Abstract
The interactions between bacteria and fungi, the main actors of the soil microbiome, remain poorly studied. Here, we show that the saprotrophic and ectomycorrhizal soil fungus Morchella crassipes acts as a bacterial farmer of Pseudomonas putida, which serves as a model soil bacterium. Farming by M. crassipes consists of bacterial dispersal, bacterial rearing with fungal exudates, as well as harvesting and translocation of bacterial carbon. The different phases were confirmed experimentally using cell counting and 13C probing. Common criteria met by other non-human farming systems are also valid for M. crassipes farming, including habitual planting, cultivation and harvesting. Specific traits include delocalization of food production and consumption and separation of roles in the colony (source versus sink areas), which are also found in human agriculture. Our study evidences a hitherto unknown mutualistic association in which bacteria gain through dispersal and rearing, while the fungus gains through the harvesting of an additional carbon source and increased stress resistance of the mycelium. This type of interaction between fungi and bacteria may play a key role in soils.
Keywords: Morchella crassipes, Pseudomonas putida, mutualism, dispersal, exudate consumption, sclerotia and melanization
1. Introduction
Soils are exceptionally complex and highly dynamic systems that have arisen from the interaction of biotic and abiotic processes over billions of years [1]. Soils are the most important reservoir of biodiversity on the planet, which is especially true for microorganisms. Those perform key processes such as decomposition and nutrient cycling, which influence soil fertility, productivity and biogeochemical cycling [2,3]. Although the importance of fungi and bacteria for ecosystem functioning is evident, the interactions between these organisms in soil are still poorly understood [4,5]. Several examples show that interactions between microorganisms can impact plant productivity (e.g. mycorrhizae [6,7] or nitrogen-fixing rhizobacteria [8]) or even metabolic processes in soil (e.g. degradation of specific substrates [9,10]). In addition, an increasing body of literature details the ability of microbes to cooperate by intra- and interspecies interactions [11–14].
Soil is a challenging system. Its opaque nature renders difficult the direct study of its biological constituents [15]. In addition, soil as a habitat poses specific constraints for the development of microbial life. Soils are highly porous materials in which approximately 50% (in a typical topsoil) comprises a mix of water and air-filled gaps that fluctuates according to prevailing environmental conditions (e.g. rainfall [1]). The structure of the pore space results in an irregular distribution of water films and substrates, affecting the living organisms within [16]. At a microscopic scale, discrete water films restrict the dispersal of individual microbial cells or populations [17]. The link between diversity and dispersal [18,19] has led to the hypothesis that bacterial diversity in soils partly reflects the limitation to free dispersal in water-unsaturated media [20,21].
Soil fungi can serve as dispersal routes for certain bacteria, which use fungal hyphae as so-called ‘fungal highways’ [22]. This facilitates dispersal and would in theory provide an important benefit for soil bacteria, as they cannot disperse efficiently otherwise [20,22,23]. Other benefits to bacteria have been proposed (i.e. exudates consumption), but experimental evidence is still missing; and no benefit to the fungus has been suggested. In this context, we analysed the mutual benefit to the saprotrophic and ectomycorrhizal soil fungus Morchella crassipes (L.) Pers. (thick-footed morel) in dispersing bacteria using its mycelial network. A strain of Pseudomonas putida constitutively expressing the green fluorescent protein (GFP) [20] was used as a model soil bacterium. Our hypothesis was that not only bacteria but also the fungus would benefit from the interaction. To test this, we designed experiments to evaluate the benefits of bacterial dispersal and availability of an additional nutrient source in the form of fungal exudates. No benefit had been reported for the fungus, therefore we evaluated various scenarios including increase in mycelial growth rate, use of bacteria as an additional nutrient source and the effect of co-culturing on survival strategies (i.e. formation of resting bodies called sclerotia [24]) or stress response (i.e. mycelia melanization [25,26]).
The results pointed towards a conceptual model where the fungus acts as a farmer of bacteria. Cultivation of crops for nourishment has evolved only a few times among eukaryotes. The most unambiguous examples include ants [27], termites [28], ambrosia beetles [29,30] and humans [31]. In the case of ants, termites and beetles, fungi are used as crops and the farming species are dependent on the crop for food. Humans started to transition to farming about 10 000 years ago, and agriculture has become critical for our survival. The defining features of insect and human farming include habitual planting or inoculation of sessile cultivars in particular habitats or substrates, cultivation (improvement of the crop's growth conditions) or protection of the crop, harvesting of the cultivar and obligate (or effectively obligate as in humans) nutritional dependence on the crop. Other features such as artificial selection or development and cultural transmission of agricultural innovations appear to be either absent or unknown except in humans [32]. Examples that do not fulfil all criteria listed above are considered animal husbandry or proto-farming and include tending by ants of honeydew-producing aphids [33], snails that feed on fungi cultured on plant wounds [34] or the dispersal and harvesting of bacteria by social amoeba [35].
Our results indicate that bacterial farming by M. crassipes includes habitual planting, cultivation and harvesting, which are common to other unambiguous farming systems. Furthermore, specific features common only to human agriculture were also observed such as the delocalization of food production (carbon translocation to sclerotia) and the separation of roles within the fungal colony. Finally, the dependence on the crop was evaluated in experiments with two other bacterial species in order to test whether such a mutualistic interaction is either a specific or a widespread phenomenon.
2. Material and methods
(a). Selection of fungal and bacterial strains
We chose M. crassipes (L.) Pers (thick-footed morel) because this fungus forms sclerotia [24,36,37] and melanizes hyphae in response to environmental stress. The NEU ML1 strain was isolated from a forest soil in Switzerland and is affiliated at 99% identity (1461 bp) to the variety crassipes (M. crassipes QFB7377) according to the ribosomal internal transcribed spacer (ITS) sequences 1 and 2, and the 5.8S rRNA gene. The ITS sequence was deposited under the accession number JX258671 in GenBank. Pseudomonas putida KT2440 was selected as a model soil bacterium often associated with the mycorhizosphere [38] and known to migrate on fungal hyphae [39,40]. Pseudomonas putida KT2440 constitutively expresses the GFP. In addition, an isogenic non-flagellated mutant (strain ΔfliM) expressing the mCherry fluorescent protein was used for the fitness experiment. Both strains were kindly provided by Dr Arnaud Dechesne (Technical University of Denmark).
(b). Experimental system
We co-inoculated M. crassipes and P. putida in 1.5% agar Petri dishes with 12 g l–1 malt extract (Mycotec, Switzerland). Unless otherwise stated, agar was technical grade (Biolife, Italy). The fungus was inoculated in the centre of the plate from a 5-day-old M. crassipes culture. The inoculum (edge of the culture) was obtained using the wider end of a Pasteur capillary pipette. To avoid passive diffusion, the bacterial inoculum consisted of a 5 µl bacterial suspension containing 108 cells µl–1 that was plated as a line 2 cm away or at various places around the fungal inoculum after 1 day of fungal incubation. Bacteria came from an overnight culture in Nutrient Agar (Biolife, Italy). After the overnight incubation, bacteria were suspended in a saline aqueous solution (0.9% w/v NaCl) to adjust the cell concentration. All the experiments were performed at 21°C.
Fungal growth was directly observed on the plates. Bacterial dispersal was assessed by observing the plates with a Nikon (Japan) SMZ1000 epifluorescence stereoscope. Dispersal to the single cell level was assessed by observing some samples in confocal laser scanning microscopy (CLSM). CLSM observations were performed with a TCS SP5X (Leica) confocal microscope attached to an upright microscope equipped with a 63× NA 1.2 water immersion objective. Excitation was performed with a white laser line at 488 nm with 50% light intensity and emissions signals were detected at 482–495 nm (reflection) and at 500–550 nm (GFP).
(c). Bacterial consumption of fungal exudates
In order to test for a carbon transfer from the fungus to bacteria during dispersal (rearing phase of farming), fungi were grown in a Schlegel mineral technical agar medium [41] supplemented with 4 g l–1 13C-glucose (99.9% 13C, Sigma–Aldrich-Supelco Chemie GmbH, Switzerland). After 5 days of incubation, all the mycelium was collected and washed three times with a saline solution (0.9% w/v NaCl) to remove un-incorporated labelled glucose. Washing consisted of vortexing for 10 s, centrifugation at 4000 r.p.m. for 2 min and removal of the supernatant solution. The labelled fungus was inoculated on unlabelled malt extract-technical agar medium. The bulk stable carbon isotope ratio (δ13C value in ‰ versus Vienna Pee Dee Belemnite standard, VPDB) of the malt extract was determined to be –27.5 ± 0.2‰ by flash combustion on a Carlo Erba 1108 (Italy) elemental analyzer connected to a Thermo Fisher Scientific Delta V (Bremen, Germany) isotope ratio mass spectrometer that was operated in the continuous helium flow mode via a Conflo III split interface (EA-IRMS). Unlabelled bacteria were inoculated as described above but 0.5 cm away from the fungal inoculum. After 5 days of incubation, the whole plate was washed with a saline solution to collect bacteria. As a control, the same experiment was performed using an unlabelled fungus. Both experiments were repeated three times. The six samples were submitted to phospholipid fatty acids extraction and stable carbon isotope analysis of bacterial biomarkers (see below). A significant 13C enrichment of these biomarkers can only be attributed to carbon transfer from the fungus.
(d). Influence of the fungus on bacterial growth
To test the direct benefits to bacteria of dispersal on fungal mycelium, the number of colony-forming units (CFUs; detection threshold of ≈10 cells sample−1) was measured on malt agar plates (technical and purified agar—Merck, Germany) in the presence or absence of a pre-established mycelial network of M. crassipes. Eight ml of medium were poured in small 50 mm Petri dishes; a 5-day-old fungal inoculum was immediately inoculated as described before. After 24 h, the agar plug with the inoculum was removed and a bacterial suspension containing 5.9 × 106 CFU ml−1 was inoculated at the centre of the plate. In a second experiment, the wild-type P. putida strain was compared with an isogenic non-flagellated mutant (ΔfliM). The two strains were mixed and inoculated from a cell suspension containing 3.0 × 104 and 4.0 × 104 cells of each strain, respectively. After 48 h of incubation, the plates were washed with a saline solution (0.9% w/v NaCl). Total cell numbers were thus determined in five replicates by CFU counting. These values were used to calculate the relative fitness of the bacterium [42] in the presence and absence of the fungus. An unpaired t-test on log-transformed CFU data was used for the analysis.
(e). Influence of bacteria on the fungus’ growth rate
The diameter of M. crassipes colonies after 3 days of incubation with and without bacteria (inoculated as described above) was measured in 11 replicates of each treatment. An unpaired t-test was used to compare the datasets.
(f). Monitoring of bacterial harvesting
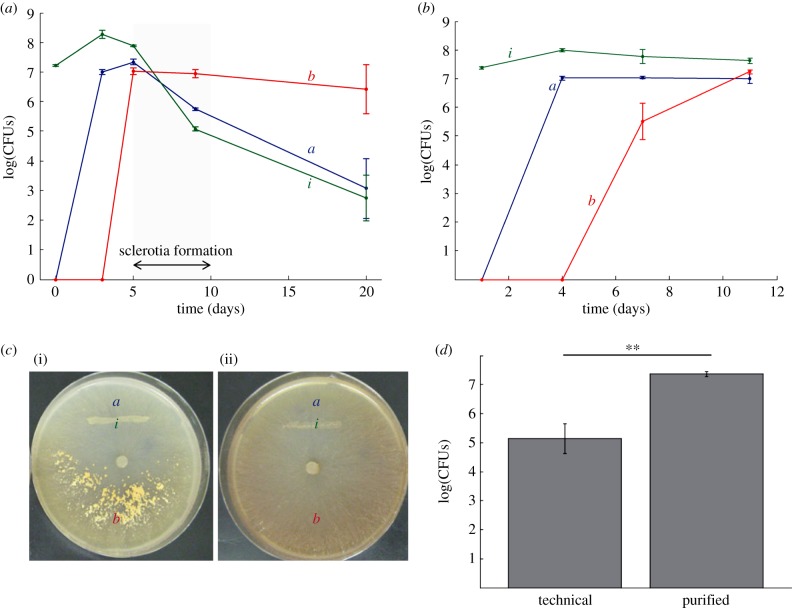
To obtain a quantitative measurement of bacterial population dynamics during co-culture with M. crassipes, the number of CFU per unit area was measured in three zones across the Petri dish: inoculation area, 1 cm from the inoculum and opposite to the inoculum. The narrow end of a Pasteur capillary pipette was used to collect a constant surface area. The resulting agar plug was washed and vortexed in a saline solution (0.9% w/v NaCl), and the suspended bacterial population assessed by CFU counting (3 independent replicates). The time points analysed were: 1 day (bacterial inoculation), 3 days, 5 days (shortly before sclerotial formation), 9 days (after sclerotial formation) and 20 days. The same experiment was carried out using purified agar. In purified agar, metal concentrations are lower than in technical agar, in particular for Mn and Zn (see electronic supplementary material, table S1). Mn, which is known to be important for mycelial growth of M. crassipes var. crassipes [43], was l3 times less concentrated in purified than in technical agar. In purified agar, the formation of sclerotia is inhibited. As in our conceptual model of fungal farming, sclerotia are the fungal storage structures for the harvested bacterial carbon; having the ability to suppress sclerotia allows us to test this point of our working hypothesis. The time points analysed for purified agar were 1, 4, 7 and 11 days. Finally, for a robust statistical comparison of the effect of sclerotia formation on the decrease of bacterial populations in the inoculum zone, an additional experiment with eight replicates each was conducted in technical (sclerotia formed) and purified (no sclerotia) agar. Bacteria were quantified after 12 days of incubation. An unpaired t-test on log-transformed data was used for the statistical analysis.
(g). Transfer of bacterial carbon during the harvesting phase
To test for carbon transfer from bacteria to fungi during the harvesting phase of farming, bacteria were grown in the same 13C-labelled medium as described above for fungal labelling. After 24 h of incubation, labelled bacteria were collected and washed three times as described for the fungus. An unlabelled fungus was inoculated at the centre of a Petri dish filled with unlabelled malt extract–technical agar medium. Labelled bacteria were inoculated 2 cm away from the fungal inoculum, 5 days after the fungal inoculation. Bacteria were inoculated as late as possible before formation of sclerotia to limit the isotopic dilution (division, dispersal and respiration) of the labelled bacterial population. Sclerotia were sampled 9 days after the fungal inoculation in 3 plates. As a control, the same experiment was repeated with unlabelled bacteria.
(h). Biomarkers extraction
The extraction of phospholipids from the bacteria and fungus was achieved by a modified one-phase extraction procedure [44]. The samples were lyophilized and stored at –20°C before analyses. The dried material was suspended with a single-phase solvent mixture (methanol/dichloromethane/phosphate buffer; 2 : 1 : 0.8; v/v/v; electronic supplementary material), vortexed for 2 min followed by sonication for 10 min. The supernatant solvent containing the extracted lipids was removed and stored in a separation funnel. The extraction was repeated four times and the supernatants combined. Dichloromethane and phosphate buffer were added to achieve a ratio of methanol/dichloromethane/phosphate buffer of 1 : 1 : 0.9 (v/v/v). Suspensions were mixed vigorously and separated for 1 h. After phase separation, the organic extract phase was removed, dried over anhydrous sodium sulfate and gently evaporated under clean N2 flow.
Fatty acid methyl esters (FAMEs) were prepared from the extracted acyl lipid mixture by acid-catalysed methanolysis [45]. The dried extracted lipids were dissolved in 0.20 ml toluene, and 1.5 ml methanol and 0.30 ml 8.0% HCl/methanol solution were added, in that order. The mixture was vortexed and incubated at 45°C for 14 h. After cooling to room temperature, 1 ml of hexane and 1 ml of reagent-grade water were added and the mixture vortexed for 1 min. The upper hexane layer containing the FAMEs was transferred to 2 ml gas chromatography vials with Teflon-lined screw caps and stored at +4°C until gas chromatography analysis (Agilent gas chromatograph 6890 coupled to an Agilent 5973 quadrupole mass selective detector (GC-MS), USA; Thermo Fisher Scientific (Bremen, Germany) Delta V isotope ratio mass spectrometer by a combustion (C) interface III (GC-C-IRMS)).
(i). Identification of bacterial and fungal biomarkers by GC-MS
As the experiments always involved the co-culture of the fungus and bacteria, to quantify the transfer of the C isotopic signature, specific lipid markers were identified from pure cultures of P. putida and M. crassipes. Chemical characterization of the lipids was performed with a GC-MS. For the analyses of FAME fractions, the system was equipped with an Agilent free fatty acids phase-fused silica capillary column (50 m length, 0.20 mm i.d.) coated with nitroterephthalic acid modified polyethylene glycol stationary phase (film thickness 0.33 µm). A sample aliquot was injected splitless at a temperature of 200°C. Helium was used as carrier gas (1 ml min−1 flow rate). After an initial period of 2 min at 100°C, the column was heated to 240°C at 5°C min−1 followed by an isothermal period of 30 min. The MS was operated in the electron impact mode at 70 eV, source temperature of 250°C, emission current of 1 mA and multiple-ion detection with a mass range from 50 to 600 amu. Compound identifications were based on comparison of standards, gas chromatography retention time and mass spectrometric fragmentation patterns. Palmitoleic acid (16 : 1ω7) and 2-hexyl cyclopropane octanoic acid (cy17 : 0) were used as bacterial biomarkers, and linoleic acid (18 : 2ω6) as fungal biomarker. This choice is consistent with other studies [46,47].
(j). Isotopic analysis by GC-C-IRMS
Compound-specific stable carbon isotope analyses (determination of the δ13C values) of the fatty acids were obtained with a GC-C-IRMS under a continuous helium flow. The GC was operated with the same type of column and temperature program used for GC-MS analyses. The stable isotope composition of carbon was reported as δ13C values (‰ versus VPDB standard).
(k). Mycelial response to an environmental stress
To evaluate the effect of co-culturing on fungal stress response, the same experimental set-up was conducted for the inoculation of the fungus and the bacteria but varying the drying period of the Petri dish. The degree of mycelial melanization was used as a proxy for the fungal stress response. The medium (25 ml) was poured at 60°C and the open plates were dried for precisely 15 or 23 min under a constant laminar flow (470 m3 h–1). A last set of plates were dried for 15 min but aged for a week before inoculation. Fungal and bacterial inoculations were performed as described before. A protocol to extract melanin was adapted from a previous report [48]. Briefly, mycelium was immersed in 3 ml 1N NaOH, sonicated for 10 min and heated in a boiling water bath for 45 min. The resulting solutions were measured for absorbance at 500 nm as indicated elsewhere [49].
(l). Quantification of the bacterial population associated to sclerotia
Bacterial populations on individual sclerotia were assessed from seven plates after 10 days of incubation. Each sclerotium was suspended in 1 ml of saline/detergent solution (0.9% w/v NaCl), Tween 80 (0.12 mM) and water (1 ml). The suspension was vortexed for 10 s and the bacterial population in the suspension assessed by CFU counting. After a washing cycle, the sclerotium was removed and washed in fresh saline/detergent solution. Washing was repeated until no additional bacteria were detected (seven times) in the washing solution. For the second-generation farming, an unwashed 10-day-old sclerotium from co-cultures with bacteria was transferred to the centre of a fresh malt extract–technical agar plate. Fungal growth and bacterial dispersal were followed as mentioned before.
3. Results and discussion
When M. crassipes was co-inoculated with P. putida KT2440, bacteria dispersed throughout the fungal network (figure 1a,b) even across an air barrier (see electronic supplementary material, figure S1). Dispersal in an unsaturated environment is a benefit for bacteria in the interaction with the fungus, as has been suggested in the past for other bacterial strains dispersing using the fungal mycelia network [22,40,50].
Figure 1.
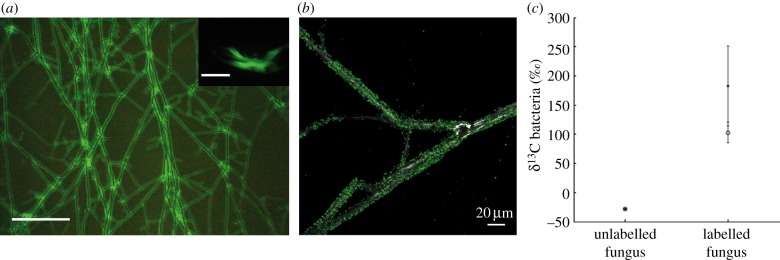
Benefits of the bacterial partner. (a) Dispersal of P. putida on the mycelium of M. crassipes. The insert shows dispersion of bacteria alone. Scale bars represent 500 µm. (b) Confocal microscopy showing maximum intensity projections of migrating GFP-labelled P. putida cells (fluorescence) on the fungal hyphae of M. crassipes (reflection). (c) Bacterial biomarkers (cy17 : 0, filled circles and 16 : 1ω7, open circles) were significantly 13C-enriched during the rearing phase compared to a control with an unlabelled fungus.
Other studies of fungus-driven bacterial dispersal suggest the consumption of fungal exudates as an additional benefit for the bacterial partner [4]. However, this had never been shown experimentally. Specific experiments were conducted to test the assimilation of fungal material during the colonization of the fungal network (i.e. rearing phase of farming). Pseudomonas putida KT2440 was co-inoculated with a fungal inoculum labelled with 13C-glucose. Bacterial biomarkers (cy17 : 0 and 16 : 1ω7; electronic supplementary material, figure S2) were significantly enriched in 13C when bacteria grew and dispersed in the fungal network (see electronic supplementary material, table S2; figure 1c). This is consistent with the observation that mycosphere bacteria are often specialized in the metabolization of fungal exudates [5,51], and we provide here, to the best of our knowledge, the first direct evidence of carbon transfer from fungi to bacteria.
We also assessed the direct effect of fungus-driven dispersal on bacterial density (table 1). When the bacterium was inoculated alone, there was no fitness effect (unpaired t-test on log CFU data, n = 8, p = 0.82 for technical and p = 0.94 for purified agar) in the co-culturing with the fungus, in both technical and purified agar. Although this was unexpected given the fact that the bacterium can colonize a larger surface of the plate in the presence of the fungus, this can be explained by a competition for resources with the latter or a shift into the utilization of a combined carbon source (malt and exudates), which can affect the growth rate of the bacterium. To take into account the role of dispersal on bacterial fitness, a second experiment was conducted comparing a non-flagellated mutant that cannot disperse with the wild-type strain. Fitness for the wild-type strain was significantly higher in the presence than in the absence of the fungus (unpaired t-test on log CFU data, n = 10, p = 0.01), with an increase of six times in relative cell density. By contrast, for the non-flagellated mutant there was no fitness effect (unpaired t-test on log CFU data, n = 8, p = 0.94). These results show that bacteria benefit from the co-culturing, in particular in a condition in which dispersal can offer an advantage, for example during competition with other bacteria that are unable to disperse using fungal hyphae.
Table 1.
Effect of co-culturing with the fungus on the fitness of P. putida. Fitness was assessed by measuring cell numbers in the presence or absence of the fungus. The inoculation was carried out in technical and purified agar, either for the wild-type strain (indicated as flagellated) alone (single inoculation) or in competition with an isogenic ΔfliM non-flagellated mutant (indicated as non-flagellated). The relative fitness of the bacterium in the presence and absence of the fungus was calculated according to [42] using the average cell numbers measured in four independent experiments. The CFU values are provided in the electronic supplementary material, table S3.
| agar | inoculation | bacterium | fitness | cell ratio (fungus/non-fungus) |
|---|---|---|---|---|
| technical | single | flagellated | 0.97 | 0.87 |
| purified | single | flagellated | 0.99 | 0.96 |
| technical | competition | flagellated | 1.31 | 6.18 |
| non-flagellated | 1.03 | 1.20 |
When we studied the benefit for the fungal partner, we observed that co-culturing with bacteria did not affect the fungus’ growth rate (ANOVA test, n = 11, F = 0.61; p > 0.1), but it did influence the formation of sclerotia. Sclerotia are hard-surface resting bodies formed by M. crassipes, which are produced as a nutrient reservoir for resisting unfavourable environmental conditions [24]. In a Petri dish inoculated only with M. crassipes, these structures formed homogeneously throughout the agar plate (figure 2a). However, when inoculated with bacteria, M. crassipes formed sclerotia only in the region opposed to the bacterial inoculum zone (figure 2b). The formation of sclerotia in this zone was not triggered by a decrease in nutrients in the area of the Petri dish supporting bacterial growth, as shown when replacing the bacterial inoculum by an agar plug without malt (figure 2c). In addition, this behaviour was observed regardless of the position of the bacterial inoculum, with the sclerotia always formed at the most distant possible position (figure 2d,e).
Figure 2.
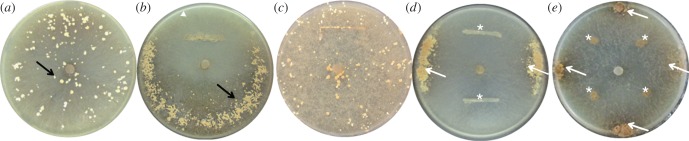
Influence of P. putida on the formation of sclerotia. (a) In the absence of bacteria, sclerotia are formed homogeneously throughout the entire surface of the Petri dish. (b) When the fungus is co-inoculated with bacteria, sclerotia are formed only in the region opposed to the bacterial inoculum zone. (c) Sclerotia are homogeneously distributed if the bacterial inoculation area is replaced by depleted agar medium. (d,e) Sclerotia (arrows) are always formed at the most distant position from the bacterial inoculum regardless of its position (stars).
A related species to M. crassipes, Morchella esculenta (L) Pers., is known to form sclerotia in a nutrient poorer region using resources extracted in a richer region [24,36,37]. Therefore, one can hypothesize that the bacterial inoculum zone can be considered a nutrient-rich pool for the fungus. To test this hypothesis, several experiments were conducted. If bacteria were farmed, the increase in bacterial populations in the area near the inoculum after the dispersal and rearing phases should be followed by a decrease during the formation of sclerotia (i.e. harvesting). To test this, bacterial populations were measured by CFU counting along a transect covering three areas of the Petri dish (inoculation, i, near the inoculum, a, and in the sclerotia-forming zone, b; figure 3). During the dispersal phase, bacteria rapidly colonized the whole plate, reaching a similar density in the three regions after 5 days (figure 3a). However, with the onset of sclerotia formation on day 7, bacterial populations at the inoculum zone (i and a) decreased dramatically (harvesting phase). By contrast, no drop in bacterial population was observed in the zone of formation of sclerotia (figure 3a), also showing that bacteria do not directly inhibit the formation of sclerotia. The decrease of bacterial numbers was contingent on sclerotia formation; blocking its formation in purified agar yielded stable bacterial populations throughout the plate (figure 3b,c; electronic supplementary material, figure S3; statistically significant reduced numbers of bacterial cells at the inoculum zone; unpaired t-test on log-transformed data, t = 12.16, p < 10–8; figure 3d). Thus, bacteria at the inoculation zone did not lose their viability over time, making it likely that the fungus harvested them when conditions for resource storage were favourable.
Figure 3.
Bacterial populations during dispersal and the formation of sclerotia. (a) Bacterial population increase and decrease are linked first to the migration on the fungal mycelium and later to the formation of sclerotia by the fungus. Bacterial populations were counted in three zones of the Petri dish (zones indicated in (c)). Bacteria colonized the entire Petri dish after 5 days, reaching a density close to 7×107 CFUs. However, bacteria in the inoculum zone disappeared during sclerotial formation. (b) In the absence of sclerotia formation, bacteria are dispersed as in previous experiments, but they are not harvested as shown by the absence of a decrease in bacterial population in the inoculum zone. (c) Comparison of fungal phenotype in the presence of bacteria but in the absence of sclerotia formation (ii). The letters indicate the regions for which bacterial populations were counted. (d) In technical agar, bacterial population in the inoculum zone is significantly lower than in purified agar (unpaired t-test on log-transformed data, n = 8, t = 12.16, p < 10−8).
We hypothesized that the decrease in bacterial populations was due to the mycelial translocation of carbon from the inoculum zone (the source) to sclerotia (the sink) during the harvesting phase of fungal farming. The direct contribution of bacterial biomass to sclerotia formation was tested by labelling P. putida with 13C-glucose and measuring the carbon isotope composition of a fungal biomarker (18 : 2ω6; electronic supplementary material, figure S2). A significant 13C enrichment of the fungus was observed during the harvesting phase (figure 4a and electronic supplementary material, table S2), demonstrating carbon transfer and for the first time, to the best of our knowledge, the consumption of bacteria and/or bacterial exudates by a fungus.
Figure 4.

Benefits of the fungal partner. (a) The fungal biomarker (18 : 2ω6) was significantly 13C-enriched during the harvesting phase with respect to the same experiment carried out with unlabelled bacteria. (b) Representative images (three replicates per condition) of M. crassipes co-inoculated or not with P. putida and showing the difference in melanization of the fungal mycelium after three weeks of growth. In the presence of P. putida (1, 2 and 3), the mycelium is less dark than in its absence (4, 5 and 6). This difference in colour is attributed to the level of hyphae melanization. 1 and 4 = one-week-old medium; 2 and 5 = 23 min drying; 3 and 6 = 15 min drying.
Bacteria were replaced by solutions with different concentrations of glucose to evaluate whether a concentrated carbon source could trigger the same effect as the bacterial inoculum. A solution as high as 10 g l−1 does not trigger the differential formation of sclerotia (see electronic supplementary material, figure S4). Although this might appear contradictory, carbon is not the only constituent of a bacterium cell and other nutrients, such as nitrogen, phosphorous, sulfur, trace elements, or other carbon sources from bacterial metabolites might also play a role in sclerotia formation and the observed effect for living bacteria. Indeed, a suspension of killed bacteria also failed to induce differential sclerotia formation (see electronic supplementary material, figure S4), suggesting that a combination of elements (e.g. bacterial density, metabolites and excretion products) is sensed by the fungus. Recently, it has been observed that bacterial communication mediated by quorum-sensing molecules can be important to regulate the interaction of bacteria and eukaryotes, as in plant–bacteria interactions [52]. To our knowledge, this has never been tested for fungi–bacteria interactions, but our results suggest the possibility that the fungus may use quorum-sensing signals between bacteria to assess bacterial presence and density in the co-culture.
In addition to the changes induced by the presence of bacteria on sclerotia formation, co-culturing with bacteria also modified the melanization of the hyphae, which is a marker of fungal stress. Fungal melanins are non-essential pigments derived from phenolic compounds [25] that can enhance survival and competitive abilities of fungi in stressful environments [25,26]. Inducing stressful conditions by changing the water content of the culture media, we observed a 24% lower melanin response in fungi associated with bacteria compared with fungi alone (for a one-week-old medium, the ratio of melanin between the mycelia with bacteria over the fungus alone was 0.76; figure 4b). We consider that both the documented carbon transfer and the reduced stress response are direct benefits to the fungal partner as part of a mutualistic interaction with P. putida.
Another aspect of farming, the storage of ‘seeds’ in structures allowing re-initiation of the culture after the end of a farming cycle, was also tested by checking for bacteria in or on sclerotia. Sequential washing steps and bacterial counting shows that over 105 bacteria are associated to each sclerotium with different strength levels of attachment. We tested the re-dispersion of bacteria by re-picking bacteria-carrying sclerotia in fresh medium (see electronic supplementary material, figure S5). Upon sclerotium germination and fungal development, bacteria dispersed and colonized the mycelium as in the case of the external bacterial inoculation. At the stage of sclerotia formation, bacteria were once more harvested in the centre of the plate and the resource translocated all over the outer rim of the Petri dish, opposite to the position of the initial inoculum (i.e. germinating sclerotia).
In other farming models, the obligate dependence of the farmers has been demonstrated by a reduced reproductive output [32]. Thus, ideally, one should measure reproductive success of the fungus in the presence versus the absence of bacteria. However, this could not be measured as fructification and spore formation in M. crassipens are difficult to induce in vitro. Hence, to test whether M. crassipes farming was restricted to P. putida, two other bacterial strains were included. Pseudomonas knackmusii B13 and Cupriavidus necator JMP289 could both disperse on M. crassipes hyphae (see electronic supplementary material, figure S6). However, only P. knackmusii B13 modified the formation of sclerotia and was likely to be farmed by M. crassipes in a similar way to P. putida. This indicates that farming is neither restricted to P. putida KT2440 nor universal to other bacteria that could migrate on M. crassipes hyphae, suggesting that specific interspecies signalling might also be involved.
The emergence of agriculture is one of the factors explaining the ecological success of humans [31]. Moreover, farming behaviour is also present in other animals like ants [27], termites [28], beetles [29,30] and damselfish [53]. This non-human agriculture can be the product of long-term coevolution and adaptation, for example in farming insects [32]. However examples that involve fewer adaptations are also known and include bacterial husbandry by social amoeba [35] or fungal farming in snails [34]. Our system fulfils some criteria that have been previously identified as shared by insect fungiculture and human farming [32]. These include habitual planting, which corresponds to bacterial dispersal along the mycelial network; cultivation, comparable to the rearing of bacteria with fungal exudates and the fitness benefit of dispersal depending on the conditions; and harvesting, consisting in decrease in bacterial numbers and translocation of C. Another criterion, the obligated nature of the interaction, is difficult to assess in our system because we are working with an artificial farming pair. Therefore it would be ideal to try to identify similar interactions directly in soils. In this context, a definition of proto-farming could be applied to our system. However, in some aspects, farming by M. crassipes differed from previously described non-human agriculture, which would suggest that the interaction observed is more complex than husbandry or proto-farming. First and similar to human agriculture, we found a delocalization of food production and consumption, with a division of labour within a single organism rather than within a society. Also, the evolution of farming has been largely associated to sociality in the past because of the multigenerational benefits in established kin groups [35]. However, in the case of M. crassipes farming, kin selection is not involved as the re-dispersion and harvesting of bacteria colonizing a survival structure will be used by a next generation of the same farmer, thus yielding only direct fitness benefits.
The appearance of fungi in terrestrial ecosystems must have had a strong impact on the evolution of soil bacteria, creating a myriad of possible interactions ranging from competition to mutualism [4]. The case of farming by a fungus shown in this study is an example of a mutualistic behaviour in which fungi first coexist with their bacterial partner, and once the medium is depleted, take advantage of a self-created resource. Mutualism is a crucial factor in shaping ecological communities [54]. Given the eminent role of dispersal for the dynamics of species communities [18,55], the effects of bacterial dispersal and farming by fungi on soil microbial communities merit detailed attention.
Acknowledgements
We acknowledge critical discussions and comments from T. Junier and D. Johnson on earlier versions of this manuscript. We thank T. Goetschi for technical assistance as well as Dr T. R. Neu and U. Kuhlicke for their assistance in using Laser microscopy.
References
- 1.O'Donnell AG, Young IM, Rushton SP, Shirley MD, Crawford JW. 2007. Visualization, modelling and prediction in soil microbiology. Nat. Rev. Microbiol. 5, 689–699 (doi:10.1038/nrmicro1714) [DOI] [PubMed] [Google Scholar]
- 2.Torsvik V, Ovreas L, Thingstad TF. 2002. Prokaryotic diversity: magnitude, dynamics, and controlling factors. Science 296, 1064–1066 (doi:10.1126/science.1071698) [DOI] [PubMed] [Google Scholar]
- 3.Gans J, Wolinsky M, Dunbar J. 2005. Computational improvements reveal great bacterial diversity and high metal toxicity in soil. Science 309, 1387–1390 (doi:10.1126/science.1112665) [DOI] [PubMed] [Google Scholar]
- 4.de Boer W, Folman LB, Summerbell RC, Boddy L. 2005. Living in a fungal world: impact of fungi on soil bacterial niche development. FEMS Microbiol. Rev. 29, 795–811 (doi:10.1016/j.femsre.2004.11.005) [DOI] [PubMed] [Google Scholar]
- 5.Nazir R, Warmink JA, Boersma H, van Elsas JD. 2010. Mechanisms that promote bacterial fitness in fungal-affected soil microhabitats. FEMS Microbiol. Ecol. 71, 169–185 (doi:10.1111/j.1574-6941.2009.00807.x) [DOI] [PubMed] [Google Scholar]
- 6.Mohammadi K, Khalesro S, Sohrabi Y, Heidari G. 2011. A review: beneficial effects of the mycorrhizal fungi for plant growth. J. Appl. Environ. Biol. Sci. 1, 310–319 [Google Scholar]
- 7.Johansson JF, Paul LR, Finlay RD. 2004. Microbial interactions in the mycorrhizosphere and their significance for sustainable agriculture. FEMS Microbiol. Ecol. 48, 1–13 (doi:10.1016/j.femsec.2003.11.012) [DOI] [PubMed] [Google Scholar]
- 8.van der Heijden MG, Bardgett RD, van Straalen NM. 2008. The unseen majority: soil microbes as drivers of plant diversity and productivity in terrestrial ecosystems. Ecol. Lett. 11, 296–310 (doi:10.1111/j.1461-0248.2007.01139.x) [DOI] [PubMed] [Google Scholar]
- 9.Martin G, Guggiari M, Bravo D, Zopfi J, Cailleau G, Aragno M, Job D, Verrecchia E, Junier P. 2012. Fungi, bacteria and soil pH: the oxalate-carbonate pathway as a model for metabolic interaction. Environ. Microbiol. 14, 2960–2970 (doi:10.1111/j.1462-2920.2012.02862.x) [DOI] [PubMed] [Google Scholar]
- 10.Harms H, Schlosser D, Wick LY. 2011. Untapped potential: exploiting fungi in bioremediation of hazardous chemicals. Nat. Rev. Microbiol. 9, 177–192 (doi:10.1038/nrmicro2519) [DOI] [PubMed] [Google Scholar]
- 11.Keller L, Surette MG. 2006. Communication in bacteria: an ecological and evolutionary perspective. Nat. Rev. Microbiol. 4, 249–258 (doi:10.1038/nrmicro1383) [DOI] [PubMed] [Google Scholar]
- 12.West SA, Griffin AS, Gardner A, Diggle SP. 2006. Social evolution theory for microorganisms. Nat. Rev. Microbiol. 4, 597–607 (doi:10.1038/nrmicro1461) [DOI] [PubMed] [Google Scholar]
- 13.Ingham CJ, Kalisman O, Finkelshtein A, Ben-Jacob E. 2011. Mutually facilitated dispersal between the nonmotile fungus Aspergillus fumigatus and the swarming bacterium Paenibacillus vortex. Proc. Natl Acad. Sci. USA 108, 19 731–19 736 (doi:10.1073/pnas.1102097108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Raymond B, West SA, Griffin AS, Bonsall MB. 2012. The dynamics of cooperative bacterial virulence in the field. Science 337, 85–88 (doi:10.1126/science.1218196) [DOI] [PubMed] [Google Scholar]
- 15.Young IM, Crawford JW. 2004. Interactions and self-organization in the soil–microbe complex. Science 304, 1634–1637 (doi:10.1126/science.1097394) [DOI] [PubMed] [Google Scholar]
- 16.Or D, Smets BF, Wraith JM, Dechesne A, Friedman SP. 2007. Physical constraints affecting bacterial habitats and activity in unsaturated porous media: a review. Adv. Water Resour. 30, 1505–1527 (doi:10.1016/j.advwatres.2006.05.025) [Google Scholar]
- 17.Crawford JW, Harris JA, Ritz K, Young IM. 2005. Towards an evolutionary ecology of life in soil. Trends Ecol. Evol. 20, 81–87 (doi:10.1016/j.tree.2004.11.014) [DOI] [PubMed] [Google Scholar]
- 18.Venail PA, MacLean RC, Bouvier T, Brockhurst MA, Hochberg ME, Mouquet N. 2008. Diversity and productivity peak at intermediate dispersal rate in evolving metacommunities. Nature 452, 210–214 (doi:10.1038/nature06554) [DOI] [PubMed] [Google Scholar]
- 19.Venail PA, Maclean RC, Meynard CN, Mouquet N. 2010. Dispersal scales up the biodiversity–productivity relationship in an experimental source-sink metacommunity. Proc. R. Soc. B 277, 2339–2345 (doi:10.1098/rspb.2009.2104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dechesne A, Wang G, Gulez G, Or D, Smets BF. 2010. Hydration-controlled bacterial motility and dispersal on surfaces. Proc. Natl Acad. Sci. USA 107, 14 369–14 372 (doi:10.1073/pnas.1008392107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Treves DS, Xia B, Zhou J, Tiedje JM. 2003. A two-species test of the hypothesis that spatial isolation influences microbial diversity in soil. Microb. Ecol. 45, 20–28 (doi:10.1007/s00248-002-1044-x) [DOI] [PubMed] [Google Scholar]
- 22.Kohlmeier S, Smits THM, Ford RM, Keel C, Harms H, Wick LY. 2005. Taking the fungal highway: mobilization of pollutant-degrading bacteria by fungi. Environ. Sci. Technol. 39, 4640–4646 (doi:10.1021/es047979z) [DOI] [PubMed] [Google Scholar]
- 23.Wang G, Or D. 2010. Aqueous films limit bacterial cell motility and colony expansion on partially saturated rough surfaces. Environ. Microbiol. 12, 1363–1373 (doi:10.1111/j.1462-2920.2010.02180.x) [DOI] [PubMed] [Google Scholar]
- 24.Amir R, Levanon D, Hadar Y, Chet I. 1995. Factors affecting translocation and sclerotial formation in Morchella esculenta. Exp. Mycol. 19, 61–70 (doi:10.1006/emyc.1995.1007) [Google Scholar]
- 25.Bell AA, Wheeler MH. 1986. Biosynthesis and function of fungal melanins. Annu. Rev. Phytopathol. 24, 411–451 (doi:10.1146/annurev.py.24.090186.002211) [Google Scholar]
- 26.Butler MJ, Day AW. 1998. Fungal melanins: a review. Can. J. Microbiol. 44, 1115–1136 (doi:10.1139/w98-119) [Google Scholar]
- 27.Currie CR, Wong B, Stuart AE, Schultz TR, Rehner SA, Mueller UG, Sung G-H, Spatafora JW, Straus NA. 2003. Ancient tripartite coevolution in the attine ant–microbe symbiosis. Science 299, 386–388 (doi:10.1126/science.1078155) [DOI] [PubMed] [Google Scholar]
- 28.Aanen DK, Eggleton P, Rouland-Lefevre C, Guldberg-Froslev T, Rosendahl S, Boomsma JJ. 2002. The evolution of fungus-growing termites and their mutualistic fungal symbionts. Proc. Natl Acad. Sci. USA 99, 14 887–14 892 (doi:10.1073/pnas.222313099) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hulcr J, Cognato AI. 2010. Repeated evolution of crop theft in fungus-farming Ambrosia beetles. Evolution 64, 3205–3212 (doi:10.1111/j.1558-5646.2010.01055.x) [DOI] [PubMed] [Google Scholar]
- 30.Biedermann PH, Taborsky M. 2011. Larval helpers and age polyethism in ambrosia beetles. Proc. Natl Acad. Sci. USA 108, 17 064–17 069 (doi:10.1073/pnas.1107758108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Diamond J. 2002. Evolution, consequences and future of plant and animal domestication. Nature 418, 700–707 (doi:10.1038/nature01019) [DOI] [PubMed] [Google Scholar]
- 32.Mueller UG, Gerardo NM, Aanen DK, Six DL, Schultz TR. 2005. The evolution of agriculture in insects. Annu. Rev. Ecol. Evol. 36, 563–595 (doi:10.1146/annurev.ecolsys.36.102003.152626) [Google Scholar]
- 33.Styrsky JD, Eubanks MD. 2007. Ecological consequences of interactions between ants and honeydew-producing insects. Proc. R. Soc. B 274, 151–164 (doi:10.1098/rspb.2006.3701) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Silliman BR, Newell SY. 2003. Fungal farming in a snail. Proc. Natl Acad. Sci. USA 100, 15 643–15 648 (doi:10.1073/pnas.2535227100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brock DA, Douglas TE, Queller DC, Strassmann JE. 2011. Primitive agriculture in a social amoeba. Nature 469, 393–396 (doi:10.1038/nature09668) [DOI] [PubMed] [Google Scholar]
- 36.Amir R, Levanon D, Hadar Y, Chet I. 1992. Formation of sclerotia by Morchella esculenta: relationship between media composition and turgor potential in the mycelium. Mycol. Res. 96, 943–948 (doi:10.1016/S0953-7562(09)80595-9) [Google Scholar]
- 37.Amir R, Levanon D, Hadar Y, Chet I. 1994. The role of source–sink relationships in translocation during sclerotial formation by Morchella esculenta. Mycol. Res. 98, 1409–1414 (doi:10.1016/S0953-7562(09)81071-X) [Google Scholar]
- 38.Bonfante P, Anca IA. 2009. Plants, mycorrhizal fungi, and bacteria: a network of interactions. Annu. Rev. Microbiol. 63, 363–383 (doi:10.1146/annurev.micro.091208.073504) [DOI] [PubMed] [Google Scholar]
- 39.Furuno S, Pazolt K, Rabe C, Neu TR, Harms H, Wick LY. 2010. Fungal mycelia allow chemotactic dispersal of polycyclic aromatic hydrocarbon-degrading bacteria in water-unsaturated systems. Environ. Microbiol. 12, 1391–1398 (doi:10.1111/j.1462-2920.2009.02022.x) [DOI] [PubMed] [Google Scholar]
- 40.Wick LY, Remer R, Wurz B, Reichenbach J, Braun S, Schafer F, Harms H. 2007. Effect of fungal hyphae on the access of bacteria to phenanthrene in soil. Environ. Sci. Technol. 41, 500–505 (doi:10.1021/es061407s) [DOI] [PubMed] [Google Scholar]
- 41.Aragno M, Schlegel HG. 1991. The mesophilic hydrogen-oxidizing (Knallgas) bacteria. In The prokaryotes (eds Balows A, Trüper HG, Dworkin M, Harder W, Schleifer) KH, pp. 344–384 New York, NY: Springer [Google Scholar]
- 42.Kerr B, Riley MA, Feldman MW, Bohannan BJM. 2002. Local dispersal promotes biodiversity in a real-life game of rock–paper–scissors. Nature 418, 171–174 (doi:10.1038/nature00823) [DOI] [PubMed] [Google Scholar]
- 43.Robbins WJ, Hervey A. 1965. Manganese, calcium and filtrate factor for Morchella crassipes. Mycologia 57, 262–274 (doi:10.2307/3756827) [Google Scholar]
- 44.Guckert JB, Antworth CP, Nichols PD, White DC. 1985. Phospholipid, ester-linked fatty-acid profiles as reproducible assays for changes in prokaryotic community structure of estuarine sediments. FEMS Microbiol. Ecol. 31, 147–158 (doi:10.1016/0378-1097(85)90016-3) [Google Scholar]
- 45.Ichihara K, Fukubayashi Y. 2010. Preparation of fatty acid methyl esters for gas–liquid chromatography. J. Lipid Res. 51, 635–640 (doi:10.1194/jlr.D001065) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tunlid A, White DC. 1992. Biochemical analysis of biomass, community structure, nutritional status, and metabolic activities of microbial communities in soil. In Soil biochemistry (eds Ballog JM, Stotzky G.), pp. 229–262 New York, NY: Marcel Dekker [Google Scholar]
- 47.Vestal JB, White DC. 1989. Lipid analysis in microbial ecology. BioScience 39, 535–541 (doi:10.2307/1310976) [PubMed] [Google Scholar]
- 48.Singaravelan N, Grishkan I, Beharav A, Wakamatsu K, Ito S, Nevo E. 2008. Adaptive melanin response of the soil fungus Aspergillus niger to UV radiation stress at ‘Evolution Canyon’, Mount Carmel, Israel. PLoS ONE 3, e2993 (doi:10.1371/journal.pone.0002993) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ozeki H, Ito S, Wakamatsu K, Hirobe T. 1995. Chemical characterization of hair melanins in various coat-color mutants of mice. J. Invest. Dermatol. 105, 361–366 (doi:10.1111/1523-1747.ep12320792) [DOI] [PubMed] [Google Scholar]
- 50.Warmink JA, Nazir R, Corten B, van Elsas JD. 2011. Hitchhikers on the fungal highway: the helper effect for bacterial migration via fungal hyphae. Soil Biol. Biochem. 43, 760–765 (doi:10.1016/j.soilbio.2010.12.009) [Google Scholar]
- 51.Boersma FGH, Otten R, Warmink JA, Nazir R, van Elsas JD. 2010. Selection of Variovorax paradoxus-like bacteria in the mycosphere and the role of fungal-released compounds. Soil Biol. Biochem. 42, 2137–2145 (doi:10.1016/j.soilbio.2010.08.009) [Google Scholar]
- 52.Gonzalez JF, Venturi V. 2013. A novel widespread interkingdom signaling circuit. Trends Plant Sci. 18, 167–174 (doi:10.1016/j.tplants.2012.09.007) [DOI] [PubMed] [Google Scholar]
- 53.Hata H, Kato M. 2006. A novel obligate cultivation mutualism between damselfish and Polysiphonia algae. Biol. Lett. 2, 593–596 (doi:10.1098/rsbl.2006.0528) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Douglas AE. 2010. The symbiotic habit, p. 202 Princeton, NJ: Princeton University Press [Google Scholar]
- 55.Lindstrom ES, Ostman O. 2011. The importance of dispersal for bacterial community composition and functioning. PLoS ONE 6, e25883 (doi:10.1371/journal.pone.0025883) [DOI] [PMC free article] [PubMed] [Google Scholar]