Abstract
Introduction
Thoracic malignancies and human breast cancer (HBC) continue to be aggressive solid tumors that are poor responders to the existing conventional standard chemotherapeutic approaches. Malignant pleural mesothelioma (MPM) is an asbestos-related tumor of the thoracic pleura that lacks effective treatment options. Altered ubiquitin proteasome pathway is frequently encountered in many malignancies including HBC and MPM and thus serves as an important target for therapeutic intervention strategies. Although proteasome inhibitor Velcade (Bort-ezomib) has been under clinical investigation for a number of cancers, limited preclinical studies with this agent have thus far been conducted in HBC and MPM malignancies.
Purpose
To study the biological and molecular responses of MPM and HBC cells to Velcade treatments, and to identify mechanisms involved in transducing growth inhibitory effects of this agent.
Methods
Flow-cytometric analyses coupled with western immunoblotting and gene-array methodologies were utilized to determine mechanisms of Velcade-dependent growth suppression of five MPM (H2595, H2373, H2452, H2461, and H2714) and two breast cancer (MDA MB-468, SKBR-3) cell lines.
Results
Our data revealed significant reduction in cell growth properties that were dose and time dependent. Velcade treatment resulted in G2M phase arrest, increased expression of cyclin-dependent kinase inhibitor p21 and pro-apoptotic protein Bax. Pretreatment of mesothelioma cells with Velcade showed synergistic effect with cisplatin combination regimens. High-throughput gene expression profiling among Velcade treated and untreated mesothelioma cell lines resulted in identification of novel transducers of apoptosis such as CARP-1, XAF1, and Troy proteins.
Conclusions
Velcade targets cell cycle and apoptosis signaling to suppress MPM and HBC growth in part by activating novel transducers of apoptosis. This pilot study has paved way for further in-depth analysis of the downstream target molecules associated with presensitization of mesothelioma cells in finding effective therapeutic treatment options for both mesothelioma and recalcitrant breast cancers.
Keywords: Malignant pleural mesothelioma, Bortezomib (Velcade), Apoptosis, Gene expression
Introduction
Malignant pleural mesothelioma (MPM) is an aggressive pulmonary cancer, which, if untreated, has a median survival of less than 8 months [1]. Its incidence is still increasing with about 3,000 cases per year in the United States and 250,000 deaths expected in Western Europe in the next 30 years [2]. MPM is resistant to various treatment options available at present [3]. Most chemotherapeutic agents are not very effective against MPM, with typical single-agent response rates of ≤ 20% [4–6]. Therefore, the new therapies for MPM are desperately needed that may offer hope for improved palliation, prolonged survival and even potential cure for MPM.
The ubiquitin proteasome pathway is essential for many fundamental cellular processes, including the cell cycle, apoptosis, angiogenesis, and differentiation [7]. The proteasome plays a central role in this process. It has been identified as an excellent target for cancer therapy because of its critical metabolic function [8]. Velcade (already know as PS-341, Bortezomib) is the first proteasome inhibitor to have progressed to clinical trails [9]. It was approved for the treatment of patients with relapsed or refractory multiple-myeloma in May 2003 by the US Food and Drug Administration [10], and is currently being evaluated alone and in combination with other chemotherapy in a number of solid tumors, such as lung cancer [11].
In this study, we investigated effects of Velcade on in vitro growth of MPM cells derived from untreated patients and established breast cancer cell lines. Consistent with previous observations, Velcade treatments resulted in G2M phase arrest as well as elevated apoptosis, and a combination of Velcade and Cisplatin demonstrated superior efficacy in inhibiting MPM cell growth in vitro. Although, Velcade caused increased expression of cyclin-dependent kinase inhibitor p21 and proapoptotic proteins such as Bax, our gene-array-based studies revealed that Velcade targeted novel transducers of apoptosis such as CARP-1/CCAR1, XIAP-associated factor 1 (XAF1) and Troy (TNFRSF-19) proteins [12–14]. These studies are expected to facilitate exploitation of ubiquitin proteasome pathway to inhibit MPM and the development of more effective therapeutic strategies for this disease.
Materials and methods
Cells and reagents
Five MPM patient derived cell lines [H2373, H2452, H2595, H2461 and H2714] established in our laboratory and characterized in detail [15] were cultured in RPMI 1640 (Mediatech Inc., Herndon, VA) supplemented with 100 units/ml of penicillin, 100 µg/ml streptomycin, 4 mM L-glutamine, and 10% fetal calf serum. The human breast cancer (HBC) cells (MDA-MB-468 and SKBR-3) were obtained from ATCC (Manassas, VA) and cultured according to ATCC guidelines. Cells were incubated at 37°C in a humidified atmosphere of 5% CO2 in air and were passaged weekly. Bortezomib (Velcade) was obtained from Millennium (Cambridge, MA, USA). Anti-PARP antibodies were purchased from Biomed, while anti-XAF1 and anti-Troy antibodies were obtained from Abcam Inc., (Cambridge, MA) and Alexis Biochemicals (San Diego, CA, USA), respectively. Anti-Bax, anti-caspase-3, anti-p21, anti-p27 antibodies were purchased from Cell Signaling Technology (Beverly, MA, USA), anti-cyclin B were obtained from Santa Cruz Biotechnology Inc (Santa Cruz, CA, USA), and anti-actin were from Sigma Biologicals, St. Louis, MO. Generation and characterization of the anti-CARP-1/ CCAR1 rabbit polyclonal antibodies have been described before [12].
Cell growth inhibition studies by MTT assay
MPM (H2373 and H2595) or HBC (MDA-MB-468 and SKBR-3) cells (5 × 103) were seeded in a 96-well culture plate and subsequently were treated with indicated agents for 48 h. Control cells were treated with 0.1% dimethyl sulfoxide (DMSO) in culture medium. After treatment, the cells were incubated with MTT reagent (1 mg/mL; Sigma Chemical Company) at 37°C for 4 h and then MTT was removed and 100 µL of DMSO was added, followed by colorimetric analysis using a multilabel plate reader at 560 nm (Victor3; PerkinElmer (Wellesley, MA, USA)). Results were plotted as the mean from triplicate experiments.
Proteasomal chymotrypsin-like activity assay in cell extracts
Whole cell extracts (10 µg) derived from untreated or cells treated were incubated for 60 min at 37°C in 100 µL of assay buffer (50 mM Tris-HCl, pH 7.5) with 40 µM of fluo-rogenic peptide substrate Suc-Leu-Leu-Val-Tyr-AMC (for the proteasomal chymotrypsin-like activity). After incubation, production of hydrolyzed AMC groups was measured using a Wallac Victor3™ multilabel counter with an excitation filter of 355 nm and an emission filter of 460 nm.
Caspase-3 activity assay
Cell-free caspase-3 activities were determined by measuring the release of the AMC groups from a caspase-3 specific substrate Ac-Asp-Glu-Val-Asp-AMC. Briefly, H2373 and H2595 cells were treated under various conditions, followed by preparation of whole cell extracts. The cell extract (30 µg) was then incubated in 100 µl of the assay buffer (50 mM Tris-HCl, pH 7.5) along with 40 mM of caspase-3 substrate in a 96-well plate. The reaction mixture was incubated at 37°C for 3 h and the hydrolyzed fluorescent AMC groups were quantified as described above.
Apoptosis assay
Apoptosis levels were determined by utilizing DNA fragmentation-based ELISA kit (Roche Diagnostics, Indianapolis, IN, USA) essentially following manufacturer suggested protocols [12]. For apoptosis assay, 4–5 × 102 cells were seeded in 96-well plates and treated essentially as indicated in the MTT assay above. Untreated as well as treated cells were lysed, and levels of mono and oligo-nucleosomal DNA fragments in the lysates were determined by measuring optical density of each sample at 405 and 495 nm wavelengths. The “enrichment factor” indicating level of apoptosis was calculated essentially by the manufacturer suggested formula.
Western blot analysis
Cells were treated with various agents for indicated times, and cell lysates were prepared by allowing cells to incubate on ice for 30 min in 400 µl of lysis buffer (1% Nonidet P-40, 150 mM NaCl, 20 mM Tris, pH 8.0, 0.5 mM phenyl-metylsulfonylfluoride (PMSF), 1 µg/ml each: pepstatin, aprotinin and leupeptin). The lysates were centrifuged at 14,000 rpm at 4°C for 15 min to remove debris. Supernatant proteins, 50 µg from each sample were separated by SDS-12% polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to polyvinylidene difluoride (PVDF) membrane (Bio-rad, Hercules, CA, USA) by standard procedures. Proteins were visualized by treatment with the chemiluminescence detection reagent (Pierce) according to manufacturer’s instructions, followed by exposure to film (Kodak X-Oat). The same membrane was reprobed with the anti-β action antibody, which was used as an internal control for protein loading.
Flow cytometry and cell cycle analysis
The cell cycle was analyzed by flow cytometry. Briefly, 1 × 106 cells were harvested and washed in PBS, then fixed in 70% alcohol for 30 min at 4°C. After washing in cold PBS thrice, cells were resuspended in 1 mL of PBS solution with 40 µg of propidium iodide and 100 µg of RNase A for 30 min at 37°C. Samples were then analyzed for their DNA content by FACSCalibur (Becton-Dickinson, Mountain View, CA, USA).
Isolation of RNA
Total RNA was extracted from untreated or Velcade-treated H2373 and H2595 MPM cells. Although, both the cell lines were significantly growth inhibited by 20–40 nM doses of Velcade (see below in “Results”), treatments with suboptimal doses of 10 and 20 nM for H2373 and H2595, respectively, for 48 h period were chosen to adequately capture early changes in transcriptional alterations in cellular RNA levels while minimizing interference from the apoptotic end-point. At the end of treatments, the untreated and treated cells were harvested and total RNAs isolated using RNA-STAT solution (Tel Test, Friendswood, TX, USA) according to the manufacturer’s instruction. The total RNAs were next treated with DNase I to remove contaminating genomic DNA, subsequently purified using RNA-easy Mini Kit (Qiagen, Valencia, CA, USA).
Microarray analysis
Velcade-dependent changes in gene expression in MPM cells were performed at the Genomic Core Facility, Karmanos Cancer Institute utilizing Illumina BeadChip® Arrays essentially according to manufacturer’s instruction (Illumina). Briefly, 0.5 µg total RNA was biotin-labeled and hybridized with BeadChips. The signal was detected with streptovadin-Cy3 according to manufacturer’s instruction (Illumina). The imaging of the BeadChips was conducted using a Bead Array Reader in conjunction with Bead Studio software (Illumina). Normalization of the data was carried out using a quantile based approach which transforms the raw data so that the resulting normalized expression values of each sample have the same distribution [16]. An unsupervised cluster analysis was performed to detect similarities among samples based on gene expression profiles. The genes retained to perform the clustering were those varying the most regardless their group membership as described elsewhere [17]. Significance of the differentially expressed genes among various groups was tested using a moderated t test to allow for p value computation for the significance of gene changes. The p values were then adjusted using the False Discovery Rate method [17] to derive corrected p values. The p values of <0.5 were considered significant provided that the fold change in expression was also equal to or larger than twofold.
Results
Velcade inhibits MPM and HBC cell growth
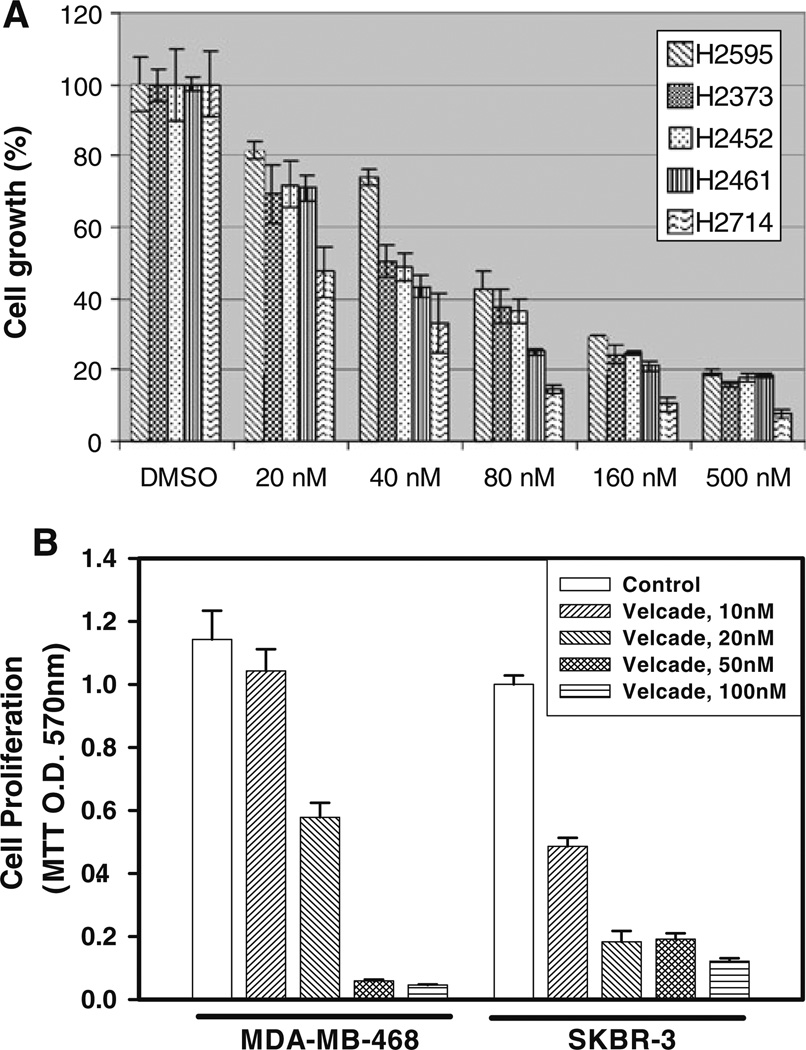
Velcade is a proteasome inhibitor that has been approved in clinics to treat multiple-myeloma and several other solid tumors [8, 9]. Since precise mechanism(s) of cell growth inhibition by this agent have yet to be elucidated, here we utilized a number of MPM and HBC cells to investigate their growth inhibition by Velcade and the extent pathways/mechanisms of cell growth inhibition by this agent overlap in these tumor types. Elucidation of overlapping similarities among the growth inhibitory mechanisms, if proven correct, will have potential to allow for development of additional strategies for effective utilization of this agent to treat a range of malignancies including MPM and breast carcinomas. As a first step towards this goal, we determined the effects of Velcade on the growth of human MPM (H2373, H2452, H2595, H2461 and H2714) as well as HBC (MDA-MB-468 and SKBR-3) cell lines. Each of the cells was separately treated with medium containing various doses of Velcade for 48 h followed by measurements of their viabilities by conducting MTT assays as detailed in methods. Velcade inhibited growth of both the MPM and HBC cells in a dose-dependent manner. Although, Velcade was cytotoxic to the MPM and HBC cells, the IC50 of ∼40 and 10–20 nM were noted for MPM and HBC cells, respectively (Fig. 1).
Fig. 1.
Antiproliferative effect of Velcade on MPM a and HBC b cells. Cells were treated with vehicle (Control) or indicated doses of Velcade for 48 h. Determination of viable/live cells was carried out by MTT assay. Columns in each of the histograms represent means of 3–4 independent experiments; bars SE
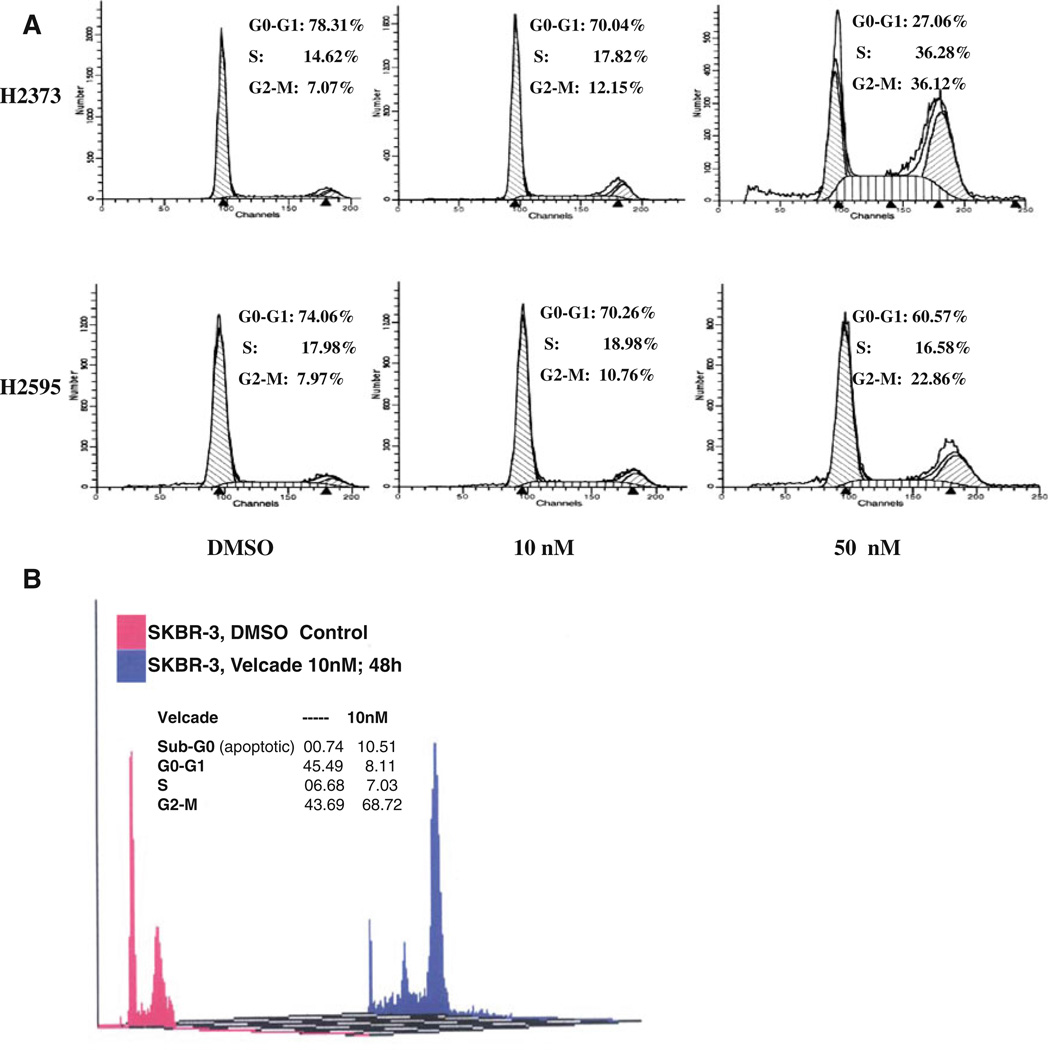
To investigate the growth inhibitory mechanisms utilized by Velcade, we conducted cell cycle analyses of untreated and Velcade-treated MPM cells using propidium iodide staining and flow cytometry. Consistent with our earlier studies [18] and observations from other laboratories [19], our current analyses revealed a G2-M arrest in a concentration-dependent manner in H2373 and H2595 MPM cells (Fig. 2; Table 1). The H2373 MPM cells also accumulated in the S in a Velcade concentration-dependent manner. Whether Velcade treatments induced S or G2M phase arrests was further investigated in the HBC cells. Although treatment of SKBR-3 HBC cells with 50 nM dose of Velcade for 48 h caused extensive apoptosis, exposure of these cells to 10 nM dose of Velcade for 48 h resulted in their significantly higher accumulation in the G2M phase when compared with their DMSO-treated counterparts (Fig. 2b).
Fig. 2.
Velade induces G2M cell cycle arrest in MPM cells. MPM a or HBC b cells were treated with vehicle (Control) or indicated doses of Velcade for 48 h. Cells were stained with propidium iodide followed by their cell cycle distribution analysis by FACS as in methods. Percent of cells in different cell cycle phases is indicated as inset with each histogram
Table 1.
Flow cytometric analysis of Velcade treated cells
| G0–G1 (%) | S(%) | G2–M (%) | |
|---|---|---|---|
| H2373 | |||
| DMSO | 78.1 ±0.3 | 14.715 ±0.1 | 7.2 ±0.2 |
| 10 nM | 69.9 ± 0.1 | 18.8 ±1.4 | 11.3 ±1.3 |
| 50 nM | 26.1 ± 2.1 | 37.7 ± 2.0 | 36.2 ±0.1 |
| H2595 | |||
| DMSO | 72.3 ± 2.4 | 18.8 ±1.2 | 8.9 ±1.2 |
| 10 nM | 70.2 ± 0.1 | 18.8 ±0.2 | 11.0 ±0.3 |
| 50 nM | 62.0 ± 2.0 | 15.6 ±1.4 | 22.4 ± 0.6 |
MPM cells were treated with Velcade for 48 h, followed by FACS analysis. DMSO was used as vehicle control. The mean values (±SD) represent the percentage of cells in the indicated phase of the cell cycle from two independent experiments
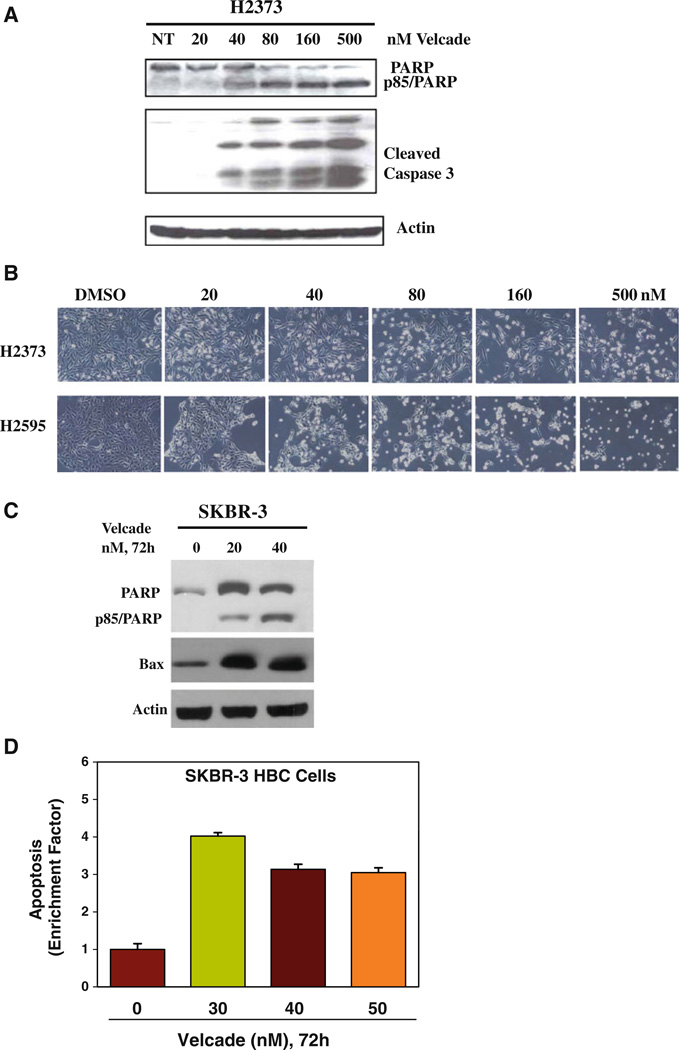
Moreover, Velcade-treated HBC cells had significant accumulation in the apoptotic (sub G0) fractions while undergoing a drastic reduction in cell numbers in the G0/G1 phase (Fig. 2b). Together with previous studies from us and others [18, 19], our data in large part support Velcade targeting of cell cycle progression in MPM and HBC cells involving their arrest in G2M phase. We next determined whether Velcade inhibition of MPM and HBC cell growth involved induction of apoptotic cell death. Cell lysates derived from untreated or Velcade-treated H2373 and SKBR-3 cells were subjected to Western blot analysis for measurement of PARP, an indicator of caspase activation and apoptosis induction [20], Caspase-3 cleavage, and expression of pro-apoptotic protein Bax. Velcade caused cleavage of PARP, activation of caspase 3, and elevated expression of Bax in a dose-dependent manner in both the cell types (Fig. 3a, c). To further confirm the apoptosis-inducing ability of Velcade, we measured apoptosis-associated cellular morphologic changes following exposure of H2373 MPM cells to various doses of Velcade. Cellular morphology changes (i.e., spherical and detached changes) were visualized by phase-contrast microscopy. The apoptotic cellular changes (Fig. 3b) were observed only in the cells treated with Velcade, but not with DMSO. In addition, lysates from untreated or Velcade-treated SKBR-3 HBC cells were subjected to DNA fragmentation-based ELISA assay to measure the level of apoptosis as detailed in methods. A three- to fourfold increase in apoptosis was noted in HBC cells treated with various doses of Velcade (Fig. 3d). Collectively, these results strongly support the conclusion that Velcade-dependent MPM and HBC cell growth suppression involved apoptosis induction.
Fig. 3.
Velcade inhibition of cell growth involves apoptosis induction. Cells were treated with vehicle (Control) or indicated doses of Velcade for 48 h a, b or 72 h b, d. Cell lysates were analyzed by western blotting with anti-PARP or caspase 3 antibodies in a, and with anti-PARP or anti-Bax antibodies in c. The membranes in a and c were then probed with anti-actin antibodies to ascertain equal protein loading, b Apoptosis-associated changes in cellular morphology documented by the phase-contrast photomicrographs of the vehicle or Velcade-treated MPM cells. d The histogram shows apoptosis levels in untreated and treated HBC cells as measured by ELISA-based DNA fragmentation assay. Columns, means of two independent experiments; bars SE
Velcade enhances cisplatin-induced MPM cell growth inhibition
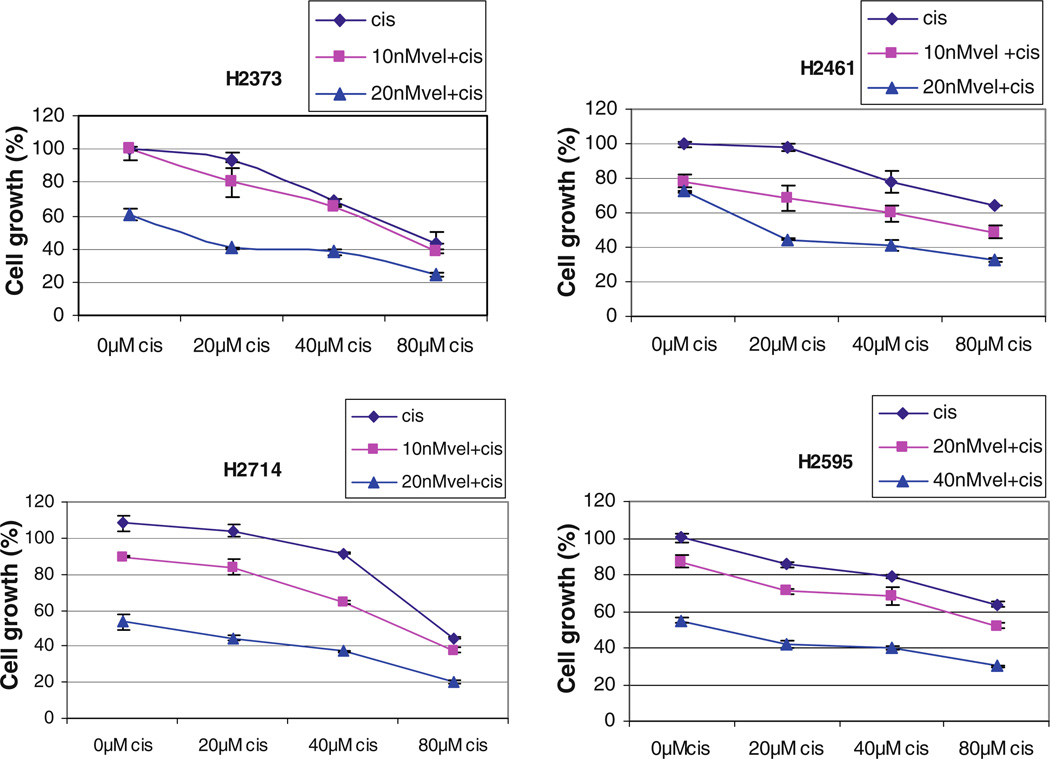
We performed survival studies to establish a baseline cytotoxicity profile for MPM cell lines exposed to cisplatin alone. The results showed that the 4 MPM cells were relatively resistant to cisplatin with the IC50 values around 80 µM (Fig. 4). These IC50 values were approximately 2,000-fold higher than those for Velcade. Since ∼50% inhibition of H2373, H2461, H2714, and H2595 MPM cell growth was noted in the presence of 40,40, 20, and 80 nM doses, respectively, of Velcade (Fig. 1a), we utilized sub-optimal doses of this agent to examine its potential to enhance activity of cisplatin. MPM cells, with the exception of H2595, were pretreated with either 10 or 20 nM of Velcade for 24 h followed by treatments with 20, 40, or 80 µM cisplatin for additional 24 h. Since H2595 cells elicited higher IC50 for Velcade (∼80 nM, Fig. 1), these cells were pretreated with 20 or 40 nM dose of Velcade for 24 h followed by exposure to cisplatin as above. Of note is the fact that all the MPM cells were treated with different doses of cisplatin alone for 24 h periods. MPM cell viabilities were measured by MTT assay as above. As shown in Fig. 4, Velcade treatment consistently enhanced the growth-inhibitory activity of cisplatin in all the MPM cells.
Fig. 4.
Velcade enhances cisplatin efficacy. MPM cells were either treated with different concentrations of cisplatin (cis) for 24 h, or pretreated with noted doses of Velcade (vel) for 24 h followed by addition of indicated concentrations of cisplatin for further 24 h. Determination of viable/live cells from 2 to 3 independent experiments was carried out by MTT assay as detailed in “Methods”
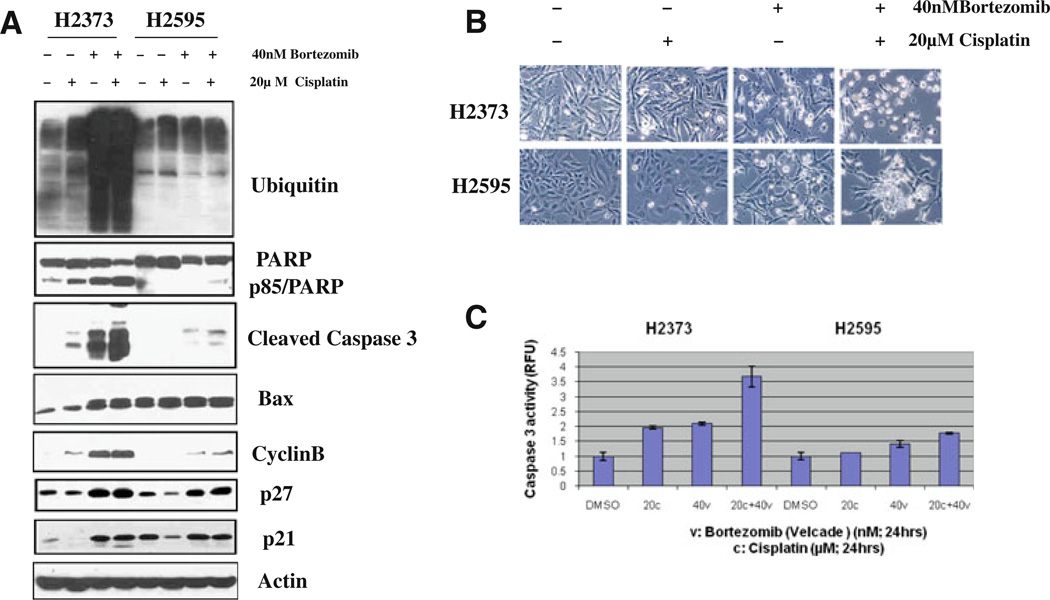
In an attempt to explore the mechanism of enhanced apoptosis of the MPM cells by combined treatments of Velcade and Cisplatin, we assessed the levels of ubiquitin, PARP, caspase-3, and Bax, cyclin B and p27/Kipl cyclin-dependent kinase inhibitor proteins that are known to be involved in diverse apoptosis signaling pathways. H2373 and H2595 cells were treated with either Velcade, cisplatin, or a combination for 48 h, followed by western blot analysis of the cell lysates in conjunction with respective antibodies for the above noted proteins. Treatment with Velcade or a combination of Velcade and cisplatin caused significant accumulation of ubiquitin proteins in H2373 MPM cells (Fig. 5a). Velcade alone or in combination with cisplatin also resulted in a significant increase in expression of pro-apoptotic protein Bax, cleaved caspase-3 and PARP proteins in H2373 cells, while a modest increase in levels of Bax and cleavage of caspase-3 and PARP proteins was noted in similarly treated H2595 cells (Fig. 5a). In addition, treatment of both the MPM cells with Velcade alone or in combination with cisplatin caused significant induction of cyclin B, CDKI p27 and p21 expression (Fig. 5a). We also measured cellular apoptotic morphologic changes (condensation and fragmentation) and apoptotic cell death by caspase-3 activity levels in both cells. Consistent with our western blot analyses, a combination treatment caused increased cellular apoptotic morphological changes (Fig. 5b), as well as significantly higher caspase-3 activities in MPM cells (Fig. 5c). Collectively, the data in Fig. 5 strongly suggest that inhibition of proteasome causes MPM cell growth inhibition in part by inducing cell cycle arrest and apoptosis, and thus provide a mechanistic basis of MPM chemosensitization by Velcade pretreatment.
Fig. 5.
Velade pretreatment enhances cisplatin-induced apoptosis. MPM cells were treated with vehicle (DMSO), indicated doses of Vel-cade (indicated as Bortezomib) or cisplatin for 48 h. For combination study, cells were pretreated with Velcade for 24 h followed by addition of cisplatin for further 24 h. Cell lysates were utilized for western blot analyses with indicated antibodies in a while apoptosis-associated cellular morphologic changes in the treated cells were documented by phase-contrast microscopy in b. In c, apoptosis levels were quantitated by caspase-3 activation assay as described in “Methods”. Histogram columns, means of two independent experiments; bars SE
Velcade targets novel apoptosis transducers
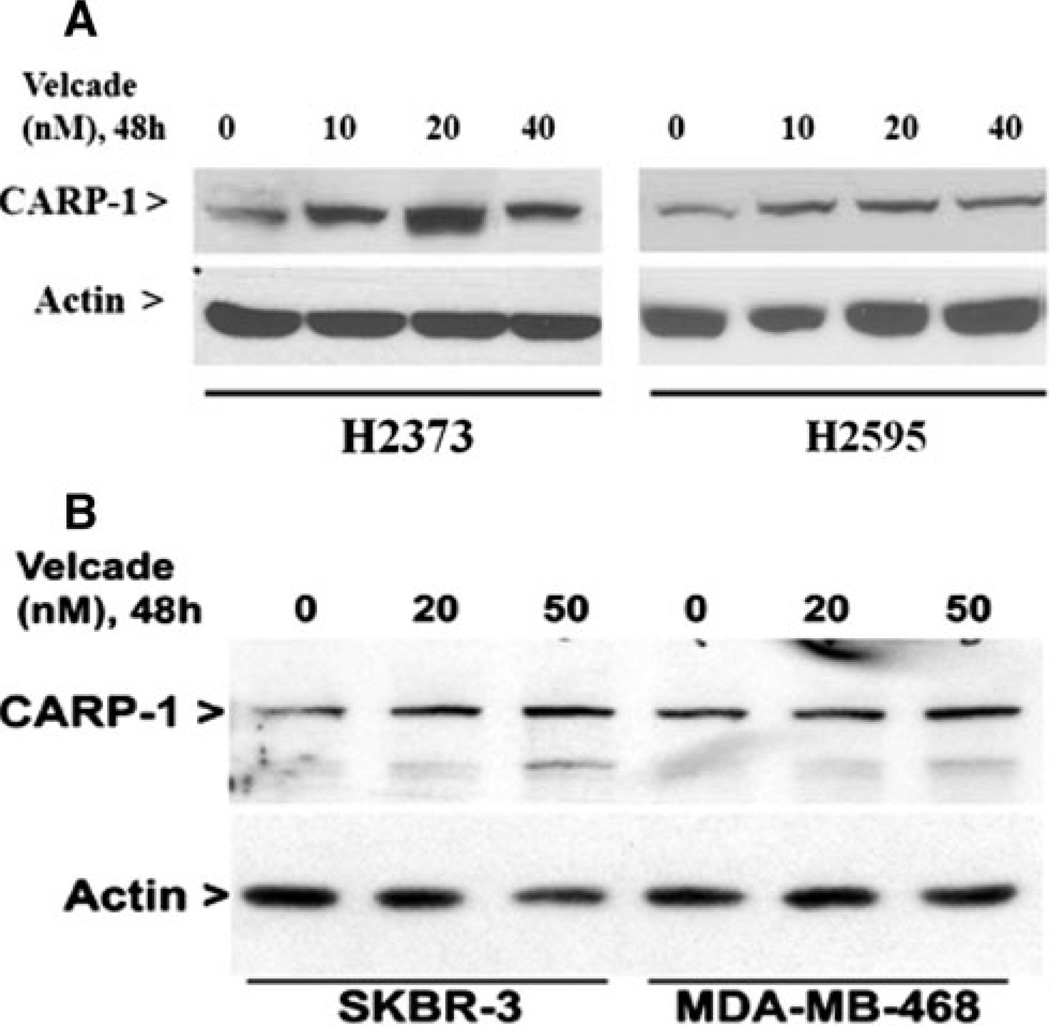
The molecular complexity of cancers and therapy-associated side effects often limit effectiveness of many anti-cancer modalities, and warrant identification of novel cancer cell growth inhibitory targets/pathways for potential exploitation in devising efficacious therapeutic strategies. CARP-1/CCAR1, a novel signaling transducer, was previously identified as a target of select chemotherapy such as adria-mycin, etoposide, and Iressa to inhibit HBC cells growth [12, 21]. CARP-1 has recently been shown to also function in a biphasic manner in regulation of cell growth by functioning as a co-activator of steroid/thyroid nuclear receptors, β-catenin, as well as p53 tumor suppressor protein [22, 23]. Here we investigated whether Velcade-dependent MPM and HBC cell growth inhibition involved elevated CARP-1 expression. Cells were treated with various doses of Velcade followed by western blot analysis of lysates for CARP-1 expression. Our data in Fig. 6 shows that Velcade treatments caused increased expression of CARP-1 in MPM and HBC cells.
Fig. 6.
Velcade induces pro-apoptotic CARP-1 expression. MPM and HBC cells were treated with vehicle (DMSO, notes as 0) or indicated doses and time of Velcade. Cell lysates were utilized for western blot analyses with anti-CARP-1 (alpha 2) antibodies followed by probing of membranes with anti-actin antibodies
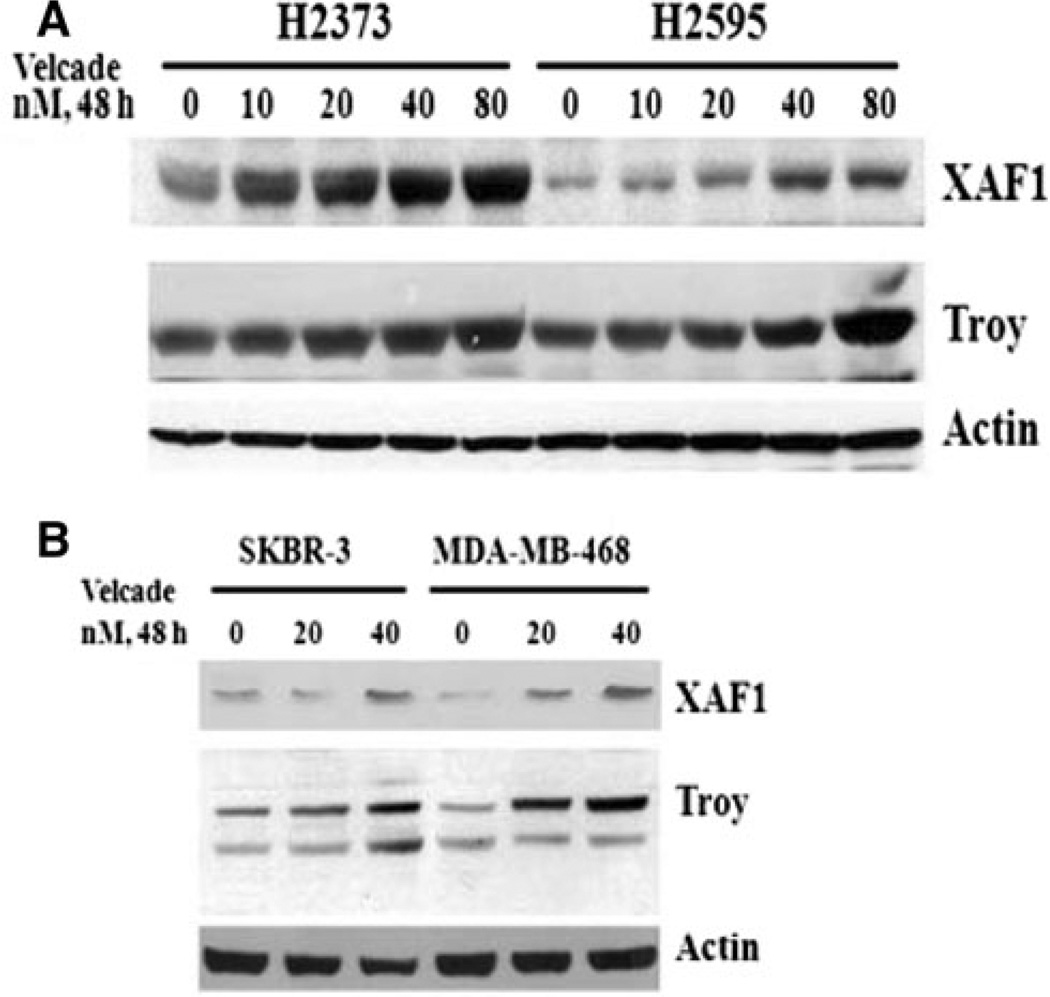
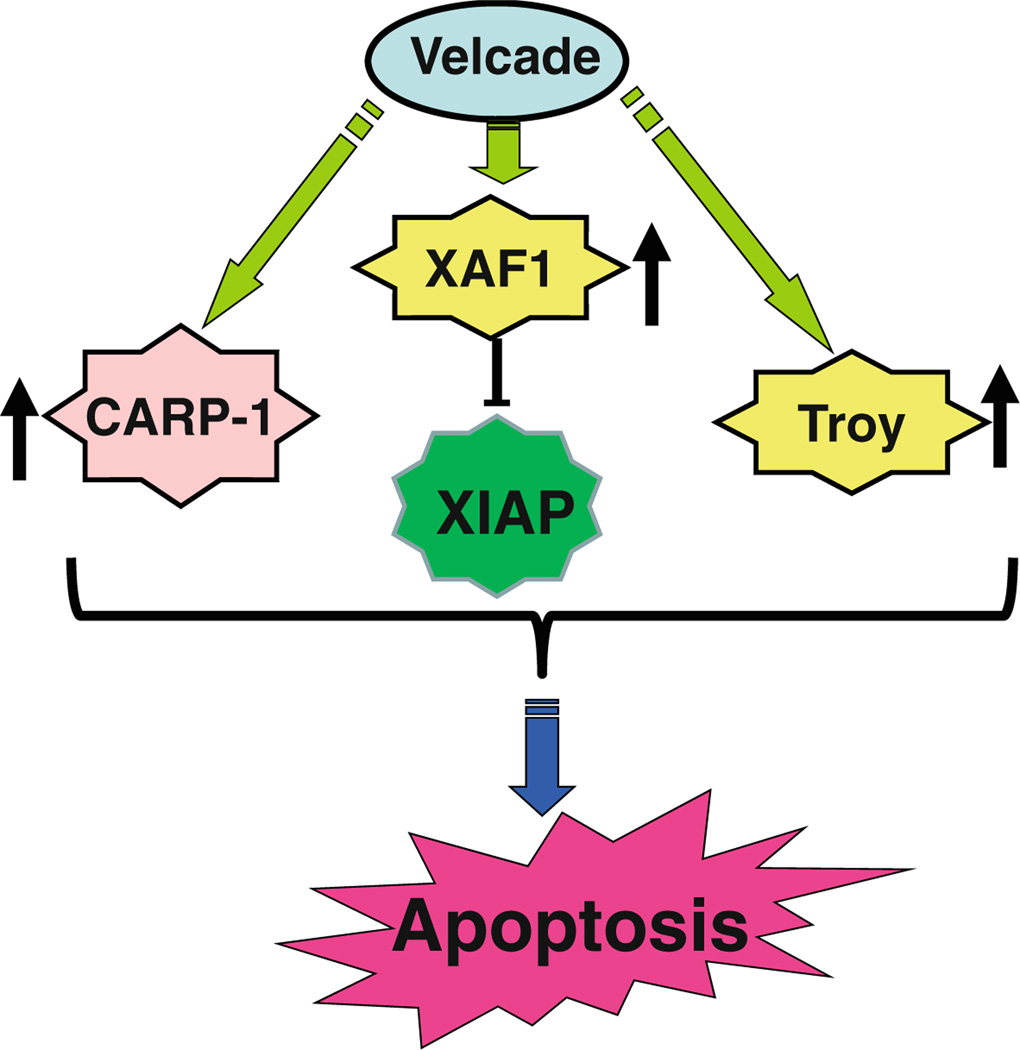
To further investigate mechanisms of MPM cell growth regulation by Velcade, a gene-array-based strategy was employed. H2373 and H2595 MPM cells were either untreated or separately treated with two doses of Velcade as indicated in methods. The RNAs from each group were hybridized with gene-array chips, and the data computed to identify genes that had a significant twofold or higher altered expression following Velcade treatments of both the MPM cells. Next, a subset of genes that had similar Vel-cade-dependent altered expression in both the MPM cells were derived from this data, and a select list of these genes is indicated in Table 2. Of note is the fact that Velcade treatment caused down-regulation of several growth-promoting genes. These included stratifin, peroxisome proliferator-activated receptor gamma (PPARG), v-fos FBJ murine osteosarcoma viral oncogene homolog c-fos, matrix metalloproteinase 3 (MMP3, stromelysin 1, progelatinase) Rho family GTPAse 3 (RND3), ubiquitin protein ligase E3 component n-recognin 4 (UBR4), UBR1, and ubiquitin family domain containing 1 (UBFD1). On the other hand, Velcade caused elevated expression of several growth inhibitory genes. These group of genes included XIAP-associated factor 1 (XAF1), tumor necrosis factor receptor superfamily member 19 (TNFRSF19, TROY), TNFRSF6B, PYD and CARD domain containing protein (PYCARD), claudin 1 (CLDN1), CLDN11, retinoic acid receptor responder (tazarotene induced) 1 (RARRES1), RARRES2, and tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 1 (TANC1), and growth arrest specific 6 (GAS6). Although, previous studies have reported Velcade inhibition of expression of several anti-apoptotic genes in a manner dependent as well as independent of TNF-α [24–27], our gene array experiments reveal that Velcade targets expression of several members of TNF receptor superfamily, as well as XAF1 that is a novel antagonist of pro-survival XIAP protein. Our western blot analyses further demonstrate that treatments of HBC and MPM cells with Velcade results in elevated expression of XAF1 and Troy proteins (Fig. 7). Together, data in Figs. 6 and 7, and Table 2 strongly suggest that Velcade-dependent inhibition of HBC and MPM cancer cells is accomplished, in part, by targeting novel transducers of apoptosis signaling such as CARP-1/ CCAR1, XAF1, and Troy proteins as summarized in Fig. 8.
Table 2.
List of select Velcade regulated genes in MPM cells
| ID | adj.P.Val | FoldChange | Direction | Symbol | Name | Entrez |
|---|---|---|---|---|---|---|
| 1940497 | 0.02984924 | 2.641078795 | DOWN | EPS15L1 | Epidermal growth factor receptor pathway Substrate 15-like 1 | 58,513 |
| 4390747 | 0.2854652 | 2.300179562 | DOWN | HK1 | Hexokinase 1 | 3,098 |
| 3990647 | 0.3404443 | 2.149919083 | DOWN | CLK3 | CDC-like kinase 3 | 1,198 |
| 5720497 | 0.58914674 | 2.294300429 | DOWN | PSMD2 | Proteasome (prosome, macropain) 26S subunit, non-ATPase, 2 | 5,708 |
| 3190593 | 0.58914674 | 2.106476252 | DOWN | UBR4 | Ubiquitin protein ligase E3 component n-recognin 4 | 23,352 |
| 5340592 | 0.58914674 | 2.056172196 | DOWN | AKAP8L | A kinase (PRKA) anchor protein 8-like | 26,993 |
| 6560482 | 0.58914674 | 2.569074114 | UP | CIRBP | Cold inducible RNA binding protein | 1,153 |
| 6180192 | 0.70227989 | 2.769594163 | DOWN | PSMC4 | Proteasome (prosome, macropain) 26S subunit, ATPase, 4 | 5,704 |
| 5690553 | 0.81019895 | 2.549852512 | DOWN | NDRG1 | N-myc downstream regulated gene 1 | 10,397 |
| 4260441 | 0.86383241 | 2.03400233 | DOWN | UBR1 | Ubiquitin protein ligase E3 component n-recognin 1 | 197,131 |
| 1980458 | 0.87893475 | 3.343181361 | DOWN | RND3 | Rho family GTPase 3 | 390 |
| 5670139 | 0.95625714 | 2.305873036 | DOWN | IER5 | Immediate early response 5 | 51,278 |
| 1940711 | 0.96254178 | 2.135861801 | DOWN | UBFD1 | Ubiquitin family domain containing 1 | 56,061 |
| 2260445 | 0.99968708 | 2.994567021 | DOWN | BAG3 | BCL2-associated athanogene 3 | 9,531 |
| 1940048 | 0.99997172 | 3.560655456 | DOWN | OSGIN1 | Oxidative stress induced growth inhibitor 1 | 29,948 |
| 1990433 | 0.99997172 | 2.270710674 | UP | TFAP2A | Transcription factor AP-2 alpha (activating enhancer binding protein 2 alpha) |
7,020 |
| 3850242 | 0.99997172 | 2.142991697 | DOWN | RASSF1 | Ras association (RalGDS/AF-6) domain family member 1 | 11,186 |
| 2680291 | 0.99997172 | 2.396075856 | DOWN | HRK | Harakiri, BCL2 interacting protein (contains only BH3 domain) |
8,739 |
| 5340754 | 0.99997172 | 2.820934606 | UP | ANTXR1 | Anthrax toxin receptor 1 | 84,168 |
| 5810471 | 0.99997172 | 2.431797077 | UP | DRAM | Damage-regulated autophagy modulator | 55,332 |
| 5260026 | 0.99997172 | 2.273615266 | UP | ASS1 | Argininosuccinate synthetase 1 | 445 |
| 1710376 | 0.99997172 | 2.549056416 | UP | TANC1 | Tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 1 |
85,461 |
| 3370167 | 0.99997172 | 2.222452114 | UP | TNFRSF6B | Tumor necrosis factor receptor superfamily, member 6b, decoy |
8,771 |
| 7400408 | 0.99997172 | 2.242145554 | DOWN | SFN | Stratifin | 2,810 |
| 5690370 | 0.99997172 | 2.478611601 | UP | IGFBP2 | Insulin-like growth factor binding protein 2, 36 kDa | 3,485 |
| 3140239 | 0.99997172 | 2.322796252 | DOWN | GADD45A | Growth arrest and DNA-damage-inducible, alpha | 1,647 |
| 3190239 | 0.99997172 | 3.166443653 | UP | CLDN1 | Claudin 1 | 9,076 |
| 2230131 | 0.99997172 | 2.255362514 | UP | PTX3 | Pentraxin-related gene, rapidly induced by IL-1 beta | 5,806 |
| 870358 | 0.99997172 | 2.167596068 | UP | PYCARD | PYD and CARD domain containing | 29,108 |
| 7160711 | 0.99997172 | 2.165129419 | DOWN | PPARG | Peroxisome proliferator-activated receptor gamma | 5,468 |
| 1070403 | 0.99997172 | 2.159964952 | UP | GAS6 | Growth arrest-specific 6 | 2,621 |
| 2600739 | 0.99997172 | 2.115975024 | UP | RARRES1 | Retinoic acid receptor responder (tazarotene induced) 1 | 5,918 |
| 1740341 | 0.99997172 | 2.142555768 | UP | XAF1 | XIAP associated factor 1 | 54,739 |
| 4250379 | 0.99997172 | 2.433203999 | DOWN | FOS | v-fos FBJ murine osteosarcoma viral oncogene homolog | 2,353 |
| 4480019 | 0.99997172 | 2.013663595 | UP | TNFRSF19 | Tumor necrosis factor receptor superfamily, member 19 | 55,504 |
| 3850730 | 0.99997172 | 2.311574067 | UP | CLDN11 | Claudin 11 | 5,010 |
| 6550634 | 0.99997172 | 2.390854775 | UP | RARRES2 | Retinoic acid receptor responder (tazarotene induced) 2 | 5,919 |
| 2970767 | 0.99997172 | 2.424118771 | DOWN | MMP3 | Matrix metallopeptidase 3 (stromelysin 1, progelatinase) | 4,314 |
Fig. 7.
Velcade induces expression of pro-apoptotic XAF1 and Troy proteins. MPM and HBC cells were treated with vehicle (DMSO, notes as 0) or indicated doses and time of Velcade. Cell lysates were utilized for western blot analyses with anti-XAFl or Troy antibodies followed by probing of membranes with anti-actin antibodies
Fig. 8.
Schematic diagram of apoptosis-inducing pathway by Velcade
Discussion
Number of studies has revealed a critical role for ubiquitin proteasome pathway (UPP) in regulating processes of cellular growth and differentiation as well as carcinogenesis. Many cancers harbor abnormalities/mutations of the UPP that affect functions of diverse effectors of cell growth and apoptosis, and with regard to MPM, UPP has been implicated as an important biomarker of poor prognosis [27]. Targeting of UPP and its regulatory components, therefore, is an attractive strategy for MPM inhibition. Velcade (Bort-ezomib, PS341) is a potent and reversible inhibitor of 20S proteasome that exhibits tumor-selective toxicity [28]. In this context, molecular profiling studies have indicated a potential utility for Velcade in targeting MPM either as a monotherapy or in combination with current platinum-based cytotoxic agents [27]. Nevertheless, the treatment options for MPM remain limited, and further investigation of therapeutic strategies is warranted. The fact that alterations in apoptosis signaling pathways often contribute to MPM growth and survival [25], and data from several studies including our current investigation suggest that Velcade suppresses MPM growth in part by stimulating apoptosis, we undertook an in vitro and gene-array-based profiling to identify novel transducers of apoptosis signaling that may be targeted by Velcade. Our studies revealed that Velcade not only inhibited growth of MPM and HBC cells, and sensitized MPM cells to inhibition by cisplatin, it also caused elevated expression of multiple novel apoptosis transducers such as CCAR1/CARP-1, XAF1, and Troy.
The proteasome activity is tightly regulated during various phases of cell cycle, and a timely removal of different cyclins is orchestrated by proteasome-mediated pathways to facilitate orderly progression of the cell cycle. Our current observations of G2-M phase arrest of MPM cells in the presence of Velcade (Fig. 2) are consistent with previous studies [24], and together with increased expression of CDKIs p21WAFl and p27Kipl (Fig. 5a) strongly suggest that Velcade treatments cause cell growth inhibition in part by blocking cell cycle progression. Although, various cyclins together with their cognate cyclin-dependent kinases positively regulate cell cycle, loss of the cyclin(s) and/or inhibition of CDKs results in inhibition of cell cycle and consequent reduction in cell growth. Consistent with the fact that proteasome activity targets cyclin B1 turnover, Velcade exposure caused consistent increase in cyclin B1 in both MPM cell lines (Fig. 5a). The available literature indicates that while a precipitous loss of cyclin B1 will often result in a mitotic catastrophe and cell death, prolonged presence of cyclin B1 beyond the window of the G2M transit will prevent cells from successfully exiting the G2M phase. Thus it is likely that Velcade treatments facilitate G2M accumulation of MPM cells by increasing cyclin B1 levels that will interfere with their orderly exit from the G2M phase.
Our current data and several previous studies [24–28] amply demonstrate that Velcade treatments induced apoptosis in a variety of cancer cells types, including MPM and HBC. Although, a robust cleavage of PARP, caspase 3 and induction of Bax was noted in Velcade-treated H2373 MPM and SKBR-3 HBC cells, a somewhat modest increase in PARP cleavage and cleaved caspase 3 levels without increase in Bax levels was noted in H2595 MPM cells (Figs. 3a, c, 5a), These sobservations suggest that although Velcade inhibits MPM growth by inducing growth arrest and apoptosis, its effects are likely transduced in part by targeting specific mediator(s) in a cell-type dependent manner. Our gene-array-based analyses (Table 2) together with western blot data in Figs. 6 and 7 convincingly reveal a new group of apoptosis signal transducing genes that are consistently activated by Velcade in MPM and HBC cells. Since, CCAR1/CARP-1 is an emerging and novel target of diverse cell growth and apoptosis signaling pathways [12, 21–23] and is not involved in cisplatin-dependent HBC growth inhibition [12], our identification of novel Velcade targets such as CCAR1/CARP-1 has potential to allow for design of effective anti-MPM strategies for optimizing Velcade utility in a range of MPM including the drug (Cisplatin)-resistant phenotype. Moreover, Velcade induction of XAF1 (Fig. 7), an inhibitor of XIAP, is consistent with its targeting of key effectors of apoptosis signaling. In light of the studies by Gordon et al. [25] demonstrating loss of XIAP expression following treatments of MPM cells with Velcade, the increased expression of XAF1 in our current study further underscores effective targeting of XIAP-dependent cell survival pathway by this agent.
The gene array data also revealed a number of interesting genes that may be involved in regulating UPP-depen-dent cell growth and survival pathways. Of note is the down-regulation of two members of the claudin family of transmembrane proteins (claudin 1, 11; Table 2). Claudins play crucial roles in formation and maintenance of tight junctions and their loss could potentially lead to cellular disorientation and detachment [29, 30], which is commonly seen in neoplasia. Decreased expression of claudin 1 has been found to correlate with recurrence status in breast cancers [29]. Although, Velcade inhibition of MPM and HBC cell growth involves enhanced cells cycle arrest and apoptosis, Velcade induction of claudin 1 and/or 11 expression, if proven correct, would suggest its targeting of novel pathways regulating cellular adhesion and perhaps metastasis.
Acknowledgments
This work was supported by the department of Veterans Affairs Merit Review grants to AKR and AW, Susan G. Komen for the Cure grant to AKR, and MARF support to AW.
Contributor Information
Ying Wang, John D. Dingell VA Medical Center, Karmanos Cancer Institute, Wayne State University, VAMC, 4646 John R., Detroit, MI 48201, USA.
Arun K. Rishi, John D. Dingell VA Medical Center, Department of Internal Medicine, Karmanos Cancer Institute, Wayne State University, Room B4334, VAMC, 4646 John R., Detroit, MI 48201, USA Rishia@Karmanos.org
Vineshkumar T. Puliyappadamba, John D. Dingell VA Medical Center, Karmanos Cancer Institute, Wayne State University, Room B4325, VAMC, 4646 John R., Detroit, MI 48201, USA
Sunita Sharma, John D. Dingell VA Medical Center, Department of Surgery, Karmanos Cancer Institute, Wayne State University, VAMC, 4646 John R, Detroit, MI 48201, USA.
Huanjie Yang, Department of Pathology, Karmanos Cancer Institute, Wayne State University, 4100 John R., Detroit, MI 48201, USA.
Adi Tarca, Karmanos Cancer Institute, Wayne State University, 4100 John R, Detroit, MI 48201, USA.
Q. Ping Dou, Department of Pathology, Karmanos Cancer Institute, Wayne State University, 4100 John R., Detroit, MI 48201, USA.
Fulvio Lonardo, Karmanos Cancer Institute, Wayne State University, 4100 John R, Detroit, MI 48201, USA.
John C. Ruckdeschel, Nevada Cancer Center, Las Vegas, Nevada, USA
Harvey I. Pass, Division of Cardiothoracic Surgery, New York University Cancer Center, New York, USA
Anil Wali, John D. Dingell VA Medical Center, Department of Surgery, Karmanos Cancer Institute, Wayne State University, Room B4325, VAMC, 4646 John R., Detroit, MI 48201, USA.
References
- 1.Antman KH, Pass HI, Schiff PB. Benign and malignant mesothelioma. In: DeVita VTJ, Hellman S, Rosenberg SA, editors. Cancer: principles and practice of oncology. 6th edn. Philadelphia: Lippincott-Raven; 2001. pp. 1943–1970. [Google Scholar]
- 2.Peto J, Decaril A, La Vecchia C, Levi F, Negri E. The European mesothelioma epidemic. Br J Cancer. 1999;79:666–672. doi: 10.1038/sj.bjc.6690105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sterman DH, Kaiser LR, Albelda SM. Advances in the treatment of malignant pleural mesothelioma. Chest. 1999;116:504–520. doi: 10.1378/chest.116.2.504. [DOI] [PubMed] [Google Scholar]
- 4.Ryan CW, Herndon J, Vogelzang NJ. A review of chemotherapy trials for malignant mesothelioma. Chest. 1998;113:66S–73S. doi: 10.1378/chest.113.1_supplement.66s. [DOI] [PubMed] [Google Scholar]
- 5.Goudar RK. New therapeutic options for mesothelioma. Curr Oncol Rep. 2005;7(4):260–265. doi: 10.1007/s11912-005-0048-3. [DOI] [PubMed] [Google Scholar]
- 6.Krug LM. An overview of chemotherapy for mesothelioma. Hematol Oncol Clin North Am. 2005;19(6):1117–1136. doi: 10.1016/j.hoc.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 7.Orlowski RZ, Dees EC. Applying drugs that affect the ubiq-uitin-proteasome pathway to the therapy of breast cancer. Breast Cancer Res. 2003;5:1–7. doi: 10.1186/bcr460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Richardson PG, Hideshima T, Anderson KC. Bortezomib (PS-341): a novel, first-in-class proteasome inhibitor for the treatment of multiple myeloma and other cancers. Cancer Control. 2003;10(5):361–369. doi: 10.1177/107327480301000502. Review. [DOI] [PubMed] [Google Scholar]
- 9.Dou QP, Goldfarb RH. Bortezomib/P341 (millennium pharmaceuticals) (invited review) IDrugs. 2002;5:828–834. [PubMed] [Google Scholar]
- 10.Kane RC, Bross PF, Farrell AT, Pazdur R. Velcade: U.S. FDA approval for the treatment of multiple myeloma progressing on prior therapy. Oncologist. 2003;8:508–513. doi: 10.1634/theoncologist.8-6-508. [DOI] [PubMed] [Google Scholar]
- 11.Adams J. The proteosome: a suitable antineoplastic target. Nat Rev Cancer. 2004;4:349–360. doi: 10.1038/nrc1361. [DOI] [PubMed] [Google Scholar]
- 12.Rishi AK, Zhang L, Boyanapalli M, Wali A, Mohammad RM, Yu Y, Fontana JA, Hatfield JS, Dawson MI, Majumdar APN, Reichert U. Identification and characterization of a Cell-Cycle and Apoptosis Regulatory Protein (CARP)-1 as a novel mediator of apoptosis signaling by Retinoid CD437. J Biol Chem. 2003;278:33422–33435. doi: 10.1074/jbc.M303173200. [DOI] [PubMed] [Google Scholar]
- 13.Shao Z, Browning JL, Lee X, Scott ML, Shulga-Morskaya S, AUaire N, Thill G, Levesque M, Sah D, McCoy JM, Murray B, Jung V, Pepinsky RB, Mi S. TAJ/TROY, an orphan TNF receptor family member, binds Nogo-66 receptor 1 and regulates axonal regeneration. Neuron. 2005;45:353–359. doi: 10.1016/j.neuron.2004.12.050. [DOI] [PubMed] [Google Scholar]
- 14.Liston P, Fong WG, KeUy NL, Toji S, Miyazaki T, Conte D, Tamai K, Craig CG, McBurney MW, Korneluk RG. Identification of XAF1 as an antagonist of XIAP anti-Caspase activity. Nat Cell Biol. 2001;3(2):128–133. doi: 10.1038/35055027. [DOI] [PubMed] [Google Scholar]
- 15.Pass HI, Lott D, Lonardo F, Harbut M, Liu Z, Tang N, Carbone M, Webb C, Wali A. Asbestos exposure, pleural mesothelioma, and serum osteopontin levels. N Engl J Med. 2005;353(15):1564–1573. doi: 10.1056/NEJMoa051185. [DOI] [PubMed] [Google Scholar]
- 16.Smyth GK, Limma L. Liner models for microarray data. In: Gentleman R, Carey V, Duoit S, Irizarry R, Huber W, editors. Bioin-formatics and computational biology solutions using R and bio-conductor. New York: Springer; 2005. pp. 397–120. [Google Scholar]
- 17.Tarca AL, Carey VJ, Chen XW, Romero R, Draghici S. Machine learning and its applications to biology. PLoS Comput Bio. 2007;l3:ell6. doi: 10.1371/journal.pcbi.0030116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wali A, Morin PJ, Hough CD, Lonardo F, Seya T, Carbone M, Pass HI. Identification of intelectin overexpression in malignant pleural mesothelioma by serial analysis of gene expression (SAGE) Lung Cancer. 2005;48(l):19–29. doi: 10.1016/j.lungcan.2004.10.011. [DOI] [PubMed] [Google Scholar]
- 19.Sartore-Bianchi A, Gasparri F, Galvani A, Nici L, Darnowski JW, Barbone D, Fennell DA, Gaudino G, Porta C, Mutti L. Bortezomib inhibits nuclear factor-kappaB dependent survival and has potent in vivo activity in mesothelioma. Clin Cancer Res. 2007;13(19):5942–5951. doi: 10.1158/1078-0432.CCR-07-0536. [DOI] [PubMed] [Google Scholar]
- 20.Lazebnik YA, Kaufmann SH, Desnoyers S, Poirier GG, Earnshaw WC. Cleavage of poly(ADP-ribose) polymerase by a proteinase with properties like ICE. Nature. 1994;371:346–347. doi: 10.1038/371346a0. [DOI] [PubMed] [Google Scholar]
- 21.Rishi AK, Zhang L, Yu Y, Jiang Y, Nautiyal J, Wali A, Fontana JA, Levi E, Majumdar APN. Cell cycle and apoptosis regulatory protein (CARP)-1 is involved in apoptosis signaling by epidermal growth factor receptor. J Biol Chem. 2006;281:13188–13198. doi: 10.1074/jbc.M512279200. [DOI] [PubMed] [Google Scholar]
- 22.Kim JH, Yang CK, Heo K, Roeder RG, An W, StaUcup MR. CCAR1, a key regulator of mediator complex recruitment to nuclear receptor transcription complexes. Mol Cell. 2008;31:510–519. doi: 10.1016/j.molcel.2008.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ou CY, Kim JH, Yang CK, Stallcup MR. Requirement of cell cycle and apoptosis regulator 1 for target gene activation by Wnt and {beta}-catenin and for anchorage-independent growth of human colon carcinoma cells. J Biol Chem. 2009;284(31):20629–20637. doi: 10.1074/jbc.M109.014332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yuan BZ, Chapman JA, Reynolds SH. Proteasome inhibitor MG132 induces apoptosis and inhibits invasion of human malignant pleural mesothelioma cells. Transl Oncol. 2008;1(3):129–140. doi: 10.1593/tlo.08133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gordon GJ, Mani M, Mukhopadhyay L, Dong L, Yeap BY, Sugarbaker DJ, Bueno R. Inhibitor of apoptosis proteins are regulated by tumour necrosis factor-alpha in malignant pleural mesothelioma. J Pathol. 2007;211(4):439–446. doi: 10.1002/path.2120. [DOI] [PubMed] [Google Scholar]
- 26.Gordon GJ, Mani M, Maulik G, Mukhopadhyay L, Yeap BY, Kindler HL, Salgia R, Sugarbaker DJ, Bueno R. Preclinical studies of the proteasome inhibitor bortezomib in malignant pleural mesothelioma. Cancer Chemother Pharmacol. 2008;61(4):549–558. doi: 10.1007/s00280-007-0500-1. [DOI] [PubMed] [Google Scholar]
- 27.Borczuk AC, Cappellini GC, Kim HK, Hesdorffer M, Taub RN, Powell CA. Molecular profiling of malignant peritoneal mesothelioma identifies the ubiquitin-proteasome pathway as a therapeutic target in poor prognosis tumors. Oncogene. 2007;26(4):610–617. doi: 10.1038/sj.onc.1209809. [DOI] [PubMed] [Google Scholar]
- 28.Fennell DA, Chacko A, Mutti L. BCL-1 family regulation by the 20S proteasome inhibitor bortezomib. Oncogene. 2008;27:1189–1197. doi: 10.1038/sj.onc.1210744. [DOI] [PubMed] [Google Scholar]
- 29.Morohashi S, Kusumi T, Sato F, Odagiri H, Chiba H, Yoshihara S, Hakamada K, Sasaki M, Kijima H. Decreased expression of claudin-1 correlates with recurrence status in breast cancer. Int J Mol Med. 2007;20:139–143. [PubMed] [Google Scholar]
- 30.Tsukita S, Furuse M, Itoh M. Multifunctional strands in tight junctions. Nat Rev Mol CeU Biol. 2001;2:285–293. doi: 10.1038/35067088. [DOI] [PubMed] [Google Scholar]