Abstract
We investigated the gene flow of the common marine diatom, Skeletonema marinoi, in Scandinavian waters and tested the null hypothesis of panmixia. Sediment samples were collected from the Danish Straits, Kattegat and Skagerrak. Individual strains were established from germinated resting stages. A total of 350 individuals were genotyped by eight microsatellite markers. Conventional F-statistics showed significant differentiation between the samples. We therefore investigated whether the genetic structure could be explained using genetic models based on isolation by distance (IBD) or by oceanographic connectivity. Patterns of oceanographic circulation are seasonally dependent and therefore we estimated how well local oceanographic connectivity explains gene flow month by month. We found no significant relationship between genetic differentiation and geographical distance. Instead, the genetic structure of this dominant marine primary producer is best explained by local oceanographic connectivity promoting gene flow in a primarily south to north direction throughout the year. Oceanographic data were consistent with the significant FST values between several pairs of samples. Because even a small amount of genetic exchange prevents the accumulation of genetic differences in F-statistics, we hypothesize that local retention at each sample site, possibly as resting stages, is an important component in explaining the observed genetic structure.
Keywords: oceanographic connectivity, Bacillariophyceae, microsatellites, Skeletonema marinoi
1. Introduction
Studies during the past decade have repeatedly revealed high genetic diversity within populations of various microeukaryote taxa [1] and patterns of genetic structure and differentiation between populations of aquatic protists [2]. However, little is known about the causes of spatial and temporal patterns of genetic variation or how genetic variation influences population dynamics (e.g. algal blooms) and biogeochemical cycles. On one hand, there is support for largely unstructured populations, such as the diatom Pseudo-nitzschia pungens that spans a 200 km region of the North Sea [3]. By contrast, there is evidence from other diatom species that populations less than 100 km apart are genetically different despite the absence of apparent dispersal barriers [1,4]. Oceanographic barriers caused by currents and density gradients are known to restrict the transport of pelagic organisms [5]. Recently, correlations between genetic differentiation and oceanographic barriers have also been shown for populations of phytoplankton over larger geographical scales, i.e. marine basins [6].
Connectivity between two populations is dependent on the organisms’ traits and the permeability of the environment. In the marine environment, the speed and direction of ocean currents together with temperature and salinity are the main features. On global geographical scales, dispersal probability may be well correlated with the Euclidean distance, leading to classic IBD population differentiation [7]. However, this may fail on regional scales where complex oceanographic circulation can lead to connectivity patterns that are poorly explained by geographical distance [8]. Therefore, gene flow in holo- or meroplanktonic marine organisms often yields significant IBD correlations on a global scale, but attempts to correlate genetic and geographical distance may fail over regional distances [9]. By contrast, efforts to correlate gene flow with oceanographic connectivity have offered more promising explanations for the genetic structures observed on local scales [10]. For instance, frequency of larval exchange and empirical genetic differences were uncorrelated between sites using Euclidean distance, but when transformed into oceanographic distance, the frequency of larval exchange explained nearly 50% of the variance in genetic differences among sites over scales of tens of kilometres [5].
Many planktonic protists produce resting stages when conditions in the water column are unfavourable. These can act as either a short- or long-term survival mechanism, with cells remaining viable in the sediment for several decades [11]. Resting stages in the sediment are of ecological importance, as they provide a seed bank of genetic material for future years when resuspended in the water column [12]. It has previously been proposed that the ability to form resting stages increases the potential for dispersal and extends a species’ or a population's geographical range [13]. However, recent studies indicate that resting stages are perhaps even more important for anchoring protist populations within a specific habitat [14], and studies of genetic structure indicate a strong link between cells in the planktonic and benthic community within a restricted area [4]. Thus, counterintuitively, resting stage formation in free-living marine protists may promote, rather than inhibit, the formation of discrete populations.
In this study, we used the chain-forming marine diatom Skeletonema marinoi as a model organism. Skeletonema is a cosmopolitan genus and there are 11 known species [15], but in Scandinavian marine waters only one species, S. marinoi, has been reported [16]. Skeletonema marinoi is a common species year round, but during the spring bloom, in February to March, it often dominates the plankton community in the Skagerrak and Kattegat [17]. Provided a plentiful nutrient supply, the cells proliferate asexually in the photic zone at a growth rate of one division per day [18]. The predominant means of propagation is through vegetative division, but auxospore formation and sexual reproduction have been documented in Skeletonema species [19]. Skeletonema marinoi has a benthic resting stage, and in Scandinavian sediments up to 50 000 propagules per gram of sediment can be found [11]. Additionally, S. marinoi is easy to collect, isolate and maintain in culture and the survival of monoclonal cultures after single cell isolation is almost 100% [20].
Here, we report on the genetic structure of this common diatom from sampling sites located along the Swedish west coast. We tested the null hypothesis of panmixia using conventional F-statistics. Spatial patterns in our data were discovered, and thereafter we applied analyses for IBD and a seascape approach. Patterns of oceanographic circulation, such as intensity and direction, are often seasonally dependent, and this variability affects the genetic structure of mero- and holoplanktonic marine species [21]. We therefore examined how well estimates of local oceanographic connectivity can explain the gene flow between different sample sites of S. marinoi on a seasonal basis.
2. Material and methods
(a). Study site, sample collection and establishment of clonal cultures
The seven sampling sites were located in the Skagerrak, Kattegat and Öresund (figure 1a and table 1). Two major current systems affect the Swedish west coast; the low saline surface Baltic current running northward parallel to the coast, and the central Skagerrak water circulation pattern resulting in an inflow of more saline North Atlantic water [22]. Hence, the water is permanently stratified in terms of salinity and a pronounced halocline (average depth 10–15 m) is present.
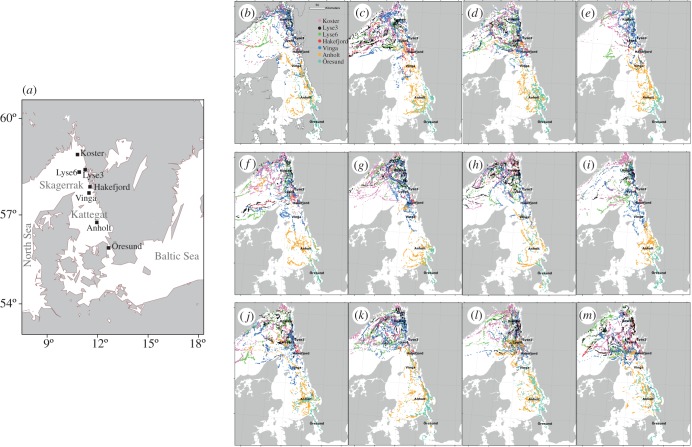
Figure 1.
(a) Southern Scandinavia. Strains of S. marinoi were established from sediment samples collected from inshore and offshore sites in the Skagerrak, Kattegat and Öresund. (b–m) Oceanographic trajectories for the seven sampling stations for each month of the year. The trajectories for each sampling station are colour coded according to the legend in (b). Connectivity is based on trajectories released from 15 grid cells per site. The total numbers of trajectories released at each site over the period 1995–2002 was 5880. (b) January, (c) February, (d) March, (e) April, (f) May, (g) June, (h) July, (i) August, (j) September, (k) October, (l) November, (m) December.
Table 1.
Details of sediment samples from which monoclonal cultures of S. marinoi were established.
| location | position | depth (m) | no. initial isolates | no. isolates that survived | no. isolates that resulted in successful DNA extraction | no. genotyped isolates |
|---|---|---|---|---|---|---|
| Koster | 58°51.0′ N, 10°45.7′ E | 102 | 56 | 51 | 43 | 42 |
| Lyse3 | 58°20.35′ N, 11°21.43′ E | 29 | 86 | 58 | 58 | 57 |
| Lyse6 | 58°15.2′ N, 11°03.5′ E | 101 | 61 | 55 | 48 | 46 |
| Hakefjord | 57°57.58′ N, 11°42,92′ E | 41 | 68 | 60 | 58 | 57 |
| Vinga | 57°33.0′ N, 11°31.5′ E | 78 | 57 | 54 | 50 | 45 |
| Anholt | 56°40.0′ N, 12°07.0′ E | 54 | 56 | 51 | 44 | 42 |
| Öresund | 55°59.16′ N, 12°44.02′ E | 14 | 61 | 61 | 61 | 61 |
| total | 350 |
Sediment samples were collected once (spring 2009) at each location using a box corer. The top (less than 0.5 cm) of the sediment cores was retained and, before further processing, kept dark and cool (4°C) for several months. Inference from nearby geographical sites indicates that 0.5 cm corresponds to one year of accumulation [23]. Approximately 1 g of sediment from each of the samples was distributed into smaller aliquots and inoculated in 24-well NUNC plates. The wells were filled with f/2 medium, 26 PSU [24]. The sediment slurries were kept at 10°C in a 12 L : 12 D cycle at an irradiance of 60 μmol photons m–2 s–1. Slurries were examined daily for germination and vegetative growth using an inverted microscope (Axiovert 135, Zeiss).
Following germination, one cell chain from each well was isolated by micropipetting. Each chain was transferred to a drop of sterile f/2 medium. This was repeated several times to assure that only one cell chain was isolated from each well. The cell chain was thereafter transferred to a Petri dish (Ø 50 mm) with f/2 medium and incubated under the same conditions as described above. When growth in the Petri dish was confirmed, the monoclonal culture was transferred to 50 ml NUNC flasks containing f/2 medium. Cultures in exponential growth stage were filtered onto 3 µm pore size filters (Ø 25 mm, Versapor-3000, Pall Corporation). Filters were folded, put in Eppendorf tubes and stored at −80°C.
(b). DNA extraction, PCR and microsatellite genotyping
Genomic DNA was extracted from the cultures in exponential phase following a CTAB-based protocol described in [25]. Eight microsatellite loci were amplified (S.mar1–8) [26] by PCR as described in [4]. The products were analysed in an ABI 3730 (Applied Biosystems) and allele sizes were assigned relative to the internal standard (GS600LIZ). Allele sizes for the individual loci were determined and processed using GeneMapper (ABI PrismGeneMapperSoftware v. 3.0).
(c). Population differentiation and gene flow
Genepop v. 4.0.7 [27] was used to estimate deviations from Hardy–Weinberg equilibrium (HWE, 10 000 Markov Chain dememorizations, 20 batches and 5000 iterations per batch) of each locus in each sample, genotypic linkage disequilibrium between pairs of loci in each sample (10 000 dememorizations, 100 batches and 5000 iterations per batch). Levels of statistical significance were adjusted according to sequential Bonferroni correction for multiple comparisons [28]. Identical eight-loci genotypes were identified in Microsatellite Tools for Excel [29]. The microsatellite dataset was analysed for null alleles, stuttering, and large allele drop out by means of 1000 randomizations using MicroChecker v. 2.2.3. Null allele frequencies cannot be accurately estimated in non-HWE loci unless the rate of inbreeding (or selfing) is known [30]. Despite susceptibility of heterozygote deficiency in some microsatellite loci [4], and no prior knowledge of the proportion of asexually reproducing individuals, we calculated null allele frequencies according to [31]. This allowed us to exclude the possibility that heterozygote deficiency in any locus was biased at particular sample sites.
Genetic differentiation between all pairs of samples was determined by calculating pairwise multilocus FST using Arlequin v. 3.1 [32] with 10 000 permutations. The heterozygosity-independent Jost D [33] was calculated using DEMEtics, and 1000 bootstrap replicates were used to estimate p-values [34]. Bonferroni technique was used to calculate p-values from all multiple tests [28].
We used a new approach for the estimation of directional migration from allelic frequencies in individual samples [35]. This procedure is a directional extension of D [33] and is based on a pool of migrants defined for each combination of two samples in pairwise comparisons. The allele-frequencies of the pool of migrants between two samples were calculated as the geometric means of the frequencies of the respective alleles in the two samples and consecutive normalization. The concept of using the geometric mean is that the pool of migrants only consists of alleles present in both samples. Directional D-values, Dd, were then calculated the same as regular D-values, with the exception that the samples were compared to the pool of migrants instead of to each other [33]. Consecutively, migration (m) was estimated from the directional Dd. The approximate equation for this is m ≈ μ(n−1)(1−Dd)/Dd, where μ is mutation rate and n is the number of samples [33]. We analysed only pairwise comparisons (n = 2) and one locus at a time. Therefore, the equation could be simplified to m/μ ≈ (1−Dd)/Dd. The migration rates, i.e. m/μ, between the seven different samples were normalized and varied between zero and one, yielding a relative measure of direction of migration between the different sample sites.
(d). Oceanographic connectivity
We estimated connectivity between the seven sampling sites with a biophysical model, where velocity fields from an ocean circulation model were combined with a particle-tracking routine to simulate drift trajectories at two different depth intervals to represent the dispersal of diatoms. Ocean current data from 1995 to 2002 were produced in hind-cast model using the BaltiX model. BaltiX is a regional model covering the Baltic and the North Sea and is based on the NEMO ocean engine [36]. A detailed model description with preliminary validations is given in [37] and the electronic supplementary material, text S1. The BaltiX model has a spatial resolution of approximately 3.7 km in the horizontal, with vertical layers ranging between 3 and 22 m. It has a free surface and uses z* vertical coordinates, as described by [38], which allow the grid boxes to stretch and shrink vertically to model the tides without generating empty grid cells at low tide. At the open boundaries, the model is forced with tidal harmonics, velocities and sea surface heights (SSHs) [39]. Temperature and salinity were obtained from climatology [40]. Atmospheric forcing used the ERA40 dataset, dynamically downscaled using a regional atmospheric circulation model, to fit the higher resolution grid of BaltiX. Precipitation was added every 12 h and river run-off each month. Validation shows that the BaltiX model provides a good representation of the tidal-driven SSH and wind-driven SSH in the Baltic Sea [37], which are important aspects for the circulation pattern.
The dispersal of diatoms was simulated using the Lagrangian trajectory model TracMass [41]. It is a particle-tracking model that calculates transport of particles using temporal and spatial interpolation of flow-field data from the BaltiX circulation model using a time step of 15 min. Each sample site was represented by 15 grid cells. Particles were released on the 15th day of each of the 12 months over an 8-year period and allowed to drift in surface (0–3 m) or deeper (12–14 m) water for 10 or 20 days. The choice of drift period was based on an approximation of the longevity of a Skeletonema bloom in the area. Connectivity among the seven sampling sites was estimated by calculating the proportion of particles released from site i that ended up in site j. Each sampling site was assumed to represent the 15 grid cells closest to the locations given in table 1. In total, the connectivities estimated among the seven sites were based on 1.98 million released particles. We also tested whether multi-generational dispersal [5] could explain the pattern of genetic differentiation. In this analysis, all locations in the model domain could act as stepping-stones between dispersal events that were 10 or 20 days. The dispersal probability over 10 dispersal events was calculated by multiplication of the connectivity matrix 10 times, which allowed for all possible dispersal routes.
(e). Comparing gene flow versus geographical distance and oceanographic connectivity
IBD analyses from matrices of genetic (FST/1−FST) and D versus geographical distances (Loge of nautical miles) were performed in GenePop [27]. Geographical distances were measured as linear distances between pairs of sites. The significance was assessed using 30 000 permutations.
To investigate the correlation between the observed gene flow and oceanographic connectivity, one-tailed Mantel tests (999 permutations) were performed. The Mantel test checks for significance between the matrices of migration calculated from the pairwise Dd value, and the oceanographic trajectories. We analysed the eight matrices of estimated migration (one each for locus S.mar1–8) versus oceanographic connectivity, represented by four different sets of 12 matrices each (one for each month of the year). The four sets represented (i) cells dispersed in surface water (0–3 m) drifting 10 days; (ii) cells dispersed in deeper water (12–14 m) drifting 10 days; (iii) cells dispersed in surface water (0–3 m) drifting 20 days and (iv) cells dispersed in deeper water (12–14 m) drifting 20 days. Additionally, we tested for significant correlations between the eight matrices of estimated migration (S.mar1–8) versus the two stepping-stone matrices (drift for 10 and 20 days). All Mantel tests were analysed using the software PASSaGE [42]. The migrations were normalized and the diagonal value was set to 1. The trajectories were log (x + 1) transformed and the diagonal value was set to 5. Correlations were considered significant at p < 0.05.
3. Results
On average, 88% of the isolated germinated cell chains from the sediment samples survived and monoclonal cultures were established. Genotyping success was 97% and 350 clonal isolates from seven locations were genotyped (table 1).
All loci were polymorphic. Locus S.mar3 was the least variable while S.mar5 was the most variable locus (see electronic supplementary material, table S1). Significant (p < 0.05) departures from HWE were observed for all loci in a varying number of samples. Loci S.mar1, S.mar3, S.mar5 and S.mar 7–8 displayed heterozygote deficiency in all samples. Locus S.mar4 displayed heterozygote deficiency in one out of seven samples. The numbers of loci that displayed departure from HWE varied among the samples. There was no evidence for large allele drop out or stuttering effects using MicroChecker. Based on the method Brookfield 1 for loci in HWE, estimates of null alleles frequency were low or non-existent in S.mar2, S.mar3, S.mar4 and S.mar6, moderate in S.mar7 and S.mar8, and highest in S.mar1 and S.mar5. Indications of null allele coincided with the loci displaying heterozygote deficiencies (Spearman correlation, n = 56, p < 0.01). There was no significant correlation between samples and potential null allele frequencies (two-tailed paired samples t-test, p > 0.05), and all loci were used in subsequent calculations of genetic differentiation and gene flow. No pairs of microsatellite loci were significantly linked across all samples, thus the eight loci were considered independent. Out of 350 individuals, three identical genotypes were identified. Two strains were identical in the Vinga sample, and two different pairs of strains were identical in the Koster sample.
Genetic structure was examined by estimating pairwise FST and D (table 2). Pairwise FST ranged from −0.0004 to 0.0277. Thirteen of 21 pairs were significant (p < 0.05), and five pairs remained significantly differentiated after Bonferroni correction (p < 0.0024). The Jost D values ranged between 0.015 and 0.149. Seventeen of 21 pairs were significant (p < 0.05), and nine pairs remained significantly differentiated after Bonferroni correction (p < 0.0024). Based on these results, we rejected a model based on panmixia.
Table 2.
Genetic differentiation between pairs of samples. Multilocus Jost D distances between populations above the diagonal and FST below the diagonal. Italic numbers denote significant differentiation (p < 0.05). Bold numbers denote significance after Bonferroni correction (p < 0.0024).
| Koster | Lyse3 | Lyse6 | Hakefjord | Vinga | Anholt | Öresund | |
|---|---|---|---|---|---|---|---|
| Koster | — | 0.105 | 0.101 | 0.084 | 0.073 | 0.085 | 0.124 |
| Lyse3 | 0.0217 | — | 0.015 | 0.094 | 0.149 | 0.091 | 0.050 |
| Lyse6 | 0.0214 | −0.0004 | — | 0.082 | 0.118 | 0.063 | 0.046 |
| Hakefjord | 0.0132 | 0.0213 | 0.0128 | — | 0.058 | 0.061 | 0.109 |
| Vinga | 0.0100 | 0.0277 | 0.0163 | 0.0043 | — | 0.075 | 0.104 |
| Anholt | 0.0163 | 0.0163 | 0.0056 | 0.0093 | 0.0078 | — | 0.049 |
| Öresund | 0.0241 | 0.0101 | 0.0055 | 0.0209 | 0.0138 | 0.0022 | — |
The Mantel test revealed no significant relationship between genetic distance (FST or D) and geographical distance in all pairwise combinations (p = 0.271 and p = 0.364; electronic supplementary material, figure S1).
The major migration direction, as measured by Dd, was from south to north (see electronic supplementary material, table S2a–h). Migration from inshore to offshore sampling sites (from station Öresund, Hakefjord and Lyse3) exceeded migration from offshore to inshore sampling sites. Symmetrical migration rates between sites were rare (18% of all possible migration routes). Among the stations, the northern offshore sampling stations (Koster and Lyse6) constituted population sinks, whereas the southern stations (Vinga and Öresund) constituted sources.
The dominating dispersal direction, as estimated from the oceanographic model, was from south to north, independent of season (figure 1b–m; electronic supplementary material, table S3a–l and figure S2). For the northern stations, there was a westward dispersal direction that was pronounced for the offshore stations (Lyse6, Koster and Vinga; figure 1b–m). There was no dispersal bias from inshore to offshore stations or vice versa. Local recruitment was supported by the oceanographic trajectories for all sampled stations. The northern most stations (Koster and Lyse3) were sinks i.e. the number of received trajectories exceeded the numbers dispersed. Vinga in particular, but also the southern-most sampling sites (Anholt and Öresund) were sources (see electronic supplementary material, table S3).
The analyses between the matrices of migration pattern, assessed from the pairwise directional Dd of the individual loci, and the matrices of oceanographic connectivity for each month of the year, yielded significant correlations with all dispersal sets, i.e. 10 days drift in surface or deep water, 20 days drift in surface or deep water and stepping-stone dispersal or 10 or 20 days drift. The majority of significant correlations were generated from the set with trajectories dispersed in the surface water for 10 days. The migration patterns for loci S.mar4 and S.mar5 yielded significant correlations to the connectivity in nine months (table 3). The migration patterns for S.mar2, S.mar6 and S.mar7 were significantly correlated to the connectivity for several months of the year, but for S.mar 8 only in the month of July. The migration matrices for S.mar1 and S.mar3 did not yield any significant correlation to connectivity in any month. The connectivity for individual months was significantly correlated to the migration pattern assessed by one to five individual microsatellite markers (table 3).
Table 3.
Mantel test of normalized migration calculated from directional genetic differentiation (Dd) assessed from individual locus and log10-transformed oceanographic trajectories for 10 days dispersal in surface water each month. Each cell gives the correlation between the matrices. Significant correlations are indicated in italics.
| month | microsatellite loci |
|||||||
|---|---|---|---|---|---|---|---|---|
| S.mar1 | S.mar2 | S.mar3 | S.mar4 | S.mar5 | S.mar6 | S.mar7 | S.mar8 | |
| Jan | 0.13 | 0.34* | 0.04 | 0.23** | 0.50* | 0.27 | 0.37* | 0.06 |
| Feb | 0.08 | 0.31* | 0.04 | 0.19 | 0.19 | 0.21 | 0.33 | 0.27 |
| Mar | 0.10 | 0.22 | 0.02 | 0.20* | 0.56* | 0.28 | 0.31 | 0.02 |
| Apr | 0.06 | 0.34* | 0.02 | 0.18* | 0.27 | 0.29 | 0.44* | 0.16 |
| May | 0.22 | 0.29 | 0.05 | 0.21* | 0.42* | 0.34* | 0.33 | 0.03 |
| Jun | 0.15 | 0.29 | 0.09 | 0.25** | 0.62* | 0.31 | 0.27 | 0.24 |
| July | 0.08 | 0.34* | 0.01 | 0.24* | 0.56* | 0.21 | 0.39* | 0.36* |
| Aug | 0.18 | 0.28 | 0.09 | 0.19* | 0.32 | 0.39* | 0.23 | 0.11 |
| Sep | 0.02 | 0.26 | 0.01 | 0.17 | 0.49* | 0.23 | 0.53* | 0.01 |
| Oct | 0.01 | 0.31* | 0.01 | 0.21* | 0.49* | 0.25 | 0.44* | 0.32 |
| Nov | 0.04 | 0.26 | 0.07 | 0.18 | 0.53* | 0.31* | 0.46* | 0.01 |
| Dec | 0.09 | 0.30 | 0.09 | 0.20* | 0.53* | 0.15 | 0.49* | 0.01 |
*p < 0.05, **p < 0.01.
4. Discussion
By germinating resting stages of S. marinoi from selected locations and applying microsatellite markers, we demonstrated that this bloom-forming species form a distinct population structure among oceanographically well-connected sites. The differentiated populations displayed large genetic diversity and the patterns of genetic structure were best explained by local oceanographic connectivity. We did not find any seasonal pattern in gene flow supported by oceanographic connectivity. Migration of cells and consequential gene flow was supported throughout the year. This is to our knowledge the first study showing that regional circulation patterns may structure planktonic protists on fine spatial scales (less than 100 km).
The survival rate of the strains from the germinated resting stages was high. This eliminates the risk of introducing bias towards strains that are able to survive under laboratory conditions. Ninety-eight per cent of the genotyped individuals were unique. This confirms the high clonal diversity reported earlier for this [4] and other diatom species [1,3]. Skeletonema marinoi mainly reproduces asexually, but the high levels of genotypic diversity and lack of linkage between the microsatellite loci imply occasional sexual reproduction. The frequency of sexual reproduction probably varies among different species and populations [43], and therefore the contribution of reproductive modes to diversity is difficult to estimate. Populations with mainly asexual propagation, large population sizes, high growth rates and short generation time maintain high genotypic diversity even if the proportion of sexually derived individuals is low [44]. The proportion of asexually reproducing individuals is unknown, but the populations analysed here all displayed heterozygote deficiency in several loci. The deviation from HWE is possibly due to the mode of reproduction and non-random mating. This will cause a Wahlund effect and deviation from expectations under panmixia, but could also be explained, especially in some loci, from a potential presence of null alleles.
The level of genetic structure in the Skeletonema populations examined here was weaker than the high level of differentiation previously reported for the same species and other diatoms occupying specific niches of sill fjord environment versus the open sea [1,4]. Presumably, gene flow among microscopic aquatic organisms may be affected not only by physical dispersal barriers, but also by priority effects and local adaptation [45]. Such paradoxes of reduced gene flow despite high dispersal capacities in aquatic organisms have also been recorded for multicellular animals and macrophytes in ponds and rock pools [46,47]. Effects of founder events are presumably enhanced by banks of resting stages that buffer against new immigrants [45]. However, the preservation of genetic differentiation among populations collected in the open sea at well-connected sampling sites where priority effects and local adaptation may be weaker due to stronger homogenizing effects of ocean circulation is puzzling. The pairwise FST recorded here of 1–2% indicates that dispersal between subpopulations might be very low. There are few analogues among pelagic protists on equivalent geographical scales. The genetic structure of the diatom Pseudo-nitzschia pungens in the North Sea has revealed a high level of gene flow and evidence of a single, unstructured population with no genetic differentiation among different sampling sites [3]. Pseudo-nitzschia is, like Skeletonema, a bloom-forming diatom, which seasonally can reach high densities [48], but unlike Skeletonema, Pseudo-nitzschia does not produce resting stages. A proportion of the Skeletonema resting stages will sediment locally, and when re-suspended they continue to contribute to the local gene pool and support the formation of discrete populations.
Another factor that may be important is their respective means of propagation. A distinctive property of the diatom life cycle is a progressive reduction in cell size during the asexual phase. This is caused by the way diatom cells divide, and the only way to restore maximum cell size and avoid death for Pseudo-nitzschia and most other diatom species, is by sexual reproduction [49]. A few genera, including Skeletonema, have evolved vegetative cell enlargement to escape miniaturization [50]. The possibility to restore cell size without sexual reproduction thus account for a larger proportion of asexually reproducing individuals in populations of Skeletonema. If the newly arrived strains can be maintained for longer periods by asexual propagation, the gene flow is impeded. Contrary, alleles arriving from a neighbouring population will faster become integrated in the local gene pool in an obligate sexual organism. Thus, a larger proportion of asexually reproducing individuals and the ability to form resting stages anchoring Skeletonema to particular sites, may account for the observation that this genus displays a reduced level of gene flow and maintains genetic structure, also in the open sea.
The dispersal trajectories modelled here support the explanation that retention of individuals and local recruitment of the populations may lead to the observed population structure in Skeletonema. Deposition of locally produced resting stages is possible with the predicted circulation pattern, especially at the inshore stations. The modelled dispersal may even underestimate the local retention because the simulated dispersal in the surface layer yielded the highest number of significant correlations, and this is where current velocities are highest. Thus, the oceanographic data are consistent with the significant FST values. Small amount of genetic exchange is enough to prevent the accumulation of genetic differences in F-statistics. Therefore, the local seeding of a greater proportion of the population at each sample site is probably important for explaining the genetic structure.
Significant IBD patterns most commonly indicate restrictions to gene flow over broad scales [51]. Thus, the absence of a significant pattern among the examined populations over the relatively small geographical area was not surprising. Patterns of IBD have been observed in sea stars with planktonic larvae spanning different basins in the Pacific and Indian Oceans, but within east Asia, this pattern was not significant [9]. In smaller areas, or in areas of high oceanographic complexity, population genetic models of panmixia and IBD may be too simplistic to describe the barriers caused by current-induced gradients or fronts of salinity and temperature differences. For elucidating barriers or zones of low gene flow, seascape approaches have proved more useful for describing observed population structures among marine holo- and meroplanktonic organisms [52].
The oceanographic connectivity of the studied region offered a seascape genetic assessment of the gene flow among the sampling sites. In particular, the strong south to north component of the migration is certainly consistent with the oceanographic connectivity simulations. However, certain patterns of gene flow could not be detected from the matrices of oceanographic connectivity. Gene flow from the inshore to the offshore sites was more common than the opposite, but the same was not obvious from oceanographic trajectories. Tentatively, cells originating near the coast are transported westward, form resting cells which subsequently sink to the sediment at offshore sites. The number of stations investigated here are perhaps a minimum given the complexity of the oceanographic circulation, but the directional gene flow might be due to a proportionally larger number of migrating cells during the spring bloom relative to the rest of the year. The spring bloom progresses from coastal to offshore waters. The initial stratification, necessary for bloom initiation, is due to outflow of fresh water from the coastal zone. Therefore, the blooms start near the coast and propagate to offshore regions [53]. In northern temperate seas, this event dominates the annual phytoplankton productivity cycle. The spring bloom contributes half of the annual carbon fixed. Owing to the mismatch between the timing of the spring bloom and the growth of grazers, the majority of the fixed carbon sinks out of the euphotic layer and sediments [54,55]. In the Öresund–Kattegat–Skagerrak, the spring bloom is dominated by Skeletonema. Cell density is highest at this time of the year (10 000 cells ml−1), and presumably this event is responsible for a large part of the resting stages accumulation. Hypothetically, the seed banks produced by S. marinoi during the spring bloom are by far the richest, and the proportion of advected cells from inshore to offshore sites is more important for the migration patterns than analyses of oceanographic connectivity reveals.
As Skeletonema dominate the phytoplankton standing stock during the spring bloom period, hypothetically the resting stages produced, transported and settled during the spring bloom would dominate the genotyped populations. If so, the gene flow would display stronger correlation to the oceanographic connectivity during February to April. According to our analyses, no particular month or season favoured migration. On the contrary, the oceanographic connectivity supported migration throughout the year. Indeed, Skeletonema is present in the water column all year round but at varying densities. During spring, it can constitute more than 50% of the biomass, and in the autumn it is also common, constituting up to 10% of the recorded phytoplankton biomass, but in a more diverse plankton community. During summer and winter months, the lowest densities of Skeletonema are observed [17].
Some of the microsatellite loci were more strongly correlated to the matrices of oceanographic trajectories. Microsatellites, in general, exhibit high mutation rates, which are estimated to be in the order of 10−3 to 10−4 per locus and per human generation [56]. Mutation rates vary between different loci, and microsatellites with more core-repeats accumulate mutations faster [57]. Owing to the different characteristics of the microsatellite loci used, it is not surprising that the correlation of migration and oceanographic connectivity varies among the different loci. The loci were not linked and we assumed that they were neutral and unaffected by selective forces. However, given enough time in divergent environments, especially if extensive asexual reproduction is present, neutral microsatellites could also become differentiated. This is particularly true in markers linked to selected loci [58].
The position of the microsatellite loci in the genome, or possible linkage to genes affected by natural selection, is unknown. Two microsatellite loci showed no (S.mar3) or weak (S.mar1) correlation with oceanographic connectivity. Locus S.mar3 displayed a low level of polymorphism at any sampling site. Locus S.mar1 on the other hand, displayed a relatively high degree of polymorphism. This indicates that S.mar1 accumulates mutations, but also that the diversity is evenly distributed among the samples. S.mar1 might be inherited and linked to a coding gene of selective advantage in all seven populations. By contrast, the loci S.mar2, S.mar4 and S.mar7, which are less polymorphic, displayed migration rates that were significantly correlated to the oceanographic connectivity of the region for several months of the year. Simulated gene flow data have demonstrated stronger correlations between landscape and genetic distances when the microsatellites are more variable [59]. Therefore, with a different set of markers the correlations obtained could be slightly different.
Results presented here add to the growing evidence for significant population structure in pelagic marine protists and further highlights the extensive genetic diversity. We conclude that the geographical patterns and the genetic structure of S. marinoi cannot be explained by genetic models based on IBD, but are caused by local oceanographic connectivity promoting gene flow in a south to north direction. We therefore anticipate that wherever oceanographic data permit, biophysical modelling to test seascape genetic hypotheses can be informative in interpreting patterns of genetic differentiation.
Acknowledgements
We are thankful to Stefan Agrenius (University of Gothenburg) and Peter Göransson (Municipality of Helsingborg) for providing the sediment samples. The fragment analysis was performed at the Genomics Core Facility, University of Gothenburg, by Dr Elham Rekabdar.
Data accessibility
Microsatellite sequences: GenBank accessions EU855763, EU855769–EU855771, EU855775, EU855777, GQ250935, GQ250937. The Skeletonema marinoi strains are available from Gothenburg University's Marine Algal Culture Collection (GUMACC) and assessed through http://assemblemarine.org/the-sven-lov-n-centre-for-marine-sciences-tj-rn/.
Funding statement
This work was supported by grants to A.G. from Formas (2009-1185), European Community-RI Action ASSEMBLE grant no. 227799, Oscar and Lilli Lamms Minne, Wilhelm and Martina Lundgrens Vetenskapsfond, Wåhlströms Minnesfond, KVVS, and by grants to P.R.J. from the Linnaeus CeMEB at the University of Gothenburg, Formas (2008-115), and the Swedish Research Council (2011-3600).
References
- 1.Rynearson TA, Armburst EV. 2004. Genetic differentiation among populations of the planktonic marine diatom Ditylum brightwellii (Bacillariophyceae). J. Phycol. 40, 34–43 (doi:10.1046/j.1529-8817.2004.03089.x) [Google Scholar]
- 2.Nagai S, Nishitani G, Sakamoto S, Sugaya T, Lee C, Kim C, Itakura S, Yamaguchi M. 2009. Genetic structururing and transfer of marine dinoflagellate Cochlodinium polykrikoides in Japanese and Korean coastal waters revealed by microsatellites. Mol. Ecol. 18, 2337–2352 (doi:10.1111/j.1365-294X.2009.04193.x) [DOI] [PubMed] [Google Scholar]
- 3.Evans KM, Kühn SF, Hayes PK. 2005. High levels of genetic diversity and low levels of genetic differentiation in North Sea Pseudo-nitzschia pungens (Bacillariophyceae) populations. J. Phycol. 41, 506–514 (doi:10.1111/j.1529-8817.2005.00084.x) [Google Scholar]
- 4.Godhe A, Härnström K. 2010. Linking the planktonic and benthic habitat: genetic structure of the marine diatom Skeletonema marinoi. Mol. Ecol. 19, 4478–4490 (doi:10.1111/j.1365-294X.2010.04841.x) [DOI] [PubMed] [Google Scholar]
- 5.White C, Delkoe KA, Watson J, Siegel DA, Zacherl DC, Toonen RJ. 2010. Ocean currents help explain population genetic structure. Proc. R. Soc. B 277, 1685–1694 (doi:10.1098/rspb.2009.2214) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Casabianca S, Penna A, Pecchioli E, Jordi A, Basterretxea G, Vernesi C. 2012. Population genetic structure and connectivity of the harmful dinoflagellate Alexandrium minutum in the Mediterranean Sea. Proc. R. Soc. B 279, 129–138 (doi:10.1098/rspb.2011.0708) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wright S. 1951. The genetical structure of populations. Ann. Eugenics 15, 323–354 (doi:10.1111/j.1469-1809.1949.tb02451.x) [DOI] [PubMed] [Google Scholar]
- 8.Treml EA, Halpin PN, Urban DL, Pratson LF. 2008. Modeling population connectivity by ocean currents, a graph-theoretic approach for marine conservation. Landscape Ecol. 23, 19–36 (doi:10.1007/s10980-007-9138-y) [Google Scholar]
- 9.Yasuda N, Nagai S, Hamaguchi M, Okaji K, Gerard K, Nadaoka K. 2009. Gene flow of Acanthaster planci (L.) in relation to ocean currents revealed by microsatellite analysis. Mol. Ecol. 18, 1574–1590 (doi:10.1111/mec.2009.18.issue-8) [DOI] [PubMed] [Google Scholar]
- 10.Serra IA, Innocenti AM, Maida GD, Calvo S, Migliaccio M, Zambianchi E, Pizzigalli C, Arnaud-Haond S, Duarte CM. 2010. Genetic structure in the Mediterranean seagrass Posidonia oceanica: disentangling past vicariance events from contemporary patterns of gene flow. Mol. Ecol. 19, 557–568 (doi:10.1111/j.1365-294X.2009.04462.x) [DOI] [PubMed] [Google Scholar]
- 11.McQuoid MR, Godhe A, Nordberg K. 2002. Viability of phytoplankton resting stages in the sediments of a coastal Swedish fjord. Eur. J. Phycol. 37, 191–201 (doi:10.1017/S0967026202003670) [Google Scholar]
- 12.Hairston NG, Vanbrunt RA, Kerrns CM, Engstrom DR. 1995. Age and survivorship of diapausing eggs in a sediment egg bank. Ecology 76, 1706–1711 (doi:10.2307/1940704) [Google Scholar]
- 13.Anderson DM, Fukuyo Y, Matsuoka K. 1995. Cyst methodologies. In Manual on harmful marine microalgae (eds Hallegraeff GM, Anderson DM, Cembella AD.), pp. 229–249 IOC Manuals and Guides Paris, France: UNESCO [Google Scholar]
- 14.Tahvanainen P, Figueroa RI, Alpermann TJ, John U, Hakanen P, Nagai S, Blomster J, Kremp A. 2012. Patterns of post-glacial genetic differentiation in marginal populations of a marine microalga. PLoS ONE 7, e53602 (doi:10.1371/journal.pone.0053602) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sarno D, Kooistra W, Balzano S, Hargraves P, Zingone A. 2007. Diversity in the genus Skeletonema (Bacillariophyceae). III. Phylogenetic position and morphological variability of Skeletonema costatum and Skeletonema grevillei, with the description of Skeletonema ardens sp. nov. J. Phycol. 43, 156–170 (doi:10.1111/j.1529-8817.2006.00305.x) [Google Scholar]
- 16.Ellegaard M, Godhe A, Härnström K, McQuoid M. 2008. The species concept in a marine diatom: LSU rDNA-based phylogenetic differentiationin Skeletonema marinoi/dohrnii (Bacillariophyceae) is not reflected in morphology. Phycologia 47, 156–167 (doi:10.2216/07-79.1) [Google Scholar]
- 17.Saravanan V, Godhe A. 2010. Physiologic differentiation and genetic heterogeneity among seasonally separated clones of Skeletonema marinoi (Bacillariophyceae) in Gullmar Fjord, Sweden. Eur. J. Phycol. 45, 177–190 (doi:10.1080/09670260903445146) [Google Scholar]
- 18.Taylor R, Abrahamsson K, Godhe A, Wängberg S-Å. 2009. Seasonal variability in polyunsaturated aldehyde production potential between strains of Skeletonema marinoi (Bacillariophyceae). J. Phycol. 45, 46–53 (doi:10.1111/j.1529-8817.2008.00625.x) [DOI] [PubMed] [Google Scholar]
- 19.Migita S. 1967. Sexual reproduction of centric diatom Skeletonema costatum. Bull. Jpn Soc. Sci. Fish. 33, 392–398 (doi:10.2331/suisan.33.392) [Google Scholar]
- 20.Godhe A, McQouid M, Karunasagar I, Karunasagar I, Rehnstam-Holm A-S. 2006. Comparison of three common molecular tools for distinguishing among geographically separated clones of the diatom Skeletonema marinoi Sarno et Zingone (Bacillariophyceae). J. Phycol. 42, 280–291 (doi:10.1111/j.1529-8817.2006.00197.x) [Google Scholar]
- 21.Schunter C, Carreras-Carbonell J, Macpherson E, Tintoré J, Vidal-Vijande E, Pascual A, Guidetti P, Pascual M. 2011. Matching genetics with oceanography: directional gene flow in a Mediterranean fish species. Mol. Ecol. 20, 5167–5181 (doi:10.1111/j.1365-294X.2011.05355.x) [DOI] [PubMed] [Google Scholar]
- 22.Lindahl O, Belgrano A, Davidsson L, Hernroth B. 1998. Primary production, climatic oscillation, and physio-chemical processes: the Gullmar Fjord time-series data set (1985–1996). ICES J. Mar. Sci. 55, 723–729 (doi:10.1006/jmsc.1998.0379) [Google Scholar]
- 23.Gustafsson M, Nordberg K. 1999. Benthic foraminifera and their response to hydrography, periodic hypoxic conditions and primary production in the Koljö Fjord on the Swedish west coast. J. Sea Res. 41, 163–178 (doi:10.1016/S1385-1101(99)00002-7) [Google Scholar]
- 24.Guillard RRL. 1975. Culture of phytoplankton for feeding marine invertebrates. In Culture of marine invertebrate animals (eds Smith W, Chanley M.), pp. 29–60 New York, NY: Plenum Press [Google Scholar]
- 25.Kooistra WHCF, DeStefano M, Mann DG, Salma N, Medlin LK. 2003. Phylogenetic position of Toxarium, a pennate-like lineage within centric diatoms (Bacillariophyceae). J. Phycol. 39, 185–197 (doi:10.1046/j.1529-8817.2003.02083.x) [Google Scholar]
- 26.Almany GR, et al. 2009. Permanent genetic resources added to molecular ecology resources database 1 May 2009–31 July 2009. Mol. Ecol. Resour. 9, 1460–1466 (doi:10.1111/j.1755-0998.2009.02759.x) [DOI] [PubMed] [Google Scholar]
- 27.Raymond M, Rousset F. 1995. GENEPOP (version 1.2): population genetics software for exact tests and ecumenicism. J. Hered. 86, 248–249 [Google Scholar]
- 28.Rice WR. 1989. Analyzing tables of statistical tests. Evolution 43, 223–225 (doi:10.2307/2409177) [DOI] [PubMed] [Google Scholar]
- 29.Park SDE. 2001. Trypanotolerance in West African Cattle and the population genetic effects of selection. PhD thesis, University of Dublin, Dublin, Ireland [Google Scholar]
- 30.van Oosterhout C, Weetman D, Hutchinson WF. 2006. Estimation and adjustment of microsatellite null alleles in nonequilibrium populations. Mol. Ecol. Notes 6, 255–256 (doi:10.1111/j.1471-8286.2005.01082.x) [Google Scholar]
- 31.Brookfield JFY. 1996. A simple new method for estimating null allele frequency from heterozygote deficiency. Mol. Ecol. 5, 453–455 (doi:10.1111/j.1365-294X.1996.tb00336.x) [DOI] [PubMed] [Google Scholar]
- 32.Excoffier L, Laval G, Schneider S. 2005. Arlequin ver. 3.0: an integrated software package for population genetics data analysis. Evol. Bioinform. Online 1, 47–50 [PMC free article] [PubMed] [Google Scholar]
- 33.Jost L. 2008. GST and its relatives do not measure differentiation. Mol. Ecol. 17, 4015–4026 (doi:10.1111/j.1365-294X.2008.03887.x) [DOI] [PubMed] [Google Scholar]
- 34.Gerlach G, Jueterbock A, Kraemer P, Deppermann J, Harmand P. 2010. Calculations of population differentiation based on GST and D: forget GST but not all of statistics! Mol. Ecol. 19, 3845–3852 (doi:10.1111/j.1365-294X.2010.04784.x) [DOI] [PubMed] [Google Scholar]
- 35.Sundqvist L, Zackrisson M, Kleinhans D. 2013. Directional genetic differentiation and asymmetric migration. J. Theor. Biol. (http://arxiv.org/abs/1304.0118) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Madec G. 2010. Nemo ocean engine, v. 3.3. Technical report, IPSL, Paris, France [Google Scholar]
- 37.Hordoir R, Dieterich C, Basu C, Dietze H, Meier HEM. 2013. Freshwater outflow of the Baltic Sea and transport in the Norwegian current: a statistical correlation analysis based on a numerical experiment. Cont. Shelf Res. 64, 1–9 (doi:10.1016/j.csr.2013.05.006) [Google Scholar]
- 38.Adcroft A, Campin J-M. 2004. Re-scaled height coordinates for accurate representation of free-surface fows in ocean circulation model. Ocean Model 7, 269–284 (doi:10.1016/j.ocemod.2003.09.003) [Google Scholar]
- 39.Egbert G, Bennett A, Foreman M. 1994. Topex/poseidon tides estimated using a global inverse model. J. Geophys. Res. Oceans 99, 24821–24852 (doi:10.1029/94JC01894) [Google Scholar]
- 40.Levitus S, Boyer TP. 1994. World Ocean Atlas 1994, vol. 5: salinity. NOAA Atlas Washington, DC: US Government Printing Office [Google Scholar]
- 41.Döös K. 1995. Inter-ocean exchange of water masses. J. Geophys. Res. Oceans 100, 13 499–13 514 (doi:10.1029/95JC00337) [Google Scholar]
- 42.Rosenberg M, Anderson C. 2011. PASSaGE: pattern analysis, spatial statistics and geographic exegesis, v. 2. Method Ecol. Evol. 2, 229–232 (doi:10.1111/j.2041-210X.2010.00081.x) [Google Scholar]
- 43.Mann DG. 1993. Patterns of sexual reproduction in diatoms. Hydrobiologia 269/270, 11–20 (doi:10.1007/BF00027999) [Google Scholar]
- 44.Bengtsson BO. 2003. Genetic variation in organisms with sexual and asexual reproduction. J. Evol. Biol. 16, 189–199 (doi:10.1046/j.1420-9101.2003.00523.x) [DOI] [PubMed] [Google Scholar]
- 45.DeMeester L, Goméz A, Okamura B, Schwenk K. 2002. The monopolization hypothesis and the dispersal–gene flow paradox in aquatic organisms. Acta Oecol. 23, 121–135 (doi:10.1016/S1146-609X(02)01145-1) [Google Scholar]
- 46.Palsson S. 2000. Microsatellite variation in Daphnia pulex from both sides of the Baltic Sea. Mol. Ecol. 9, 1075–1088 (doi:10.1046/j.1365-294x.2000.00969.x) [DOI] [PubMed] [Google Scholar]
- 47.Campillo S, Garcia-Roger EM, Carmona MJ, Gomez A, Serra M. 2009. Selection on life-history traits and genetic population divergence in rotifers. J. Evol. Biol. 22, 2542–2553 (doi:10.1111/j.1420-9101.2009.01871.x) [DOI] [PubMed] [Google Scholar]
- 48.Sarno D, Zingone A, Montresor M. 2010. A massive and simultaneous sex event of two Pseudo-nitzschia species. Deep Sea Res. II 57, 248–255 (doi:10.1016/j.dsr2.2009.09.012) [Google Scholar]
- 49.Round FE, Crawford RM, Mann DG. 1990. The diatoms. Cambridge, UK: Cambridge University Press, 747 [Google Scholar]
- 50.Gallagher JC. 1983. Cell enlargement in Skeletonema costatum (Bacillariophyceae). J. Phycol. 19, 539–542 (doi:10.1111/j.0022-3646.1983.00539.x) [Google Scholar]
- 51.Casteleyn G, Leliaert F, Backeljau T, Debeer A-E, Kotakai Y, Rhodes L, Lundholm N, Sabbe K, Vyverman W. 2010. Limits to gene flow in a cosmopolitan marine planktonic diatom. Proc. Natl Acad. Sci. USA 107, 12 952–12 957 (doi:10.1073/pnas.1001380107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kenchington EL, Patwary MU, Zouros E, Bird CJ. 2006. Genetic differentiation in relation to marine landscape in a broadcast-spawning bivalve mollusc (Placopecten magellanicus). Mol. Ecol. 15, 1781–1796 (doi:10.1111/j.1365-294X.2006.02915.x) [DOI] [PubMed] [Google Scholar]
- 53.Stipa T. 2002. Temperature as a passive isopycnal tracer in salty, spiceless oceans. Geophys. Res. Lett. 29, 1953–1957 (doi:10.1029/2001GL014532) [Google Scholar]
- 54.Lignell R. 1993. Fate of a phytoplankton spring bloom: sedimentation and carbon flowin the planktonic food web in the northern Baltic. Mar. Ecol. Prog. Ser. 94, 239–252 (doi:10.3354/meps094239) [Google Scholar]
- 55.Tiselius P, Kuylenstierna M. 1996. Growth and decline of a diatom spring bloom: phytoplankton species composition, formation of marine snow and the role of heterotrophic dinoflagellates. J. Plankton Res. 18, 133–155 (doi:10.1093/plankt/18.2.133) [Google Scholar]
- 56.Ellegren H. 2004. Microsatellites: simple sequences with complex evolution. Nat. Rev. Genet. 5, 435–443 (doi:10.1038/nrg1348) [DOI] [PubMed] [Google Scholar]
- 57.Tesson SVM, Legrand C, van Oosterhout C, Montresor M, Kooistra WHCF, Procaccini G. 2013. Mendelian inheritance pattern and high mutation rates of microsatellite alleles in the diatom Pseudo-nitzschia multistriata. Protist 164, 89–100 (doi:10.1016/j.protis.2012.07.001) [DOI] [PubMed] [Google Scholar]
- 58.Thibert-Plante X, Hendry AP. 2010. When can ecological speciation be detected with neutral markers? Mol. Ecol. 19, 2301–2314 (doi:10.1111/j.1365-294X.2010.04641.x) [DOI] [PubMed] [Google Scholar]
- 59.Landguth EL, Fedy BC, Oyler-McCance SJ, Garey AL, Emel SL, Mumma M, Wagner HH, Fortin M-J, Cushman SA. 2011. Effects of sample size, number of markers, and allelic richness on the detection of spatial genetic pattern. Mol. Ecol. Resour. 12, 276–284 (doi:10.1111/j.1755-0998.2011.03077.x) [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Microsatellite sequences: GenBank accessions EU855763, EU855769–EU855771, EU855775, EU855777, GQ250935, GQ250937. The Skeletonema marinoi strains are available from Gothenburg University's Marine Algal Culture Collection (GUMACC) and assessed through http://assemblemarine.org/the-sven-lov-n-centre-for-marine-sciences-tj-rn/.