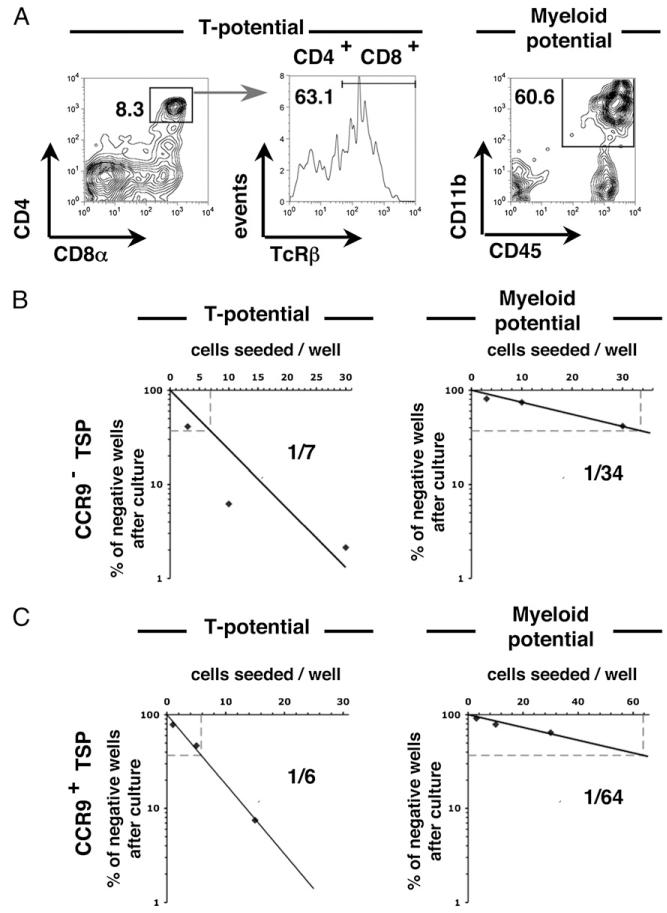
We next performed limit dilution analysis (Fig. 2A) to determine the T cell and myeloid lineage potential of CCR9+ and CCR9− TSP subsets. A strong and comparable T cell precursor frequency (one of six for CCR9+ and one of seven for CCR9−) was observed, whereas only residual myeloid potential was detected for both populations (Fig. 2B, 2C). Moreover, limit dilution analysis of CCR9− and CCR9+ TSP produced comparable numbers of T cell progeny (data not shown), indicating a similar efficacy of T cell development as well as T cell potential. In agreement with other reports (36), B lineage potential was not detected in E12 TSP preparations (data not shown). Interestingly, however, other studies have demonstrated the B cell potential of thymic progenitors and shown this to be an age-dependent process. For example, Ceredig et al. (46) demonstrated the B lineage potential of thymocytes at E14 of gestation. Whether the absence of B lineage potential shown in this study in E12 TSP is linked to gestational age, which may further highlight age-related differences in thymus colonizing cells, is currently unclear. Collectively, our data show that during the first wave of thymus colonization, the dominant F4/80− TSP fraction contains discrete subsets that can be distinguished by their differential expression of CCR9 yet are inseparable on the basis of their T/myeloid potential and lack of B cell potential.
FIGURE 2. CCR9+ and CCR9− TSP both display a strong and comparable T cell precursor frequency.
The hematopoietic potential of FACS-sorted E12 thymic CD45+F4/80−CCR9− and CD45+F4/80−CCR9+ cells was assessed in vitro. A, Flow cytometric analysis shows the T cell progeny (CD4+CD8α+TcRβ+ cells) and the myeloid cell progeny (CD45highCD11b+ cells) obtained after culture on OP9-DL1 and OP9, respectively. The T cell (left panels) and myeloid (right panels) frequencies are shown for CD45+F4/80−CCR9− (B) and CD45+F4/80−CCR9+ (C) cells.
Differential requirement for FoxN1+ thymic epithelial cells in the recruitment of CCR9+ and CCR9− TSP to the E12 thymic anlagen
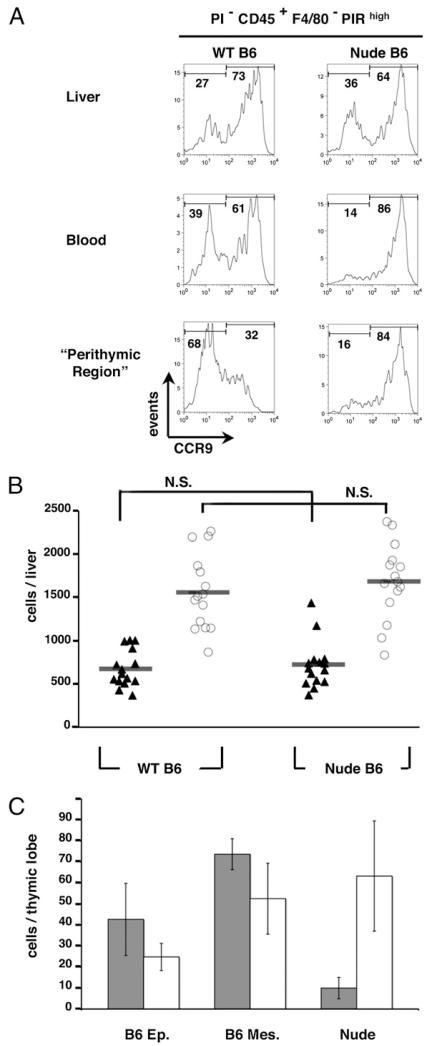
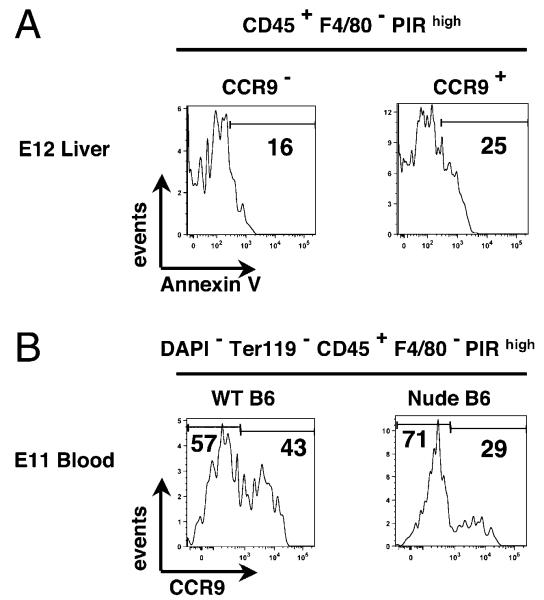
The identification of CCR9+ and CCR9− TSP subsets with comparable developmental potential at E12 suggests that, although the majority of T cell progenitors recruited to the early thymus havethe same functional potential, they display heterogeneity with regard to their mechanisms of thymus recruitment and colonization. To investigate this further, we next analyzed CCR9 expression within the CD45+F4/80−PIRhigh subset in extrathymic sites and in nude mouse embryos in which intact thymic epithelial microenvironments fail to form because of the absence of FoxN1 gene expression (47, 48). In line with the notion that CCR9 defines heterogeneity within T cell progenitors that are already evident by the time they reach the thymus, CCR9+ and CCR9− subsets of the CD45+F4/80−PIRhigh population were readily detectable in the E12 fetal liver of WT embryos (Fig. 3A, 3B). Interestingly, although a similar distribution of CCR9+ and CCR9− subsets within CD45+F4/80−PIRhigh cells was observed in both E12 WT and nude fetal liver preparations (Fig. 3A, 3B), differences were observed in peripheral blood obtained from WT and nude embryos. Thus, blood from WT and nude embryos both contained CD45+ F4/80−PIRhigh cells, the latter finding being in agreement with an earlier study identifying T lineage-restricted progenitors in the blood of nude mouse embryos (14). Interestingly, however, in contrast to WT embryos, which contained readily detectable CCR9+ and CCR9− subsets, CD45+F4/80-PIRhigh blood-borne progenitors in nude embryos were predominantly (83.2 ± 4.2%) CCR9+, with few CCR9−CD45+F4/80-PIRhigh cells detected (Fig. 3A). We next analyzed hemopoietic progenitors recruited to the WT and nude thymus anlagen by comparing cells within physically separated perithymic mesenchyme of WT thymus with those obtained from the nude thymic anlagen, where cells are recruited to the perithymic mesenchyme but fail to enter the FoxN1-deficient epithelial microenivironment (49). At E12 of gestation, unlike the numbers of CCR9+CD45+F4/80−PIRhigh cells, which were comparable in WT perithymic mesenchyme and nude thymus preparations (Fig. 3A, 3C), the number of CCR9−CD45+F4/80−PIRhigh cells recruited to the E12 nude thymus anlage was reduced dramatically (Fig. 3A, 3C). Importantly, when we next analyzed the apoptotic levels of the two CCR9− and CCR9+ TSP subsets, no major differences were observed (Fig. 4A), suggesting that the depletion of CCR9− TSP is not simply due to more stringent survival requirements of CCR9− TSP as compared with CCR9+ TSP.
FIGURE 3. Recruitment of CCR9− TSP requires signals from FoxN1+ thymic epithelial cells.
A shows flow cytometric analysis of CCR9 expression by CD45+F4/80−PIRhigh progenitors in fetal liver, blood, and perithymic mesenchyme from WT and nude E12 embryos. B, The number of PIRhiCCR9− (filled triangles) and PIRhiCCR9+ cells (open circles) in fetal liver preparations of WT and nude embryos was calculated, with each shape representing a single embryo. C shows quantitation of PIRhiCCR9− (gray bars) and PIRhiCCR9+ (open bars) progenitors recovered from WT thymic epithelial rudiments, WT perithymic mesenchyme, or nude thymic anlagen. N.S., nonsignificant according to the one tail t test for a degree of freedom = 50 and α = 0.0005.
FIGURE 4. Both CCR9− and CCR9+ TSP are detectable in the circulation of E11 nude mouse embryos.
A shows flow cytometric analysis of Annexin V staining in CD45+F4/80−PIRhigh CCR9− and CCR9+ cells from freshly isolated E12 WT C57BL/6 fetal liver. The number on the histogram indicates the percentage of cells positive for Annexin V staining. B shows flow cytometric analysis of CCR9 expression by CD45+F4/80− PIRhigh progenitors in the peripheral blood isolated from WT and nude E11 embryos.
To further investigate the dramatic reduction in CCR9− TSP in the circulation of nude mouse embryos, we analyzed the TSP compartment in the blood at an earlier stage, E11 of gestation. Interestingly, and in contrast to our findings at E12 of gestation (Fig. 3A), both CCR9− and CCR9+ TSP subsets were readilydetectable in both E11 nude and WT mouse blood (Fig. 4B). Collectively, these findings indicate that both CCR9− and CCR9+ T cell progenitor subsets are detectable in extrathymic tissues such as fetal liver, and both subsets are capable of exiting the liver and entering the circulation in the absence of FoxN1+ thymic epithelial cells.
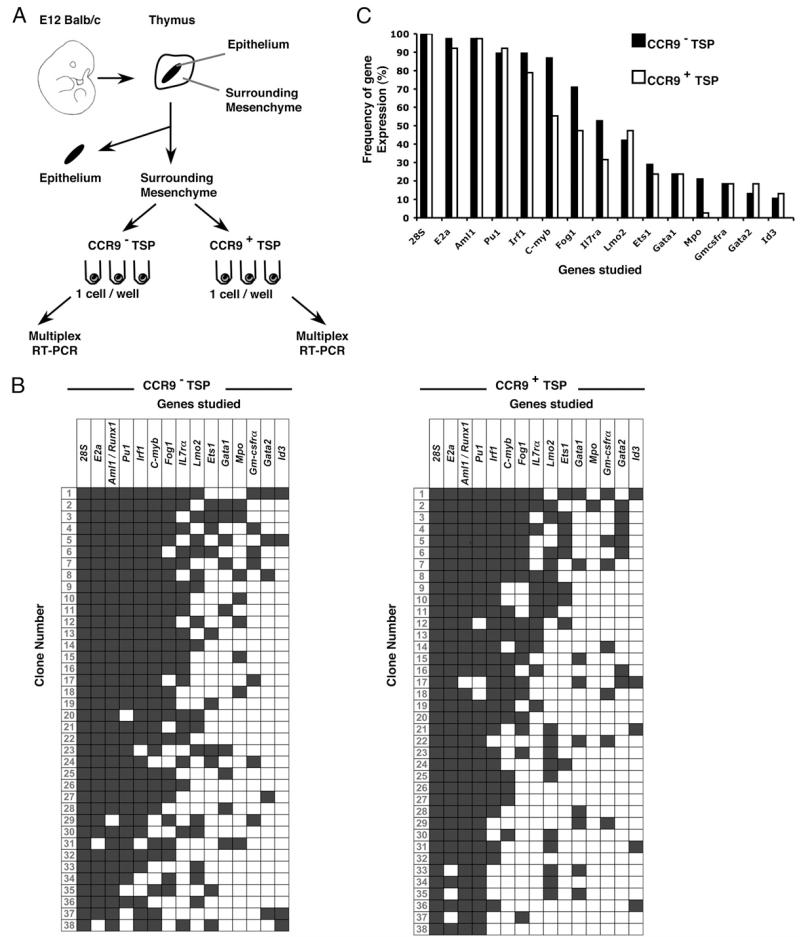
Multiplex clonal PCR analysis identifies a molecular consensus in TSP
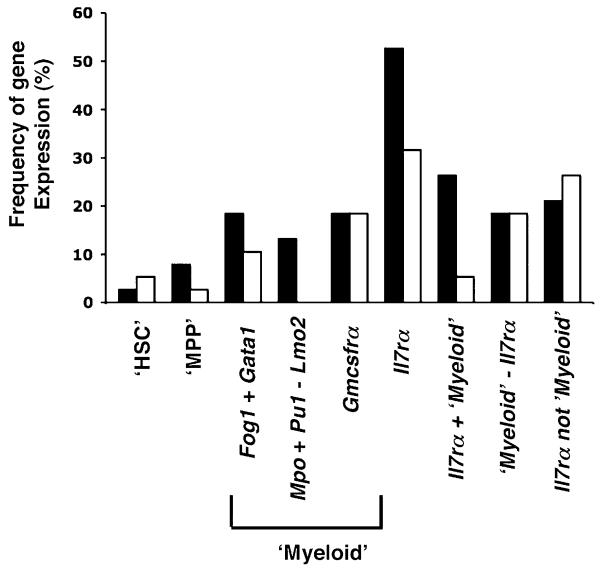
Although the above data demonstrate that CCR9+ and CCR9− CD45+F4/80−PIRhigh TSP subsets share similar developmental potential, the observed differences in CCR9 expression and differential dependency on FoxN1+ thymic epithelium for their thymic recruitment suggested that additional intrinsic differences might also exist. To investigate this possibility, we adopted a multiplex single-cell RT-PCR approach to examine, within single CCR9+ and CCR9− TSP, the simultaneous expression of 14 genes involved in hematopoietic cell development (38). Individual CCR9+ and CCR9− TSP were isolated from the perithymic mesenchyme of the E12 WT thymus anlage by MoFlo cell sorting and subjected to Multiplex PCR (Fig. 5A) as previously described (38), with expression of 28S mRNA used as a positive control housekeeping gene. The panel of genes analyzed included several groups of molecular regulators including transcription factors (Pu1, C-myb, and Gata1) and growth factor receptors (Il-7ra and Gm-csfra). Fig. 5B shows a summary of coexpression data obtained from 38 individual CCR9+ TSP and 38 individual CCR9− TSP. Interestingly, a homogeneous profile of gene expression was revealed in individual cells contained within each TSP subset with E2a, Aml1/Runx1, and Pu1 being examples of genes expressed by essentially all CCR9+ and CCR9− TSP analyzed, whereas Gata2, Mpo, and Id3 represent examples of genes that were absent in the vast majority of CCR9+ as well as CCR9− cells (Fig. 5B). Moreover, by performing a comparative analysis of gene expression in CCR9+ and CCR9− TSP subsets, in which the percentage of cells expressing any given gene is determined, a striking similarity pattern is revealed (Fig. 5C), suggesting that despite differences in chemokine receptor expression, the initial cohort of TSP to colonize the embryonic thymus display uniformity in their expression of hemopoietic regulators. By clustering the transcript expression of certain genes into distinct groups, we also determined the “signature” of defined progenitor populations, including hematopoietic stem cells (HSC: coexpression of E2a, Aml1/Runx1, Irf1, C-myb, Lmo2, Gata2, and Fog1 but not Gata1 and Il7ra) and myeloid committed cells (coexpression of Fog1 and Gata1, coexpression of Mpo and Pu1 without Lmo2, or expression of Csf2ra) (38). We also identified lymphoid committed cells using Il7ra expression, which has recently been used in fate-mapping studies to identify T lineage-restricted progenitors (32). As expected, few (<6%) of the CCR9+ and CCR9− TSP have an HSC-like signature (Fig. 6), whereas 11 and 18% of CCR9+ and CCR9− TSP coexpress Fog1 and Gata1 transcripts. Interestingly, Mpo and Pu1 transcript coexpression in the absence of Lmo2 expression, indicative of macrophage/granulocyte commitment (38, 50), was only observed in 13% of CCR9− TSP, whereas Csf2ra mRNA expression was detected in 18% of both CCR9+ and CCR9− subsets. Finally, analysis of expression of Il7ra mRNA, characteristic of progenitors primed toward the lymphoid fate (16, 32, 51), suggests an enrichment for lymphoid committed cells being detected in 32 and 53% of CCR9+ and CCR9− TSP, respectively (Fig. 6), which is in agreement with earlier studies and data shown in this study, indicating a T lineage bias of thymus-colonizing cells. Collectively, clonal analysis of embryonic TSP reveals a genetic signature that is indicative of a bias toward lymphoid-restricted progenitors and identifies homogeneity within and between CCR9+ and CCR9− TSP subsets.
FIGURE 5. Clonal multiplex coexpression analysis reveals genetic similarities between, and within, CCR9+ and CCR9− TSP.
The schema in A summarizes isolation of CD45+CCR9− and CD45+CCR9+ TSP from perithymic mesenchyme surrounding the E12 thymus, which were studied individually by multiplex RT-PCR for the expression of 15 genes. B shows a summary of gene expression of each clone, as determined by multiplex RT-PCR. ■, Positive gene expression; □, an absence of expression. Cells were scored as positive by the expression of 28S rRNA. C shows a summary of the frequency of gene expression in CD45+CCR9− (■) and CD45+CCR9+ (□) cells.
FIGURE 6. The TSP genetic signature of TSP is distinct to other hemopoietic progenitors.
The gene expression profile of individual CD45+ CCR9− TSP (■) and CD45+CCR9+ TSP (□) from E12 perithymic mesenchyme was compared with gene coexpression profiles characteristic of a panel of hemopoietic cell subsets and lineages. The frequencies of the HSC signature (‘HSC’: coexpression of E2a, Aml1, Irf1, C-myb, Lmo2, Gata2, and Fog1; lack of expression of Gata1 and Il7ra); the MPP/early thymic progenitors signature (‘MPP’: E2a, Aml1, Irf1, C-myb, Lmo2, Pu1 and Mpo), the myeloid signatures (positive for Fog1 and Gata1; positive for Mpo and Pu1 but negative for Lmo2; positive for Gmcsfra), and lymphoid signature (Il7ra) determined by multiplex RT-PCR are shown.
Discussion
The continued production of functionally competent T cells requires the constant input of hemopoietic progenitors to the thymus, a process that begins in early embryonic life, and represents a key stage in the development of the self-tolerant peripheral T cell pool. In this study, we analyzed the phenotype, developmental potential, and gene expression profile of cells representing the earliest TSP (19) that colonize the embryonic thymus. Our data indicate that initially the developing thymus anlage is colonized by hemopoietic cells that consist largely of T lineage-biased PIRhighc-Kit+ progenitors but also include a smaller subset of PIR−F4/80+ cells, the latter likely representing previously reported myeloid committed cells (42, 52). Regarding PIRhighc-Kit+ TSP, we provide evidence that these cells can be subdivided into distinct subsets on the basis of the homeostatic chemokine receptor CCR9. This finding contrasts with a previous study analyzing CCR9-GFP reporter mice (23), in which all thymus-colonizing cells at E12 of gestation were reported to be GFP+, which was taken as evidence that these cells are uniformly CCR9+. The presence of CCR9− and CCR9+ cell subsets within PIR+ progenitors in fetal liver shown in this study, representing stages prior to thymus colonization, argues against the notion that CCR9− TSP are detected because of CCR9 downregulation following thymic homing. Perhaps critically, unlike in our study, levels of cell surface CCR9 protein expression were not analyzed in CCR9-GFP knockin mouse embryos (23), which may explain the discrepancy as GFP expression in this system measures gene promoter activity that may not always correlate directly with protein expression. For example, it may be the case that, as shown by Cooper et al. (53) in other GFP-reporter systems (e.g., using GFP to monitor Rag2 gene expression), the protein of interest (in this case CCR9) and the GFP protein may not always be synchronously expressed.
In further experiments, we show that CCR9+ and CCR9− TSP, which are both present in fetal liver, show a differential requirement for FoxN1-expressing epithelial cells for their recruitment to the E12 thymus anlage. Importantly, however, despite these differences, functional analysis indicates that both CCR9+ and CCR9− TSP subsets share a strong and common bias toward T cell-committed progenitors. This conclusion is also supported by an analysis of gene expression in individual CCR9+ and CCR9− TSP, which shows that CCR9+ and CCR9− TSP share a gene expression profile that predominantly lacks HSC and myeloid signatures, but is indicative of a bias toward lymphoid-restricted progenitors. Interestingly, however, single-cell gene coexpression analysis revealed that a fraction of TSP within the E12 perithymic mesenchyme coexpress transcripts indicative of lymphoid (Il7ra) as well as myeloid (Fog1 and Gata1, Mpo and Pu1, and Csf2ra) potential, which correlate with the residual myeloid potential revealed in our limiting dilution assays. Furthermore, when analysis of clonal transcriptional activity was recently performed in various subsets of adult bone marrow and thymic hematopoietic progenitors, Aml1/Runx1, Irf1, C-myb, Lmo2, Pu1, and Mpo, expression was shown to define MPP/early thymic progenitors (38). Interestingly, in the current study on embryonic thymus, we noted that both CCR9+ and CCR9− TSP subsets recruited to the E12 thymus anlage contain a high frequency of cells coexpressing E2a, Aml1/Runx1, Irf1, C-myb, and Pu1 but not Lmo2 and Mpo, underlining the notion that fetal and adult progenitor populations recruited to the thymus are distinct.
Collectively, these data suggest that the cells giving rise to the initial T cell cohorts represent T cell-biased progenitors that colonize the thymus via multiple mechanisms. How this heterogeneity in thymus recruitment relates to our current understanding of the roles of CCR7 and CCR9 is currently unclear. Interestingly, however, in early fetal life, progenitor recruitment to the thymic region has been shown to be a shared function of the parathyroid and the thymus anlagen, which are closely associated at these stages (54). Moreover, the parathyroid provides a source of the CCR7 and CCR9 ligands CCL21 and CCL25, respectively (18). Interestingly, although CCL21 and CCL25 expression remains intact in the parathyroid of nude mouse embryos, CCL25 is absent from the nude thymus anlagen (18, 55). We have shown previously that the E12 CCR9+ thymic progenitor subset contains CCR7 expressing cells, whereas CCR9− thymic progenitors lack CCR7 expression (41). Thus, we propose that in the absence of FoxN1-expressing thymic epithelial cells, CCR9+ TSP—as a result of their dual expression of CCR7 and CCR9 (41)—are recruited to the thymic region via chemokine production from the parathyroid. In contrast, CCR9− TSP—which also lack CCR7 (41)—are unable to respond to parathyroid-derived signals and are dependent on functionally intact FoxN1+ thymic epithelial cells for their recruitment. On the basis of these observations, it would be interesting to determine the anatomical location of CCR9− and CCR9+ TSP in confocal analysis of the combined thymus/parathyroid anlagen. However, in our hands, currently available anti-CCR9 Abs are incompatible with labeling of frozen tissue sections (data not shown). Interestingly, analysis of TSP in the circulation at E11 of gestation identifies subsets of CCR9− and CCR9+ TSP in both nude and WT mouse embryos, suggesting that the presence of FoxN1+ thymic epithelial cells is not required for the initial exit of either of these TSP subsets from the fetal liver. However, the dramatic loss of CCR9− TSP in the circulation of nude mouse embryos at E12 of gestation, coupled with their absence from the nude thymus rudiment at this stage, suggests that the failure of CCR9− TSP to be recruited out of the circulation by FoxN1+ thymic epithelial cells results in the induction of cell death in this population between E11 and E12 of gestation. On the basis of these findings, we suggest that although both CCR9− and CCR9+ TSP can exit the liver independently of normal thymus development, the recruitment of the CCR9− TSP subset from the circulation to the thymus anlagen is thymus dependent but in a manner that is independent of CCL25. Interestingly, in CCR7/CCR9 double-deficient mouse embryos, small numbers of thymus-colonizing cells are still detectable around the early thymic anlagen (18), which may represent recruitment of CCR9− TSP. Moreover, although other studies have reported the recruitment of T cell progenitors to the nude thymus anlage (49), our current data demonstrating a loss of CCR9− TSP around E12 nude thymus anlage suggests that these cells do not fully represent an intact wave of initial thymus-colonizing cells. In summary, our study defines the TSP that give rise to the initial cohorts of mature T cells and suggests that T cell-primed progenitors can colonize the developing thymus anlage via multiple mechanisms.
Acknowledgments
We thank Juan Carlos Zuniga-Pflucker for providing OP9 and OP9DL1 cell lines, Roger Bird for expert cell sorting, and Biomedical Services Unit staff for animal husbandry.
This work was supported by a Medical Research Council Programme grant (to G.A. and E.J.J.) and by the Medical Research Council Center Grant core facilities of the Medical Research Council Center for Immune Regulation.
Abbreviations used in this article
- c-Kit–L
c-Kit ligand
- CLP
common lymphoid progenitor
- E12
embryonic stage 12
- Flt3-L
Flt3 ligand
- HSC
hematopoietic stem cell
- MPP
multipotent progenitor
- PIR
paired IgR
- TSP
thymus-settling progenitor
- WT
wild-type
Footnotes
Disclosures: The authors have no financial conflicts of interest.
References
- 1.Rodewald HR. Thymus organogenesis. Annu. Rev. Immunol. 2008;26:355–388. doi: 10.1146/annurev.immunol.26.021607.090408. [DOI] [PubMed] [Google Scholar]
- 2.Petrie HT, Zúñiga-Pflücker JC. Zoned out: functional mapping of stromal signaling microenvironments in the thymus. Annu. Rev. Immunol. 2007;25:649–679. doi: 10.1146/annurev.immunol.23.021704.115715. [DOI] [PubMed] [Google Scholar]
- 3.Anderson G, Jenkinson EJ. Lymphostromal interactions in thymic development and function. Nat. Rev. Immunol. 2001;1:31–40. doi: 10.1038/35095500. [DOI] [PubMed] [Google Scholar]
- 4.Griffith AV, Fallahi M, Nakase H, Gosink M, Young B, Petrie HT. Spatial mapping of thymic stromal microenvironments reveals unique features influencing T lymphoid differentiation. Immunity. 2009;31:999–1009. doi: 10.1016/j.immuni.2009.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wallis VJ, Leuchars E, Chwalinski S, Davies AJ. On the sparse seeding of bone marrow and thymus in radiation chimaeras. Transplantation. 1975;19:2–11. doi: 10.1097/00007890-197501000-00002. [DOI] [PubMed] [Google Scholar]
- 6.Donskoy E, Goldschneider I. Thymocytopoiesis is maintained by blood-borne precursors throughout postnatal life: a study in parabiotic mice. J. Immunol. 1992;148:1604–1612. [PubMed] [Google Scholar]
- 7.Petrie HT. Cell migration and the control of post-natal T-cell lympho-poiesis in the thymus. Nat. Rev. Immunol. 2003;3:859–866. doi: 10.1038/nri1223. [DOI] [PubMed] [Google Scholar]
- 8.Bhandoola A, von Boehmer H, Petrie HT, Zúñiga-Pflücker JC. Commitment and developmental potential of extrathymic and intrathymic T cell precursors: plenty to choose from. Immunity. 2007;26:678–689. doi: 10.1016/j.immuni.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 9.Chi AW, Bell JJ, Zlotoff DA, Bhandoola A. Untangling the T branch of the hematopoiesis tree. Curr. Opin. Immunol. 2009;21:121–126. doi: 10.1016/j.coi.2009.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Adolfsson J, Månsson R, Buza-Vidas N, Hultquist A, Liuba K, Jensen CT, Bryder D, Yang L, Borge OJ, Thoren LA, et al. Identification of Flt3+ lympho-myeloid stem cells lacking erythro-megakaryocytic potential a revised road map for adult blood lineage commitment. Cell. 2005;121:295–306. doi: 10.1016/j.cell.2005.02.013. [DOI] [PubMed] [Google Scholar]
- 11.Lai AY, Kondo M. Identification of a bone marrow precursor of the earliest thymocytes in adult mouse. Proc. Natl. Acad. Sci. USA. 2007;104:6311–6316. doi: 10.1073/pnas.0609608104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schwarz BA, Sambandam A, Maillard I, Harman BC, Love PE, Bhandoola A. Selective thymus settling regulated by cytokine and chemokine receptors. J. Immunol. 2007;178:2008–2017. doi: 10.4049/jimmunol.178.4.2008. [DOI] [PubMed] [Google Scholar]
- 13.Scimone ML, Aifantis I, Apostolou I, von Boehmer H, von Andrian UH. A multistep adhesion cascade for lymphoid progenitor cell homing to the thymus. Proc. Natl. Acad. Sci. USA. 2006;103:7006–7011. doi: 10.1073/pnas.0602024103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rodewald HR, Kretzschmar K, Takeda S, Hohl C, Dessing M. Identification of pro-thymocytes in murine fetal blood: T lineage commitment can precede thymus colonization. EMBO J. 1994;13:4229–4240. doi: 10.1002/j.1460-2075.1994.tb06743.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kawamoto H, Ohmura K, Katsura Y. Direct evidence for the commitment of hematopoietic stem cells to T, B and myeloid lineages in murine fetal liver. Int. Immunol. 1997;9:1011–1019. doi: 10.1093/intimm/9.7.1011. [DOI] [PubMed] [Google Scholar]
- 16.Umland O, Mwangi WN, Anderson BM, Walker JC, Petrie HT. The blood contains multiple distinct progenitor populations with clonogenic B and T lineage potential. J. Immunol. 2007;178:4147–4152. doi: 10.4049/jimmunol.178.7.4147. [DOI] [PubMed] [Google Scholar]
- 17.Carlyle JR, Zúñiga-Pflücker JC. Requirement for the thymus in ab T lymphocyte lineage commitment. Immunity. 1998;9:187–197. doi: 10.1016/s1074-7613(00)80601-9. [DOI] [PubMed] [Google Scholar]
- 18.Liu C, Saito F, Liu Z, Lei Y, Uehara S, Love P, Lipp M, Kondo S, Manley N, Takahama Y. Coordination between CCR7- and CCR9-mediated chemokine signals in prevascular fetal thymus colonization. Blood. 2006;108:2531–2539. doi: 10.1182/blood-2006-05-024190. [DOI] [PubMed] [Google Scholar]
- 19.Zlotoff DA, Sambandam A, Logan TD, Bell JJ, Schwarz BA, Bhandoola A. CCR7 and CCR9 together recruit hematopoietic progenitors to the adult thymus. Blood. 2010;115:1897–1905. doi: 10.1182/blood-2009-08-237784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Svensson M, Marsal J, Uronen-Hansson H, Cheng M, Jenkinson W, Cilio C, Jacobsen SE, Sitnicka E, Anderson G, Agace WW. Involvement of CCR9 at multiple stages of adult T lymphopoiesis. J. Leukoc. Biol. 2008;83:156–164. doi: 10.1189/jlb.0607423. [DOI] [PubMed] [Google Scholar]
- 21.Krueger A, Willenzon S, Lyszkiewicz M, Kremmer E, Förster R. CC chemokine receptor 7 and 9 double-deficient hematopoietic progenitors are severely impaired in seeding the adult thymus. Blood. 2010;115:1906–1912. doi: 10.1182/blood-2009-07-235721. [DOI] [PubMed] [Google Scholar]
- 22.Uehara S, Grinberg A, Farber JM, Love PE. A role for CCR9 in T lymphocyte development and migration. J. Immunol. 2002;168:2811–2819. doi: 10.4049/jimmunol.168.6.2811. [DOI] [PubMed] [Google Scholar]
- 23.Benz C, Bleul CC. A multipotent precursor in the thymus maps to the branching point of the T versus B lineage decision. J. Exp. Med. 2005;202:21–31. doi: 10.1084/jem.20050146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wurbel MA, Malissen B, Campbell JJ. Complex regulation of CCR9 at multiple discrete stages of T cell development. Eur. J. Immunol. 2006;36:73–81. doi: 10.1002/eji.200535203. [DOI] [PubMed] [Google Scholar]
- 25.Bajoghli B, Aghaallaei N, Hess I, Rode I, Netuschil N, Tay BH, Venkatesh B, Yu JK, Kaltenbach SL, Holland ND, et al. Evolution of genetic networks underlying the emergence of thymopoiesis in vertebrates. Cell. 2009;138:186–197. doi: 10.1016/j.cell.2009.04.017. [DOI] [PubMed] [Google Scholar]
- 26.Lei Y, Liu C, Saito F, Fukui Y, Takahama Y. Role of DOCK2 and DOCK180 in fetal thymus colonization. Eur. J. Immunol. 2009;39:2695–2702. doi: 10.1002/eji.200939630. [DOI] [PubMed] [Google Scholar]
- 27.Gossens K, Naus S, Corbel SY, Lin S, Rossi FM, Kast J, Ziltener HJ. Thymic progenitor homing and lymphocyte homeostasis are linked via S1P-controlled expression of thymic P-selectin/CCL25. J. Exp. Med. 2009;206:761–778. doi: 10.1084/jem.20082502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rossi FM, Corbel SY, Merzaban JS, Carlow DA, Gossens K, Duenas J, So L, Yi L, Ziltener HJ. Recruitment of adult thymic progenitors is regulated by P-selectin and its ligand PSGL-1. Nat. Immunol. 2005;6:626–634. doi: 10.1038/ni1203. [DOI] [PubMed] [Google Scholar]
- 29.Drake PM, Stock CM, Nathan JK, Gip P, Golden KP, Weinhold B, Gerardy-Schahn R, Bertozzi CR. Polysialic acid governs T-cell development by regulating progenitor access to the thymus. Proc. Natl. Acad. Sci. USA. 2009;106:11995–12000. doi: 10.1073/pnas.0905188106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.David-Fung ES, Butler R, Buzi G, Yui MA, Diamond RA, Anderson MK, Rowen L, Rothenberg EV. Transcription factor expression dynamics of early T-lymphocyte specification and commitment. Dev. Biol. 2009;325:444–467. doi: 10.1016/j.ydbio.2008.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yui MA, Feng N, Rothenberg EV. Fine-scale staging of T cell lineage commitment in adult mouse thymus. J. Immunol. 2010;185:284–293. doi: 10.4049/jimmunol.1000679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schlenner SM, Madan V, Busch K, Tietz A, Läufle C, Costa C, Blum C, Fehling HJ, Rodewald HR. Fate mapping reveals separate origins of T cells and myeloid lineages in the thymus. Immunity. 2010;32:426–436. doi: 10.1016/j.immuni.2010.03.005. [DOI] [PubMed] [Google Scholar]
- 33.Wada H, Masuda K, Satoh R, Kakugawa K, Ikawa T, Katsura Y, Kawamoto H. Adult T-cell progenitors retain myeloid potential. Nature. 2008;452:768–772. doi: 10.1038/nature06839. [DOI] [PubMed] [Google Scholar]
- 34.Bell JJ, Bhandoola A. The earliest thymic progenitors for T cells possess myeloid lineage potential. Nature. 2008;452:764–767. doi: 10.1038/nature06840. [DOI] [PubMed] [Google Scholar]
- 35.Schlenner SM, Rodewald HR. Early T cell development and the pitfalls of potential. Trends Immunol. 2010;31:303–310. doi: 10.1016/j.it.2010.06.002. [DOI] [PubMed] [Google Scholar]
- 36.Harman BC, Jenkinson WE, Parnell SM, Rossi SW, Jenkinson EJ, Anderson G. T/B lineage choice occurs prior to intrathymic Notch signaling. Blood. 2005;106:886–892. doi: 10.1182/blood-2004-12-4881. [DOI] [PubMed] [Google Scholar]
- 37.Schmitt TM, Zúñiga-Pflücker JC. Induction of T cell development from hematopoietic progenitor cells by delta-like-1 in vitro. Immunity. 2002;17:749–756. doi: 10.1016/s1074-7613(02)00474-0. [DOI] [PubMed] [Google Scholar]
- 38.Gautreau L, Boudil A, Pasqualetto V, Skhiri L, Grandin L, Monteiro M, Jais JP, Ezine S. Gene coexpression analysis in single cells indicates lymphomyeloid copriming in short-term hematopoietic stem cells and multi-potent progenitors. J. Immunol. 2010;184:4907–4917. doi: 10.4049/jimmunol.0902184. [DOI] [PubMed] [Google Scholar]
- 39.Peixoto A, Monteiro M, Rocha B, Veiga-Fernandes H. Quantification of multiple gene expression in individual cells. Genome Res. 2004;14(10A):1938–1947. doi: 10.1101/gr.2890204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shakib S, Desanti GE, Jenkinson WE, Parnell SM, Jenkinson EJ, Anderson G. Checkpoints in the development of thymic cortical epithelial cells. J. Immunol. 2009;182:130–137. doi: 10.4049/jimmunol.182.1.130. [Published erratum appears in 2009 J. Immunol. 182: 4488.] [DOI] [PubMed] [Google Scholar]
- 41.Jenkinson WE, Rossi SW, Parnell SM, Agace WW, Takahama Y, Jenkinson EJ, Anderson G. Chemokine receptor expression defines heterogeneity in the earliest thymic migrants. Eur. J. Immunol. 2007;37:2090–2096. doi: 10.1002/eji.200737212. [DOI] [PubMed] [Google Scholar]
- 42.Kawamoto H, Ohmura K, Katsura Y. Presence of progenitors restricted to T, B, or myeloid lineage, but absence of multipotent stem cells, in the murine fetal thymus. J. Immunol. 1998;161:3799–3802. [PubMed] [Google Scholar]
- 43.Masuda K, Kubagawa H, Ikawa T, Chen CC, Kakugawa K, Hattori M, Kageyama R, Cooper MD, Minato N, Katsura Y, Kawamoto H. Prethymic T-cell development defined by the expression of paired immunoglobulin-like receptors. EMBO J. 2005;24:4052–4060. doi: 10.1038/sj.emboj.7600878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rutishauser U. Polysialic acid in the plasticity of the developing and adult vertebrate nervous system. Nat. Rev. Neurosci. 2008;9:26–35. doi: 10.1038/nrn2285. [DOI] [PubMed] [Google Scholar]
- 45.Drake PM, Nathan JK, Stock CM, Chang PV, Muench MO, Nakata D, Reader JR, Gip P, Golden KP, Weinhold B, et al. Polysialic acid, a glycan with highly restricted expression, is found on human and murine leukocytes and modulates immune responses. J. Immunol. 2008;181:6850–6858. doi: 10.4049/jimmunol.181.10.6850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ceredig R, Bosco N, Rolink AG. The B lineage potential of thymus settling progenitors is critically dependent on mouse age. Eur. J. Immunol. 2007;37:830–837. doi: 10.1002/eji.200636728. [DOI] [PubMed] [Google Scholar]
- 47.Nehls M, Pfeifer D, Schorpp M, Hedrich H, Boehm T. New member of the winged-helix protein family disrupted in mouse and rat nude mutations. Nature. 1994;372:103–107. doi: 10.1038/372103a0. [DOI] [PubMed] [Google Scholar]
- 48.Blackburn CC, Augustine CL, Li R, Harvey RP, Malin MA, Boyd RL, Miller JF, Morahan G. The nu gene acts cell-autonomously and is required for differentiation of thymic epithelial progenitors. Proc. Natl. Acad. Sci. USA. 1996;93:5742–5746. doi: 10.1073/pnas.93.12.5742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Itoi M, Kawamoto H, Katsura Y, Amagai T. Two distinct steps of immigration of hematopoietic progenitors into the early thymus anlage. Int. Immunol. 2001;13:1203–1211. doi: 10.1093/intimm/13.9.1203. [DOI] [PubMed] [Google Scholar]
- 50.Rosmarin AG, Yang Z, Resendes KK. Transcriptional regulation in myelopoiesis: hematopoietic fate choice, myeloid differentiation, and leukemogenesis. Exp. Hematol. 2005;33:131–143. doi: 10.1016/j.exphem.2004.08.015. [DOI] [PubMed] [Google Scholar]
- 51.Krueger A, von Boehmer H. Identification of a T lineage-committed progenitor in adult blood. Immunity. 2007;26:105–116. doi: 10.1016/j.immuni.2006.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Morris L, Graham CF, Gordon S. Macrophages in haemopoietic and other tissues of the developing mouse detected by the monoclonal antibody F4/80. Development. 1991;112:517–526. doi: 10.1242/dev.112.2.517. [DOI] [PubMed] [Google Scholar]
- 53.Cooper CJ, Orr MT, McMahan CJ, Fink PJ. T cell receptor revision does not solely target recent thymic emigrants. J. Immunol. 2003;171:226–233. doi: 10.4049/jimmunol.171.1.226. [DOI] [PubMed] [Google Scholar]
- 54.Gordon J, Bennett AR, Blackburn CC, Manley NR. Gcm2 and Foxn1 mark early parathyroid- and thymus-specific domains in the developing third pharyngeal pouch. Mech. Dev. 2001;103:141–143. doi: 10.1016/s0925-4773(01)00333-1. [DOI] [PubMed] [Google Scholar]
- 55.Bleul CC, Boehm T. Chemokines define distinct microenvironments in the developing thymus. Eur. J. Immunol. 2000;30:3371–3379. doi: 10.1002/1521-4141(2000012)30:12<3371::AID-IMMU3371>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]