Abstract
This study was carried out to investigate the protective effects of chitosan nanoparticles (CNP) against hydrogen peroxide (H2O2)-induced oxidative damage in murine macrophages RAW264.7 cells. After 24 h pre-incubation with CNP (25–200 μg/mL) and chitosan (CS) (50–200 μg/mL, as controls), the viability loss in RAW264.7 cells induced by H2O2 (500 μM) for 12 h was markedly restored in a concentration-dependent manner as measured by MTT assay (P < 0.05) and decreased in cellular LDH release (P < 0.05). Moreover, CNP also exerted preventive effects on suppressing the production of lipid peroxidation such as malondialdehyde (MDA) (P < 0.05), restoring activities of endogenous antioxidant including superoxide dismutase (SOD), and glutathione peroxidase (GSH-Px) (P < 0.05), along with increasing total antioxidant capacity (T-AOC) (P < 0.05). In addition, pre-incubation of CNP with RAW264.7 cells for 24 h resulted in the increase of the gene expression level of endogenous antioxidant enzymes, such as MnSOD and GSH-Px (P < 0.05). At the same concentration, CNP significantly decreased LDH release and MDA (P < 0.05) as well as increased MnSOD, GSH-Px, and T-AOC activities (P < 0.05) as compared to CS. Taken together, our findings suggest that CNP can more effectively protect RAW264.7 cells against oxidative stress by H2O2 as compared to CS, which might be used as a potential natural compound-based antioxidant in the functional food and pharmaceutical industries.
Keywords: chitosan nanoparticles, antioxidant activity, stress injury, RAW264.7 cells
1. Introduction
Antioxidants may have a positive effect on human health since they can protect human body against deterioration by reactive oxygen species (ROS), which attack biomolecules such as proteins, lipid, and DNA, and consequently, lead to many health disorders including aging, cancer, diabetes, neurodegenerative, cardiovascular, and inflammation disease. Cells possess cellular defense systems including nonenzymatic and enzymatic substances, such as superoxide dismutase (SOD) and glutathione peroxidase (GSH-Px) to scavenge radicals and protect major cellular biomolecules from oxidative damage to [1,2,3]. When the cellular defense mechanisms are unable to cope with excessive generation of ROS, subsequently oxidative stress will happen. Oxidative stress has been shown to be involved in various pathogenic processes including aging, cancer, wrinkle formation, rheumatoid arthritis, and inflammation. Therefore, antioxidant might theoretically retard spreading of cell damage and improve intracellular antioxidant enzymatic and nonenzymatic systems.
Many studies have been carried out to seek novel antioxidative compounds from marine resources, such as chitin, chitosan, and their derivates. Chitosan, a partially deacetylated polymer of N-acetyl glucosamine, is prepared by alkaline deacetylation of chitin. It exhibits a wide variety of biological activities, including antitumor activities [4], immunostimulating effects [5], cholesterol-lowering effects [6], antimicrobial effects [7], wound-healing effects [8], antifungal activities, and free radical scavenging activities [9]. Xie, Xu, and Liu (2001) reported that the scavenging of hydroxyl radicals by chitosan inhibits the lipid peroxidation of phosphatidylcholine and linoleate liposomes [10]. Santhosh, Sini, Anandan, and Mathew (2006) reported that the administration of chitosan to rats, treated with isoniazid or rifampicin, prevented the oxidation of hepatotoxic lipids [11]. Similarly, chitosan, when injected, inhibited glycerol-induced renal oxidative damage in rats [12]. Owing to its many antioxidant studies in vitro and in vivo, chitosan has attracted considerable attention from researchers.
In the previous studies, the antioxidant activity of chitosan, and its oligosaccharides, was studied in vitro and in vivo. The results showed that chitosan could significantly reduce serum free fatty acid (FFA) and MDA concentrations and elevate SOD, CAT, and GSH-Px activities, the latter being the major antioxidant enzyme in the body, indicating that chitosan regulated the antioxidant enzyme activities and reduced lipid peroxidation [13]. The unique character of nanoparticles could make chitosan nanoparticles (CNP) exhibit more superior activities than chitosan, CNP have been reported to have heightened immune-enhancing effect [14], anticancer activity [15], and antimicrobial activity [16], of chitosan (CS). However, other biological activities are unknown, such as the antioxidant activity of CNP, has received less attention. Therefore, we hypothesized that CNP may have a potential to augment the antioxidant activity of chitosan. Herein, we investigated the protective effects of CNP against hydrogen peroxide (H2O2)-induced oxidative damage in murine macrophages RAW264.7 cells.
2. Results
2.1. Morphology, Size, and Zeta Potential of CNP
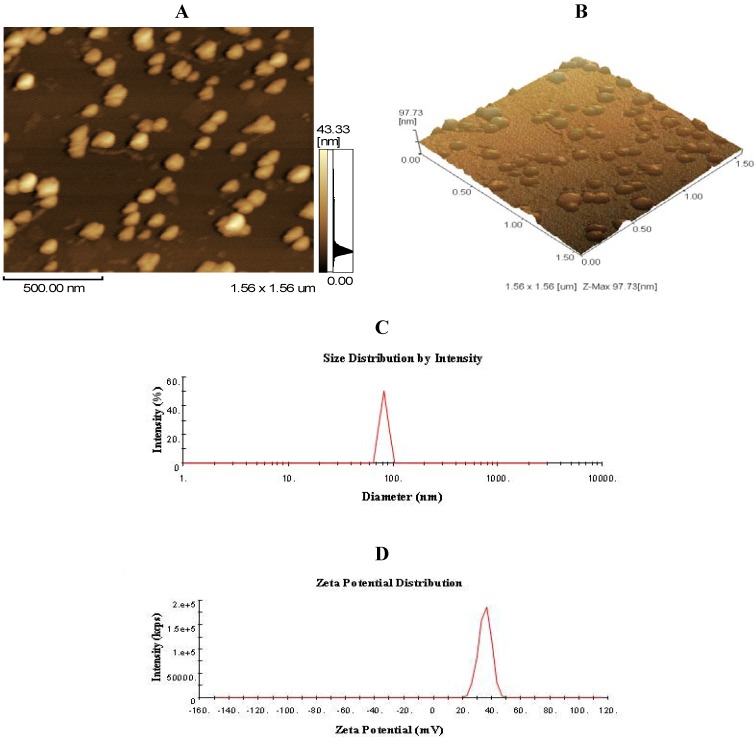
As shown in Figure 1A,B, chitosan nanoparticles regularly formed and well distributed in the acetic acid (HOAc)/sodium tripolyphosphate solution (pH 5.5) used in this study. The mean size and size distribution of each batch of nanoparticle suspension was analyzed using the Zetasizer analysis (Figure 1C,D). The size distribution profile represents a typical batch of nanoparticles with a mean diameter of 83.66 nm and a narrow size distribution ranging from 63.16 to 101.70 nm (polydispersity index < 1), and shows that the surfaces of chitosan nanoparticles have a positive surface charge of about 35.43 mV. These excellent characteristics are of benefit to the stabilization and penetration capability of CNP.
Figure 1.
Morphology of chitosan nanoparticles (CNP). (A,B) atomic force micrographs (AFMs) of CNP; (C) the size distribution by intendity of CNP, the size of CNP ranges from 63.16 to 101.70 nm, and the mean of size is about 83.66 nm; (D) Zeta potential distribution of CNP, CNP exhibit a zeta potential range from 20.04 to 51.13 mV and have a mean charge with 35.43 mV.
2.2. Time-Dependent and Concentration-Dependent Viability Losses in RAW264.7 Cells Induced by H2O2
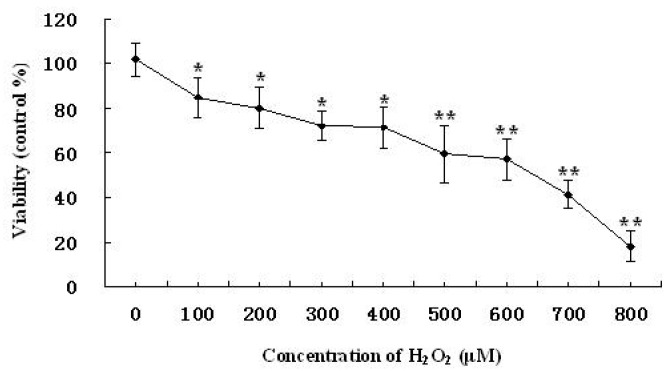
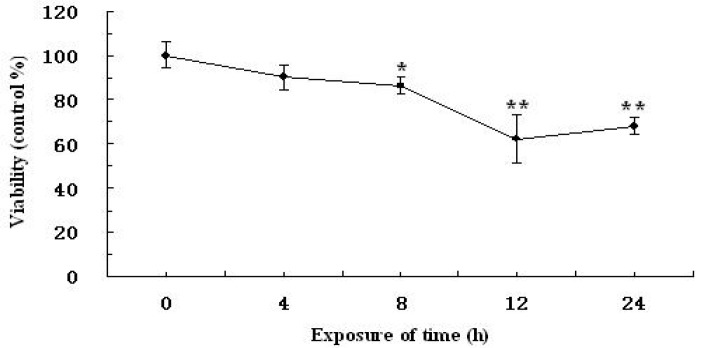
The time-dependent and concentration-dependent studies of viability losses were investigated in RAW264.7 cells induced by H2O2. After treatment with increasing concentrations of H2O2 for 12 h, cell viability was then examined by MTT method. As shown in Figure 2, gradual losses of cell viability were observed with increasing concentrations of H2O2. The degree of cell injury was maximum at 800 μM, among the cell viability was close to 18.0% ± 7.0% as compared with the vehicle-treated control group. The results of time-response study in which cells were exposed to 500 μM of H2O2 up to 24 h (Figure 3), the concentration of H2O2 started to increase oxidative injury in RAW264.7 cells 4 h after treatment with H2O2. The magnitude of cell injury peaked at 12 h after H2O2 exposure, and the cell viability was about 59.1% ± 12.6% (P < 0.05). Based on these results, RAW264.7 cells were treated with 500 μM of H2O2 for 12 h, or vehicle as control, in the extensive studies.
Figure 2.
Viability losses in RAW264.7 cells induced by various concentration of H2O2. Values are presented as means ± SD. Mean with different asterisks are statistically different (P < 0.05).
Figure 3.
Viability losses in RAW264.7 cells induced by H2O2 for different time. Values are presented as means ± SD. Mean with different asterisks are statistically different (P < 0.05).
2.3. Effect of CNP on the Viability of H2O2-Induced RAW264.7 Cells
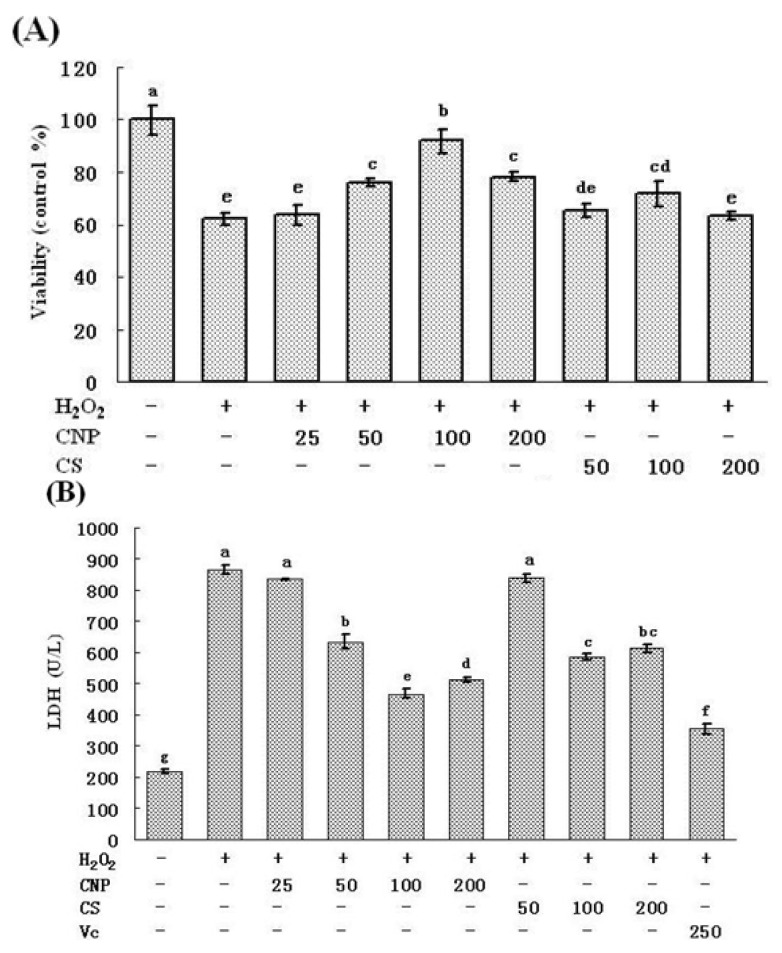
Effects of CNP on the viability of H2O2-induced RAW264.7 cells were evaluated by MTT analysis. The results in Figure 4A show that the survival rate of RAW264.7 cells was decreased markedly (P < 0.05) after exposure to 500 μM of H2O2 for 12 h. However, pre-incubation of RAW264.7 cells with different concentrations of CNP (50, 100, 200 μg/mL) for 24 h significantly increased the vialibity of H2O2-induced RAW264.7 cells in a dose-dependent manner (P < 0.05). In addition, no difference was found in cell viability between cells treated with CNP (25–200 μg/mL) alone and vehicle-treated controls (data not shown). Apparently, CNP were effective for the protection of RAW264.7 cells against H2O2-induced injury.
Figure 4.
Protective effects of CNP on viability losses in RAW264.7 cells induced by H2O2 (500 μM). Values are presented as means ± SD. Bars with different letters are statistically different (P < 0.05).
To further investigate the protective effects of CNP, LDH assay, another indicator of cell toxicity, was performed. As shown in Figure 4B, LDH release in RAW264.7 cells was minimal in the vehicle-treated control group and a dramatic increase was observed after 12 h exposure to 500 μM of H2O2 Contrary to this, pre-treatment with CNP for 24 h at concentrations above 50 μg/mL attenuated, markedly, the H2O2-induced increase in LDH release as well as CS (100, 200 μg/mL), while at the same concentration, CNP significantly decreased LDH release as compared to CS (P < 0.05). The results was consistent with that determined by MTT assay.
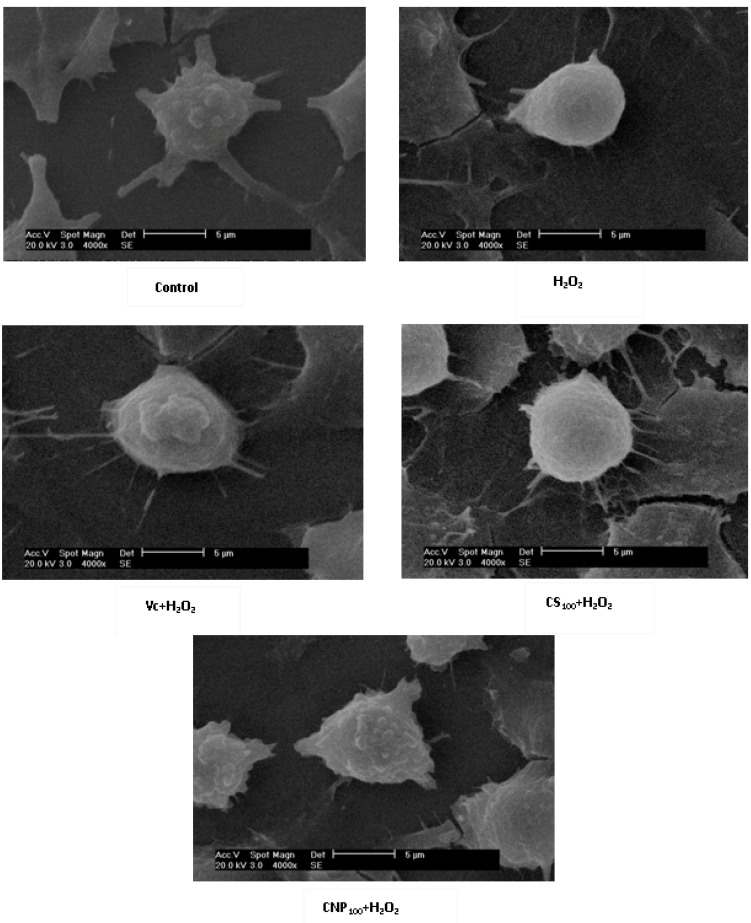
2.4. Effects of CNP on Morphology of RAW264.7 Cells
As shown in Figure 5, RAW264.7 cells treated with CNP (100 μg/mL), Vitamin C, and the vehicle-treated control group exhibited a typical macrophage-like morphology, indicated by the presence of surface ruffles and numerous extending pseudopodia, however, RAW264.7 cells with 500 μM of H2O2 for 12 h were observed with morphological differences that were indicated by the presence of surface smooth, shrink, and a small number of pseudopodia compared to the vehicle-treated control group.
Figure 5.
Effects of CNP on morphology of RAW264.7 cells observed with Scanning Electron Microscope (SEM) (4000×).
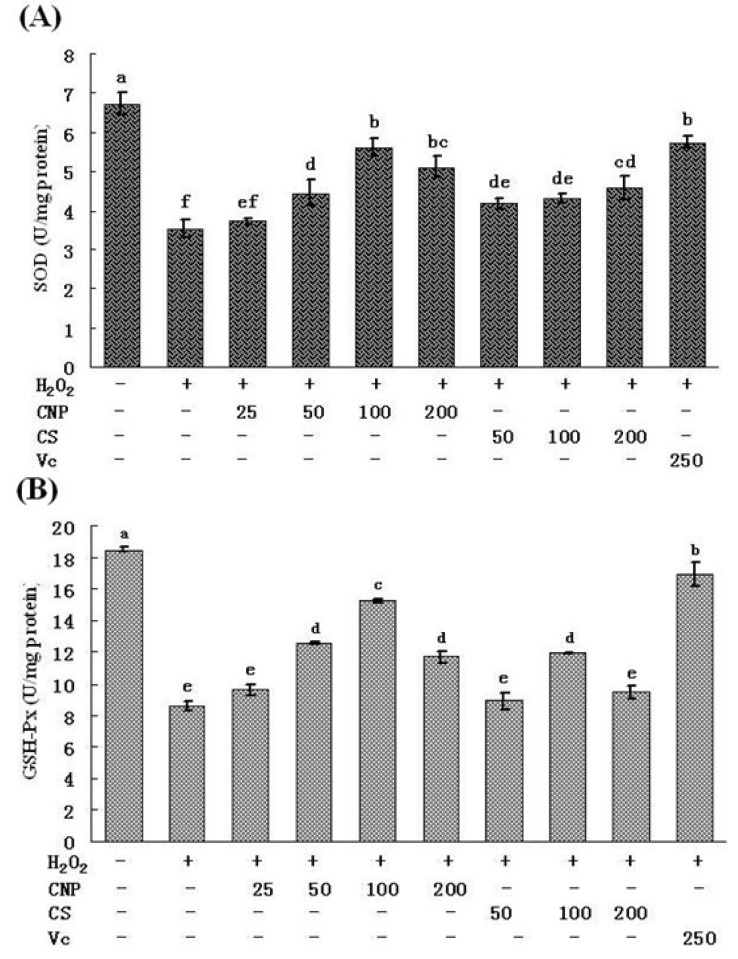
2.5. Measurement of SOD, GSH-Px, GSH, and T-AOC Activities as Well as MnSOD and GSH-Px mRNA Expression Levels
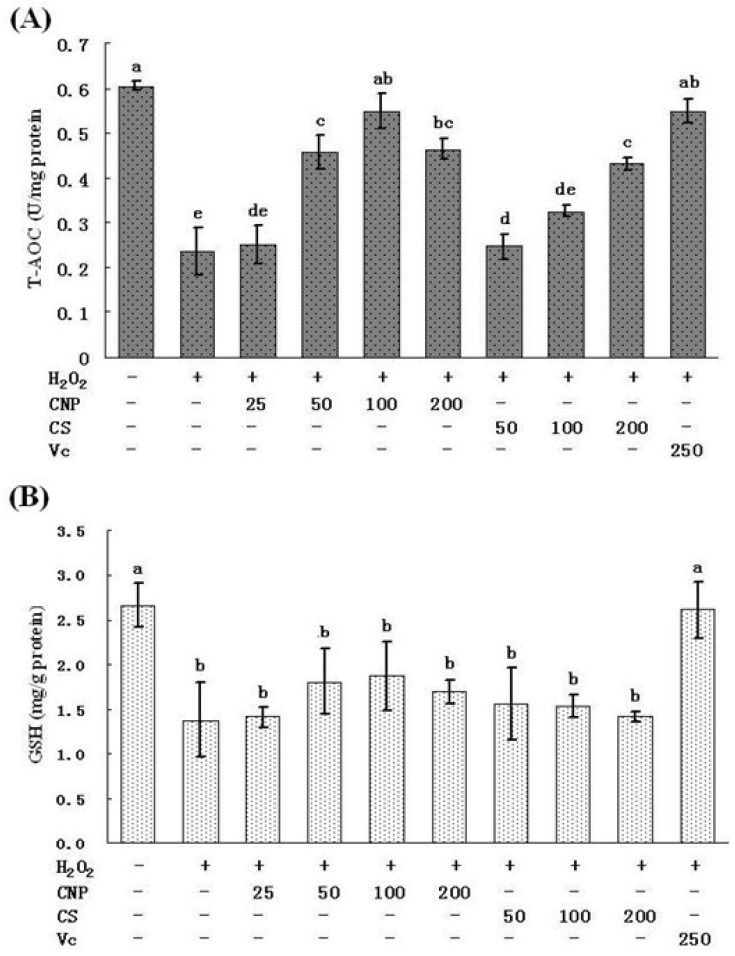
Treatment of RAW264.7 cells with 500 μM of H2O2 for 12 h caused the decrease in the activities of SOD and GSH-Px, respectively (P < 0.05). However, pre-incubation with CNP (50–200 μg/mL) significantly attenuated the changes of SOD and GSH-Px activities (Figure 6A,B). At 100 μg/mL of CNP, the H2O2-induced decrease in SOD and GSH-Px activities were restored respectively, which were the highest among the concentration of CNP and close to that recovered by Vitamin C at 250 μg/mL. CNP and CS did not affect (P > 0.05) the concentration of GSH (Figure 7B) in RAW264.7 cells with 500 μM of H2O2 for 12 h. However, CNP and CS increased significantly the activity of T-AOC (P < 0.05) in a dose-dependent manner as compared to the H2O2 alone control group (Figure 7A). At the same concentration, CNP can more effectively increase SOD, GSH-Px, and T-AOC activities, as compared to CS (P < 0.05).
Figure 6.
Effects of CNP and antioxidant Vitamin C (Vc) on the production of SOD (A), GSH-Px (B) in RAW264.7 cells. Values are presented as means ± SD. Bars with different letters are statistically different (P < 0.05).
Figure 7.
Effects of CNP and antioxidant Vitamin C (Vc) on the production of T-AOC (A), GSH (B) in RAW264.7 cells. Values are presented as means ± SD. Bars with different letters are statistically different (P < 0.05).
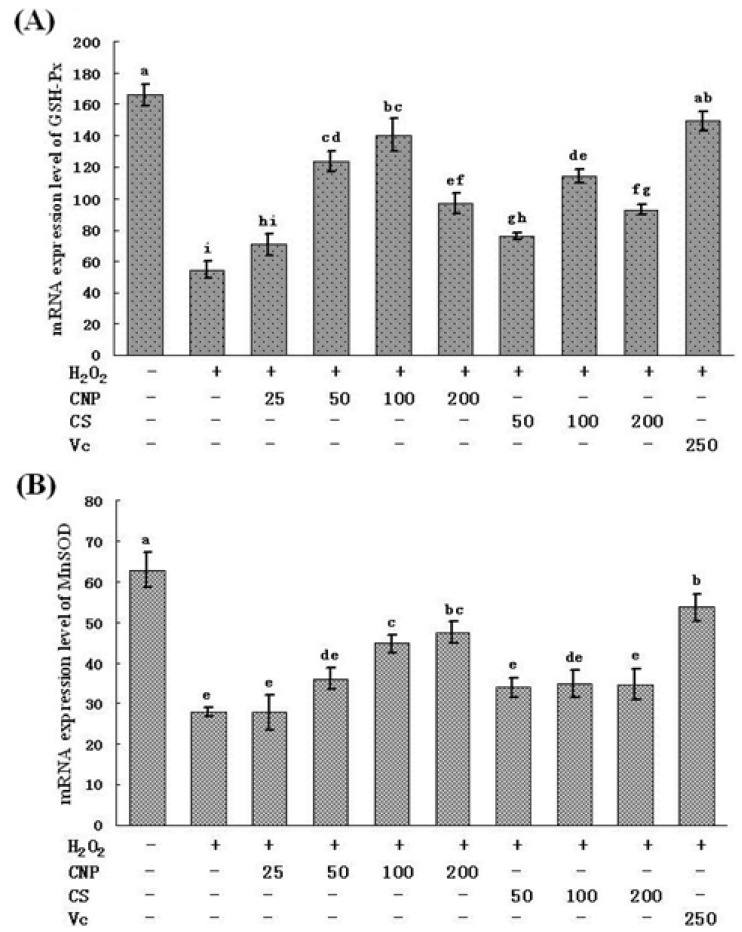
In parallel, as shown in Figure 8, treatment of RAW264.7 cells with 500 μM of H2O2 for 12 h caused the decrease in the mRNA expression of MnSOD and GSH-Px. Moreover, after RAW264.7 cells with pre-incubated with CNP (50–200 μg/mL), CS (50–100 μg/mL), or Vitamin C (250 μg/mL) for 24 h, the mRNA expression of SOD and GSH-Px were augmented consistently with intracellular SOD and GSH-Px levels.
Figure 8.
mRNA expression of GSH-Px and MnSOD. Values are presented as means ± SD. Bars with different letters are statistically different (P < 0.05).
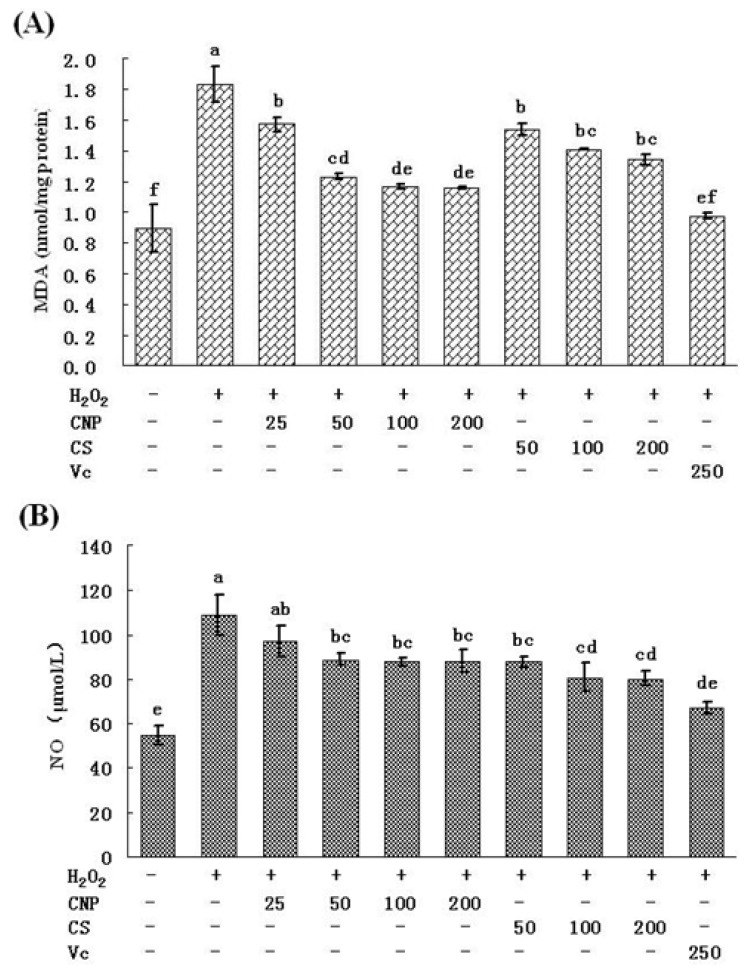
2.6. Detection of Nitric Oxide (NO) Release in Cell Culture Medium as Well as MDA Content
RAW264.7 cells treated with 500 μM of H2O2 for 12 h caused an marked increase in the intracellular MDA level (P < 0.05) compared with others, while pre-incubation of cells with CNP (25–200 μg/mL), CS (50–200 μg/mL), or Vitamin C (250 μg/mL) significantly decreased the intracellular MDA level (P < 0.05) (Figure 9A), at concentrations (50–200 μg/mL), CNP can more effectively decrease the intracellular MDA level as compared to CS (P < 0.05).
Figure 9.
Effects of CNP and antioxidant Vitamin C (Vc) on the production of MDA (A), NO (B) in RAW264.7 cells. Values are presented as means ± SD. Bars with different letters are statistically different (P < 0.05).
In addition, the intracellular NO content in RAW264.7 cells was also assayed according to protocol of commercial kits. As shown in Figure 9B, NO content was increased significantly at the vehicle-treated control group after treatment with 500 μM of H2O2 for 12 h. In addition pre-incubation with CNP (50–200 μg/mL), CS (50–100 μg/mL), or Vitamin C (250 μg/mL), before H2O2 exposure, significantly decreased the NO level as compared to H2O2 treatment alone (P < 0.05).
3. Discussion
Oxidative stress has been implicated in the pathogenesis of many disease states, such as aging, atherosclerosis, carcinogenesis, ischemia-reperfusing tissue injury, and acute and chronic inflammatory disorders [17,18]. Oxidative stress can be defined as the imbalance between cellular oxidant species production and antioxidant capability. Toxic reactive oxygen species (ROS), including the superoxide and hydrogen peroxide (H2O2), generated from normal cellular respiration by aerobic metabolism or exogenous oxidants, can cause cellular damage by oxidizing nucleic acids, proteins, and membrane lipids [18]. In biological systems, equilibrium between oxidant formation and endogenous antioxidant defense mechanisms exists to protect cellular biomolecules against oxidation by reactive oxygen species (ROS). ROS are generated excessively in oxidative stress and cause cell or tissue injury, leading to cell death, if this balance is disturbed [19]. Furthermore, ROS have direct or indirect relationships with oxidative process of cellular components and play an important role in various diseases such as cancer, arthritis, inflammation, Alzheimer’s, hypertension, diabetes, and aging [20,21,22].
In this study, we used CNP to pretreat cells before hydrogen peroxide treatment, and cells were washed by PBS, then hydrogen peroxide was used to induce oxidative injury, the uptake of chitosan nanoparticles and hydrogen peroxide were not simultaneous, thus, chitosan nanoparticles did not affect the uptake of hydrogen peroxide. Therefore, we think attenuation of oxidative injury did not depend on inhibition of the uptake of hydrogen peroxide on the cell membrane, maybe due to nanoparticle enhancing efficiency of cellular internalization of chitosan [23]. In addition, there are the previous studies showing that chitosan reduced the oxidative stress in vivo and in vitro [24,25,26]. Macrophages participate in host defense and are main targets for action of pro-oxidants. H2O2 is commonly used on macrophage cells for investigation of apoptosis or oxidative stress-mediated cell injury [27,28]. We confirmed that 500 μM H2O2 and 12 h incubation is sufficient enough to test oxidant-induced cell injury. In this study, we aimed to investigate whether treatment of CNP or CS prior to acute oxidative stress (H2O2) in macrophage cell can afford to cytoprotection and, if so, if the protection effects of CNP or CS are due to their abilities to enhance cell antioxidant activities. Thus, we examined viability, morphology, the MnSOD, and GSH-Px expression, enzyme and non-enzyme antioxidant activity by CNP on RAW264.7 cells with 500 μM of H2O2 for 12 h. Current cell culture experiments demonstrated cell viability and morphology were protected significantly by the presence of CNP (50–200 μg/mL). Lactate dehydrogenase (LDH) is a crucial biomarker for assessing cell viability of proliferation [29]. Using LDH as a marker for cell viability and membrane interity, in keeping with viability (MTT assay) and morphology results, this study revealed that pre-treatment of RAW264.7 cells with CNP or CS (50–200 μg/mL) markedly decreased LDH release compared to H2O2 -induction alone. RAW264.7 cells treated with CNP (100 μg/mL), Vitamin C, and the vehicle-treated control group exhibited a typical macrophage-like morphology, indicated by the presence of surface ruffles and numerous extending pseudopodia, however, RAW264.7 cells with 500 μM of H2O2 for 12 h were observed morphological differences that indicated by the presence of surface smooth, shrink, and a small number of pseudopodia compared to the vehicle-treated control group. Based on viability and morphology, our findings suggest RAW264.7 cells treated with CNP can effectively attenuate H2O2-induced cell injury.
Lipid peroxidation is one of the primary events in free radical-mediated cell injury [30]. GSH-Px and SOD are the major antioxidant enzymes, which can effectively eliminate free radicals (FR), and have antioxidative stress function, and they not only prevent the damage of oxygen free radicals, but also have an intrinsic protective effect, therefore maintaining a healthy balance between oxidants and antioxidants [31]. MDA is the direct products of lipid peroxidation after unsaturated fatty acids attacked by radicals on cell membrane and is widely used as a biomarker of oxidative stress [32]. MDA content serves as an indicator of the extent of lipid peroxidation and an indirect reflection of the extent of cell damage. MDA may destruct cell membrane structure, cause DNA fragmentation, rearrangement, cross-linking, and accelerate apoptosis, and is one of the most important elements of phlegmnosis or tumorigenesis [33]. Therefore, determination of MDA, SOD, and GSH-Px can reflect the level of metabolism of oxygen free radicals. On the other hand, cells are often equipped with several antioxidants for the prevention of free-radical damage. SOD and GSH-Px, along with other enzymatic and non-enzymatic antioxidants, play pivotal roles in preventing cellular damage caused by ROS [34]. The MnSOD isoform (SOD2) is primarily located in mitochondria and is considered the primary isoform in relation to oxidative stress [35]. Therefore, the intracellular ROS can be effectively eliminated by the combined action of SOD, GSH-Px, and other endogenous antioxidants, which provide a repairing mechanism for oxidized membrane components. In the present study, significant decreases in SOD and GSH-Px were observed in RAW264.7 cells after exposure to H2O2, indicating the impairment in antioxidant defenses. Moreover, the mRNA expression of SOD and GSH-Px were found also marked decreases. In addition, an obvious elevation of MDA production was associated with an increase of LDH release. Nonetheless, when RAW264.7 cells were pre-incubated with CNP, these H2O2-induced cellular events were blocked to a great extent. Our studies also show a similar efficiency in protecting RAW264.7 cells against H2O2-induced oxidative injury between CNP (50–200 μg/mL) and Vitamin C (250 μg/mL). These results jointly suggest that enhancement of endogenous antioxidant preservation and attenuation of lipid peroxidation may represent a major mechanism of cellular protection by CNP.
NO is one of the most important mediators in the regulation of cell functions [36]. NO stimulates H2O2 production from the mitochondrial respiratory chain, and NO directly inhibits catalase [37], while peroxynitrite oxidizes glutathione and inactivates glutathione reductase [38]. Thus, NO can raise H2O2 levels in cells. H2O2 and NO can react on superoxide dismutase to produce peroxynitrite [39]. Thus, high NO levels in cell can, in the presence of oxygen, give rise to a potent mixture of ROS and reactive nitrogen species (RNS), and the most significant results of this are oxidation of protein cysteine residues, lipid oxidation, and DNA mutation [40]. In this study, we found CNP could significantly decrease NO level as compared to the H2O2 alone control group, the result suggest that CNP can attenuate RAW264.7 cells injury by H2O2-induction, which may be through decrease in NO content.
4. Experimental Section
4.1. Chemicals and Reagents
Dulbecco’s modified Eagle’s medium (DMEM), penicillin/streptomycin, and the other materials required for culture of cells were purchased from Gibco BRL, Life Technologies (Grand Island, NY, USA). H2O2, dimethylsulfox-ide (DMSO), 3-(4,5-dimethylthiazol-2-yl) 2,5-diphenyltetrazolium bromide (MTT), Vitamin C, and bovine serum albumin (BSA) were obtained from Sigma (St. Louis, MO, USA). The reagent kits for measurement of levels of LDH, NO, MDA, SOD, GSH-Px, GSH, and T-AOC were purchased from the Nanjing Institute of Jiancheng Bioengineering (Nanjing, China). Trizol was from Invitrogen (Carlsbad, CA, USA), revert Aid™ M-MuLV reverse transcriptase was from Fermentas (Amherst, NY, USA), diethylpyrocarbonate (DEPC) and ribonuclease inhibitor were from Biobasic, Canada, oligo (dT)18 were from Sangon, China. All other chemicals were of analytical grade or of the highest grade available commercially.
4.2. Cell Culture and Treatment
Mouse macrophages RAW 264.7 cell line was obtained from the Shanghai Institute of Cell Biology (Shanghai, China) and maintained in DMEM, supplemented with heat-inactivated 10% fetal bovine serum, 100 U/mL penicillin, 100 U/mL streptomycin in a humidified atmosphere of 5% CO2 at 37 °C. When the cells reached sub-confluence, they were pretreated for 24 h with culture medium containing different concentrations of CNP (25, 50, 100, and 200 μg/mL), CS (50, 100, and 200 μg/mL), or Vitamin C (250 μg/mL) that were tested in the experiments. Following this, the culture supernatant was collected and cells were washed three times with phosphate-buffered saline (PBS, pH 7.2). Subsequently, the cells were exposed to H2O2 (500 μM) diluted in culture medium for 12 h in a humidified atmosphere of 5% CO2 at 37 °C until further assay.
4.3. Preparation and Characterization of CNP
Chitosan (CS) was obtained from the Chitosan Company of Pan’an (Zhejiang, China). The degree of deacetylation was about 95%, as determined by elemental analysis, and the average molecular weight of the chitosan was 220 kDa, as determined by viscometric methods. Chitosan nanoparticles were prepared and characterized as described previously [14,16]. Briefly, Chitosan was dissolved at 0.5% (w/v) with 1% (v/v) acetic acid (HOAc) and then raised to pH 4.6–4.8 with 10 N NaOH. CNP were formed by coacervation between positively charged chitosan (0.5%, w/v) and negatively charged sodium tripolyphosphate (0.25%, w/v). Nanoparticles with different mean size were obtained by adjusting the volume ratio of chitosan to tripolyphosphate solution. Nanoparticles were purified by centrifugation at 9000× g for 30 min. Supernatants were discarded and chitosan nanoparticles were extensively rinsed with distilled water to remove any NaOH residue, and freeze dried before further use or analysis. The freeze-dried chitosan nanoparticles were suspended in water for characterization or use for other experiments. Particle size distribution and the zeta potential of chitosan nanoparticles were determined using Zetasizer Nano-ZS90 (Malvern, Worcestershire, UK). The analysis was performed at a scattering angle of 90° at a temperature of 25 °C using samples diluted to different intensity concentration with de-ionized distilled water. Atomic force microscopy (AFM, SPM-9500J3) was used for visualization of the chitosan nanoparticles deposited on silicon substrates operating in the contact mode. AFM imaging was performed using Si3N4 probes with a spring constant of 0.06 N/m.
4.4. Cell Viability Measurement
RAW 264.7 cells were seeded at density of 1 × 106 cells/mL in 6-well plates and the cell viability was measured using the MTT assay [41]. Briefly, at the indicated time after the treatment as before, the culture supernatant was removed. The cells were washed with PBS and incubated with MTT (5 mg/mL) in culture medium at 37 °C for another 4 h. After MTT removal, the colored formazan was dissolved in 150 μL of DMSO. The absorption values were measured at 490 nm using a SpectraMax M5 Microplate Reader (Molecular Devices, MDS Analytical Technologies, Sunnyvale, CA, USA). The viability of RAW 264.7 cells in each well was presented as percentage of control cells.
4.5. Morphology of RAW264.7 Cells
RAW264.7 cells (1 × 106 per coverslip) were seeded onto sterile coverslips and treated as the same as “4.2. Cell Culture and Treatment” in a humidified CO2 incubator at 37 °C. Following incubation, cells were prepared for scanning electron microscopy. Briefly, coverslip cultures were rinsed with PBS, fixed in 2.5% glutaraldehyde at 4 °C overnight, and post-fixed in 1% osmium tetroxide for 1 h. Coverslip cultures, then, were washed in 0.1 M sodium cacodylate, dehydrated in a graded series of ethanol 50%, 70%, 80%, and 95%, 3×, 10 min each; followed by 100%, 3×, 20 min each and immersed in hexamethyldisilazane (HMDS, 3×, 15 min each). Coverslip cultures were air-dried, mounted, and coated with gold. Cells were examined using a XL30-ESEM Scanning Electron Microscope operating at an accelerating voltage of 20 kV.
4.6. Preparation of Cell Lysates
The cells were seeded at a density of 1 × 106 cells/mL in 6-well plates. When the cells reached sub-confluence, they were pretreated for 24 h with culture medium containing different concentrations of CNP (25, 50, 100, and 200 μg/mL), CS (50, 100, and 200 μg/mL), or Vitamin C (250 μg/mL). Upon completion of the incubation studies, the culture supernatant was collected for analysis of LDH and NO release. The cells were scraped from the plates into ice-cold 1% Triton X-100 lysis buffer and protein concentration was determined by the bicinchoninic acid (BCA) method, using BSA as a reference standard. Aliquots were stored at −80 °C until detection for MDA, SOD, GSH-Px, GSH, and T-AOC.
4.7. Measurement of LDH and Nitric Oxide (NO) Release
LDH, an indicator of cell injury, was detected after the exposure to H2O2 with an assay kit according to the manufacturer’s protocol. The activity of enzyme was expressed as units per liter and the absorbance was read at 450 nm. The concentration of nitriles (NO2−) and nitrates (NO3−), stable end products of nitric oxide (NO), were determined by the reagent kits from the Nanjing Institute of Jiancheng Bioengineering (Nanjing, China). NO production was determined by measuring the optical density at 550 nm and expressed as units per liter.
4.8. Assay for Intracellular Contents of SOD, GSH-Px, and MDA
The activities of SOD and GSH-Px, as well as the concentration of MDA, were all determined by using commercially available kits. All procedures completely complied with the manufacturer’s instructions. The activities of enzymes were expressed as units per milligram protein. The assay of SOD activity was based on its ability to inhibit the oxidation of hydroxylamine by O2− produced from the xanthine-xanthineoxiase system. One unit of SOD activity was defined as the amount that reduced the absorbance at 450 nm by 50%. The assay of GSH-Px activity was assayed by quantifying the rate of oxidation of the reduced glutathione to the oxidized glutathione by H2O2 catalyzed by GSH-Px. One unit of GSH-Px was defined at 412 nm as the amount that reduced the level of GSH by 1 μM in 1 min/mg protein. MDA was measured at a wavelength of 532 nm by reacting with thiobarbituric acid (TBA) to form a stable chromophoric production. Values of MDA level were expressed as nanomoles per milligram protein.
4.9. Measurement of the mRNA Expression Levels of MnSOD and GSH-Px by Real-Time PCR
Cells were lysed in 0.8 mL of Trizol reagent (Invitrogen™, Carlsbad, CA, USA) and the total RNA was isolated according to the manufacture’s protocol. The concentration of total RNA was quantified by determining the optical density at 260 nm. The total RNA was used and reverse transcription was performed by mixing 2 µg of RNA with 0.5 µg oligo (dT)18 primer in a DEPC-treated tube. Nuclease-free water was added giving a final volume of 12.5 µL. This mixture was incubated at 70 °C for 5 min and chilled on ice for 2 min. Following this, a solution containing 4 µL of M-MuLV 5× reaction buffer, 2 µL of 10 mM dNTP, 20 U of ribonuclease inhibitor, and DEPC-treated water was added, giving a final volume of 19 μL, and the tubes were incubated for 5 min at 37 °C. The tubes then received 200 U of M-MuLV reverse transcriptase and were incubated for 60 min at 42 °C. Finally, the reaction was stopped by heating at 70 °C for 10 min. The samples were stored at −20 °C until further use.
As shown in Table 1, the primers were used to amplify cDNA fragments (238-bp GSH-Px fragment, 79-bp MnSOD fragment, and 94-bp 18S fragment). Amplification was carried out in total volume of 20 µL containing 1 µL (5 µM) of each target and 18S specific primers, 2 µL of cDNA template, 2 µL of 10× PCR buffer, 2 μL of MgCl2 (25 mM), 2 μL of dNTPs (2.5 mM), 2 μL of transcribed cDNA, and 0.4 μL of Taq DNA polymerase. Reaction conditions were the standard conditions for the iQTM5 PCR (Bio-Rad, Hercules, CA, USA) (10 s denaturation at 95 °C, 25 s annealing at 60 °C (GSH-Px, MnSOD), 62 °C (18S)) with 45 PCR cycles. Ct values were obtained automatically using software (Bio-Rad, USA). The comparative Ct method (2−ΔΔCt method) [42] was used to analyze the expression levels of MnSOD and GSH-Px.
Table 1.
Real-Time PCR Primers and Conditions.
| Gene | Genbank Accession | Primer sequence | Product size (bp) | Annealing (°C) |
|---|---|---|---|---|
| GSH-Px | NM_008160 | 5′ ACAGTCCACCGTGTATGCCTTC 3′ | 238 | 60 |
| 5′ CTCTTCATTCTTGCCATTCTCCTG 3′ | ||||
| MnSOD | X04972 | 5′ CTGTGGGAGTCCAAGGTTCA 3′ | 79 | 60 |
| 5′ GAGCAGGCAGCAATCTGTAAG 3′ | ||||
| 18S | NR_003278 | 5′ CGGACACGGACAGGATTGACA 3′ | 94 | 62 |
| 5′ CCAGACAAATCGCTCCACCAACTA 3′ |
4.10. Effect of CNP on the Antioxidant Capacity of H2O2-Induced RAW264.7 Cells
Levels of GSH were determined colorimetrically at 405 nm with the spectrophotometer following reaction with 5,5-dithiobes (2-ni-trobenzoic acid) (DTNB), and was expressed as mg gpro−1. T-AOC was measured according to protocol of commercial kits. In the reaction mixture ferric ion was reduced by antioxidant reducing agents and to form the complex ferrous-tripyridyltriazine, which can be estimated by colorimetric assay to reflect T-AOC, which was expressed as U mgpro−1.
4.11. Statistical Analysis
Data were expressed as mean ± standard deviations (S.D.) and examined for their statistical significance of difference with ANOVA and a Tukey post hoc test by using SPSS 16.0. P-values of less than 0.05 were considered statistically significant.
5. Conclusions
The present studies clearly show that chitosan nanoparticles (CNP) can attenuate H2O2-induced stress injury in RAW264.7 cells, and the unique character of nanoparticles could make CNP exhibit more superior antioxidative activities than chitosan (CS). We explored the protective mechanism of action of CNP against H2O2-induced RAW264.7 cell injury, which are partly contributed to restoring the activities endogenous antioxidants, along with enhancement of their gene expression. These data propose a possibility that CNP can be used as a potent natural antioxidant in the treatment of oxidative-related diseases. Since oxidative stress-induced RAW264.7 cell injury plays a key role in inflammation disease, our findings suggest a novel application for CNP in the treatment of chronic inflammation.
Acknowledgments
This work was supported by grant from Science and Technology Project of Zhoushan Municipal Bureau of China (No. 2012C33004 and 2011C22044), Zhejiang Provincial Natural Science Foundation of China (No. LQ13C200002 and Y2111223) and the National Natural Science Foundation of China (NSFC, No. 81273429 and 31301982).
Conflict of Interest
The authors declare that there are no conflicts of interest.
References
- 1.Butterfield D.A., Abdul H.M., Opii W., Newman S.F., Joshi G., Ansari M.A., Sultana R. Pin1 in Alzheimer’s disease. J. Neurochem. 2006;98:1697–1706. doi: 10.1111/j.1471-4159.2006.03995.x. [DOI] [PubMed] [Google Scholar]
- 2.Dhalla N.S., Temsah R.M., Netticadan T. Role of oxidative stress in cardiovascular diseases. J. Hypertens. 2000;18:655–673. doi: 10.1097/00004872-200018060-00002. [DOI] [PubMed] [Google Scholar]
- 3.Seven A., Guzel S., Aslan M., Hamuryudan V. Lipid, protein, DNA oxidation and antioxidant status in rheumatoid arthritis. Clin. Biochem. 2008;41:538–543. doi: 10.1016/j.clinbiochem.2008.01.029. [DOI] [PubMed] [Google Scholar]
- 4.Suzuki K., Mikami T., Okawa Y., Tokoro T., Suzuki S., Suzuki M. Antitumor effect of hexa-N-acetylchitohexaose and chitohexaose. Carbohyd. Polym. 1986;151:403–408. doi: 10.1016/s0008-6215(00)90359-8. [DOI] [PubMed] [Google Scholar]
- 5.Jeon Y.J., Kim S.K. Potential immuno-stimulating effect of antitumoral fraction of chitosan oligosaccharides. J. Chtin Chitosan. 2001;6:163–167. [Google Scholar]
- 6.Schipper N.G., Varum M.K.M., Stenberg P., Cklind G.O., Lennernas H., Artursson P. Chitosans as absorption enhancers of poorly absorbable drugs: 3. Influence of mucus on absorption enhancement. Eur. J. Pharm. Sci. 1999;8:335–343. doi: 10.1016/S0928-0987(99)00032-9. [DOI] [PubMed] [Google Scholar]
- 7.Park P.J., Je J.Y., Byun H.G., Moon S.H., Kim S.K. Antimicrobial activity of hetero-chitosans and their oligosaccharides with different molecular weights. J. Mol. Microb. Biotechnol. 2004;14:317–323. [Google Scholar]
- 8.Porporatto C., Bianco I.D., Riera C.M., Correa S.G. Chitosan induces different l-arginine metabolic pathways in resting and inflammatory macrophages. Biochem. Biophys. Res. Commun. 2003;304:266–272. doi: 10.1016/S0006-291X(03)00579-5. [DOI] [PubMed] [Google Scholar]
- 9.Anraku M., Kabashima M., Namura H., Maruyama T., Otagiri M., Gebicki J.M., Furutani N., Tomida H. Antioxidant protection of human serum albumin by chitosan. Int. J. Biol. Macromol. 2008;43:159–164. doi: 10.1016/j.ijbiomac.2008.04.006. [DOI] [PubMed] [Google Scholar]
- 10.Xie W., Xu P., Liu Q. Antioxidant activity of watersoluble chitosan derivatives. Bioorg. Med. Chem. Lett. 2001;11:1699–1701. doi: 10.1016/S0960-894X(01)00285-2. [DOI] [PubMed] [Google Scholar]
- 11.Santhosh S., Sini T.K., Anandan R., Mathew P.T. Effect of chitosan supplementation on antitubercular drugs-induced hepatotoxicity in rats. Toxicology. 2006;219:53–59. doi: 10.1016/j.tox.2005.11.001. [DOI] [PubMed] [Google Scholar]
- 12.Yoon H.J., Moon M.E., Park H.S., Kim H.W., Im S.Y., Lee J.H., Kim Y.H. Effects of chitosan oligosaccharide (COS) on the glycerol-induced acute renal failure in vitro and in vivo. Food Chem. Toxicol. 2008;46:710–716. doi: 10.1016/j.fct.2007.09.111. [DOI] [PubMed] [Google Scholar]
- 13.Liu J.N. Ph.D. Thesis. Jiangnan University; Wuxi, China: 2008. Study on the hypolipidemic mechanism of chitosan. [Google Scholar]
- 14.Wen Z.S., Xu Y.L., Zou X.T., Xu Z.R. Chitosan nanoparticles act as an adjuvant to promote both Th1 and Th2 immune responses induced by ovalbumin in mice. Mar. Drugs. 2011;9:1038–1055. doi: 10.3390/md9061038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qi L., Xu Z., Chen M. In vitro and in vivo suppression of hepatocellular carcinoma growth by chitosan nanoparticles. Eur. J. Cancer. 2007;43:184–193. doi: 10.1016/j.ejca.2006.08.029. [DOI] [PubMed] [Google Scholar]
- 16.Qi L., Xu Z., Jiang X., Hu C., Zou X. Preparation and antibacterial activity of chitosan nanoparticles. Carbohyd. Res. 2004;339:2693–2700. doi: 10.1016/j.carres.2004.09.007. [DOI] [PubMed] [Google Scholar]
- 17.Droge W. Free radicals in the physiological control of cell function. Physiol. Rev. 2002;82:47–95. doi: 10.1152/physrev.00018.2001. [DOI] [PubMed] [Google Scholar]
- 18.Griendling K.K., Sorescu D., Lassegue B., Ushino-Fukai M. Modulation of protein kinase activity and gene expression by reactive oxygen species and their role in vascular physiology and pathophysiology. Arterioscl. Throm. Vas. 2000;20:2175–2183. doi: 10.1161/01.ATV.20.10.2175. [DOI] [PubMed] [Google Scholar]
- 19.Kang K.B., Lee K.H., Chae S.W., Zhang R., Jung M.S., Lee Y.K., Kim S.Y., Kim H.S., Joo H.G., Park J.W., et al. Eckol isolated from Ecklonia cava attenuates oxidative stress induced cell damage in lung fibroblast cells. FEBS Lett. 2005;579:6295–6304. doi: 10.1016/j.febslet.2005.10.008. [DOI] [PubMed] [Google Scholar]
- 20.Calabrese V., Lodi R., Tonon C., D’Agata V., Sapienza M., Scapagnini G., Mangiameli A., Pennisi G., Giuffrida Stella A.M., Butterfield D.A. Oxidative stress, mitochondrial dysfunction and cellular stress response in Friedreich’s ataxia. J. Neurol. Sci. 2005;233:145–162. doi: 10.1016/j.jns.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 21.Je J.Y., Park P.J., Kim S.K. Free radical scavenging properties of hetero-chitooligosaccharides using an ESR spectroscopy. Food Chem. Toxicol. 2004;42:381–387. doi: 10.1016/j.fct.2003.10.001. [DOI] [PubMed] [Google Scholar]
- 22.Vajragupta O., Boonchoong P., Wongkrajang Y. Comparative quantitative structure activity study of radical scavengers. Bioorgan. Med. Chem. 2000;8:2617–2628. doi: 10.1016/S0968-0896(00)00195-4. [DOI] [PubMed] [Google Scholar]
- 23.Huang M., Khor E., Lim L.Y. Uptake and cytotoxicity of chitosan molecules and nanoparticles: effects of molecular weight and degree of deacetylation. Pharm. Res. 2004;21:344–353. doi: 10.1023/B:PHAM.0000016249.52831.a5. [DOI] [PubMed] [Google Scholar]
- 24.Anraku M., Fujii T., Furutani N., Kadowaki D., Maruyama T., Otagiri M., Gebicki J.M., Tomida H. Antioxidant effects of a dietary supplement: Reduction of indices of oxidative stress in normal subjects by water-soluble chitosan. Food Chem. Toxicol. 2009;47:104–109. doi: 10.1016/j.fct.2008.10.015. [DOI] [PubMed] [Google Scholar]
- 25.Khodagholi F., Eftekharzadeh B., Maghsoudi N., Rezaei P.F. Chitosan prevents oxidative stress-induced amyloid β formation and cytotoxicity in NT2 neurons: involvement of transcription factors Nrf2 and NF-κB. Mol. Cell. Biochem. 2010;337:39–51. doi: 10.1007/s11010-009-0284-1. [DOI] [PubMed] [Google Scholar]
- 26.Anraku M., Fujii T., Kondo Y., Kojima E., Hata T., Tabuchi N., Tsuchiya D., Goromaru T., Tsutsumi H., Kadowaki D., et al. Antioxidant properties of high molecular weight dietary chitosan in vitro and in vivo. Carbohyd. Polym. 2011;83:501–505. doi: 10.1016/j.carbpol.2010.08.009. [DOI] [Google Scholar]
- 27.Maeng O., Kim Y.C., Shin H.I., Lee J.O., Huh T.L., Kang K.I., Kim Y.S., Paik S.G., Lee H. Cytosolic NADP(+)-dependent isocitrate dehydrogenase protects macrophages from LPS-induced nitric oxide and reactive oxygen species. Biochem. Biophys. Res. Commun. 2004;317:558–564. doi: 10.1016/j.bbrc.2004.03.075. [DOI] [PubMed] [Google Scholar]
- 28.Thoeni G., Werner E.R., Werner-Felmayer G. Tetrahydropteridines suppress gene expression and induce apoptosis of activated RAW264.7 cells via formation of hydrogen peroxide. Free Radical Biol. Med. 2004;37:375–385. doi: 10.1016/j.freeradbiomed.2004.05.010. [DOI] [PubMed] [Google Scholar]
- 29.Gupta A., Singh S., Jamal F., Nath S., Mehrotra S., Sharma B. Synergistic effects of glutathione and Vitamin E on ROS mediated ethanol toxicity in isolated rat hepatocytes. Asian J. Biochem. 2011;6:347–356. doi: 10.3923/ajb.2011.347.356. [DOI] [Google Scholar]
- 30.Shi G.F., An L.J., Jiang B., Guan S., Bao Y.M. Alpinia protocatechuic acid protects against oxidative damage in vitro and reduces oxidative stress in vivo. Neurosci. Lett. 2006;403:206–210. doi: 10.1016/j.neulet.2006.02.057. [DOI] [PubMed] [Google Scholar]
- 31.Aguilar A., Alvarez-Vijande R., Capdevila S., Alcoberro J., Alcaraz A. Antioxidant patterns (superoxide dismutase, glutathione reductase and glutathione peroxidase) in kidneys from non-heart-beating-donors: experimental study. Transplant Proc. 2007;39:249–252. doi: 10.1016/j.transproceed.2006.10.212. [DOI] [PubMed] [Google Scholar]
- 32.Cini M., Fariello R.G., Bianchetti A., Moretti A. Studies on lipid peroxidation in the rat brain. Neurochem. Res. 1994;19:283–288. doi: 10.1007/BF00971576. [DOI] [PubMed] [Google Scholar]
- 33.Surapaneni K.M., Venkataramana G. Status of lipid peroxidation, glutathione, ascorbic acid, vitamin E and antioxidant enzymes in patients with osteoarthritis. Indian J. Med. Sci. 2007;61:9–14. doi: 10.4103/0019-5359.29592. [DOI] [PubMed] [Google Scholar]
- 34.Luo T., Xia Z. A small dose of hydrogen peroxide enhances tumor necrosis factor- alpha toxicity in inducing human vascular endothelial cell apoptosis: reversal with propofol. Anesth. Analg. 2006;103:110–116. doi: 10.1213/01.ane.0000221183.02244.80. [DOI] [PubMed] [Google Scholar]
- 35.Valdivia A., Pérez-Álvarez S., Aroca-Aguilar J.D., Ikuta I., Jordán J. Superoxide dismutases: a physiopharmacological update. J. Physiol. Biochem. 2009;65:195–208. doi: 10.1007/BF03179070. [DOI] [PubMed] [Google Scholar]
- 36.Brown G.C. Nitric oxide and neuronal death. Nitric Oxide. 2010;23:153–165. doi: 10.1016/j.niox.2010.06.001. [DOI] [PubMed] [Google Scholar]
- 37.Brown G.C. Reversible binding and inhibition of catalase by nitric oxide. Eur. J. Biochem. 1995;232:188–191. doi: 10.1111/j.1432-1033.1995.tb20798.x. [DOI] [PubMed] [Google Scholar]
- 38.Savvides S.N., Scheiwein M., Bohme C.C., Arteel G.E., Karplus P.A., Becker K., Schirmer R.H. Crystal structure of the antioxidant enzyme glutathione reductase inactivated by peroxynitrite. J. Biol. Chem. 2002;277:2779–2784. doi: 10.1074/jbc.M108190200. [DOI] [PubMed] [Google Scholar]
- 39.McBride A.G., Borutaite V., Brown G.C. Superoxide dismutase and hydrogen peroxide cause rapid nitric oxide breakdown, peroxynitrite production and subsequent cell death. Biochim. Biophys. Acta. 1999;1454:275–288. doi: 10.1016/s0925-4439(99)00046-0. [DOI] [PubMed] [Google Scholar]
- 40.Thomas D.D., Ridnour L.A., Espey M.G., Donzelli S., Ambs S., Hussain S.P., Harris C.C., DeGraff W., Roberts D.D., Mitchell J.B., et al. Superoxide fluxes limit nitric oxide-induced signaling. J. Biol. Chem. 2006;281:25984–25993. doi: 10.1074/jbc.M602242200. [DOI] [PubMed] [Google Scholar]
- 41.Mosmann T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J. Immunol. Methods. 1983;65:55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
- 42.Livak K.J., Schmittgen T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCt Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]